Abstract
Coral disease is a global problem. Diseases are typically named or described based on macroscopic changes, but broad signs of coral distress such as tissue loss or discoloration are unlikely to be specific to a particular pathogen. For example, there appear to be multiple diseases that manifest the rapid tissue loss that characterizes ‘white plague.’ PhyloChip™ G3 microarrays were used to compare the bacterial community composition of both healthy and white plague-like diseased corals. Samples of lobed star coral (Orbicella annularis, formerly of the genus Montastraea [1]) were collected from two geographically distinct areas, Dry Tortugas National Park and Virgin Islands National Park, to determine if there were biogeographic differences between the diseases. In fact, all diseased samples clustered together, however there was no consistent link to Aurantimonas coralicida, which has been described as the causative agent of white plague type II. The microarrays revealed a large amount of bacterial heterogeneity within the healthy corals and less diversity in the diseased corals. Gram-positive bacterial groups (Actinobacteria, Firmicutes) comprised a greater proportion of the operational taxonomic units (OTUs) unique to healthy samples. Diseased samples were enriched in OTUs from the families Corynebacteriaceae, Lachnospiraceae, Rhodobacteraceae, and Streptococcaceae. Much previous coral disease work has used clone libraries, which seem to be methodologically biased toward recovery of Gram-negative bacterial sequences and may therefore have missed the importance of Gram-positive groups. The PhyloChip™data presented here provide a broader characterization of the bacterial community changes that occur within Orbicella annularis during the shift from a healthy to diseased state.
Introduction
Coral disease is now recognized as one of the major causes of reef degradation and coral mortality. The Caribbean has been particularly hard hit, with disease mortality reducing coral cover by an order of magnitude in some areas [2], [3]. A frustrating aspect of trying to manage or prevent disease is that corals have a limited number of ways to visually manifest distress (e.g., discoloration, tissue loss) and as such, the macroscopic signs that ‘define’ a disease actually have minimal diagnostic value. Using tissue loss to characterize a coral disease is on the order of characterizing a human disease solely by the presence of a fever. Being unable to clearly distinguish one disease from another, or link macroscopic signs to specific microbial pathogens, makes it difficult, if not impossible, to develop a plan for containment or prevention.
White plague is a perfect example. This disease is visibly ‘identified’ by a sharp boundary between pigmented tissue and bare skeleton that is bright white due to recent tissue loss [4], [5]. In the Caribbean, three distinct ‘types’ (WPI, WPII, WPIII) have been described, differentiated only by the progression speed of the lesion [4], [6]. One problem is that most microbial studies have collected samples at a single time point without any information about the progression speed. Therefore, each could be describing a different ‘type,’ while combining the microbial results under the single disease name. To further underscore the point that this macroscopic symptom is not diagnostic enough, Red Sea corals that appeared to have white plague were found by microbiology and histology to be suffering from an atypical black band disease [7]. Even when confined to Atlantic corals, there appear to be numerous white plague-like diseases [8]–[13]. Fortunately, researchers are beginning to realize the confusion this has created and it is becoming more common to either distinguish a disease as ‘white plague-like’ if no progression data exists, or give each outbreak a more descriptive name (e.g., “Tortugas multispecies rapid tissue loss disease” [8]) following the recommendations offered by Work and Aeby [14].
Although causative bacterial pathogens have been identified for two white plague diseases (Aurantimonas coralicida, for WPII in the Caribbean [15] and Thalassomonas loyana for white plague-like disease in the Red Sea [16]), it is increasingly believed that coral diseases arise from environmental stress triggering opportunistic pathogens rather than from primary pathogens [17], [18]. Therefore, the causative agent of a disease or syndrome may not always be the same, especially in different geographic areas. Many previous studies of coral disease have been culture-based [10], [19], [20], [21] or relied on clone libraries of a few hundred sequences [12], [22], [23], [24], both of which give a limited characterization of the microbial community diversity. Within the last five years, there has been a shift towards techniques that provide a more comprehensive view of the coral-associated microbial community in healthy and diseased corals, such as PhyloChip™ DNA microarrays [25], [26] and pyrosequencing [27]–[29]. The objective of this work was to use PhyloChip™ G3 microarrays to examine changes in the coral-associated bacterial communities between healthy Orbicella annularis (formerly of the genus Montastraea [1]) coral colonies and those affected by white-plague-like disease. The PhyloChip™ G3 (generation 3) microarray [30] provides a broad taxonomic overview of microbial communities. This microarray is printed with 1.1 million 16S rRNA gene probes representing nearly 60,000 operational taxonomic units (OTUs) that fit hierarchically into taxa from domain to species. This extensive coverage of known bacterial taxa allows detection of more bacterial richness than traditional clone library analysis. The large but standardized OTU inventory allows for easier analysis than an open-ended pyrosequencing dataset. Samples were compared from the Dry Tortugas and Virgin Islands to test whether the disease patterns clustered based on geographic areas, to look for diagnostic taxonomic shifts between healthy and diseased samples, and to see if either location’s disease was associated with Aurantimonas coralicida, the causative agent identified for White Plague Type II.
Materials and Methods
Ethics Statement
These collections were made under permits VIIS-2008-SCI-0033 (study VIIS-08033) and DRTO-2009-SCI-0018 (study DRTO-00074), granted to the first author by the Virgin Islands National Park and Dry Tortugas National Park, respectively. No ethical approval was required for the experimental research described here.
Sample Sites and Collections
Healthy and white plague-affected Orbicella annularis (lobed star coral) samples were collected in two National Parks during the summer of 2009. Five healthy and six diseased colonies were sampled at Tektite Reef in Great Lameshur Bay (18°18′46 N, 64°43′22 W), St. John, Virgin Islands National Park (VIIS) on July 16–17 (Table 1). Samples from five healthy and three diseased colonies were also collected from the ‘Little Africa’ patch reef (24°38′06 N, 82°55′15 W) in the Dry Tortugas National Park (DRTO) on August 5 (Table 1). Water temperature was 31–32°C with a salinity of 35 ppt at DRTO and 29°C with a salinity of 34 ppt at VIIS. Healthy corals were normally pigmented with no lesions or tissue loss. White plague-affected corals exhibited a sharp interface between normal tissue and freshly exposed skeleton.
