Abstract
Congenital abnormalities of the kidney and urinary tract (CAKUT) constitute the most frequent cause of chronic kidney disease in children, accounting for ~50% of all cases. Although many forms of CAKUT are likely caused by single-gene defects, only few causative genes have been identified. To identify new causative genes many candidate genes need to be analyzed due to the broad genetic locus heterogeneity of CAKUT. We therefore applied our newly developed approach of DNA pooling with consecutive massively parallel exon resequencing to overcome this problem. We pooled DNA of 20 individuals and amplified by PCR all 313 exons of 30 CAKUT candidate genes. PCR products were then subjected to massively parallel exon resequencing. Mutation carriers were identified using Sanger sequencing. We repeated the experiment to cover 40 patients in total (29 with unilateral renal agenesis and 11 with other CAKUT phenotypes). We detected 5 heterozygous missense mutations in 2 candidate genes that were not previously implicated in non-syndromic CAKUT in humans, 4 mutations in the FRAS1 gene and 1 in FREM2. All mutations were absent from 96 healthy control individuals and had a PolyPhen score of >1.4 (“possibly damaging”). Recessive truncating mutations in FRAS1 and FREM2 were known to cause Fraser syndrome in humans and mice, whereas a phenotype in heterozygous carriers has not been described. We hereby identify heterozygous missense mutations in FRAS1 and FREM2 as a new cause of non-syndromic CAKUT in human.
INTRODUCTION
Congenital abnormalities of the kidney and urinary tract (CAKUT) occur in about 3–6 per 1,000 live births and constitute 20–30% of all anomalies identified in the neonatal period1, 2. They constitute the most frequent cause of end-stage renal disease (ESRD) in children, accounting for approximately 50% of all cases3. To date there is no prophylaxis for CAKUT available.
CAKUT cover a wide range of structural and functional malformations that result from defects in the morphogenesis of the kidney and/or the urinary tract. CAKUT may occur in a large number of conditions of various primary or secondary causes. The hypothesis that CAKUT may be caused in many instances by single-gene defects is supported by the fact that monogenic forms of CAKUT in humans and mouse models of CAKUT have been published4–7. Such single-gene mutations may cause a wide phenotypic spectrum of CAKUT that ranges from vesico-ureteral reflux to renal agenesis, because the induction of the metanephrogenic mesenchyme by the ureteric bud influences morphogenesis of all three tissue groups of the excretory system: the ureterovesical junction, the ureter, and the kidney8.
Evidence from humans and mouse models suggests the existence of monogenic causes of CAKUT (Supplementary Table 1). Although many forms of CAKUT in humans are very likely caused by single-gene defects, only few causative genes have been identified so far. In our multiethnic cohort of 538 patients from 456 different families with non-syndromic CAKUT, we detected mutations of 4 known dominant CAKUT genes TCF2/HNF1β, PAX2, UMOD and EYA1 in only 12 families (1.9%)9. Thus, most of the individuals in this cohort remained unsolved regarding their potential molecular genetic cause of CAKUT.
We therefore developed a novel approach of high throughput mutation analysis to be performed in pooled genomic DNA of 20 individuals and 30 candidate genes simultaneously using exon polymerase chain reaction (PCR) and massively parallel exon resequencing10. The CAKUT candidate genes were genes that have been implicated in the severe developmental phenotype of unilateral renal agenesis in humans or mice.
RESULT
We pooled two times 20 DNA samples of individuals with CAKUT (29 with unilateral renal agenesis, 11 with other forms of CAKUT). We performed exon PCR in 30 candidate genes (Supplementary Table 2). Massively parallel exon resequencing of 342 different exon PCR products generated was carried out on an Illumina/Solexa GAII platform (one pool per lane of a flow cell) delivering about 5.3 million short reads of 39 bases (5.22 million in pool #1 and 5.37 million in pool #2). Following gapped alignment to the human normal reference sequence, 75% of all reads mapped back to one of the 342 target sequence that contained the exons plus 100 bp adjacent intronic sequence. The sequence concatenation of all 342 amplicons amounts to a total length of 175.7 kb, 68.2 kb of which were exonic coding regions. The median coverage depth for the coding regions was 1,228x (mean 1,402x). On average, about 95% of nucleotides in targeted coding regions met at least 400-fold coverage depth. This translates into a depth of at least 20-fold per patient, which is sufficient to call a heterozygous change.
