Abstract
OBJECTIVE
Patients with diabetic neuropathy (DN) demonstrate variable degrees of nerve regeneration and degeneration. Our aim was to identify risk factors associated with sural nerve degeneration in patients with DN.
RESEARCH DESIGN AND METHODS
Demographic, anthropometric, biochemical, and anatomical data of subjects with DN from a 52-week trial of acetyl-L-carnitine were retrospectively examined. Based on the change in sural nerve myelinated fiber density (ΔMFD%), subjects were divided into three groups: regenerator (top 16 percentiles, n = 67), degenerator (bottom 16 percentiles, n = 67), and intermediate (n = 290), with dramatically increased, decreased, and steady ΔMFD%, respectively. ANOVA, Fisher exact test, and multifactorial logistic regression were used to evaluate statistical significance.
RESULTS
ΔMFD%s were 35.6 ± 17.4 (regenerator), −4.8 ± 12.1 (intermediate), and −39.8 ± 11.0 (degenerator). HbA1c at baseline was the only factor significantly different across the three groups (P = 0.01). In multifactorial logistic regression, HbA1c at baseline was also the only risk factor significantly different between regenerator (8.3 ± 1.6%) and degenerator (9.2 ± 1.8%) (odds ratio 0.68 [95% CI 0.54–0.85]; P < 0.01). Support Vector Machine classifier using HbA1c demonstrated 62.4% accuracy of classifying subjects into regenerator or degenerator. A preliminary microarray experiment revealed that upregulated genes in the regenerator group are enriched with cell cycle and myelin sheath functions, while downregulated genes are enriched in immune/inflammatory responses.
CONCLUSIONS
These data, based on the largest cohort with ΔMFD% information, suggest that HbA1c levels predict myelinated nerve fiber regeneration and degeneration in patients with DN. Therefore, maintaining optimal blood glucose control is likely essential in patients with DN to prevent continued nerve injury.
Twenty-five million Americans, or >8% of the population, have diabetes, and 1.9 million new cases were diagnosed in 2010 (1). In the U.S., the total cost for management of diabetes in 2007 was 218 billion USD (1). Complications of diabetes, including diabetic neuropathy (DN), nephropathy, and retinopathy, often have a significant impact on quality of life. DN is the most common diabetes complication; 60–70% of diabetic patients develop DN (2). DN is responsible for >60% of nontraumatic lower-limb amputations (1,3). Management of DN-related complications accounts for an estimated 27% of the total cost of diabetes treatment (3).
The most common type of DN is distal symmetric polyneuropathy. It affects the longest axons in the extremities first and progresses proximally in a stocking-glove pattern with increasing severity and duration of diabetes (2). The sural nerve is one of the most frequently affected nerves in DN. Although overall sural myelinated fiber density (MFD) decreases with age, the nerve itself can regenerate, making the grafting of sural nerves into other injured nerves possible (4). Axonal regeneration is a natural response of the body to compensate for damage caused by diabetes, but incomplete or unsuccessful regeneration may constitute a critical component in DN progression (5).
Our laboratory maintains a unique repository of human sural nerve biopsies harvested as part of a double-blind placebo-controlled clinical trial testing acetyl-L-carnitine (ALC) efficacy for DN (6,7). ALC treatment alleviated pain symptoms but had no effect on sural nerve conduction velocities (NCVs), amplitudes, or MFD (6). Our initial demographic analyses of these participants revealed that elevated serum triglycerides measured at trial onset correlated with DN progression after correcting for baseline DN severity and clinical factors, such as sex, age, duration and types of diabetes, insulin treatment, ALC treatment, and HbA1c (7). A subsequent study identified 532 differentially expressed genes (DEGs) between progressive and nonprogressive DN, highly enriched with immune response and lipid metabolism (8). Our previous studies focused on the loss of absolute MFD over the course of a 52-week clinical trial, resulting into two groups of patients (a progressor group with ≥500 fibers/mm2 MFD loss and a nonprogressor group with ≤100 fibers/mm2 MFD loss). While reexamining these data, we observed that ~43% of the subjects gained MFD over 52 weeks. Although modest regeneration has been documented in DN (9), no study has investigated critical factors affecting nerve regeneration in DN.
