Abstract
Ischemia reperfusion (I/R) injury is an unavoidable event occurring during heart transplantation, leading to graft failures and lower long-term survival rate of the recipient. Several studies have demonstrated that microRNAs (miRNAs) are vital regulators of signalling pathways involved in I/R injury. The present study aims to quantify the altered expression levels of miRNA and mRNA upon I/R injury in a mouse heart transplantation model, and to investigate whether these miRNA can regulate genes involved in I/R injury. We performed heterotopic heart transplantation on mouse models to generate heart tissue samples with I/R and non-I/R (control). The expression levels of miRNAs as well as genes were measured in heart grafts by microarray and real time RT-PCR. miRNA alteration in cardiomyocytes exposed to hypoxia was also detected by qRT-PCR. We observed significant alterations in miRNA and gene expression profile after I/R injury. There were 39 miRNAs significantly downregulated and 20 upregulated up to 1.5 fold in heart grafts with I/R injury compared with the grafts without I/R. 48 genes were observed with 3 fold change and p<0.05 and 18 signalling pathways were enriched using Keggs pathway library. Additionally, hypoxia/reperfusion induced primary cardiomyocyte apoptosis and altered miRNA expression profiles. In conclusion, this is the first report on miRNA expression profile for heart transplantation associated with I/R injury. These findings provide us with an insight into the role of miRNA in I/R injury in heart transplantation.
Introduction
Since the 1970s, heart failure (HF) prevalence has been increasing in the world as a consequence of a decline in coronary artery and cerebrovascular disease mortality [1]. Although there are many treatments available for HF patients, heart transplantation remains the best option for long-term survival for end-stage HF patients [2]. However, this effective treatment for heart failure is severely affected by ischemia reperfusion (I/R) injury occurring during transplantation.
Despite major achievements in heart transplantation, I/R injury is a major contributing factor in graft failure and longer ischemia time has shown to lower the long-term survival rate, especially for older patients [3]. In addition, due to a shortage of donors, physicians are forced to enlarge the donor pool by accepting marginal organs, which include organs from elderly or ill patients, and thus they are more susceptible to I/R injury [3]. Currently, there are no effective treatments against ischemia reperfusion injury. It is important to explore new alternative mechanisms involved in I/R injury during heart transplantation.
miRNAs are endogenous, short, non-coding single-stranded RNAs that are approximately 20 nucleotides in length. miRNAs have emerged as a key player in physiology as well as pathophysiology attributable to its ability to downregulate gene expression through mRNA destabilization/degradation and translation repression by binding onto either 3′ UTR or 5′UTR of the mRNA [4]. Several studies have shown that miRNAs have the ability to regulate the expression profiles of genes in signalling pathways associated with heart diseases, including heart failure, hypertrophy, and ischemia reperfusion injury [5]. Therefore, it is crucial to examine the role of miRNA in heart transplantation and its implications for I/R signalling pathways. In this study, for the first time, we report miRNA expression profiles in I/R injured heart grafts and also investigated mRNA expression profiles that may be affected by miRNAs.
Materials and Methods
Animals
Eight weeks old C57BL/6 mice were purchased from Charles River Laboratory (Canada). All procedures involving mouse breeding and surgery were performed according to the guidelines of the Canadian Council of Animal Care and were approved by the Animal Use Subcommittee at the University of Western Ontario, Canada.
Induction of Cold Ischemia Reperfusion Injury and Heart Transplantation
C57BL/6 mice were anesthetized with ketamine/protophin and injected with 1 ml heparin. Donor hearts were excised from mice and heterotopically implanted into the peritoneal cavity with the donor aorta anastomosed to the recipient abdominal aorta and the pulmonary artery connected to the inferior vena cava. For induction of cold I/R injury, donor hearts were preserved with University of Wisconsin (UW) solution at 4°C for 18 hours before implantation. Meanwhile, the rest of the excised heart was immediately implanted into the recipient to generate non cold ischemia injury heart (non-I/R) as controls. At the endpoint of experiments, mice were sacrificed by injection over dose of ketamine/protophin and heart grafts were harvested for future studies.
Histological Analysis
At 24 hours post-transplantation, heart grafts were collected from mice and tissue slices were fixed in 10% formalin and processed for histology examination using standard techniques. Formalin tissue was embedded in paraffin and 5 µm sections were stained with hematoxylin and eosin stain (H&E).
Myeloperoxidase (MPO) Activity
To detect neutrophil infiltration, MPO activity was detected in heart tissues. Paraffin tissue sections were stained with MPO antibody (Santa Cruz, San Diego, CA) following the manufacturer’s instruction. Each slide was examined by light microscopy at 200× magnification.
Terminal Deoxynucleotidyl Transferase-mediated dUTP Nick End Labelling (TUNEL) Assay
To detect cell apoptosis in heart grafts, TUNEL assay was performed on paraffin tissue sections using an in situ cell death detection kit according to the manufacturer’s instruction (Roche, Mississauga, ON, Canada). Sections were counter-stained with hematoxylin. Each slide was examined by light microscopy at 200× magnification.
microRNA/RNA Extraction
Fresh heart tissues or primary cardiomyocytes were collected and subjected to the extraction of miRNA/RNA. miRNAs were extracted using a miRNeasy mini Kit (Qiagen, Ontario, Canada). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA) and the RNA 6000 Nano kit (Caliper Life Sciences, Mountain View, CA). One microgram of total RNA was used to synthesize cDNA for measuring miRNA expression using miRScript II kit (Qiagen, Ontario, Canada) according to the manufacturer’s manual.
miRNA Microarray
All sample labeling and GeneChip processing were performed at the London Regional Genomics Centre (Robarts Research Institute, London, Ontario, Canada; http://www.lrgc.ca). One microgram of total RNA was labeled using the Flash Tag Biotin HSR kit from Genisphere (http://www.genisphere.com/array_detection_flashtag_biotin.html). Samples were then hybridized to Affymetrix miRNA 3.0 arrays for 16 hours at 48°C. All washing steps were performed by a GeneChip Fluidics Station 450 and GeneChips were scanned with the GeneChip Scanner 3000 7 G (Affymetrix, Santa Clara, CA) using Command Console v3.2.4. Partek was used to determine ANOVA p-values and fold changes for miRNAs. Species annotations were added and used to filter only those miRNA found in Mus musculus. miRNA expression data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE50885.
Gene Expression Microarray
Single stranded complementary DNA (sscDNA) was prepared from 200 ng of total RNA as per the Ambion WT Expression Kit for Affymetrix GeneChip Whole Transcript WT Expression Arrays (http://www.ambion.com/techlib/prot/fm_4411973.pdf, Applied Biosystems, Carlsbad, CA). Total RNA was first converted to cDNA, followed by in vitro transcription to make cRNA. 5.5 µg of single stranded cDNA was synthesized; end labeled and hybridized, for 16 hours at 45°C, to Mouse Gene 1.0 ST arrays. All washing steps were performed by a GeneChip Fluidics Station 450 and GeneChips were scanned with the GeneChip Scanner 3000 7 G (Affymetrix, Santa Clara, CA) using Command Console v3.2.4. Partek was used to determine ANOVA p-values and fold changes for genes. Gene expression data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE 50884.
Quantitative Reverse Transcriptase-polymerase Reaction (q-PCR) for miRNA Expression
miRNAs were extracted from I/R and non-I/R heart tissues using miRNeasy mini Kit (Qiagen, Ontario,Canada), then cDNA were synthesized using miRScript II RT Kit (Qiagen, Ontario). Synthesized cDNA samples were then subjected to real time RT-PCR (Stratagene mx3005P) using miRScript SYBR Green PCR Kit (Qiagen, Ontario) according to instruction of the kit supplier. Thermal profiling for the real time PCR was an initial activation step at 95°C for 10 mins, followed by 40 cycles of: 95°C for 15 s, 55°C for 30 s, 70°C for 30 s. Expression levels between I/R and non-I/R were quantitatively compared using the ΔΔCt method with SNorD6 as the endogenous control for miRNA expression.