Mutation carrier identification by Sanger sequencing
Massively parallel exon resequencing of all PCR products of 40 individuals revealed initially a total of 114 variants from normal reference sequence within the coding regions and splice sites of the 30 candidate genes analyzed, 47 of which were known SNPs. Eight additional variants were present in a cohort of 96 Caucasian healthy control individuals and are thought to be either as yet unannotated SNPs or false calls due to software base calling or alignment artifacts. Of the remaining 59 variants, 11 were predicted to truncate the protein products and 16 others had a PolyPhen score higher than 1.4 (predicted to be “possibly damaging”). These 27 variants were assumed to affect the function of encoded proteins and thus were followed by direct Sanger sequencing to identify the mutation carrier(s) out of the respective two pools of 20 individuals each. This carrier analysis led to the confirmation of 10 of the 27 variants (“true positives”) in 11 individuals from 11 different families, whereas 17 of the 27 variants could not be confirmed (“false positives”). Segregation study was done in all confirmed cases where DNA of relatives was available and 3 out of 10 variants were not segregating.
The remaining 7 variants (Table 1, Figure 1) were heterozygous missense mutations in 4 genes. Four mutations were in FRAS1 (p.L259R, p.D998Y, p.R3273H and p.H3757Q) and 1 in FREM2 (p.T2338I). Mutations in FRAS1 and FREM2 have never been reported in non-syndromic CAKUT in humans. All 5 mutations were absent from 96 healthy control individuals of Caucasian origin and 270 control individuals from the “1,000 Genomes project”. Two mutations in FRAS1 (p.D998Y and p.H3757Q) and one mutation in FREM2 (p.T2338I), which were detected in individuals of Arabic and Indian origin respectively, were additionally screened and were absent in 141 ethnically matched healthy controls. Since FRAS1 and FREM2 were known to inherit recessively, we amplified and sequenced all other coding exons in respective mutation carriers to exclude the presence of the second mutation. No further mutations were found except in A2381 II-2 (FRAS1; R3273H) where an additional nonsense mutation was detected (R2621X). The R2621X change was 2,349-fold covered with frequency of 4.5% but had been systematically masked out. The 2 mutations (R3273H and R2621X) were in trans as R3273H was heterozygously present only in the mother while R2621X was heterozygously present only in the father. The elder female sibling of A2381 II-2 had died in utero of bilateral renal agenesis and absent bladder but the DNA was not available for study.
Table 1.
Genotypes and phenotypes of 8 individuals with unilateral renal agenesis (8 families) with 8 different heterozygous missense mutations in one of 30 candidate genes.