In the current study, we reexamined this DN cohort with MFD data available to identify critical factors that may impact sural nerve regeneration, focusing on subjects with the greatest gain or loss of MFD. As patients at different ages and duration of diabetes tend to have different levels of baseline MFD (4), we opted to investigate MFD percent change (ΔMFD%), rather than an absolute change in MFD to identify the clinical factors closely associated with MFD change over the specific duration of the disease course. With use of this classification approach, biomarkers and differential gene expression distinguishing patients exhibiting regeneration were examined and identified.
RESEARCH DESIGN AND METHODS
For all subjects, demographic, anthropometric, biochemical, and anatomical data included in the current study were reported previously (6,7). Briefly, human sural nerve biopsies were obtained during a double-blind, placebo-controlled, 52-week trial of ALC. The trial included both type 1 and 2 diabetic patients—all with existing neuropathy. A sural nerve biopsy (week 0, baseline) and a blood sample were collected at the time of patient enrollment, and the following measures were recorded: HbA1c, hematocrit, serum triglycerides, cholesterol, and albumin. After 52 weeks of treatment, measures of DN were reassessed and a second sural nerve biopsy was harvested (week 52) from the opposite leg. All the harvested biopsies were processed at the Nerve Biopsy Laboratory, University of Michigan, according to the published protocols (10). No blood sample was collected at the end of the trial. Among the 748 participants in the trial, 427 participants had two sural nerve biopsies and complete blood chemistry analyses.
Outcome measures
The primary outcome measure was ΔMFD% at week 52. Three outlier subjects with >200% increase in MFD were excluded from any further analyses. In the remaining 424 subjects (ΔMFD% range −78.6 to 87.7%), 183 subjects (42.9%) demonstrated positive ΔMFD%s. Based on ΔMFD%, the subjects were divided into three groups: regenerator (top 16 percentiles equivalent to beyond 1 SD from the mean); degenerator (bottom 16 percentiles), and intermediate (remaining subjects).
Neuropathy evaluations
Electrophysiological measurements, including bilateral sural NCV and amplitude, peroneal NCV, and amplitude on the dominant side and median motor and sensory NCV and amplitude on the nondominant side, were performed the baseline and completion of the trial to generate an O’Brien neuropathy score (6). These measurements were done in triplicate, and the median value was used.
Computational classifier for regenerator and degenerator
Computational classifiers of regenerator and degenerator were generated and evaluated using ORANGE (http://orange.biolab.si/), an open-source, component-based data-mining and machine learning software suite (11). Seven classification algorithms (Naïve Bayes, Logistic Regression, k Nearest Neighbors, Classification Tree, CN2 rules, Support Vector Machine [SVM], and Random Forest) available for binary class prediction were used with 20-fold cross-validation sampling to classify the subjects as regenerator or degenerator based on the demographic, anthropometric, and biochemical data.
Microarray data analysis
Based on the new grouping, we reanalyzed the previously published microarray data set (8) and an additional batch of unpublished microarray data (n = 35 and n = 33, respectively). These 68 microarrays included samples from 14 degenerators, 7 regenerators, and 45 intermediates. Microarrays were normalized using Robust Multiarray Average (12), and the batch effect was corrected using the distance-weighted discrimination method (13). Intensity-based moderated T statistics (14) was used to determine the DEGs among the groups. Owing to the small number of available microarrays (7 regenerators), a nominal P value of 0.05 without multiple testing corrections was used as the cutoff for DEGs.
The identified DEGs were further analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (15,16), to determine overrepresented biological functions in terms of Gene Ontology (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/). LRpath (http://lrpath.ncibi.org/), a logistic regression-based gene set enrichment testing tool, was also used in our analysis. LRpath accepts statistical significance values from all genes on the tested array and does not require a predefined DEG set (17).
Correlation between regeneration cluster density and MFD change
Electron microscopy (EM) was performed on the baseline and/or 52-week biopsies of approximately half of the subjects (n = 219) immediately after the termination of the 52-week trial. The number of regenerating nerve clusters were counted, and the density of regenerating clusters was calculated (6). In the current study, we correlated the ΔMFD% over 52 weeks with changes in the density of regenerating clusters for subjects with both baseline and 52-week biopsies examined by EM (n = 168).