Primary Neonatal Cardiomyocytes Culture
Neonatal ventricular cardiomyocytes were cultured as described by the Feng group [6]. Briefly, ventricular tissues from C57BL/6 mice were isolated and minced within 24 h after birth. Subsequently, cardiomyocytes were dispersed by incubation in a D-Hank’s buffer supplemented with 0.5 mg/ml Liberase (Roche, Worthington Biochemical, Lakewood, NJ) and the cellular suspension was filtered through a polypropylene macro porous filter (mesh opening 105 µm, Spectra Mesh; Spectrum Medical Industries). The suspension was then centrifuged at 200 g for 5 min, and the cellular pellet was suspended in medium 199 (M199) with 10% FBS and penicillin-streptomycin (50 µg/ml; GIBCO-BRL). The cellular suspension was pre-plated for 1.5 h at 37°C in 5% CO2 to remove any non-cardiomyocytes. Cell density was adjusted to 106 cells/ml using M199 supplemented with 10% FBS, and cells were seeded (5 × 105) in polystyrene, nonpyrogenic24-well culture plates (Becton Dickinson, Franklin Lakes, NJ) precoated with 1% gelatin. Cells were incubated in 5% CO2 at 37°C.
Hypoxia/Reperfusion Model for Cell Culture
Isolated primary cardiomyocytes were cultured in a 24 well plate pre-coated with 1% gelatine for overnight. Culture medium were replaced by 250 µl deoxygenated DMEM medium without FBS and antibiotics and then placed in a chamber with 2% O2 at 37°C for 45 min. After hypoxia, cells were reoxygenated by adding 250 µl of complete medium supplemented with 10% FBS and cultured at 5% CO2, 37°C, for 24 h.
Annexin-V Staining and Flow Cytometry
Cardiomyocytes were harvested using Trypsin and suspended in PBS solution with 2% FBS. Cells were double stained with Annexin-V and PI using Annexin-V kit (ebioscience, San Diego, CA) according to the manufacturer’s instruction. The fluorescence from the stained cells was measured by flow cytometry (BD bioscience, San Jose, CA).
Potential miRNA Targets
For selected miRNA that were significantly altered by I/R injury in heart transplantation, online computational algorithm analyses (Targetscan and FINDTAR3) were used to predict potential target genes. Based on scientific literature, target genes that are known to be involved in I/R injury pathways were selected for further analysis.
Gene Expression
Total RNA isolations were performed using the TRIzol reagent (Invitrogen), and cDNA was synthesized using oligo-(dT) primer and reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Synthesized cDNA samples were then subjected to real time RT-PCR (Stratagene mx3005P and Bio-Rad CFX) with SYBR Green and a final primer concentration of 80 nM. Thermal profiling for the real time PCR was the initial activation step at 95°C for 10 mins, followed by 40 cycles of: 95°C for 30 s, 58°C for 30 s, 72°C for 30 s. Expression levels between I/R and non-I/R were quantitatively compared using the ΔCt method with mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the endogenous control for target gene expression.
Western Blotting
Heart tissues were homogenized with RIPA buffer containing 5 µg/ml PMSF as the manufacturer’s instruction (Invitrogen, Life Tech., Ontario, Canada) and centrifuged at 10,000 g for 10 mins. Protein concentration was measured using a Bradford assay (Bio-Rad, Mississauga, Ontario, Canada). 40 µg of total proteins were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes. After blocking for 1 h in TBS supplemented with 5% milk and 0.1% Tween20, membranes were blotted with rabbit anti mouse polyclone angiopoietin 1 Ab (used at 1∶500 dilution, Abcam. Toronto, ON, Canada) and developed using HRP-conjugated anti-rabbit IgG (1∶5,000 dilution) (Santa Cruz, Dallas, Texas) and enhanced chemiluminescence (Bio-Rad).
Statistical Analysis
All data are presented as means ± SEM. Statistical comparisons between two groups were performed using student’s t-test and ANOVA was applied for the microarray data. Statistical significance was determined as P<0.05.
Results
I/R Induced Cardio Graft Damage
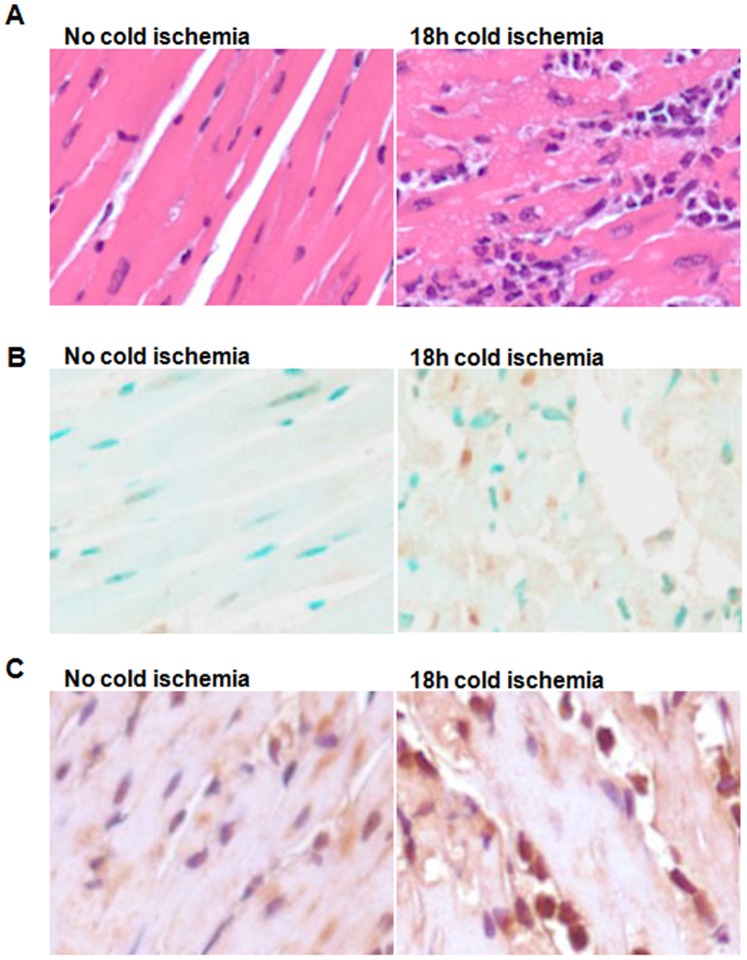
Currently during organ transplantations, donor organs are preserved using a hypothermal static method to reduce ischemia reperfusion injury. Thus, I/R injury in organ transplantation is usually referred to as cold I/R, differentiating it from that in non-transplanted. Accordingly, we used this hypothermal static method to preserve donor organs for a variety of periods in order to induce cold ischemia-reperfusion injury. Histopathological staining (H&E, apoptosis and neutrophil infiltration) were applied to assess the severity of I/R injury in grafts. We found that 18 h cold ischemia induced I/R injury - supported by results from H&E, TUNEL assay, and MPO assay. H&E staining (Figure 1A) shows disrupted architecture, necrosis, and degeneration in I/R groups compared to the control (non-I/R) group. TUNEL assay (Figure 1B) shows an increased number of apoptotic cells in I/R groups. As shown by MPO assay (Figure 1C), increased myeloperoxidase activity was observed in the I/R group, indicating there are neutrophils infiltrating into grafts.
Figure 1. 18 h cold ischemia-reperfusion caused I/R injury in heart grafts.
Donor hearts were isolated from C57BL/6 mice, infused with UW solution and preserved in UW solution at 4°C for 18 h. After 18 h preservation, donor hearts were implanted into syngeneic recipient C57BL/6 mice. At day 2 after transplantation, the heart grafts were harvested and fixed in 10% formalin. The paraffin heart sections were subjected to HE staining, TUNEL assay, and MPO assay. (A) HE staining. (B) TUNEL assay for apoptosis. (C) MPO assay. Representative images were from experiments.
I/R Injury Altered miRNA Expression Profiles in Heart Grafts
In order to characterize the miRNA expression profile that regulates genes involved in I/R injury in heart transplantation, we performed a microarray assay using Affymatrix: GeneChip miRNA 3.0 Array that contains 1111 mouse probe sequences. Heart grafts (n = 3/group) with I/R or with non-I/R (control) were collected to extract miRNA at day 2 post transplantation. Microarray assays showed that miRNA were expressed differentially in heart grafts. A total of 59 miRNA were significantly altered with the criteria of 1.5 fold change with P<0.05 (Table 1). Out of the 59 altered miRNAs, 39 were downregulated in heart grafts with I/R injury compared with the grafts without I/R, while 20 miRNA were upregulated (Table 1). As shown in a pie graph of miRNA distribution based on their fold changes in expression (Figure 2A), the majority of altered miRNA (49 out of 59) fell into the range of 1.5 to 3 fold up or downregulation. Only ten miRNAs (five up-regulated and another five down-regulated) displayed over three fold changes between two groups. Each individual altered miRNA with >3 fold change is shown in Figure 2B, indicating that the miRNA changes fit a Poisson distribution. Additionally, signalling pathway enrichment analysis was conducted based on miRNAs with 1.5 fold change and p<0.05, 71 signalling pathways were selected (enrichment P<0.05) using the Kegg pathway library.