| Family | Phenotype | Pattern of inheritance |
Origin | Genea | Nucleotide change (zygosity state)b |
Amino acid change |
PolyPhen PSIC scorec |
Count/coverage (frequency) |
Segregation | |
|---|---|---|---|---|---|---|---|---|---|---|
| A2435 | II-1 | URA | sporadic | Albania | FRAS1 | c.776T>G (h) | p.L259R | 1.692 | 16/648 (2.5%) | n/a |
| A1808 | II-1 | URA | sporadic | Kuwait | FRAS1 | c.2992G>T (h) | p.D998Y | 2.336 | 32/1,388 (2.3%) | n/a |
| A2381 | II-2 | URA | familiald | Austria | FRAS1 | c.9815G>A (h) | p.R3273H | 1.695 | 32/1,085 (2.9%) | Maternal |
| FRAS1 | c.7861C>T (h) | p.R2621X | N/A | 105/2349 (4.5%) | Paternal | |||||
| A1425 | II-1 | URA | sporadic | Lebanon | FRAS1 | c.11268C>A (h) | p.H3757Q | 2.011 | 16/1,320 (1.2%) | n/a |
| A1023 | II-1 | URA | sporadic | India | FREM2 | c.7013C>T (h) | p.T2338I | 2.050 | 29/1,067 (2.7%) | n/a |
| A1077 | II-1 | URA | sporadic | Kuwait | RET | c.2110G>T (h) | p.V704F | 1.537 | 8/494 (1.6%) | n/a |
| A1143 | II-1 | URA | sporadic | Albania | BMP4 | c.362A>G (h) | p.H121R | 2.001 | 26/826 (3.1%) | n/a |
| A1184 | II-1 | URA | sporadic | Macedonia | BMP4 | c.362A>G (h) | p.H121R | 2.001 | 800/826 (3.1%) | Paternal |
h, heterozygous; n/a, not available; ns, not segregating with CAKUT phenotype in the family; URA, unilateral renal agenesis
Mutation numbering is based on the cDNA position in reference sequences. All changes were absent from 96 healthy control (HRC-1) DNA pool, 270 individuals of the “1,000 Genomes project” (http://www.1000genomes.org/page.php) and from additional ethnically matched healthy controls.
PSIC score above 1.4 is considered “probably damaging” (http://genetics.bwh.harvard.edu/pph/).
A2381 II-1 (older female sibling, died in utero) had bilateral renal agenesis with absent urinary bladder. DNA is not available for study.
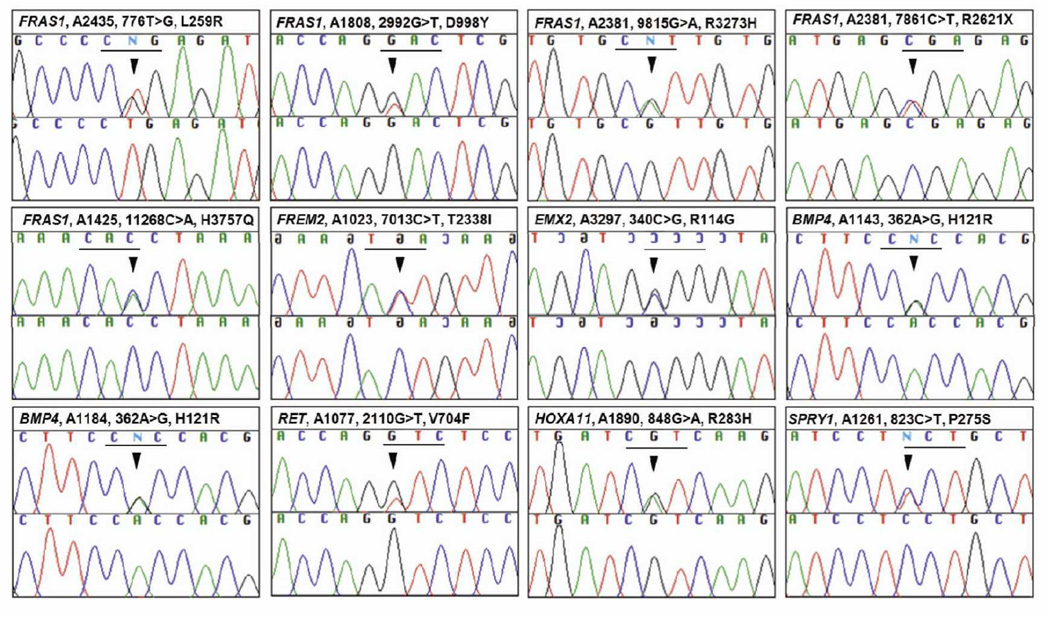
Figure 1.
Sequence chromatograms of 11 confirmed variants identified in 11 individuals with unilateral renal agenesis. Gene name, patient identifier, nucleotide change, and inferred amino acid alteration are given above sequence traces. The mutation position is indicated with an arrow. Wild type sequence chromatograms are shown below mutated sequences. Reading frames are underlined.