Statistical analysis
Variable differences between the groups were analyzed with the Fisher exact test for categorical variables and ANOVA with Bonferroni post hoc tests for continuous variables. Multifactorial logistic regression between the regenerator and degenerator groups was performed to evaluate the effect of multiple factors including age, sex, ALC treatment, diabetes type, diabetes duration, HbA1c, insulin treatment, BMI, triglyceride, cholesterol, albumin, hematocrit, and O’Brien neuropathy rank-sum score (7,18). The statistical significance level was set at 0.05. For statistical analyses, R, version 2.15.2 (http://cran.r-project.org/), was used. Data are means ± SD or percentage unless otherwise stated.
RESULTS
Group classification
Based on the ΔMFD%, subjects were divided into three groups: regenerator (n = 67), degenerator (n = 67), and intermediate (n = 290). The mean ΔMFD%s ± SD were 35.6 ± 17.4 (regenerator), −4.8 ± 12.1 (intermediate), and −39.8 ± 11.0 (degenerator). Table 1 summarizes the demographic, anthropometric, and biochemical characteristics for all subjects, comparing subjects from regenerator, degenerator, and intermediate groups by ANOVA and Fisher exact tests.
Table 1.
Characteristics of subjects and statistical evaluation results
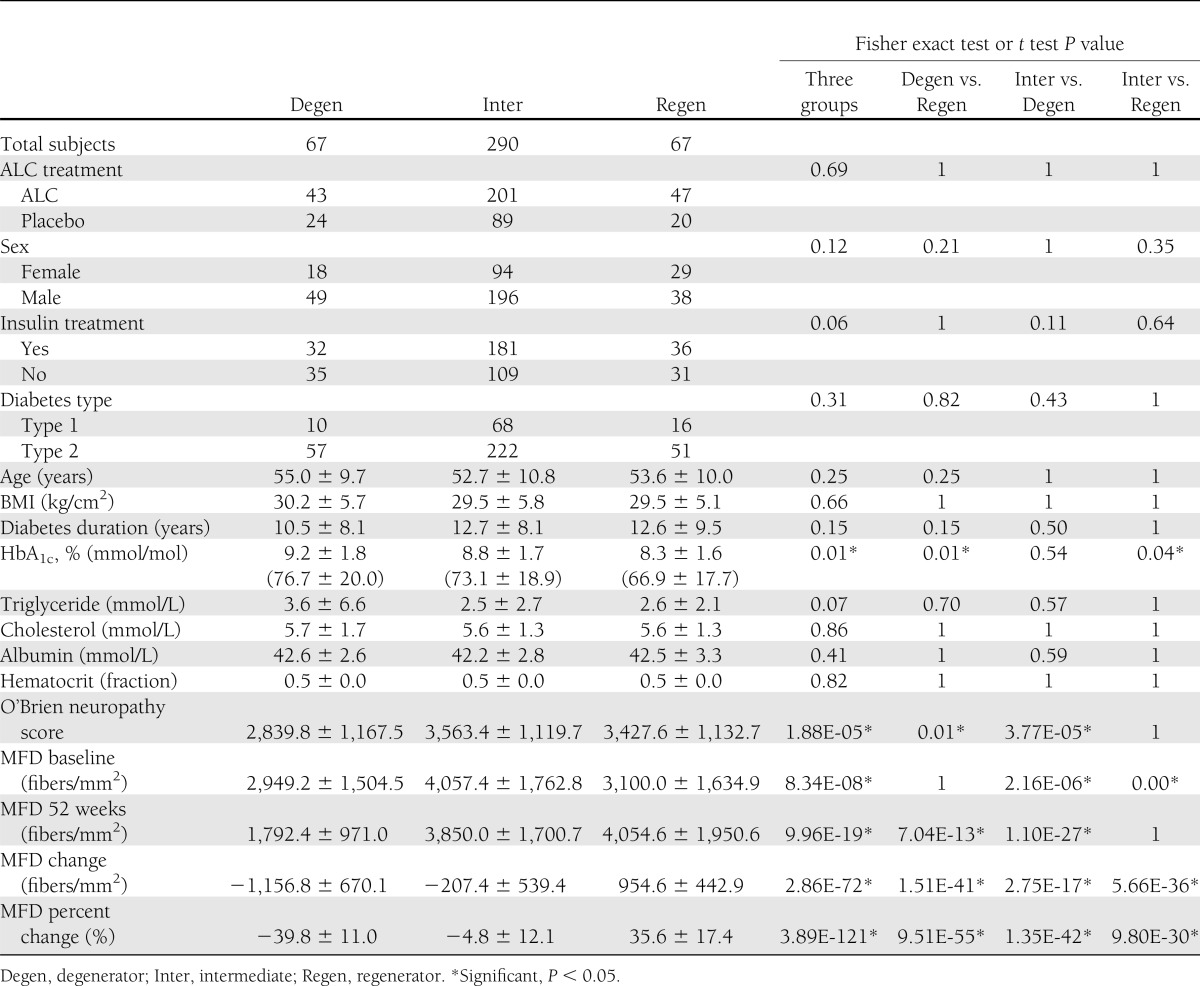
At baseline, there were no differences between groups in age, sex, diabetes duration or type, BMI, triglyceride, or total cholesterol. There were also no significant differences between groups in the number of subjects randomized to ALC or subjects treated with insulin. In addition, there were no differences between MFD at baseline between degenerator and regenerator, but MFD was significantly higher