Table 1. Orbicella annularis samples collected for microarray analysis.
| Sample ID | Location | Date Collected | Health State | Depth (m) |
| VIISMAP01 | Virgin Islands National Park | July 16, 2009 | white plague | 12 |
| VIISMAP02 | Virgin Islands National Park | July 16, 2009 | white plague | 12 |
| VIISMAP03 | Virgin Islands National Park | July 16, 2009 | white plague | 13 |
| VIISMAP04 | Virgin Islands National Park | July 17, 2009 | white plague | 8 |
| VIISMAP05 | Virgin Islands National Park | July 17, 2009 | white plague | 6 |
| VIISMAH06 | Virgin Islands National Park | July 17, 2009 | healthy | 9 |
| VIISMAH07 | Virgin Islands National Park | July 17, 2009 | healthy | 8 |
| VIISMAH09 | Virgin Islands National Park | July 17, 2009 | healthy | 5 |
| VIISMAH10 | Virgin Islands National Park | July 17, 2009 | healthy | 6 |
| VIISMAP11 | Virgin Islands National Park | July 17, 2009 | white plague | 7 |
| DRTOMAP01 | Dry Tortugas National Park | Aug 5, 2009 | white plague | 3 |
| DRTOMAP02 | Dry Tortugas National Park | Aug 5, 2009 | white plague | 2 |
| DRTOMAP03 | Dry Tortugas National Park | Aug 5, 2009 | white plague | 2 |
| DRTOMAH05 | Dry Tortugas National Park | Aug 5, 2009 | healthy | 2 |
| DRTOMAH06 | Dry Tortugas National Park | Aug 5, 2009 | healthy | 3 |
| DRTOMAH07 | Dry Tortugas National Park | Aug 5, 2009 | healthy | 3 |
| DRTOMAH08 | Dry Tortugas National Park | Aug 5, 2009 | healthy | 3 |
| DRTOMAH09 | Dry Tortugas National Park | Aug 5, 2009 | healthy | 3 |
A healthy sample designated VIISMAH08 was collected but no 16S rRNA gene PCR product was obtained, so it was not included in this table since it does not appear in any subsequent figures.
Note that in spite of extensive searching, the three samples of white plague-like disease collected in DRTO were the only ones encountered.
Corals were sampled using a bleach-sterilized metal punch of approximately 2-cm diameter to collect two biopsies of tissue with minimal skeletal material attached. Diseased corals were sampled at the lesion interface. Coral tissue samples were immediately placed into sterile Whirl-pak bags. The punch was cleaned and re-bleached between samples to prevent any transfer of microbes or contamination. Back on shore, the coral samples were briefly rinsed with sterile-filtered seawater to remove loosely associated microbes, wrapped in sterile aluminum foil, placed into a fresh Whirl-pak bag, and flash-frozen in liquid nitrogen. Samples were transported from the field to the USGS St. Petersburg Science Center and then transferred to a −80°C freezer for storage.
DNA Extraction
In the laboratory, each pair of frozen tissue samples from a single coral colony was ground to powder using a sterile mortar and pestle on dry ice and then combined. The microbial community DNA was extracted in duplicate from each combined sample using the MO BIO PowerPlant® DNA Isolation Kit with some modification as previously described by Sunagawa et al. [31]. At the end of the protocol, the replicate extractions were combined, resulting in a single microbial DNA extraction representing each coral colony sampled.
16S rRNA Gene Amplification
Amplification was carried out as previously described [32]. Primers targeting 16S bacterial ribosomal RNA genes were used: 27F with the degenerate base removed, sometimes also referred to as 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) [33] and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [34]. Eight replicate 25 µl amplifications were performed for each DNA extraction over a temperature gradient of 48–58°C. At each temperature, 2 µl of template DNA was used. Final concentrations for each 25 µl reaction were: 1× Ex Taq Buffer with 2 mM MgCl2, 300 nM each primer (27F and 1492R), 200 µM each dNTP, 25 µg bovine serum albumin and 0.625 units of Ex Taq polymerase. The reactions were amplified using an iCycler and under the following thermocycling conditions: 95°C for 3 min for initial denaturation, 30 cycles of 95°C for 30 sec, 48–58°C for 30 sec, and 72°C for 2 min, and then final extension for 10 min at 72°C. PCR products from each annealing temperature for a sample were combined and concentrated using Amicon Ultra-0.5, Ultracel-30 Membrane, 30 kDa centrifugal filter units and quantified before application to the PhyloChip™. The concentrated 1 µl PCR product was quantified on a 2% agarose E-gel using the Low Range Quantitative DNA Ladder as reference. In some cases the primers amplified both coral and bacterial DNA [35], requiring gel extraction of the bacterial amplicons for all samples to be consistent. The difference in size between the two products (i.e., coral and bacterial amplicons) was roughly 200 base pairs. To allow clear separation of the target (bacterial) amplicon, gels were run twice as long as usual before being stopped to excise the 16S rRNA amplicon.
PhyloChip™ G3 Analysis
Details about probe selection, probe scoring, data acquisition and preliminary data analysis are according to Hazen et al. [30]. For each sample, 500 ng of PCR product was applied to an individual PhyloChip™ G3 DNA microarray, with the exception of one microarray (representing sample VIISMAH06), which received 450 ng because of less robust amplification. To summarize the procedure, the sample 16S rRNA PCR products and a mixture of PCR products at known concentrations (referred to as ‘spike-mix’) were combined, fragmented using DNAseI, and biotin-labeled using the recommended protocol for Affymetrix Prokaryotic Arrays. These labeled products were hybridized on the PhyloChip™ G3 overnight at 48°C while rotating at 60 rpm. The microarrays were washed, stained, and scanned as previously described [30]. Briefly: Fluorescent images were capture with the GeneChip Scanner 3000 7G. Each 25mer probe array feature occupied approximately 8×8 pixels in the image file. The central 9 pixels were ranked by intensity, and the 75th percentile was used as the summary intensity for the feature. Array fluorescence intensities could range from 1 to 65,536. The hybridization score (HybScore, a.k.a., OTU intensity) for an OTU was calculated as the mean intensity of the perfectly matching probes exclusive of the maximum and minimum. HybScore data obtained from the CEL files (produced from GeneChip Microarray Analysis Suite, version 5.1) were scaled by setting the spike-mix control intensity to 10,000 to correct for slight differences in scan intensities or potential uneven hybridization efficiencies across microarrays.