Table 1. miRNA altered in heart transplantation.
| miRNA | Fold-Change(Exp vs. Control) | SS(Error) | Fold-Change(Exp vs Control) | p-value(Exp vs. ontrol) | |
| mmu-miR-490-5p_st | −6.24593 | 1.23515 | Down | 0.0203434 | |
| mmu-miR-326_st | −5.24883 | 0.284485 | Down | 0.00340912 | |
| mmu-miR-490-3p_st | −3.65425 | 1.19612 | Down | 0.0477261 | |
| mmu-miR-1843b-5p_st | −3.2084 | 0.636683 | Down | 0.0280223 | |
| mmu-miR-345-3p_st | −3.1504 | 0.691868 | Down | 0.0325233 | |
| mmu-miR-671-3p_st | −2.92478 | 0.667747 | Down | 0.0368904 | |
| mmu-miR-24-1-star_st | −2.85064 | 0.446847 | Down | 0.0232897 | |
| mmu-miR-181a-2-star_st | −2.7874 | 0.358902 | Down | 0.0183911 | |
| mmu-miR-1943_st | −2.62098 | 0.223826 | Down | 0.0113917 | |
| mmu-miR-425-star_st | −2.37163 | 0.416558 | Down | 0.0351811 | |
| mmu-miR-181c-star_st | −2.3374 | 0.0854605 | Down | 0.00415107 | |
| mmu-miR-185-star_st | −2.32242 | 0.115686 | Down | 0.00655456 | |
| mmu-miR-491_st | −2.2317 | 0.456292 | Down | 0.0473795 | |
| mmu-miR-328_st | −2.20344 | 0.412421 | Down | 0.0434945 | |
| mmu-miR-2183_st | −2.11913 | 0.0630796 | Down | 0.00381541 | |
| mmu-miR-3058_st | −2.01364 | 0.183612 | Down | 0.0208451 | |
| mmu-miR-128_st | −1.98828 | 0.0715253 | Down | 0.00590299 | |
| mmu-miR-874-star_st | −1.977 | 0.110185 | Down | 0.0111359 | |
| hp_mmu-mir-181a-2_st | −1.96297 | 0.169814 | Down | 0.0207353 | |
| mmu-miR-423-3p_st | −1.93968 | 0.261325 | Down | 0.0381555 | |
| mmu-miR-324-3p_st | −1.91486 | 0.201859 | Down | 0.0287916 | |
| mmu-miR-532-3p_st | −1.89947 | 0.18219 | Down | 0.0260086 | |
| mmu-miR-181a-1-star_st | −1.86983 | 0.121223 | Down | 0.0160835 | |
| mmu-miR-467a-star_st | −1.822 | 0.238043 | Down | 0.0435423 | |
| mmu-miR-433_st | −1.78743 | 0.166247 | Down | 0.0299398 | |
| mmu-miR-351-star_st | −1.75 | 0.0465557 | Down | 0.00574918 | |
| mmu-miR-5123_st | −1.74629 | 0.132341 | Down | 0.024712 | |
| hp_mmu-mir-181a-2_x_st | −1.74163 | 0.175927 | Down | 0.0362254 | |
| mmu-miR-669a-3p_st | −1.69692 | 0.198551 | Down | 0.0475402 | |
| mmu-miR-295-star_st | −1.68704 | 0.195156 | Down | 0.047832 | |
| mmu-miR-470_st | −1.64899 | 0.159001 | Down | 0.0414325 | |
| mmu-miR-139-3p_st | −1.64413 | 0.0621587 | Down | 0.012078 | |
| hp_mmu-mir-128-2_x_st | −1.61738 | 0.0527945 | Down | 0.0105619 | |
| mmu-miR-362-5p_st | −1.60748 | 0.155159 | Down | 0.0457828 | |
| hp_mmu-mir-133a-1_x_st | −1.59055 | 0.125522 | Down | 0.0371386 | |
| mmu-miR-190b-star_st | −1.55534 | 0.030848 | Down | 0.00627936 | |
| mmu-miR-324-5p_st | −1.53951 | 0.0934193 | Down | 0.0306476 | |
| mmu-miR-210_st | −1.52691 | 0.0256112 | Down | 0.00543635 | |
| mmu-let-7i-star_st | −1.51651 | 0.0849839 | Down | 0.0297218 | |
| hp_mmu-mir-128-2_st | 1.5022 | 0.112589 | up | 0.045064 | |
| hp_mmu-mir-5112_st | 1.50223 | 0.0196402 | up | 0.00415309 | |
| hp_mmu-mir-466c-1_x_st | 1.62092 | 0.162286 | up | 0.0463585 | |
| hp_mmu-mir-101a_x_st | 1.66217 | 0.129606 | up | 0.0306606 | |
| hp_mmu-mir-466b-4_x_st | 1.7585 | 0.158651 | up | 0.0303406 | |
| mmu-miR-705_st | 1.89157 | 0.269444 | up | 0.0436911 | |
| hp_mmu-mir-5130_st | 1.96821 | 0.127422 | up | 0.013874 | |
| mmu-miR-1949_st | 2.02064 | 0.0914098 | up | 0.00783574 | |
| mmu-miR-5099_st | 2.15647 | 0.30996 | up | 0.0324799 | |
| mmu-miR-328-star_st | 2.46223 | 0.335471 | up | 0.0237448 | |
| mmu-miR-5130_st | 2.57367 | 0.653544 | up | 0.0492977 | |
| mmu-miR-744_st | 2.59709 | 0.0620819 | up | 0.00185233 | |
| mmu-miR-714_st | 2.65014 | 0.567341 | up | 0.0383118 | |
| mmu-miR-346-star_st | 2.87902 | 0.78242 | up | 0.0466927 | |
| mmu-miR-1896_st | 2.91753 | 0.789354 | up | 0.0457703 | |
| mmu-miR-1982-star_st | 3.12965 | 0.771611 | up | 0.0379458 | |
| mmu-miR-2137_st | 3.14534 | 0.670176 | up | 0.0313255 | |
| mmu-miR-3104-5p_st | 3.44502 | 1.09737 | up | 0.048136 | |
| mmu-miR-1893_st | 4.59124 | 0.681606 | up | 0.01495 | |
| mmu-miR-711_st | 9.14388 | 3.24 | up | 0.0435551 | |
Note: Exp: grafts with 18 h cold-ischemia and reperfusion; Control: grafts without 18 h cold ischemia.
Figure 2. miRNA expression in heart grafts.
Donor hearts were treated and transplanted into syngeneic recipient C57BL/6 mice as described in Figure 1. miRNAwere extracted and detected by miRNA array or qPCR at day 2 after transplantation. (A) A heat map of miRNA microarrays. (B)A pie graph of miRNA distribution. (C). Altered miRNA with its fold change.