at baseline in the intermediate group (P = 8.34E-08). The groups differed in the O’Brien neuropathy rank-sum score, based on the electrophysiological measurements, with the lowest score observed in the degenerator group (2,839.8 ± 1,167.5) and the highest score in the regenerator (3,427.6 ± 1,132.7) and intermediate (3,563.4 ± 1,119.7) groups (P = 1.88E-05). Among the other evaluated risk factors shown in Table 1, HbA1c at baseline was the only risk factor significantly different across the three groups: regenerator (8.3 ± 1.6%), degenerator (9.2 ± 1.8%), and intermediate (8.8 ± 1.7%) (P = 0.01).
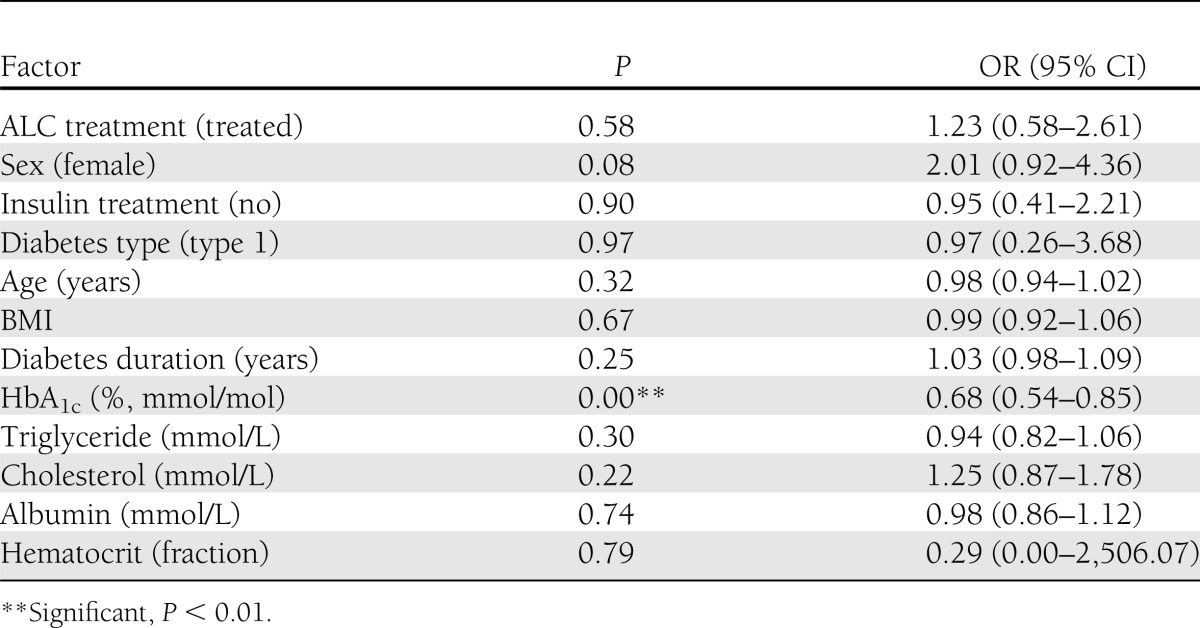
To further understand potential reasons that would drive regeneration or degeneration, we next compared the two extreme groups (degenerator vs. regenerator). Again, HbA1c level was the only significantly different biochemical factor (Bonferroni-corrected P = 0.01) between the two groups, while all other variables listed were not significantly different (Table 1). The multivariable logistic regression analysis also confirmed that the HbA1c level at baseline was the only significantly different factor (odds ratio [OR] 0.68 [95% CI 0.54–0.85]; P < 0.01) with other variables adjusted (Table 2). The regenerator group included more females and more patients with type 1 diabetes; however, the differences were not statistically significant. Interestingly, although these two groups had similar baseline MFD (2,949.2 ± 1,504.5 [degenerator] and 3,100.0 ± 1,634.9 [regenerator]; P = 1), the baseline O’Brien score was significantly lower in the degenerator group compared with the regenerator group (Table 1) (2,839.8 ± 1,167.5 vs. 3,427.6 ± 1,132.7 respectively, P = 0.01).