Data Analyses
An updated taxonomy [36] was used for this dataset (updated since [32]). Stage 1 of PhyCA analysis was performed to remove OTUs not present in any of the samples using cutoff values: q1 = 0.5, q2 = 0.93, q3 = 0.98). This reduced the total number of OTUs subject to analysis from roughly 60,000 down to 13,441 (Table 2). The microarray data has been archived online at Greengenes, accessible by the following dedicated URL: (http://greengenes.lbl.gov/Download/Microarray_Data/Orbicella_Kellogg/). Intensity values were log2 transformed before hybridization data were further analyzed.
Table 2. Enumeration of taxonomic rank members detected across replicate PhyloChips™ from healthy and diseased coral samples (after Sunagawa et al, 2009).
| All OTUs | ||||||||
| TaxonomicRank | Healthy(abs.) | Diseased(abs.) | PercentDecrease | UniqueHealthy (%) | UniqueDiseased (%) | Shared Healthy &Diseased (%) | Total | |
| OTUs | 11,869 | 6,346 | 46.5 | 7,095 (52.8) | 1,572 (11.7) | 4,774 (35.5) | 13,441 | |
| Species | 627 | 388 | 38.0 | 307 (44.2) | 67 (9.6) | 321 (46.2) | 695 | |
| Genera | 775 | 569 | 26.6 | 246 (30.2) | 40 (4.9) | 529 (64.9) | 815 | |
| Families | 333 | 280 | 15.9 | 61 (17.9) | 8 (2.3) | 272 (79.8) | 341 | |
| Orders | 185 | 165 | 10.8 | 25 (13.2) | 5 (2.6) | 160 (84.2) | 190 | |
| Classes | 126 | 109 | 13.5 | 23 (17.4) | 6 (4.6) | 103 (78.0) | 132 | |
| Phyla | 73 | 61 | 16.4 | 12 (16.4) | 0 (0) | 61 (83.6) | 73 | |
| Singletons Removed | ||||||||
| Taxonomic Rank | Healthy (abs.) | Diseased (abs.) | Percent Decrease | Unique Healthy (%) | Unique Diseased (%) | Shared Healthy & Diseased (%) | Total | |
| OTUs | 4,599 | 3,555 | 28.0 | 1,519 (22.6) | 428 (6.4) | 4,774 (71.0) | 6,721 | |
| Species | 281 | 229 | 18.5 | 66 (17.0) | 20 (5.1) | 303 (77.9) | 389 | |
| Genera | 470 | 407 | 13.4 | 33 (6.0) | 18 (3.3) | 497 (90.7) | 548 | |
| Families | 257 | 230 | 10.5 | 19 (6.7) | 2 (0.7) | 263 (92.6) | 284 | |
| Orders | 152 | 142 | 6.6 | 11 (6.5) | 2 (1.2) | 155 (92.3) | 168 | |
| Classes | 94 | 94 | 0 | 2 (1.9) | 2 (1.9) | 101 (96.2) | 105 | |
| Phyla | 59 | 54 | 8.5 | 5 (7.7) | 1 (1.5) | 59 (90.8) | 65 | |
Top panel includes all OTUs. Bottom panel excludes singletons (defined as OTUs present only in a single sample).
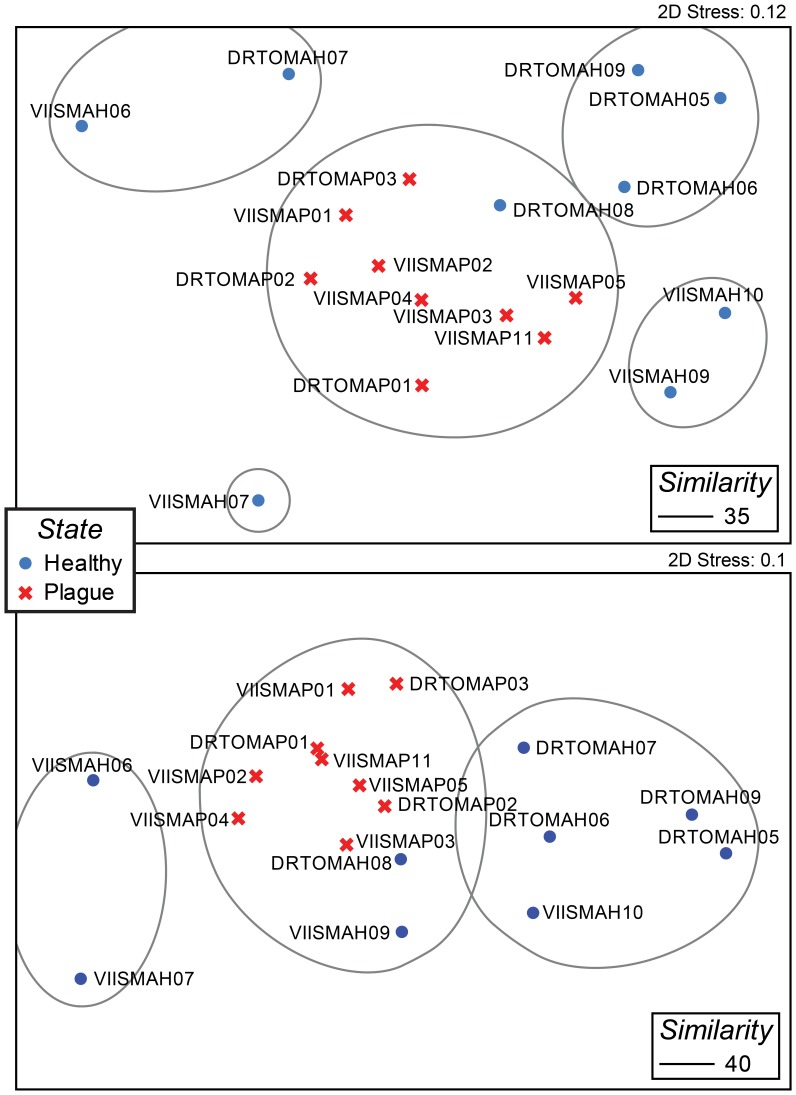
PRIMER 6 version 6.1.13 software [37] was used to generate cluster analyses and nonmetric multidimensional scaling (NMDS) plots to examine sample grouping (Figure 1). When using binary presence/absence data, no transformation was performed prior to computing the Bray Curtis similarities. Square-root transformed intensity data were used to calculate the two-way crossed analyses of similarity (ANOSIM) to test for significant differences in the bacterial community composition between predefined sample sets (i.e., based on collection location and health state).
Figure 1. Non-metric multidimensional scaling (NMDS) plot of samples, based on Bray-Curtis similarity of presence/absence OTU data.