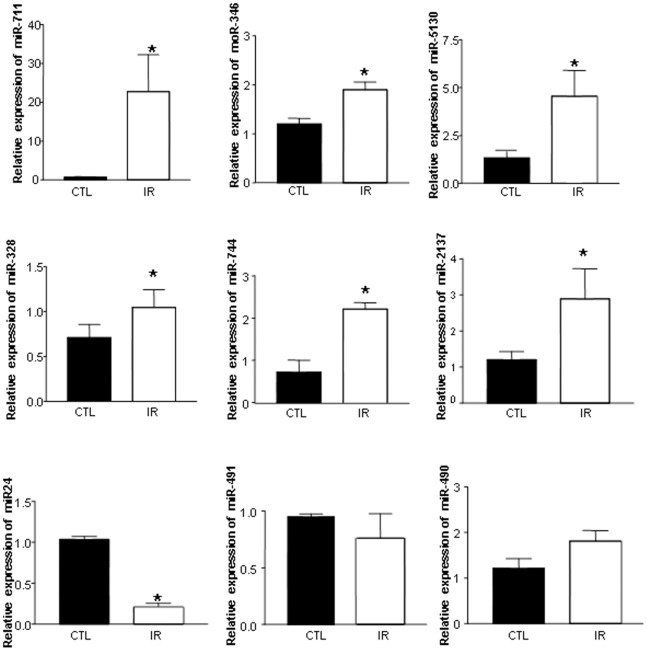
We further confirmed the expression of miRNA selected from miRNA microarray results by quantitative PCR (Figure S1). The expressions of miR-711, miR-714, miR-744, miR-2137, miR-5130, miR-1892, miR-328, miR-346, miR-5099, and miR-705 were significantly upregulated in I/R injured heart grafts, while miR-490, miR-491, miR-210, miR-362, miR-24, miR-423, miR-128, miR-328, miR -181, and miR-532 were downregulated. According to intensity of fluorescence detected on miRNA microarray and Ct values of qPCR, the expression levels of these miRNA in heart tissues varied to a great extent between each other. For example, miR-2137, miR-5130 and miR-5112 were highly expressed in heart tissues; miR-490, miR-491, miR-181, miR-362, miR-425, and miR-3104 were expressed at quite a low level (Ct value ∼ over 30), whereas 32 out of those 58 altered miRNA were expressed at an extremely low level in hearts and there were almost no Ct value detected by qPCR.
miRNA Expression Profile was Dynamically Changed in Heart Grafts
Literature has reported that miRNA expression changes dynamically [7]. Accordingly, we investigated whether miRNA expression changes over time in hearts post transplantation. We extracted grafts with I/R and with non-I/R at day 7 after transplantation and detected the expression level of miRNAs (miR-711, miR -714, miR-744, miR -2137, miR -5130, miR -346, miR -490, miR -491, miR -24, and miR -328). As shown in Figure 3, the expressions of miR-711, miR-714, miR-744, miR -2137, miR -5130, miR -346, and miR -328 was still upregulated in the I/R group, but the expression of miR-24 and miR-490 were downregulated, compared to the control group grafts. The expression of miR-491 was slightly but not significantly upregulated in I/R injured grafts. Compared with grafts taken out on day 2 post-transplantation, the expression of miR-2137, miR-714, miR-744, miR -2137, miR -5130, miR -346, and miR -328 were slightly decreased, whilst the expression of miR-711 continued its upregulation in the I/R injured grafts.
Figure 3. miRNA expression in heart grafts on day 7 post transplantation.
Donor hearts were treated and transplanted into syngeneic recipient C57BL/6 mice as described in Figure 1. miRNA were extracted and detected by qPCR on day 7 after transplantation.
Hypoxia/Reperfusion Incurred Cardiomyocyte Apoptosis and miRNA Expression Change In Vitro
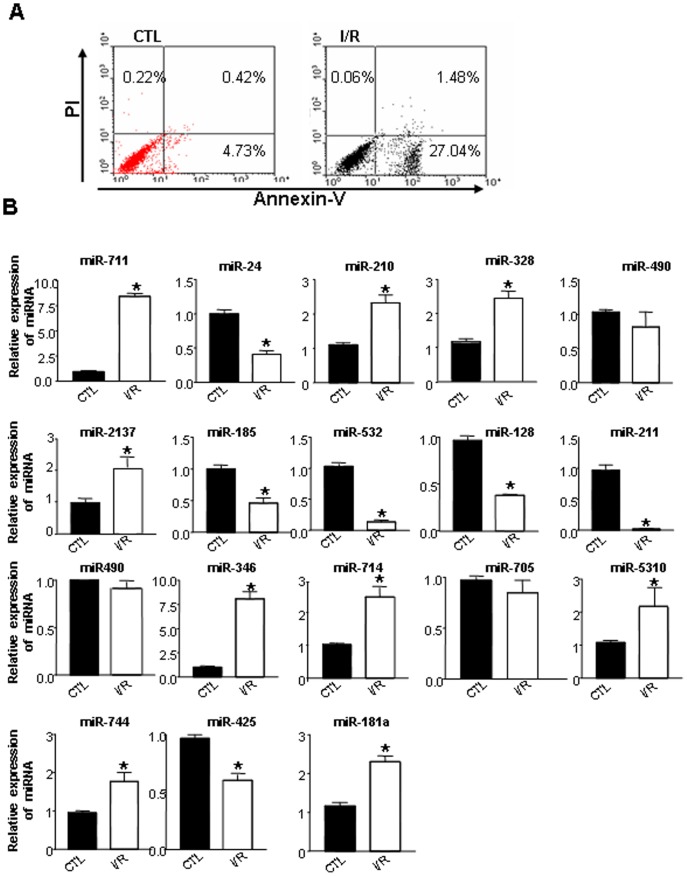
Cardiomyocyte is a main component of heart tissue. Given that prolonged cold ischemia changed miRNA expression profiles in heart grafts, we proposed to determine what the miRNA profile would be in cardiomyocytes in response to hypoxia/reperfusion stress in vitro and whether it was in line with that from heart tissues in vivo. We isolated and cultured primary cardiomyocytes from neonatal mice and subjected them to hypoxia/reperfusion stress by exposing them to a 2% O2 chamber for 45 mins and then reoxygenation for 24 h by incubation of cells in a 5% CO2 and 95% O2 incubator. We first measured cell apoptosis/death by double staining with Annexin-V and PI to confirm cell injury induced by hypoxia/reperfusion. Results showed that 45 mins hypoxia/reperfusion induced cardiomyocyte apoptosis evidenced by the increased percentage of apoptotic cells (Figure 4A) in hypoxia/reperfusion treated cells as compared with cells without hypoxia stress.
Figure 4. Hypoxia induced apoptosis of primary cardiomyocytes and altered miRNA expression in primary cardiomyocytes.
Primary cardiomyocytes were isolated from neonatal C57BL/6 mice cultured in vitro and then subjected to a hypoxia environment. Cell apoptosis was detected by staining with Annexin–V and followed by flow cytometry analysis 24 h after hypoxia. miRNAs were extract from above cells and the expression of miRNA were detected by qPCR.
Next, we measured the expression of miRNA in hypoxia/reperfusion -treated cardiomyocytes by qPCR. As compared with cells under normxia, miR-711, miR-714, miR-328, miR-346, miR-210, miR-744, miR-5130, miR-181a and miR-2137 were significantly over-expressed in hypoxia/reperfusion treated cardiomyocytes, while the expression of miR-491, miR-211, miR-532, miR-185, miR-425, miR-128, miR-24 was down-regulated (Figure 4B). There was no significant difference in the expression of miR-490 between the two groups (Figure 4B). As expected, miR-2137, miR-210, miR-5130, and miR-328 were highly expressed in cardiomyocytes, while miR-490, miR-491, and miR-211 were expressed at a low level.
Prolonged Cold I/R Changed Gene Expression Profile and Signalling Pathways in Heart Grafts
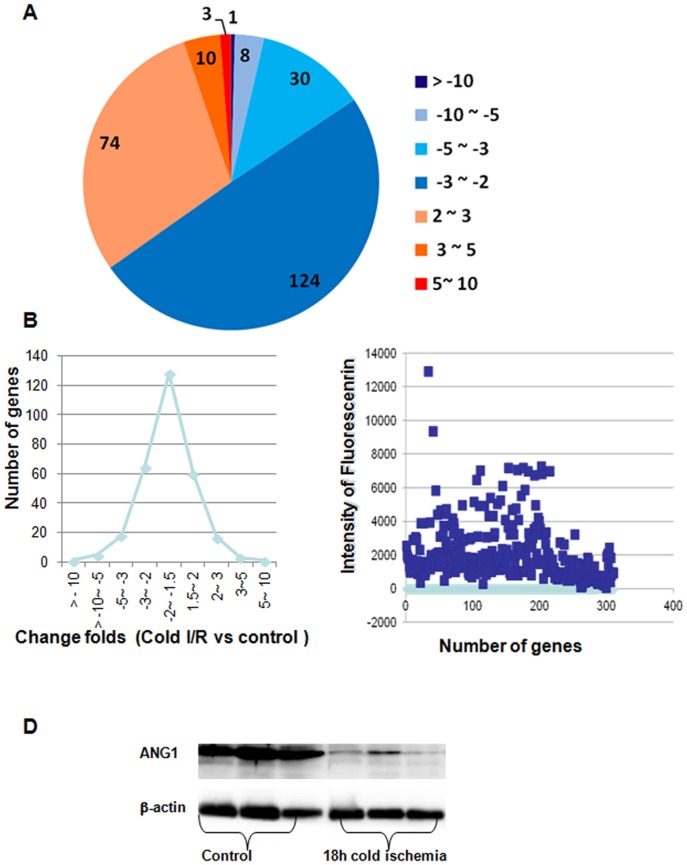
miRNA functions as a negative regulator of gene expression. Similarly, using microarray assay, we investigated the global gene expression changes in I/R injured heart grafts. Heart grafts with or without prolonged ischemia were harvested at day 2 after implantation and subjected to gene expression microarray assay (Table S1). I/R greatly altered gene expression profiles in heart grafts as shown in a heat map. The expression for the majority of significant altered genes (P<0.05) was increased or decreased 1.5–2 fold between two groups and also fits in a Poisson distribution (Figure 5A and 5B). Most of the altered genes were expressed in heart tissue at a moderate level with fluorescence intensities of around 1000–2000 units in microarray (Figure 5C). Given a three-fold change and P<0.05 (up and down) in differential expression as a cut-off, the number of altered genes was reduced to 48; 36 of them were downregulated, and 12 genes were up regulated (Table 2). Among them, Angiopoietin 1(ANG1) decreased to the greatest degree, which was confirmed by the Western blot (Figure 5D).