Two other variants were novel mutations in known CAKUT genes; RET (p.V704F) and BMP4 (p.H121R) (Table 1, Figure 1). Family A1143 and A1184 (both Macedonian) shared the same novel change in BMP4 (p.H121R). Both mutations were absent from 96 healthy control individuals and 270 control individuals from the “1,000 Genomes project”.
DISCUSSION
We here identified 7 different mutations of 4 different genes in 7 out of 40 patients with CAKUT using massively parallel exon resequencing. Two heterozygous mutations were novel mutations in known human CAKUT-causing genes: BMP4 and RET. Five heterozygous mutations were in 2 candidate genes that have never been reported in non-syndromic CAKUT, FRAS1 and FREM2. McGregor and Vrontou et al. identified recessive mutations in FRAS1 using linkage analysis in families that were affected with Fraser syndrome, a rare multi-organ disorder characterized by cryptophthalmos, cutaneous syndactyly and renal agenesis11, 12. Fras1 encodes Fras1, a protein containing repeats of the chondroitin sulfate proteoglycan (CSPG) core domain whose function is to maintain epithelial cell integrity. Fras1 protein was detected in several developing tissues such as limb, lung, gut and kidney. In metanephros, Fras1 was detected in the extracellular matrix underlying the basal surface of outgrowing ureter and the basement membrane of collecting tubule11, 12. McGregor et al. detected the homozygous Fras1 mutation that leads to premature stop codon (S2200X) in blebbed (bl) mice, the long known mouse strain with phenotype resembling human Fraser syndrome but in addition also in utero skin blisters11. Both bl/bl mice and Fras1−/− mice developed limb defects and renal agenesis or dysplasia11, 13. Pitera et al. demonstrated that the ureteric bud of bl/bl mice either failed to sprout or to enter the metanephric mesenchyme13.
All FRAS1 mutations that have been reported in Fraser syndrome individuals were either homozygous or compound heterozygous indicating a recessive genetic mechanism. Most mutations were truncating11, 14–16. In contrast, we here observe single heterozygous missense mutations of FRAS1 in non-syndromic unilateral renal agenesis. Although the disease phenotypes of recessive diseases are generally known to be absent in heterozygous carriers, there is evidence of phenotypic expression in heterozygotes of some diseases. Heterozygous mutations of SALL1 in human cause Townes-Brocks syndrome, which comprises of facial and limb defects, renal and urinary tract abnormalities and anorectal malformations17. In mice, while heterozygous Sall1 mutants mimic severe human phenotypes, Sall1-null mice only manifest renal agenesis without other organ involvement18, 19. Kiefer et al. demonstrated that the truncated Sall1 protein could interact with all Sall protein and have deleterious effects on all Sall functions in the cell18. The other example is Eya1 where both heterozygous and homozygous mutant mice have renal abnormalities and hearing defects similar to human branchio-oto-renal syndrome20.
In an individual with compound heterozygous mutations (R3273H and R2621X), since this individual has only unilateral renal agenesis, not full-spectrum Fraser syndrome, it was possible that only the truncating allele caused the phenotype. On the other hand, based on the presence of more severe CAKUT phenotypes in the deceased sibling (died in utero, no data on other phenotypes) and that the mutations were in trans, this might be compound heterozygous inheritance. Variability in expression is uncommon in recessive diseases but was reported in autosomal recessive polycystic kidney disease/congenital hepatic fibrosis, hereditary hemochromatosis and familial hypobetalipoproteinemia.
Frem2, FRAS1-related extracellular matrix protein 2 gene, was identified as segregating in the mouse myUcl strain which showed phenotypes similar to that of Fras1−/− mutant mice. Frem2 was expressed in mesonephric and metanephric epithelium, especially in the ureteric bud21. Frem2 shares several structural domains with Fras1 including the core region of 12 CSPGs. Jadeja et al. detected a homozygous mutation in FREM2 in 2 families with Fraser syndrome not linked to FRAS121. To date, there have been only 2 different homozygous mutations of FREM2 reported, representing 3 unrelated families with Fraser syndrome21, 22. Homozygous FREM2 mutations occurred in syndromic form of Fraser syndrome, whereas we found heterozygous FREM2 mutations in individual with non-syndromic unilateral renal agenesis.