Panel A is based on total OTUs (13,441). Panel B is based on the subset of OTUs that include at least one coral-associated sequence (313).
A custom PERL script was created (QueryOTU.pl, File S1) to screen the results for unique OTUs. The input file consisted of a modified OTU table (File S2) of binary presence/absence data with one extra column added labeled “Outgroup” which had zeros in all the real OTU rows and one arbitrary OTU numbered 999999. Running the PERL script on the.csv file generates a sequentially numbered list of the samples, followed by a prompts to choose the samples to include in Group A and then Group B. The program then compares the two groups, excluding any OTUs found in both, to provide a list of OTUs unique to each. The output includes the total number of OTUs per group, a list of the OTU numbers, and which samples within the group contain each OTU. The outgroup OTU and sample were included to allow determinations of total OTUs including those shared by other real samples. For a more detailed description of how this program was used to obtain the data in Table 2, please see File S3.
A QIIME-formatted OTU table based on the presence/absence data and a mapping file were created and run through QIIME [38]. The script ‘summarize_taxa_through_plots.py’ was used to generate taxa summary plots of OTU abundance at the phylum through family levels. This script was also used to compare grouped healthy samples versus grouped diseased samples by passing the –c option.
Results
There was a significant difference between healthy and diseased samples (Figure 1A; ANOSIM Global R = 0.316, p = 0.025) and the Global R value supports moderately distinct microbial communities in the diseased and healthy coral tissues. Although the depth range of collections was from 2 to 13 meters, there does not appear to be any structuring based on depth (Table 1, Figure 1). Healthy samples showed a greater taxonomic diversity compared to diseased samples (Table 2). Out of the 7,095 OTUs unique to healthy samples, 106 (0.01%) were present in greater than 50% of the healthy replicates (Table 3). The majority (94/106) consist of Gram-positive bacteria from the phyla Actinobacteria and Firmicutes.
Table 3. Operational taxonomic units (OTUs) unique to healthy samples and present in greater than 50% (five or more) healthy replicates.
| OTUs | Phylum | Class | Order | Family | Genus |
| 1 | Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Cryobacterium |
| 1 | Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | Arthrobacter |
| 8 | Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | Micrococcus |
| 1 | Actinobacteria | Actinobacteria | Actinomycetales | Propionibacteriaceae | Propionibacterium |
| 1 | Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | unclassified |
| 1 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flammeovirgaceae | Candidatus Cardinium |
| 81 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 1 | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium |
| 1 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Coprococcus |
| 9 | Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | Vibrio |
| 1 | Verrucomicrobia | Spartobacteria | Spartobacteriales | Spartobacteriaceae | MC18 |
| 106 | |||||
One of the major upgrades of the PhyloChip™ G3 over the previous generation (G2) is the representation of many coral-associated sequences in the additional OTUs. Through keyword searches followed by manual screening of the results, we determined that there are 545 total OTUs on the PhyloChip™ G3 that include at least one coral-associated sequence (Table S1). The bulk of these coral-associated sequences come from five studies [23], [26], [31], [39], [40]. Our Orbicella annularis dataset included 313 of these coral-associated OTUs. Out of the 1,572 OTUs unique to diseased samples (Table 2), only 38 (0.02%) were present in greater than 50% of the replicates (Table 4). These were pretty evenly split between Gram-positive and Gram-negative bacteria, with the main phyla being Firmicutes and Proteobacteria. Eighteen percent (7/38) of those OTUs represent sequences previously associated with corals (Table S1).
Table 4. Operational taxonomic units (OTUs) unique to diseased samples and present in ≥50% (five or more) diseased replicates.
| OTUs | Phylum | Class | Order | Family | Genus |
| 5 | Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | Corynebacterium |
| 1 | Chloroflexi | Anaerolineae | Anaerolineales | Anaerolinaceae | A4b |
| 1 | Cyanobacteria | Chloroplast | Chlorophyta | Ulvophyceae | unclassified |
| 1 | Cyanobacteria | Oscillatoriophycideae | Chroococcales | Microcystaceae | Microcystis |
| 1 | Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus |
| 1 | Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | unclassified |
| 1 | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus |
| 3 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus |
| 2 | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium |
| 1 | Firmicutes | Clostridia | Clostridiales | Incertae Sedis | Sedimentibacter |
| 1 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia |
| 2 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia |
| 2 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | unclassified |
| 1 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Faecalibacterium |
| 1 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Cohaesibacteraceae | Cohaesibacter |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Loktanella |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Paracoccus |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Rhodovulum |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Shimia |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Thalassobius |
| 3 | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | unclassified |
| 1 | Proteobacteria | Alphaproteobacteria | Rhodospirillales | Acetobacteraceae | Asaia |
| 1 | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Novosphingobium |
| 1 | Proteobacteria | Betaproteobacteria | Burkholderiales | Aquabacteriaceae | Aquabacterium |
| 1 | Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio |
| 1 | Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Desulfuromonadaceae | Desulfuromonas |
| 1 | Proteobacteria | Gammaproteobacteria | Oceanospirillales | Alteromonadaceae | nsmpVI18 |
| 38 | |||||
Given such high representation of coral-associated sequences in the subset of OTUs unique to diseased samples, we reran the NMDS plot using only the 313 coral-associated OTUs that had been identified within the total dataset of 13,441 OTUs (Figure 1B). The consistency of the disease samples clustering together (compare Figure 1 panels A and B) indicates that these coral-associated sequences are important in distinguishing the health state of the samples.