Figure 5. Gene expression in heart grafts detected by microarray assays.
Donor hearts were treated and transplanted into syngeneic C57BL/6 recipient mice. At day 2 post transplantation, total RNAs were extracted from grafts and gene expression in grafts were detected by microarray assays. (A) A Poisson distribution of gene expression. (B) Fluorescence intensities of altered genes detected by microarray assays. (C) Altered genes with two fold changes. (D) Angiopoietin 1 expression by Western Blotting.
Table 2. Annotated genes regulated in mouse heart transplants.
| Gene | p-value | Fold-Change (Exp vs. Control) | Function annotation |
| Angpt1 | 0.008529 | −19.3306 | receptor binding |
| Asb15 | 0.0182505 | −7.53081 | |
| Gpr22 | 0.0024004 | −6.50553 | signal transducer activity |
| Lrrc10 | 0.0475174 | −5.97936 | |
| Klhl38 | 0.0359302 | −5.9631 | |
| Grm1 | 0.0210762 | −5.86236 | PLC activating G-protein coupled glutamate receptor activity |
| Il15 | 0.0122715 | −5.71981 | cytokine activity |
| Iigp1 | 0.0378235 | −5.64458 | nucleotide binding |
| Art1 | 0.0219665 | −5.48431 | NAD+ ADP-ribosyltransferase activity |
| Ucp3 | 0.0087787 | −4.93265 | oxidative phosphorylation uncoupler activity |
| Adh1 | 0.0416643 | −4.91006 | nucleotide binding |
| Mylk3 | 0.00744 | −4.62612 | nucleotide binding |
| Fam40b | 0.0144888 | −4.56915 | |
| Csdc2 | 0.0223918 | −4.44072 | nucleic acid binding |
| Tmtc1 | 0.0141671 | −4.27416 | |
| Kcnj3 | 0.0285984 | −4.2726 | ion channel activity |
| Car14 | 0.0368218 | −4.11809 | carbonate dehydratase activity |
| Asb2 | 0.0086771 | −4.10112 | |
| Tmem100 | 0.0122261 | −4.00128 | |
| Slit2 | 0.0351409 | −3.99089 | GTPase inhibitor activity |
| Tnni3k | 0.01648 | −3.90863 | nucleotide binding |
| Gm4951 | 0.0269588 | −3.81868 | no |
| Lpin1 | 0.0219087 | −3.66845 | RNA polymerase II transcription factor binding |
| Filip1l | 0.0181653 | −3.62773 | |
| Ppp1r3a | 0.0463544 | −3.49446 | protein serine/threonine phosphatase activity |
| Hfe2 | 0.0235755 | −3.47549 | protein binding |
| Slc2a4 | 0.0300101 | −3.47473 | transporter activity |
| Ppargc1a | 0.0280838 | −3.30582 | nucleotide binding |
| Mapk10 | 0.0245692 | −3.29025 | nucleotide binding |
| Slc38a3 | 0.0377629 | −3.24102 | L-histidine transmembrane transporter activity |
| Adck3 | 0.0251863 | −3.2227 | nucleotide binding |
| Pnmt | 0.0497843 | −3.13239 | phenylethanolamine N-methyltransferase activity |
| Fuca2 | 0.0231706 | −3.10097 | catalytic activity |
| Asb11 | 0.0384006 | −3.07244 | |
| Ube2ql1 | 0.0294376 | −3.04395 | nucleotide binding |
| Hfe | 0.0433132 | −3.00196 | protein binding |
| Trim16 | 0.0428384 | 3.10174 | DNA binding |
| Sfn | 0.0394142 | 3.14658 | protein binding |
| Tnfrsf23 | 0.0115847 | 3.26603 | receptor activity |
| Apln | 0.0152681 | 3.36778 | receptor binding |
| Cyr61 | 0.0336177 | 3.38045 | integrin binding |
| Pgf | 0.0303102 | 3.53196 | growth factor activity |
| Loxl4 | 0.0059025 | 3.71021 | protein-lysine 6-oxidase activity |
| Prnd | 0.0159526 | 4.75449 | copper ion binding |
| Gdf15 | 0.0093257 | 4.8554 | growth factor activity |
| Fosl1 | 0.0168812 | 5.48284 | DNA binding |
| Plk2 | 0.0104977 | 5.50407 | nucleotide binding |
| Crct1 | 0.01037 | 7.60055 | protein binding |
Note: Exp: grafts with 18h cold-ischemia and reperfusion;Control: grafts without 18 h cold ischemia.
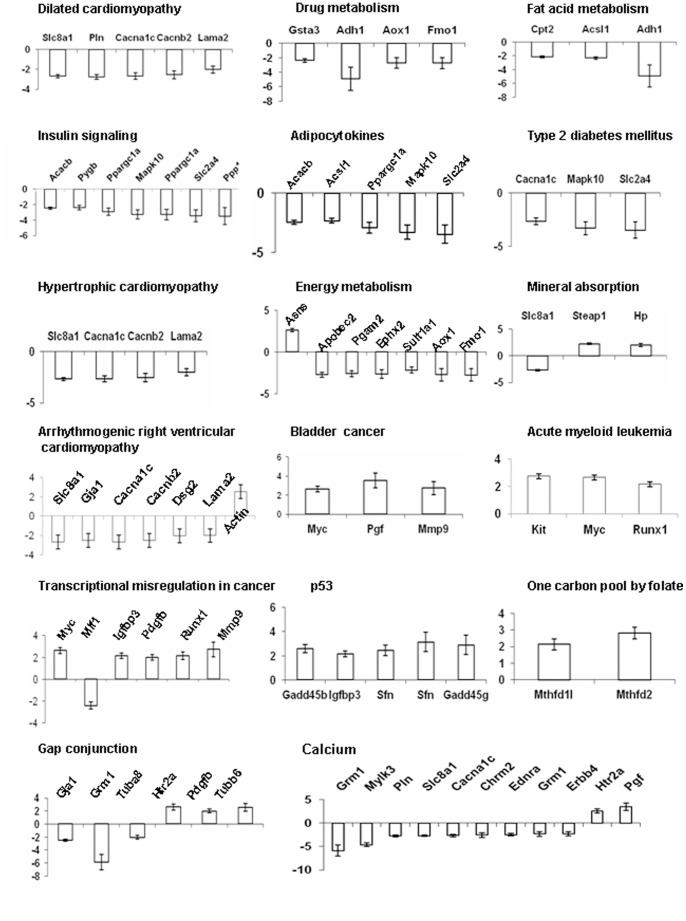
To clarify which signalling pathways were affected by I/R in heart transplantation, we applied the KEGG library and performed enrichment analysis for microarray data. Eighteen signalling pathways were enriched with the criteria of 2 fold changes and p<0.05 (Table 3), which include the Insulin, Tyrosine metabolism, cell cycle, gap junction, calcium, energy metabolism, p53, cardio function associated signalling pathways and some cancer signalling pathways. Less than 10% genes listed on those pathways significantly changed with more than 2 folds. Figure 6 shows the information of these genes from the 18 altered signalling pathways listed on Table 3. Most genes from those pathways were downregulated, except genes in the p53 and one carbon pool by folate, bladder cancer, acute myeloid leukemia and transcriptional misregulation in cancer pathways were upregulated. The Gap junction pathway evenly consisted of both up and downregulated genes.