Table 2.
Factor analysis using multivariate logistic regression
Computational classifier for regenerator and degenerator
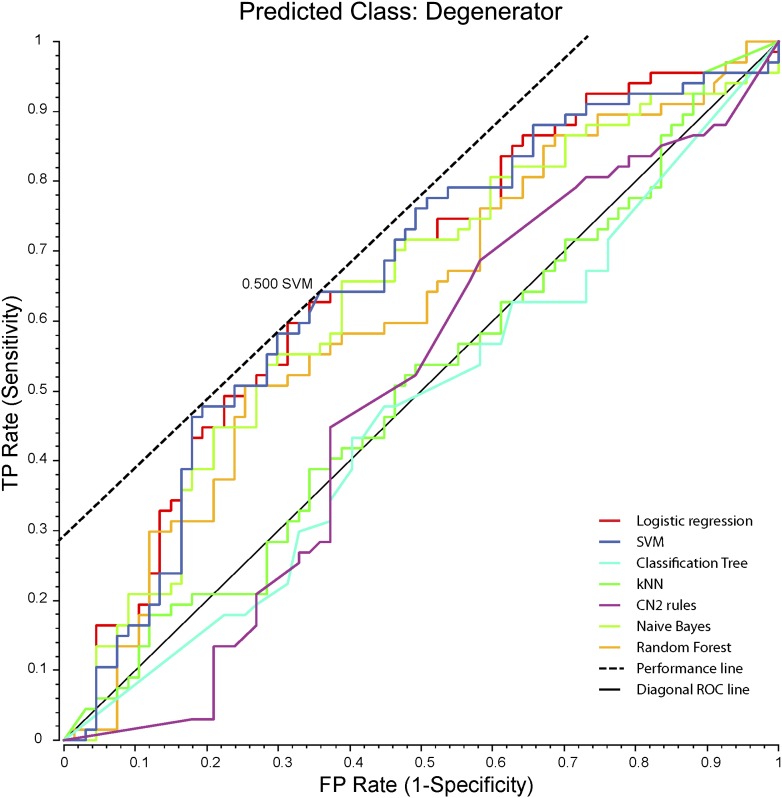
A machine-learning approach was used to test whether risk factors may predict the classification category outcome of participants with DN. Among the seven machine learning algorithms using a 20-fold cross-validation, two algorithms (SVM and logistic regression) achieved a classification accuracy (CA) >60%, with logistic regression being the best classifier (CA = 62.7%). Figure 1 illustrates receiver operating characteristic curves of the evaluated classifiers and indicates that SVM and logistic regression are the best classifiers. The addition of other factors resulted in degraded classification performance; however, O’Brien neuropathy score slightly improved the classifiers, with SVM achieving the highest CA of 64.2%.
Figure 1.
Receiver operating characteristic of the classifiers. The seven machine learning classification algorithms available in ORANGE were evaluated for classifying DN patient regenerator and degenerator groups. Classifiers were trained on the HbA1c levels of the subjects from these two groups, and testing was performed in 20-fold cross-validation. CN2, Clark Niblett 2; FP, false positive; kNN, k-nearest neighbor; TP, true positive.
Microarray data analysis
To examine the gene expression profiles that are significantly different between the two extreme groups, two batches of human sural nerve microarray datasets were combined (one published [8]). Intensity-based moderated T statistics identified a total of 490 DEGs between regenerator (n = 7) and degenerator (n = 15) at a nominal P value of 0.05 without multiple testing corrections. Supplementary Table 1 lists the 10 most upregulated and 10 most downregulated DEGs. Multiple immune-related genes such as CD177 molecule (CD177) (19), human leukocyte antigen (HLA) complex group 4 (HCG4), and chemokine (C-X-C motif) ligand 10 (CXCL10) (20) are upregulated in regenerator, indicating possible activation of multiple immune cell types such as neutrophils (19) and natural killer cells (20).