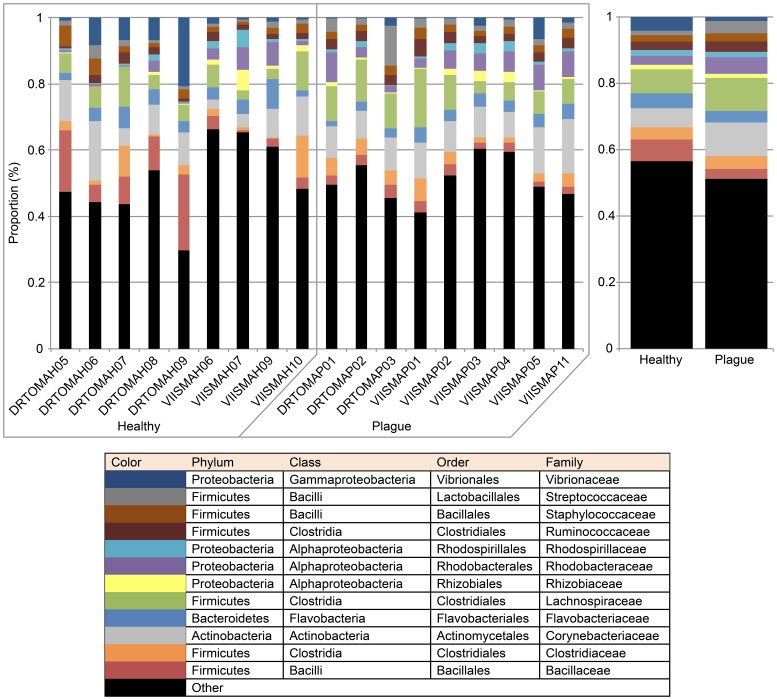
A large number of taxa are shared by healthy and diseased samples at all phylogenetic levels (Table 2), but some patterns can be seen at the family level (Figure 2). When all healthy and plague samples are combined such that the most common families can be compared, there are differences in several groups (Figure 2). There are more Vibrionaceae and Bacillaceae OTUs in healthy samples than diseased samples, which correlate well with the presence of a number of healthy-specific OTUs from those families (Table 3). Similarly, the families Corynebacteriaceae, Lachnospiraceae, Rhodobacteraceae, and Streptococcaceae are better represented in diseased samples, echoing the presence of disease-specific OTUs (Table 4).
Figure 2. Relative diversity within bacterial families associated with healthy and white plague-like diseased corals.
The 12 families shown were those representing greater than 5% of at least one sample. The remaining families are collectively represented by the ‘other’ category. The size of each color block (assigned to families in the key below) represents the number of OTUs detected in the family relative to the total number of OTUs detected in that sample. Individual samples are presented on the left and cumulative healthy and diseased profiles on the right.
When comparing entire bacterial communities, the white plague-like disease samples all clustered together and there was no significant difference between the two sampling locations (ANOSIM Global R = 0.111, p = 0.187; Figure 1). This clustering indicates that the diseased samples have similar bacterial community patterns, regardless of geography.
With the PhyloChip™ G3 microarray, the presence of each OTU is reported based on the responses of multiple probes assigned to the OTU. An OTU may encompass a single unique sequence or several hundred from a reference database. The OTU that includes Aurantimonas coralicida WP1 (the causative agent for WPII; [15]) is not exclusive to that sequence, and so a positive result, while suggesting the presence of this bacterium, could nonetheless indicate the presence of another related bacterium. Only three samples were positive for this OTU at the cutoff values used: VIISMAP04, VIISMAH06 and VIISMAH07. All three samples were from the Virgin Islands, from one diseased colony and two visibly healthy colonies.
Discussion
Using presence/absence of OTUs in our PhyloChip™results, we constructed non-metric multidimensional scaling plots that showed strong clustering of all the white plague-like samples, regardless of collection site (Figure 1). A PhyloChip™ G3 study that examined two coral species affected by a white plague-like disease on a single reef in the Indo-Pacific found similarity in the disease community irrespective of host [25]. These patterns of similar bacterial communities associated with disease states that are conserved across hosts [25] or locations (this study), suggest that the PhyloChip™ G3 may be a useful tool to differentiate and track specific coral diseases. This is in contrast to a study that examined Orbicella annularis from the Bahamas using length heterogeneity PCR (LH-PCR) to generate the OTUs [10]. In that case, all coral samples clustered together regardless of disease state and ANOSIM confirmed no significant differences between the healthy and white plague bacterial communities [10]. The larger number of informational OTUs (13,441) that the PhyloChip™ G3 provided was likely important to resolving the health state; however, as shown in Figure 1B, a subset of 313 OTUs containing coral-associated sequences was sufficient to produce a similar outcome.
Many studies, including the two prior studies of coral disease employing PhyloChips™ [25], [26], have shown increased diversity in diseased corals compared to healthy [12], [41], [42]. In this study we found the opposite, with greater numbers of OTUs both present and unique to healthy samples compared to diseased (Table 2). At first we thought that the difference in diversity detected by the PhyloChip™ G3 compared to the PhyloChip™ G2 might be partly due to the presence of the coral-specific OTUs on the G3 chip (Table S1), since they may reveal more microbial community diversity in healthy corals than was possible on the previous chip. Also, it seemed logical that the increased diversity observed with the G3 chip might simply be due to the greater taxonomic coverage: using the G2 PhyloChip, Sunagawa et al. [26] detected 1,702 out of a possible total of 8,741 OTUs versus our G3 data reflecting 13,441 out of a possible 59,222 bacterial OTUs. This is an almost eightfold increase in OTUs and therefore a substantial amount of additional information. However, both those arguments fail when comparing between two G3 studies. In our case, much of the diversity in healthy samples was due to taxa that were represented by a single OTU on the PhyloChip™ G3 (i.e., uncommon families, genera, and species). Further, of the more than 7,000 OTUs unique to healthy samples (Table 2, top panel), most were not shared by more than a few samples (e.g., greater than 4,000 OTUs were only called present in a single coral sample). The Roder et al. study [25] did not consider any OTUs that were not present in more than one sample (analogous to removing singletons from pyrosequencing datasets). For comparative purposes, we recalculated our rankings after removing all singletons (Table 2, bottom panel). While fully half (6,720) of our OTUs were singletons, their removal did not change our observation of higher diversity in healthy samples compared to diseased. It did cause the percentages of shared taxa to increase dramatically (Table 2, bottom panel).
Out of the 13,441 OTUs present across all samples, 106 OTUs were unique to healthy samples and present in more than half of the healthy replicates (Table 3). Of note are the three groups of multiple OTUs within the same genera: 8 for Micrococcus, 81 for Bacillus, and 9 for Vibrio. This matches well with Figure 2 where both families Bacillaceae and Vibrionaceae were represented by a greater proportion of OTUs in healthy samples compared to diseased. A greater abundance (based on relative intensities) of Bacillaceae was also found in healthy corals compared to white plague-like diseased corals by the PhyloChip™ G3 study in the Indo-Pacific [25]. In soil studies, some strains of Bacillus have been shown to fix nitrogen, while others may stimulate nitrogen-fixing activities of other bacteria [43], [44]. This might be a functional role for coral-associated Bacillus sp. in healthy corals that is disrupted by the transition to a diseased state. Recent studies of white plague-like disease in other coral species give conflicting results in terms of Vibrios: a lower number of Vibrionales sequences was seen in diseased Diploria strigosa and Siderastrea siderea (both Caribbean species) [28], versus a higher abundance of Vibrionaceae in diseased Pavona duerdeni and Porites lutea (Pacific species) [25]. While Vibrio species have been implicated in several coral diseases [29], [45], [46], they have also been shown to be a normal part of the coral microbiome [47]–[50].