Criteria that established causality for rare genetic variants are usually, i) absence of the allele from >50 ethnically matched control individuals, ii) “allelic strength” as supported by the finding that the variant leads to truncation or absence of the gene product (mutation of start codon, obligatory splice site, stop-codon mutations, frame-shift mutations), iii) previous publication of the allele as disease causing, and iv) functional studies in cell based systems or animal models, if available. In our study, many of these criteria apply, because, i) we confirmed the absence of all mutations in 96 healthy control individuals of European origin, 270 control individuals from the “1,000 Genomes project” and additional 141 ethnically matched healthy controls, ii) though most mutations are not truncating, they are well conserved and are predicted to be “damaging” using the prediction software, and iii) mutations in these genes were known to cause CAKUT as part of Fraser syndrome.
To evaluate the frequency of FRAS1 and FREM2 variants in normal population, we searched in “1,000 Genomes” dataset and found 81 and 20 exonic SNPs in FRAS1 and FREM2 respectively, of which ~50% (40 in FRAS1 and 9 in FREM2) are synonymous variants. Among non-synonymous variants, all 11 changes in FREM2 and 36 out of 41 in FRAS1 were annotated in dbSNP132 with frequency above 1%, leaving only 5 variants in FRAS1 unannotated. In 4 probands with FRAS1 mutations, there are 3 additional FRAS1 exonic variants with Polyphen scores between 1.435–1.575. These 3 variants with ‘possibly damaging’ Polyphen score were annotated as known SNPs with allele frequency between 5–44.7%, which very likely excludes them as functional alleles.
Here we emphasize the advantages of using DNA pooling and massively parallel exon resequencing over the conventional method of exon PCR in single individuals, which is expensive, highly labor intensive and hardly feasible in diseases with broad genetic heterogeneity. The estimated cost of next generation sequence analysis in our laboratory was reduced to be about $50–60 per individual for 313 exons23. In this study we were able to identify potential CAKUT-causing mutations in 8 out of 40 individuals (20%). In Otto et al. the exact same approach using pooled DNA was applied to 18 patients with known mutations in nephronophthisis associated ciliopathy genes. After massively parallel exon resequencing of 376 different pooled PCR products derived from the DNA pool of 18 different mutation carriers, the analysis revealed 22 out of 24 known mutations (92%) or 8% false negative24. This weakness may partially be overcome by sequencing more than one lane upon Illumina™ sequencing of pooled PCR products. Another weakness may lie in the fact that confirmation of changes by Sanger sequencing is cumbersome. This limitation can now be addressed by using the array-based Integrated Fluidic Circuit by Fluidigm™ platform with 48-sample throughput25. We also might miss mutations in areas of low coverage (<400-fold), mutations in promoter or intronic exon splice enhancers, insertions or deletions larger than 2 bp in length, and complete exon deletions. In order to overcome the relatively high false positive error rate delivered by massively parallel exon resequencing, we prioritized our analysis to detect mutations that are predicted to be damaging. The improvement in coverage might solve this problem in future project.
METHODS
Human subjects
Blood or DNA samples were obtained from 40 individuals diagnosed with CAKUT from 38 different families. The diagnosis was made by pediatric nephrologists on the basis of imaging studies. After informed consent was obtained, detailed clinical data and pedigree information was referred to us by the specialists through a standardized clinical questionnaire (www.renalgenes.org). The cohort of 40 individuals included 29 (72.5%) individuals diagnosed with unilateral renal agenesis, 3 (7.5%) individuals with renal hypodysplasia, 7 (17.5%) individuals with VUR and 1 (2.5%) individual with bilateral duplex systems.