Table 3. Enriched signalling pathways regulated in heart transplantation.
| Pathway Name | EnrichmentScore | p-value Enrichment | % genesa | # genesb | # genesc | # genesd | Database |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 9.7642 | 5.75E-05 | 9.58904 | 7 | 89 | 6829 | kegg |
| Calcium signaling pathway | 8.48247 | 0.000207 | 5.40541 | 10 | 86 | 6720 | kegg |
| Gap junction | 6.56392 | 0.00141 | 6.66667 | 6 | 90 | 6811 | kegg |
| Adipocytokine signaling pathway | 5.81675 | 0.002977 | 6.94444 | 5 | 91 | 6828 | kegg |
| Dilated cardiomyopathy | 4.86527 | 0.00771 | 5.55556 | 5 | 91 | 6810 | kegg |
| Energy Metabolism | 4.60156 | 0.010036 | 4 | 7 | 89 | 6727 | kegg |
| p53 signaling pathway | 4.22517 | 0.014623 | 5.7971 | 4 | 92 | 6830 | kegg |
| Insulin signaling pathway | 4.17598 | 0.01536 | 4.08163 | 6 | 90 | 6754 | kegg |
| Tyrosine metabolism | 3.99802 | 0.018352 | 7.31707 | 3 | 93 | 6857 | kegg |
| Bladder cancer | 3.81036 | 0.02214 | 6.81818 | 3 | 93 | 6854 | kegg |
| Hypertrophic cardiomyopathy(HCM) | 3.6154 | 0.026906 | 4.81928 | 4 | 92 | 6816 | kegg |
| One carbon pool by folate | 3.59657 | 0.027418 | 10.5263 | 2 | 94 | 6878 | kegg |
| Mineral absorption | 3.52912 | 0.029331 | 6.12245 | 3 | 93 | 6849 | kegg |
| Fatty acid metabolism | 3.477 | 0.0309 | 6 | 3 | 93 | 6848 | kegg |
| Drug metabolism - cytochromeP450 | 3.46478 | 0.03128 | 4.5977 | 4 | 92 | 6812 | kegg |
| Type II diabetes mellitus | 3.28047 | 0.037611 | 5.55556 | 3 | 93 | 6844 | kegg |
| Transcriptional misregulationin cancer | 3.27588 | 0.037784 | 3.31492 | 6 | 90 | 6720 | kegg |
| Acute myeloid leukemia | 3.01689 | 0.048953 | 5 | 3 | 93 | 6838 | kegg |
note: a% genes in pathway that are present;
numbers of genes in list and in the pathway;
numbers of genes in list, not in pathway;
numbers of genes not in list, not in pathway.
Figure 6. Genes in 18 altered signalling pathways.
Gene expression in the heart grafts was detected by microarrays as describe in Figure 5. Signalling pathways were enriched using the Kegg library with 2 fold change and p<0.05.
Discussion
I/R injury is recognized as a primary factor leading to graft dysfunction [8] and graft failure [9], [10]. It is important to understand molecular mechanisms of I/R injury for the development of therapies against I/R injury. The current study, for the first time, reported on the expression profile of miRNA in I/R injured heart grafts in heart transplantation. The findings of our study demonstrate that miR-711, miR-2137 miR-705, miR-5130, miR-346, miR-714, and miR-744 were significantly upregulated (>2 fold change) in I/R injured hearts, while miR-210, miR-490, miR-491, miR-425, miR-423-3p, and miR-532-3p were downregulated. The study also demonstrates that 250 genes and 18 signalling pathways were significantly altered with more than 2 fold changes by I/R injury by cDNA microarray assay.
microRNAs have emerged as a vital regulator in many physiological and pathological pathways. Deregulation of miRNAs associated with different forms of ischemia reperfusion injury [11]. A substantial number of miRNA including miR-1 [12], miR-15 [13], miR-21 [14], [15], miR-24 [16], [17], [18], miR-499 [19], and the miR-17-92 family [20], miR-124 [21], miR-15a/b [22] (2012), miR-93 [23], miR-29 family [24], [25], miR-146a [26], [27], miR-145/451 [28], miR-384-5p [29], miR-424 [30], and miR-494 [31] have been identified in I/R injury. It has been demonstrated that ischemia precondition (IPC) increases cardiac expression of miRNA-1, miRNA-21 and miRNA-24. IPC-regulated miRNA-21 reduces cell apoptosis by repressing the programmed cell death 4 (PDCD4) gene, which results in preventing the heart from I/R injury [32]. It has also been reported that inhibition of miR-15 prevents cardiac ischemia injury in a porcine coronary arterial ligation model [33]. However, in this study we did not observe the above miRNAs change in heart grafts. The reason for this might be that miRNA expression changes dynamically and miRNA expression varies between different models [34], [35].
In this study, we observed that miR-711 was significantly up-regulated both in I/R injured heart grafts and hypoxia/reperfusion treated primary cardiomyocytes. Despite only a few studies of miR-711 have been reported, available data have shown that miR-711 is expressed in many types of cells [36], [37], [38] and is upregulated under different stresses [39], [40]. For example, a chemical palmitate used to induce insulin resistance increases the expression of miR-711 in mouse muscle C2C12 cells [41]. A study has also shown that miR-711 was significantly upregulated in the myocardium with acute myocardial infarction on day 14 post ischemia [42]. Tranter et al reported shows that cardiac ischemic preconditioning (IPC) of the in vivo mouse heart results in decreased levels of miR-711 which was dependent on NF-kB, and that miR-711 post-transcriptionally suppresses Hsp70.3 [43]. A more recent study reported that Pioglitazone (an insulin sensitizing drug with cardio protective effect, it attenuates cardiac fibrosis) increased miR-711 levels in myocardial infarction rats and miR-711 directly targeted and downregulated SP1, leading to reduced collagen-I levels [44]. However, the expression level of SP1 and HSP70 was not altered in I/R injured heart tissues in this study (data not shown). It may be attributed to differences in animal model and injury. Predicted by TargetScan and FINDTAR3, Angiopoietin 1 (ANG1) is a putative target of miR-711. Our data also showed that ANG1 was significantly downregulated in I/R injured hearts and hypoxia-treated cardiomyocytes, suggesting ANG1 might be a target of miR-711. Supportively, Lee et al [45] demonstrated that ANG1 can exert cardio protective effects by preventing vascular leakage and cardiomyocyte death by inhibiting activities of Caspase 3 and Caspase 9. Further study on miR-711 function will help us understand the regulatory roles of miR-711.
Additionally, miR-2137, miR-1893, miR-744, miR-705 and miR-714 are highly expressed in heart tissue and cardiomyocytes as well, suggesting that these miRNA are important for cardiomyocytes survival and growth. However, there are no reports available on miR-2137 and 1893 regarding their functions. Performing computational analysis using TargetScan, two conserved genes, retrograde golgi transport homolog (RGP1) and pleckstrin and Sec7 domain containing (Psd), were predicted as targets of miR-2137, while another 144 genes including calmodulin binding transcription activator 1(Camta1) and furry homolog-like (Fryl) were predicted irrespective of site conservation. The data from the gene expression microarray assay showed that the Camta1 gene was significantly downregulated in I/R injured grafts. It has been shown that CAMTA 1 can activate the expression of the anti-proliferative cardiac hormone natriuretic peptide A (NPPA) in the heart and is recognised as a tumor suppressor [46]. Another family member of CAMTA, the transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases, while the loss of CAMTA2 promotes cardiomyocyte hypertrophy [47]. Taken together, the increased expression of miR-2137 may play a role in I/R injury in heart transplantation through regulating CAMTA 1.