There were eight families that occurred in diseased corals but were not present in any of the healthy samples (Table 2): BSV26/SM1H02, Chlamydiaceae, Gallionellaceae, Rarobacteraceae, Scalinduaceae, Solimonaceae, Spirulinaceae, Thermodesulfobiaceae. Unfortunately, in all but one case (Scalinduaceae, 4 OTUs), each family is only represented by a single OTU on the PhyloChip™ G3, making it difficult to assess the significance of these groups. There were only 38 OTUs that were unique to diseased samples and shared by more than half the diseased replicates (Table 4). Included are two cyanobacterial OTUs and one for Desulfovibrio, providing some evidence for the ‘atypical black band disease’ that Ainesworth et al. [7] described as having a macroscopic appearance of white plague. Pantos et al. [12] also had noted similarities in 16S rRNA gene clone library sequences between white plague and black band diseases.
There is also noticeable representation of the Rhodobacteraceae family (8 OTUs; Table 4); this family previously had been detected as enriched in white plague samples of Orbicella faveolata by Sunagawa et al. [26] using an earlier generation PhyloChip™ microarray and also in white plague-like samples of Indo-Pacific corals by PhyloChip™ G3 [25]. Rhodobacteraceae OTUs were represented by a greater proportion of OTUs in diseased samples compared to healthy (Figure 2). Sequences from the order Rhodobacterales were consistently and significantly enriched in pyrosequencing datasets from white plague diseased Diploria strigosa and Siderastrea siderea compared to healthy [28] and have also been associated with another white plague-like disease [12]. Further, a meta-analysis of coral-associated sequences [51] found Rhodobacter spp. to be associated with several coral diseases, not just white plague. This seems to corroborate the suggestion by Sunagawa et al. [26] that Rhodobacteraceae are opportunists.
In the recent PhyloChip™ G3 comparison of healthy and white plague-like diseased Indo-Pacific corals, several of the bacterial families that showed higher abundance in diseased corals were Clostridiaceae, Corynebacteriaceae, Lachnospiraceae, and Ruminococcaceae [25]. Similarly, we found a higher relative abundance of OTUs from these families in our diseased samples (Figure 2). Families that both appeared in higher relative abundance in our diseased samples (Figure 2) and were also represented by OTUs unique to disease samples (Table 4) were Corynebacteriaceae, Lachnospiraceae, and Streptococcaceae. Unlike Rhodobacteraceae, these families are all Gram-positive, and recent work on white band disease has indicated that the causative pathogen is a Gram-positive microbe [52]. It has been pointed out that it can be difficult to distinguish between white band and white plague in Acropora when surrounded by an outbreak of white plague-like disease in other coral species [13]. Further, it has been suggested that given our difficulty in identifying defining causative agents, coral diseases with this phenotype on the Great Barrier Reef should be collectively referred to as white syndromes [53] rather than distinguishing white plague from white band based solely on host identity. In general, the presence and roles of Gram-positive bacterial groups in corals (both healthy and diseased) have been understudied, likely because of a bias towards the recovery of Gram-negative bacteria in many culture media as well as clone libraries. To this point, the focus has been dominated by attention to Gram-negative Proteobacteria. For example, several white plague studies have described a shift in the bacterial community towards Alphaproteobacteria being more heavily represented in diseased corals [12], [26], [28]. We did not observe that in this study, either in terms of higher intensities or in terms of greater numbers of Alphaproteobacterial OTUs. Newer methods such as the PhyloChip™ G3 and pyrosequencing are uncovering the presence and diversity of Gram-positive bacteria in corals, and suggest we may need to look at them more closely.
Two recent coral disease studies [10], [27] have found no significant differences between the bacterial community composition of healthy and diseased corals. In one case, the comparison was based on a total of 67 bacterial OTUs [10], which likely did not provide enough depth of coverage. The large number of shared OTUs revealed by our study (Table 2) could explain this finding, particularly since the community fingerprints used in that study only identified the dominant taxa. The other study used pyrosequencing and so achieved a much greater depth of coverage, but was limited by having only one healthy and one diseased replicate per location [27]. While we expected that our diseased samples might cluster based on geographic location, indicating different pathogens or disease processes, that did not occur. Instead, the diseased samples all cluster together (Figure 1). The clustering is not tight enough to indicate a specific pathogen or even a single group of bacteria, but there is some evidence for a number of bacterial groups affecting the health state (Figure 2). It is also likely that each coral sampled was at a different stage of the disease process, and we currently have no baseline of the changes a disease lesion undergoes as it progresses over time.
Aurantimonas coralicida has been described as the causative agent of white plague type II (WPII) [15]. We found no evidence for its association with any of the diseased samples from the Dry Tortugas and only three samples from the Virgin Islands (one diseased, two healthy) were positive for the OTU that includes the A. coralicida sequence. Using the second generation (G2) PhyloChip™and clone libraries, Sunagawa et al. [26] were unable to detect A. coralicida in either healthy or white plague-affected Orbicella faveolata. More recent work using the PhyloChip™ G3 and clone libraries to examine a white plague-like disease in Pavona duerdeni and Porites lutea also did not find any indication of A. coralicida. Similarly, previous work on O. annularis using clone libraries failed to detect A. coralicida [12]. Deliberate attempts to culture or detect A. coralicida using specific PCR primers (on other Caribbean species Diploria strigosa and Siderastrea siderea) also failed [28]. However, since it was not mentioned in the methods sections of these papers that any effort was made to measure the disease progression rate (the only macroscopic differentiator between WPI, WPII, and WPIII), there is no way to be certain that the diseases being sampled were in fact WPII. It has been suggested that there are likely multiple etiologies that result in these macroscopic symptoms [5], [9], [10], [26], [28], [54]. Alternatively, recently it has been shown that popular fixatives used to preserve tissue after sampling (before DNA extraction) seem biased against A. coralicida and other bacteria with a high percentage of GC bases in their DNA, such that they are not detected in a mixed community even when present [55].