Whole genome amplification and DNA pooling
To normalize various DNA samples, whole genome amplification was performed on DNA of 40 different individuals and 96 healthy control samples using Phi29 based DNA polymerase strand displacement amplification (GenomiPhi DNA amplification kit, GE Healthcare). Subsequently, whole genome amplified DNA was purified using 96 well spin columns (Rapid 96TM PCR purification system, Marligen-Biosciences). DNA of 20 individuals were pooled at 2 µg each and diluted to 60 ng/µl. Pooling was repeated to represent 40 individuals. In addition, an equimolar DNA pool was generated by pooling 96 DNA samples derived from healthy individuals of Caucasian origin [Human Random Control DNA Panel-1 (HRC-1); European Collection of Cell Cultures, Salisbury, UK].
PCR amplification and massively parallel exon resequencing
All 313 exons (342 amplicons) of 30 candidate genes: ATGR2, BMP4, CDC5L, EMX2, EYA1, FOXC1, FRAS1, FREM2, GATA1, GDF11, GDNF, GFRA1, GREM1, HOX11, HOXA11, KAL1, PAX2, RET, ROBO2, SALL1, SIX1, SIX2, SIX4, SIX5, SLIT2, SPRY1, TCF2 (HNF1β), UPK3A and USF2 were individually amplified using exon flanking primers (Supplementary table 2) on each of the DNA pools. PCR products from the same pool were combined and enzymatically modified. The modified PCR fragment mixture were then used to construct an Illumina sequencing library using “Genomic DNA Sample Prep Kit” as previously described10. Each library was run on a single lane of a Solexa/Illumina Genome Analyzer GAII platform, generating about 15–20 million single-end sequence reads of 39 bases each.
Sequence analysis and mutation carrier identification
Sequence alignment was performed with CLC Genomics Workbench™ software (CLC-bio, Aarhus, Denmark) using imported and annotated human reference genome assembly NCBI36/hg18 as a reference. Sequence reads were mapped to exonic coding regions plus adjacent 100 bp intronic sequence of all 313 exons. Variant calls were obtained using the following filter parameters: Coverage ≥ 400-fold, variant frequency > 1%, and a minimum variant count of 5 reads. The variant analysis included coordinates of obligatory splice sites, and all variants predicted to change the amino acid sequence (missense, nonsense and coding indels). Variants present in dbSNP130, the “1,000 Genomes Project” (270 control individuals, http://www.1000genomes.org/page.php) or in the healthy control pool of 96 individuals (HRC-1) were excluded from further analysis. To prioritize for downstream analysis we scored missense variants according to the information of evolutionary conservation and the likelihood of a potential protein damaging effect using PolyPhen software predictions (http://genetics.bwh.harvard.edu/pph/)26. All variants with a predicted “probably damaging” or “possibly damaging” effect and a score above 1.4 were further analyzed. The selected variants were amplified by PCR for each individual in the corresponding DNA pool with subsequent Sanger sequencing to identify the mutation carrier.
Supplementary Material
ACKNOWLEDGEMENT
The authors wish to thank the physicians and families for participating in this study. We acknowledge Robert Lyons and the University of Michigan DNA Sequencing Core for excellent Illumina sequencing.
F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Frederick G.L. Huetwell Professor. This research was supported by grants from the National Institutes of Health to F.H. (R01-DK045345 and R01-DK088767).
Footnotes
DISCLOSURE
The authors declare that there is no conflict of interest.