miR-705, miR-714 and miR-744 have not been extensively investigated yet. It has been reported that the expression of miR-714 and miR-744 are significantly higher in mice aorta with vascular calcification [48]. miR-744 is known to be expressed in cardiac valves [49] and involved in cancer cell growth and proliferation [50], [51]. Recent studies have shown that miR-744 targets TGF-β and eukaryotic translation elongation factor 1 alpha 2(eEF1A2) which is able to promote cell growth and inhibit apoptosis [52], [53], [54]. Our data showed that eEF1A2 was decreased in I/R injured hearts, implying that there may be a causative relationship between miR-744 and eEF1A2. Predicted by TargetScan, miR-705 has 89 putative targets including Transmembrane BAX inhibitor motif containing 1 (TMBIM1) with conservative sites. TMBIM1 has been well documented as a part of the Bax Inhibitor-1(BI-1) family that has a similar anti-apoptotic function as Bcl-2 and miR-705 induced inactivation of TMBIM1 may be responsible for the increase rate of apoptosis after I/R injury [55]. Although our cDNA microarray data did not show significant changes of TMBIM1 expression at the mRNA level, we can not rule out TMBIM1 as a putative target of miR-705 because that miRNA functions via translation repression as well. Our data also showed that miR-346 was highly expressed in normal heart tissues and upregulated by I/R. There are 90 putative targets of miR-346 predicted by the TargetScan. Most studies on miR-346 have been investigated in a rheumatoid arthritis model [56], [57]. The expression of miR-346 was positively correlated with the severity of ischemic injury in a mouse hepatic ischemia/reperfusion injury model [58]. It has been demonstrated that miR-346 can target receptor-interacting protein 140 (RIP140), TNFα, Leukemia inhibitory factor (LIF), IL18 and antigen peptide transporter 1 (TAP1) [56], [59], [60]. The expression of LIF was decreased in the I/R injured heart grafts (data not shown), indicating miR-346 may negatively regulate LIF.
miR-24 has previously shown a protective effect on I/R injury [16], [17], [18]. We observed a decrease in the expression of miR-24 in the I/R injured heart grafts but it was not significant. However, we did see a significant reduction of miR-24 in hypoxia-treated primary cardiomyocytes, which is consistent with reported literature [16], [17], [18]. Since an entire heart tissue consists of multiple lineages of cells such as cardiomyocyte, endothelial cell, and fibroblast cell, the expression of miR-24 in heart is an accumulation from all cells in the heart, and other cells may buffer the reduction of miR-24 in cardiomyocytes.
The data from gene expression microarray showed that 48 genes (36 downregulated and 12 up-regulated) were significantly changed with more than three fold in prolonged cold I/R. Among them, some genes have been demonstrated to be involved in the growth and function of cardiac cells. For examples, growth of Ankyrin repeat and suppressor of cytokine signaling box-containing protein (ASB) 15 regulates myoblast differentiation [61]; G protein-coupled receptors 22 (GPCRs 22) which are highly expressed in cardiac myocytes and coronary arteries plays an essential role in the regulation of cardiac contractile function and cardiomyocyte apoptosis [27] and Leucine-rich repeat containing 10 (Lrrc10), a cardiac-specific factor, is crucial for proper cardiac development. Deletion of Lrrc10 in mice results in dilated cardiomyopathy [62]. Furthermore, uncoupling protein 3(UCP3) [63], angiopoietin 1 [45] and growth differentiation factor-15 (GDF-15) [64] are anti-apoptotic genes and show protective effects on I/R injury in non-transplantation settings. Therefore, those altered genes might be good targets for the prevention of I/R injury in heart transplantation.
In conclusion, our study demonstrates that ischemia reperfusion injury in heart tissue after transplantation is associated with an altered miRNA profile, which will help us to understand roles of miRNA in I/R injury. In addition, this study provides insights into the mechanisms involved in I/R injury and investigate specific miRNA that regulate genes associated with signalling pathways involved in I/R injury. As miRNAs hold great potential as therapeutic targets, expression levels can be modified to prevent, or revert ischemia reperfusion injury that occurs during heart transplantation - minimizing graft failure and increasing graft long-term survival rate.
Supporting Information
miRNA expression detected by qPCR. miRNA was extracted from heart grafts at day 2 post transplantation as described in Figure 2. cDNA was synthesized using miRScript II RT Kit. The expression of miRNA was detected by qPCR using SYBRGreen systems.
(TIF)
Gene expression with two fold changes and p<0.05 in the heart grafts of I/R injury vs.non-IR, detected by gene expression microarray assays.
(XLS)
Acknowledgments
We would like to thank Winnie Liu for assistance with histology sectioning and David Carter for microarray assays.
Funding Statement
This study was supported by the Heart and Stroke Foundation of Canada; Lawson Health Research Institute; and Multiple Organ Transplant Program. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Barker WH, Mullooly JP, Getchell W (2006) Changing incidence and survival for heart failure in a well-defined older population, 1970–1974 and 1990–1994. Circulation 113: 799–805. [DOI] [PubMed] [Google Scholar]
- 2. Klotz S, Scheld HH (2011) Surgical approach to end-stage heart failure. Curr Opin Anaesthesiol 24: 86–91. [DOI] [PubMed] [Google Scholar]
- 3. Subramaniam K (2012) Early graft failure after heart transplantation: prevention and treatment. Int Anesthesiol Clin 50: 202–227. [DOI] [PubMed] [Google Scholar]
- 4. Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A 104: 9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ye Y, Perez-Polo JR, Qian J, Birnbaum Y The role of microRNA in modulating myocardial ischemia-reperfusion injury. Physiol Genomics 43: 534–542. [DOI] [PubMed] [Google Scholar]
- 6. Lepic E, Burger D, Lu X, Song W, Feng Q (2006) Lack of endothelial nitric oxide synthase decreases cardiomyocyte proliferation and delays cardiac maturation. Am J Physiol Cell Physiol 291: C1240–1246. [DOI] [PubMed] [Google Scholar]
- 7. Kirklin JK, McGiffin DC (1999) Control of the inflammatory response in extended myocardial preservation of the donor heart. Ann Thorac Surg 68: 1978–1982. [DOI] [PubMed] [Google Scholar]
- 8. Slater JP, Amirhamzeh MM, Yano OJ, Shah AS, Starr JP, et al. (1995) Discriminating between preservation and reperfusion injury in human cardiac allografts using heart weight and left ventricular mass. Circulation 92: II223–227. [DOI] [PubMed] [Google Scholar]
- 9. Knight RJ, Dikman S, Liu H, Martinelli GP (1997) Cold ischemic injury accelerates the progression to chronic rejection in a rat cardiac allograft model. Transplantation 64: 1102–1107. [DOI] [PubMed] [Google Scholar]
- 10. Tanaka M, Mokhtari GK, Terry RD, Gunawan F, Balsam LB, et al. (2005) Prolonged cold ischemia in rat cardiac allografts promotes ischemia-reperfusion injury and the development of graft coronary artery disease in a linear fashion. J Heart Lung Transplant 24: 1906–1914. [DOI] [PubMed] [Google Scholar]
- 11.Fasanaro P, D’Alessandra Y, Magenta A, Pompilio G, Capogrossi MC (2013) MicroRNAs: promising biomarkers and therapeutic targets of acute myocardial ischemia. Curr Vasc Pharmacol. [DOI] [PubMed]
- 12. Pan Z, Sun X, Ren J, Li X, Gao X, et al. (2012) miR-1 exacerbates cardiac ischemia-reperfusion injury in mouse models. PLoS One 7: e50515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, et al. (2012) Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 110: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia P, Teng J, Zou J, Fang Y, Zhang X, et al.. (2013) miR-21 Contributes to Xenon-conferred Amelioration of Renal Ischemia-Reperfusion Injury in Mice. Anesthesiology. [DOI] [PMC free article] [PubMed]
- 15. Qin Y, Yu Y, Dong H, Bian X, Guo X, et al. (2012) MicroRNA 21 inhibits left ventricular remodeling in the early phase of rat model with ischemia-reperfusion injury by suppressing cell apoptosis. Int J Med Sci 9: 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brody MJ, Cho E, Mysliwiec MR, Kim TG, Carlson CD, et al. Lrrc10 is a novel cardiac-specific target gene of Nkx2–5 and GATA4. J Mol Cell Cardiol. [DOI] [PMC free article] [PubMed]
- 17. Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, et al. (2011) miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med 208: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song YR, Liu Z, Gu SL, Qian LJ, Yan QF (2011) [Advances in the molecular pathogenesis of hypertrophic cardiomyopathy]. Yi Chuan 33: 549–557. [DOI] [PubMed] [Google Scholar]