Conclusions
The PhyloChip™ G3 microarray provides a broader taxonomic overview than can be achieved by a traditionally sized clone library and also provides data that can be analyzed without a steep learning curve of bioinformatics, such as is required to handle pyrosequencing datasets. Our results show no evidence for geographically distinct disease states of coral-associated microbiota or for a single causative agent of this white plague-like disease. The higher taxonomic diversity observed in the healthy samples is likely indicative of the natural spatial heterogeneity that has been documented in coral-associated bacterial communities on O. annularis [48] as well as other species [25], [56]. The microbial communities of healthy and diseased samples are clearly different, with differential representation of the families Bacillaceae, Corynebacteriaceae, Lachnospiraceae, Rhodobacteraceae, Streptococcaceae, and Vibrionaceae indicating a need to better understand the relationships between coral and Gram-positive bacteria, which appear to have been underappreciated to date.
Supporting Information
PhyloChip™ G3 operational taxonomic units (OTUs) that contain one or more coral-associated sequences.
(XLS)
OTU sorting program, QueryOTU.pl, written in Perl.
(PL)
Comma-delineated OTU table provided as input for the QueryOTU.pl program.
(CSV)
Detailed example for using QueryOTU.pl program to sort PhyloChip™ G3 OTUs.
(DOC)
Acknowledgments
Field assistance by T. McDole, B.J. Reynolds, and V.H. Garrison is gratefully acknowledged. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Although the software program in File S1 has been used by the U.S. Geological Survey (USGS) for this study, no warranty, expressed or implied, is made by the USGS or the U.S. Government as to the accuracy and functioning of the program and related program material, nor shall the fact of distribution constitute any such warranty, and no responsibility is assumed by the USGS in connection therewith.
Funding Statement
This project was supported by the Coral Reef Ecosystems Study (CREST) of the United States Geological Survey’s Coastal and Marine Geology Program. A portion of this work was performed under the auspices of the United States Department of Energy under contract DE-AC02-05CH11231 to Lawrence Berkeley National Laboratory. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Budd AF, Fukami H, Smith ND, Knowlton N (2012) Taxonomic classification of the reef coral family Mussidae (Cnidaria: Anthozoa: Scleractinia). Zool J Linn Soc 166: 465–529. [Google Scholar]
- 2. Hughes TP (1994) Catastrophes, phase-shifts and large-scale degradation of a Caribbean coral reef. Science 265: 1547–1551. [DOI] [PubMed] [Google Scholar]
- 3. Rogers C (2009) Coral bleaching and disease should not be underestimated as causes of Caribbean coral reef decline. Proc R Soc B 276: 197–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bythell JC, Pantos O, Richardson LL (2004) White plague, white band, and other “white” diseases. In: Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin: Springer-Verlag. 351–365.
- 5. Dustan P (1977) Vitality of reef coral populations off Key Largo, Florida: recruitment and mortality. Environ Geol 2: 51–58. [Google Scholar]
- 6. Richardson LL (1998) Coral diseases: what is really known? Trends Ecol Evol 13: 438–443. [DOI] [PubMed] [Google Scholar]
- 7. Ainsworth TD, Kramarsky-Winter E, Loya Y, Hoegh-Guldberg O, Fine M (2007) Coral disease diagnostics: what’s between a plague and a band? Appl Environ Microbiol 73: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandt ME, Ruttenberg BI, Waara R, Miller J, Witcher B, et al. (2012) Dynamics of an acute coral disease outbreak associated with the macroalgae Dictyota spp. in Dry Tortugas National Park, Florida, USA. Bull Mar Sci 88: 1035–1050. [Google Scholar]
- 9. Brandt ME, Smith TB, Correa AMS, Vega Thurber R (2013) Disturbance driven colony fragmentation as a driver of a coral disease outbreak. PLOS ONE 8: e57164 10.1371/journal.pone.0057164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cook GM, Rothenberger JP, Sikaroodi M, Gillevet PM, Peters EC, et al. (2013) A comparison of culture-dependent and culture-independent techniques used to characterize bacterial communities on healthy and white plague-diseased corals of the Montastraea annularis species comlex. Coral Reefs 32: 375–388. [Google Scholar]
- 11. Francini-Filho R, Reis R, Meirelles P, Moura R, Thompson F, et al. (2010) Seasonal prevalence of white plague like disease on the endemic Brazilian reef coral Mussismilia braziliensis . Lat Am J Aquat Res 38: 292–296. [Google Scholar]
- 12. Pantos O, Cooney RP, Le Tissier MDA, Barer MR, O’Donnell AG, et al. (2003) The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis . Environ Microbiol 5: 370–382. [DOI] [PubMed] [Google Scholar]
- 13. Sánchez JA, Herrera S, Navas-Camacho R, Rodríguez-Ramírez A, Herron P, et al. (2010) White plague-like coral disease in remote reefs of the Western Caribbean. Rev Biol Trop 58: 145–154. [DOI] [PubMed] [Google Scholar]
- 14. Work TM, Aeby G (2006) Systematically describing gross lesions in corals. Dis Aquat Org 70: 155–160. [DOI] [PubMed] [Google Scholar]
- 15. Denner EBM, Smith GW, Busse H-J, Schumann P, Narzt T, et al. (2003) Aurantimonas coralicida gen. nov., sp. nov., the causitive agent of white plague type II on Caribbean scleractinian corals. Int J Syst Evol Microbiol 53: 1115–1122. [DOI] [PubMed] [Google Scholar]
- 16. Thompson FL, Barash Y, Sawabe T, Sharon G, Swings J, et al. (2006) Thalassomonas loyana sp. nov., a causative agent of the white plague-like disease of corals on the Eliat coral reef. Int J Syst Evol Microbiol 56: 365–368. [DOI] [PubMed] [Google Scholar]
- 17. Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O (2007) Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol 346: 36–44. [Google Scholar]
- 18. Muller EM, van Woesik R (2012) Caribbean coral diseases: primary transmission or secondary infection? Glob Change Biol 18: 3529–3535. [Google Scholar]
- 19. Cervino JM, Thompson FL, Gomez-Gil B, Lorence EA, Goreau TJ, et al. (2008) The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. J Appl Microbiol 105: 1658–1671. [DOI] [PubMed] [Google Scholar]
- 20. Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, et al. (2002) The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata . Proc Nat Acad Sci USA 99: 8725–8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viehman S, Mills DK, Meichel GW, Richardson LL (2006) Culture and identification of Desulfovibrio spp. from corals infected by black band disease on Dominican and Florida Keys reefs. Dis Aquat Org 69: 119–127. [DOI] [PubMed] [Google Scholar]