REFERENCES
- 1.Pohl M, Bhatnagar V, Mendosa A, et al. Toward an etiological classification of developmental disorders of the kidney and upper urinary tract. Kidney Int. 2002;61(1):10–19. doi: 10.1046/j.1523-1755.2002.00086.x. [DOI] [PubMed] [Google Scholar]
- 2.Woolf AS. A molecular and genetic view of human renal and urinary tract malformations. Kidney Int. 2000;58(2):500–512. doi: 10.1046/j.1523-1755.2000.00196.x. [DOI] [PubMed] [Google Scholar]
- 3.NAPTRCS. Annual Report. 2003. [Google Scholar]
- 4.Abdelhak S, Kalatzis V, Heilig R, et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997;6(13):2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- 5.Lu W, van Eerde AM, Fan X, et al. Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am J Hum Genet. 2007;80(4):616–632. doi: 10.1086/512735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruf RG, Xu PX, Silvius D, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101(21):8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber S, Taylor JC, Winyard P, et al. SIX2 and BMP4 mutations associate with anomalous kidney development. J Am Soc Nephrol. 2008;19(5):891–903. doi: 10.1681/ASN.2006111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichikawa I, Kuwayama F, Pope JCt, et al. Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int. 2002;61(3):889–898. doi: 10.1046/j.1523-1755.2002.00188.x. [DOI] [PubMed] [Google Scholar]
- 9.Hoskins BE, Cramer CH, 2nd, Tasic V, et al. Missense mutations in EYA1 and TCF2 are a rare cause of urinary tract malformations. Nephrol Dial Transplant. 2008;23(2):777–779. doi: 10.1093/ndt/gfm685. [DOI] [PubMed] [Google Scholar]
- 10.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2010 doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGregor L, Makela V, Darling SM, et al. Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat Genet. 2003;34(2):203–208. doi: 10.1038/ng1142. [DOI] [PubMed] [Google Scholar]
- 12.Vrontou S, Petrou P, Meyer BI, et al. Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat Genet. 2003;34(2):209–214. doi: 10.1038/ng1168. [DOI] [PubMed] [Google Scholar]
- 13.Pitera JE, Scambler PJ, Woolf AS. Fras1, a basement membrane-associated protein mutated in Fraser syndrome, mediates both the initiation of the mammalian kidney and the integrity of renal glomeruli. Hum Mol Genet. 2008;17(24):3953–3964. doi: 10.1093/hmg/ddn297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slavotinek AM, Tifft CJ. Fraser syndrome and cryptophthalmos: review of the diagnostic criteria and evidence for phenotypic modules in complex malformation syndromes. J Med Genet. 2002;39(9):623–633. doi: 10.1136/jmg.39.9.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Haelst MM, Maiburg M, Baujat G, et al. Molecular study of 33 families with Fraser syndrome new data and mutation review. Am J Med Genet A. 2008;146A(17):2252–2257. doi: 10.1002/ajmg.a.32440. [DOI] [PubMed] [Google Scholar]
- 16.van Haelst MM, Scambler PJ, Hennekam RC. Fraser syndrome: a clinical study of 59 cases and evaluation of diagnostic criteria. Am J Med Genet A. 2007;143A(24):3194–3203. doi: 10.1002/ajmg.a.31951. [DOI] [PubMed] [Google Scholar]
- 17.Kohlhase J, Wischermann A, Reichenbach H, et al. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet. 1998;18(1):81–83. doi: 10.1038/ng0198-81. [DOI] [PubMed] [Google Scholar]
- 18.Kiefer SM, Ohlemiller KK, Yang J, et al. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum Mol Genet. 2003;12(17):2221–2227. doi: 10.1093/hmg/ddg233. [DOI] [PubMed] [Google Scholar]
- 19.Nishinakamura R, Matsumoto Y, Nakao K, et al. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128(16):3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- 20.Xu PX, Adams J, Peters H, et al. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23(1):113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 21.Jadeja S, Smyth I, Pitera JE, et al. Identification of a new gene mutated in Fraser syndrome and mouse myelencephalic blebs. Nat Genet. 2005;37(5):520–525. doi: 10.1038/ng1549. [DOI] [PubMed] [Google Scholar]
- 22.Shafeghati Y, Kniepert A, Vakili G, et al. Fraser syndrome due to homozygosity for a splice site mutation of FREM2. Am J Med Genet A. 2008;146A(4):529–531. doi: 10.1002/ajmg.a.32091. [DOI] [PubMed] [Google Scholar]
- 23.Janssen S, Ramaswami G, Davis EE, et al. Mutation analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum Genet. 2010 doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2011;48(2):105–116. doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voelkerding KV, Dames S, Durtschi JD. Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12(5):539–551. doi: 10.2353/jmoldx.2010.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.