- 19. Qin H, Chen GX, Liang MY, Rong J, Yao JP, et al. The altered expression profile of microRNAs in cardiopulmonary bypass canine models and the effects of mir-499 on myocardial ischemic reperfusion injury. J Transl Med 11: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou M, Cai J, Tang Y, Zhao Q (2013) MiR-17–92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses 81: 108–110. [DOI] [PubMed] [Google Scholar]
- 21.Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, et al.. (2013) MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. [DOI] [PubMed]
- 22. Liu LF, Liang Z, Lv ZR, Liu XH, Bai J, et al. (2012) MicroRNA-15a/b are up-regulated in response to myocardial ischemia/reperfusion injury. J Geriatr Cardiol 9: 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hazarika S, Farber CR, Dokun AO, Pitsillides AN, Wang T, et al. (2013) MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation 127: 1818–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R (2013) MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS One 8: e58039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye Y, Hu Z, Lin Y, Zhang C, Perez-Polo JR (2010) Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc Res 87: 535–544. [DOI] [PubMed] [Google Scholar]
- 26. Chen Q, Kong L, Xu X, Geng Q, Tang W, et al. (2013) Down-regulation of microRNA-146a in the early stage of liver ischemia-reperfusion injury. Transplant Proc 45: 492–496. [DOI] [PubMed] [Google Scholar]
- 27. Adams JW, Wang J, Davis JR, Liaw C, Gaidarov I, et al. (2008) Myocardial expression, signaling, and function of GPR22: a protective role for an orphan G protein-coupled receptor. Am J Physiol Heart Circ Physiol 295: H509–521. [DOI] [PubMed] [Google Scholar]
- 28. Wang X, Zhu H, Zhang X, Liu Y, Chen J, et al. (2012) Loss of the miR-144/451 cluster impairs ischaemic preconditioning-mediated cardioprotection by targeting Rac-1. Cardiovasc Res 94: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bao Y, Lin C, Ren J, Liu J (2013) MicroRNA-384-5p regulates ischemia-induced cardioprotection by targeting phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta (PI3K p110delta). Apoptosis 18: 260–270. [DOI] [PubMed] [Google Scholar]
- 30. Zhao H, Wang J, Gao L, Wang R, Liu X, et al. (2013) MiRNA-424 Protects Against Permanent Focal Cerebral Ischemia Injury in Mice Involving Suppressing Microglia Activation. Stroke 44: 1706–1713. [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Zhang X, Ren XP, Chen J, Liu H, et al. MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation 122: 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yin C, Salloum FN, Kukreja RC (2009) A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res 104: 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 110: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, et al. (2009) MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J 50: 377–387. [DOI] [PubMed] [Google Scholar]
- 35. Ren XP, Wu J, Wang X, Sartor MA, Qian J, et al. (2009) MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 119: 2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DG, et al. (2010) Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol 185: 3835–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, et al. (2011) Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 118: 5891–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Opalinska JB, Bersenev A, Zhang Z, Schmaier AA, Choi J, et al. (2010) MicroRNA expression in maturing murine megakaryocytes. Blood 116: e128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sadoshima YYNNJ (2009) miR-206 Mediates Yap-induced Cardiac Hypertrophy: A Component of the Mammalian Hippo Pathway Regulating Cardiac Hypertrophy Circulation. 120: S758. [Google Scholar]
- 40. Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, et al. (2009) Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 106: 4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li ZY, Na HM, Peng G, Pu J, Liu P (2011) Alteration of microRNA expression correlates to fatty acid-mediated insulin resistance in mouse myoblasts. Mol Biosyst 7: 871–877. [DOI] [PubMed] [Google Scholar]
- 42. Shi B, Guo Y, Wang J, Gao W (2010) Altered expression of microRNAs in the myocardium of rats with acute myocardial infarction. BMC Cardiovasc Disord 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, et al. (2011) Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem 286: 29828–29837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao N, Yu H, Sun M, Zhang Y, Xu M, et al. (2013) MiRNA-711-SP1-collagen-I pathway is involved in the anti-fibrotic effect of pioglitazone in myocardial infarction. Sci China Life Sci 56: 431–439. [DOI] [PubMed] [Google Scholar]
- 45. Lee SW, Won JY, Lee HY, Lee HJ, Youn SW, et al. Angiopoietin-1 protects heart against ischemia/reperfusion injury through VE-cadherin dephosphorylation and myocardiac integrin-beta1/ERK/caspase-9 phosphorylation cascade. Mol Med 17: 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schraivogel D, Weinmann L, Beier D, Tabatabai G, Eichner A, et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J 30: 4309–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song K, Backs J, McAnally J, Qi X, Gerard RD, et al. (2006) The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125: 453–466. [DOI] [PubMed] [Google Scholar]
- 48. Gui T, Zhou G, Sun Y, Shimokado A, Itoh S, et al. (2012) MicroRNAs that target Ca(2+) transporters are involved in vascular smooth muscle cell calcification. Lab Invest 92: 1250–1259. [DOI] [PubMed] [Google Scholar]
- 49. Vacchi-Suzzi C, Hahne F, Scheubel P, Marcellin M, Dubost V, et al. (2013) Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One 8: e52442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang V, Place RF, Portnoy V, Wang J, Qi Z, et al. (2012) Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song MY, Pan KF, Su HJ, Zhang L, Ma JL, et al. (2012) Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One 7: e33608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin J, Jenkins RH, Bennagi R, Krupa A, Phillips AO, et al. (2011) Post-transcriptional regulation of Transforming Growth Factor Beta-1 by microRNA-744. PLoS One 6: e25044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vislovukh A, Kratassiouk G, Porto E, Gralievska N, Beldiman C, et al. (2013) Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer 108: 2304–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Z, Qi CF, Shin DM, Zingone A, Newbery HJ, et al. (2010) Eef1a2 promotes cell growth, inhibits apoptosis and activates JAK/STAT and AKT signaling in mouse plasmacytomas. PLoS One 5: e10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reimers K, Choi CY, Bucan V, Vogt PM (2008) The Bax Inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med 8: 148–156. [DOI] [PubMed] [Google Scholar]
- 56. Semaan N, Frenzel L, Alsaleh G, Suffert G, Gottenberg JE, et al. (2011) miR-346 controls release of TNF-alpha protein and stability of its mRNA in rheumatoid arthritis via tristetraprolin stabilization. PLoS One 6: e19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA (2012) miRNAs and related polymorphisms in rheumatoid arthritis susceptibility. Autoimmun Rev 11: 636–641. [DOI] [PubMed] [Google Scholar]
- 58. Xu CF, Yu CH, Li YM (2009) Regulation of hepatic microRNA expression in response to ischemic preconditioning following ischemia/reperfusion injury in mice. OMICS 13: 513–520. [DOI] [PubMed] [Google Scholar]
- 59. Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, et al. (2011) The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem 286: 41862–41870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsai NP, Lin YL, Wei LN (2009) MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J 424: 411–418. [DOI] [PubMed] [Google Scholar]
- 61. McDaneld TG, Spurlock DM (2008) Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 alters differentiation of mouse C2C12 myoblasts and phosphorylation of mitogen-activated protein kinase and Akt. J Anim Sci 86: 2897–2902. [DOI] [PubMed] [Google Scholar]
- 62.Brody MJ, Cho E, Mysliwiec MR, Kim TG, Carlson CD, et al.. (2012) Lrrc10 is a novel cardiac-specific target gene of Nkx2–5 and GATA4. J Mol Cell Cardiol. [DOI] [PMC free article] [PubMed]
- 63. Ozcan C, Palmeri M, Horvath TL, Russell KS, Russell RR 3rd (2013) Role of uncoupling protein 3 in ischemia-reperfusion injury, arrhythmias, and preconditioning. Am J Physiol Heart Circ Physiol 304: H1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heger J, Schiegnitz E, von Waldthausen D, Anwar MM, Piper HM, et al. (2013) Growth differentiation factor 15 acts anti-apoptotic and pro-hypertrophic in adult cardiomyocytes. J Cell Physiol 224: 120–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miRNA expression detected by qPCR. miRNA was extracted from heart grafts at day 2 post transplantation as described in Figure 2. cDNA was synthesized using miRScript II RT Kit. The expression of miRNA was detected by qPCR using SYBRGreen systems.
(TIF)
Gene expression with two fold changes and p<0.05 in the heart grafts of I/R injury vs.non-IR, detected by gene expression microarray assays.
(XLS)