- 22. Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW (2002) Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol 68: 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sekar R, Kaczmarsky LT, Richardson LL (2008) Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar Ecol Prog Ser 362: 85–98. [Google Scholar]
- 24. Pantos O, Bythell JC (2006) Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture-independent 16S rRNA techniques. Dis Aquat Org 69: 79–88. [DOI] [PubMed] [Google Scholar]
- 25.Roder C, Arif C, Bayer T, Aranda M, Daniels C, et al. (2013) Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8 August 2013, 10.1038/ismej.2013.127 [DOI] [PMC free article] [PubMed]
- 26. Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, et al. (2009) Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata . ISME J 3: 512–521. [DOI] [PubMed] [Google Scholar]
- 27. Apprill A, Hughen K, Mincer T (2013) Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol 15: 2063–2072. [DOI] [PubMed] [Google Scholar]
- 28. Cárdenas A, Rodriguez-R LM, Pizarro V, Cadavid LF, Arévalo-Ferro C (2012) Shifts in bacterial communities of two caribbean reef-building coral species affected by white plague disease. ISME J 6: 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia GD, Gregoracci GB, Santos EdO, Meirelles PM, Silva GGZ, et al. (2013) Metagenomic analysis of healthy and white plague-affected Mussismilia braziliensis corals. Microb Ecol 65: 1076–1086. [DOI] [PubMed] [Google Scholar]
- 30. Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, et al. (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208. [DOI] [PubMed] [Google Scholar]
- 31. Sunagawa S, Woodley CM, Medina M (2010) Threatened corals provide underexplored microbial habitats. PLOS ONE 5: e9554 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kellogg CA, Piceno YM, Tom LM, DeSantis TZ, Zawada DG, et al. (2012) PhyloChip™ microarray comparison of sampling methods used for coral microbial ecology. J Microbiol Meth 88: 103–109. [DOI] [PubMed] [Google Scholar]
- 33. Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucl Acids Res 17: 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stackebrandt E, Liesack W (1993) Nucleic acids and classification. In: Goodfellow M, O’Donnell AG, editors. Handbook of New Bacterial Systematics. London: Academic Press. 152–189.
- 35. Galkiewicz JP, Kellogg CA (2008) Cross-kingdom amplification using Bacteria-specific primers: complications for studies of coral microbial ecology. Appl Environ Microbiol 74: 7828–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDonald D, Price MN, Goodrich J, Nawrock EP, DeSantis TZ, et al. (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aus J Ecol 18: 117–143. [Google Scholar]
- 38. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sekar R, Mills DK, Remily ER, Voss JD, Richardson LL (2006) Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea . Appl Environ Microbiol 72: 5963–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Penn K, Wu D, Eisen JA, Ward N (2006) Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl Environ Microbiol 72: 1680–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barneah O, Ben-Dov E, Kramarsky-Winter E, Kushmaro A (2007) Characterization of black band disease in Red Sea stony corals. Environ Microbiol 9: 1995–2006. [DOI] [PubMed] [Google Scholar]
- 42. Reis AMM, Araújo SD Jr, Moura RL, Francini-Filho RB, Pappas G Jr, et al. (2009) Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis . J Appl Microbiol 106: 1378–1387. [DOI] [PubMed] [Google Scholar]
- 43. Rózycki H, Dahm H, Strzelczyk E, Li CY (1999) Diazotrophic bacteria in root-free soil and in the root zone of pine (Pinus sylvestris L.) and oak (Quercus robur L.). Appl Soil Ecol 12: 239–250. [Google Scholar]
- 44. Zlotnikov AK, Sharpovalova YN, Makarov AA (2001) Association of Bacillus firmus E3 and Klebsiella terrigena E6 with increased ability for nitrogen fixation. Soil Biol Biochem 33: 1525–1530. [Google Scholar]
- 45.Rosenberg E, Kushmaro A (2011) Microbial diseases of corals: pathology and ecology. In: Dubinsky Z, Stambler N, editors. Coral Reefs: An Ecosystem in Transition. New York: Springer. 451–464.
- 46. Arotsker L, Siboni N, Ben-Dov E, Kramarsky-Winter E, Loya Y, et al. (2009) Vibrio sp. as a potentially important member of the Black Band Disease (BBD) consortium in Favia sp. corals. FEMS Microbiol Ecol 70: 515–524. [DOI] [PubMed] [Google Scholar]
- 47. Bourne DG, Munn CB (2005) Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7: 1162–1174. [DOI] [PubMed] [Google Scholar]
- 48. Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, et al. (2011) Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis . Mar Ecol Prog Ser 426: 29–40. [Google Scholar]
- 49. Gray MA, Stone RP, McLaughlin MR, Kellogg CA (2011) Microbial consortia of gorgonian corals from the Aleutian islands. FEMS Microbiol Ecol 76: 109–120. [DOI] [PubMed] [Google Scholar]
- 50. Nithyanand P, Pandian SK (2009) Phylogenetic characterization of culturable bacterial diversity associated with the mucus and tissue of the coral Acropora digitifera from the Gulf of Mannar. FEMS Microbiol Ecol 69: 384–394. [DOI] [PubMed] [Google Scholar]
- 51. Mouchka ME, Hewson I, Harvell CD (2010) Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr Comp Biol 50: 662–674. [DOI] [PubMed] [Google Scholar]
- 52. Kline DI, Vollmer SV (2011) White band disease (type I) of endangered Caribbean acroporid corals is caused by pathogenic bacteria. Sci Rep 1: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willis BL, Page CA, Dinsdale EA (2004) Coral disease on the Great Barrier Reef. In: Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin: Springer-Verlag. 69–104.
- 54. Pollock FJ, Morris PJ, Willis BL, Bourne DG (2011) The urgent need for robust coral disease diagnostics. PLOS Pathogens 7: e1002183 10.1371/journal.ppat.1002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gray MA, Pratte ZA, Kellogg CA (2013) Comparison of DNA preservation methods for environmental bacterial community samples. FEMS Microbiol Ecol 83: 468–477. [DOI] [PubMed] [Google Scholar]
- 56. Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O (2010) Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and species-specificity of common bacterial associates. PLOS ONE 5: e10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PhyloChip™ G3 operational taxonomic units (OTUs) that contain one or more coral-associated sequences.
(XLS)
OTU sorting program, QueryOTU.pl, written in Perl.
(PL)
Comma-delineated OTU table provided as input for the QueryOTU.pl program.
(CSV)
Detailed example for using QueryOTU.pl program to sort PhyloChip™ G3 OTUs.
(DOC)