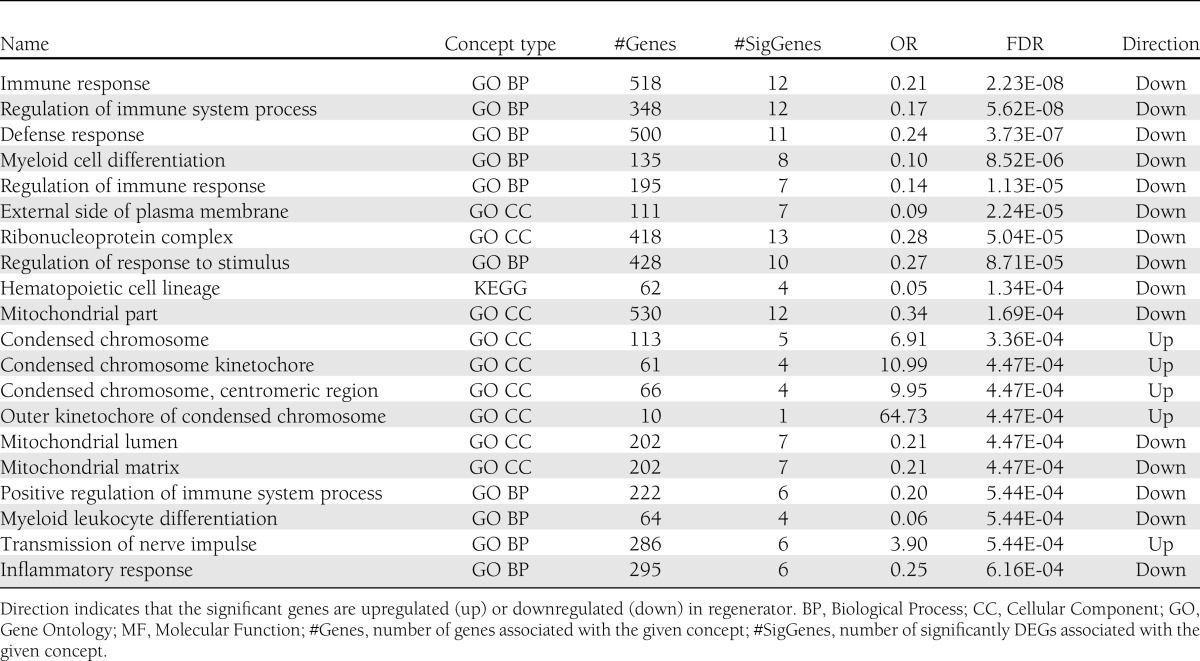
Table 3 lists the top 20 concepts (gene sets defined by biological functional terms such as Gene Ontology terms) identified by LRpath that have a significantly lower false discovery rate (FDR) for differential gene expression. Although some immune activation gene markers (CD177 and CXCL10) were markedly upregulated in regenerator, LRpath suggests that genes associated with immune response (FDR = 2.23E-08), defense response (FDR = 3.73E-07), and inflammatory response (FDR = 6.16E-04) were generally downregulated in regenerator. The top concepts upregulated in regenerator included condensed chromosome (FDR = 4.47E-04) and transmission of nerve impulse (FDR = 5.44E-04). Supplementary Table 2 lists all the significant genes in these 20 top concepts. DAVID, another gene set enrichment analysis tool, identified biological functional terms significantly overrepresented in the 490 DEG set. A heat map (Supplementary Fig. 1) was generated to summarize the most significant functions and indicates that the genes upregulated in regenerators were highly enriched with cell cycle, suggesting active regeneration.
Table 3.
Top 20 most differential biological functions between degenerator and regenerator identified by LRpath
Correlation with regeneration clusters
Approximately one-third (n = 168) of the study subjects had their baseline and 52-week biopsies examined by EM, and the Pearson correlation coefficient between ΔMFD% and the regeneration fiber density change was 0.33 (P < 0.001). When the analysis is limited only to the regenerator and degenerator groups, the correlation coefficient was 0.35 (P = 0.014 with n = 48). Although the correlation is not strong, the data suggest that the change in MFD is partially reflected in the decreased level of nerve regeneration. We also examined whether HbA1c level is correlated with the absolute density and the change of the regenerating clusters. The correlation between baseline HbA1c and the changes in regenerating clustering density was 0.02 for all 168 subjects; however, this correlation became −0.15 with the analysis limited to the regenerator and degenerator groups (n = 48). Although this does not reach the statistical significance cutoff, the negative correlation values suggest that there is a trend for decreased regeneration cluster density over 52 weeks with a higher baseline HbA1c level. However, no linear correlation was observed between the baseline HbA1c level and the baseline regeneration cluster density (correlation coefficient = 0.01 in both sets using all subjects and the two extreme sets). These results suggest that the HbA1c level at a certain time point may not be predictive of the absolute level of the regenerating cluster density but may partially predict the changes in the regenerating cluster and MFD over time.
CONCLUSIONS
Previous analyses by our group of human sural nerve biopsies harvested as part of a double-blind, placebo-controlled, 52-week trial of ALC for DN (6,7) revealed that elevated serum triglycerides measured at trial onset correlate with DN progression (7) and that the alterations in immune response and lipid metabolism genes are also associated with progressive DN (8). Further examination of these data, however, revealed that MFD improved in ~43% of the subjects, and although modest regeneration has been documented in DN (9), no study has investigated critical factors affecting nerve regeneration in DN. In the current study, we examined demographic, anthropometric, and biochemical data of these subjects to identify the potential risk factors that correlate with myelinated nerve fiber regeneration and degeneration. We found that HbA1c was the only factor significantly associated with regeneration and degeneration. In fact, the baseline HbA1c level alone was able to correctly classify 62.7% of the subjects as a regenerator or degenerator.
This study is an extension of the previously published analysis of the same cohort and was pursued in an attempt to better understand factors contributing to nerve fiber degeneration associated with DN. The previous study used the absolute loss of MFD over the course of the 52-week clinical trial as the classifier, resulting in three groups of patients (progressor group with ≥500 fibers/mm2 of MFD loss, nonprogressor group with ≤100 fibers/mm2 of MFD loss, and intermediate group for the remainders) (7,8). Therefore, the nonprogressor group in the previous study comprised all of the regenerator subjects from the current study, as well as a large portion of the intermediate subjects. Furthermore, the present subject groups were selected using the ΔMFD% as the classifier rather than absolute MFD change, as patients at different ages and with different durations of diabetes tend to have substantially variable levels of baseline MFD. Thus, the percent change, rather than absolute change of MFD, was used to evaluate the effects of several important clinical factors shown to contribute to DN progression.
Peripheral nerves undergo spontaneous regeneration upon injury; however, the risk factors in diabetes affecting nerve regeneration and degeneration are not clearly understood. Although the overall MFD decreases with age, the nerve itself may regenerate either spontaneously or in response to external stimuli (21). Axonal regeneration actively takes place as a natural compensatory response to damage caused by diabetes, but incomplete or unsuccessful regeneration may constitute a critical component of DN progression (5). We anticipated that genes related to axonal regeneration or cell growth would be more actively expressed in the regenerator compared with the degenerator group.
Microarray analyses confirmed that those genes involved in cell cycle functions are highly upregulated in regenerator compared with degenerator. Biological functions associated with neuron projection and myelin sheath were highly enriched in those genes upregulated in regenerator compared with the intermediate group. These functions were not significantly overrepresented in the DEGs between degenerator and regenerator, which is probably due to the low power of detecting differential expression with a limited number of samples. The results should be only considered as preliminary, and more samples need to be processed to increase the statistical power. It should also be noted that Schwann cells are major contributors to the mRNA in the sural nerve biopsies, with a small contribution coming from axons, epineural fibroblasts, adipocytes, vascular endothelial cells, and immune cells such as macrophages. Therefore, the gene expression changes observed by microarray are most likely to represent the changes in Schwann cells in response to diabetes.
Another interesting finding is the fact that in spite of similar MFD at baseline in the degenerator and regenerator groups, the degenerator group had a much lower O’Brien neuropathy score. The O’Brien scores are accepted methods to quantify multiple electrophysiological measurements obtained from nerve conduction studies (NCSs). NCSs assess mostly large myelinated nerve fiber function and are still considered by most as the gold standard end point for DN in clinical trials (22). Our findings suggest that nerve fiber function as assessed by NCS may be decreased before an anatomical loss of myelinated fibers and can be used in combination with other factors (such as HbA1c) to predict nerve fiber degeneration.
Fiber regeneration delays have been observed in both the tibial (largely motor) and sural (sensory) distal sciatic branches after both sciatic nerve crush injury and complete sciatic nerve transection in streptozocin-treated mice, a type 1 diabetes animal model (23). Interestingly, macrophage invasion was associated with the delay in this model, supporting a potential mechanism for impaired regeneration due to abnormal macrophage participation in nerve repair (23); however, the role of macrophages in nerve repair is still controversial (24–26). According to our microarray data, macrophage differentiation had a FDR of 0.018. Although only one gene, THO complex 5 (THOC5), was deemed a significant gene according to this concept, the overall changes of genes related to macrophage differentiation and other similar terms, such as macrophage activation, were found to be downregulated in regenerator by LRpath. More studies on the potential role of macrophages will be necessary to elucidate the exact mechanisms.
Study limitations include that blood chemistry data were only available at baseline and no subsequent measuring was done during or at the end of the trial. Therefore, controlling for in-trial changes in the covariates analyzed was not possible. It is possible that the overall HbA1c levels changed during the trial and data regarding lifestyle or diet changes were not available. In addition, even though the study cohort is the largest one available to date with ΔMFD% information, we may have lacked sufficient power to detect meaningful effects for all risk factors. Although diabetes type did not have a statistically significant effect on regenerator and degenerator classification in the current study, future separate analyses of type 1 and 2 diabetic subjects may be informative and will be pursued.
In the current study, we evaluated potential biomarkers and gene expression profiles of the sural nerve biopsies from the largest available DN patient cohort with ΔMFD% information in order to ascertain factors associated with nerve regeneration and degeneration in DN. The data suggest that HbA1c levels are significantly associated with the nerve regeneration and degeneration and may be predictive of future sural peripheral nerve regeneration. The microarray data suggest that immune and inflammatory responses may play a crucial role in nerve regeneration and degeneration. Although the exact mechanisms must still be elucidated, these data indicate that optimal blood glucose control in patients with DN is likely to impact sural nerve regeneration and that the immune response may play an important role in this process.
Acknowledgments
This work was supported by the American Diabetes Association (to E.L.F.), the Program for Neurology Research and Discovery, the A. Alfred Taubman Medical Research Institute, and a JDRF Postdoctoral Fellowship (to J.H.).
No potential conflicts of interest relevant to this article were reported.
J.H. collected and analyzed data and wrote the manuscript. K.A.S., R.P.-B., and B.C.C. contributed to discussion and reviewed the manuscript. E.L.F. obtained funding, supervised the analysis, and revised the manuscript. E.L.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented in abstract form at the 73rd Scientific Sessions of the American Diabetes Association, Chicago, Illinois, 21–25 June 2013.
The authors acknowledge Dr. Stacey Sakowski Jacoby at the University of Michigan for her expert editorial advice.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc12-2530/-/DC1.
References
- 1.National Diabetes Fact Sheet [internet], 2011. Available from http://www.cdc.gov/diabetes/pubs/pdf/ndfs_2011.pdf Accessed 5 November 2012
- 2.Edwards JL, Vincent AM, Cheng HT, Feldman EL. Diabetic neuropathy: mechanisms to management. Pharmacol Ther 2008;120:1–34 [DOI] [PMC free article] [PubMed]
- 3.Gordois A, Scuffham P, Shearer A, Oglesby A, Tobian JA. The health care costs of diabetic peripheral neuropathy in the US. Diabetes Care 2003;26:1790–1795 [DOI] [PubMed] [Google Scholar]
- 4.Wolford LM, Stevao EL. Considerations in nerve repair. Proc (Bayl Univ Med Cent) 2003;16:152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley JL, Thomas PK, King RH, et al. Myelinated nerve fibre regeneration in diabetic sensory polyneuropathy: correlation with type of diabetes. Acta Neuropathol 1995;90:403–410 [DOI] [PubMed] [Google Scholar]
- 6.Sima AA, Calvani M, Mehra M, Amato A, Acetyl-L-Carnitine Study Group Acetyl-L-carnitine improves pain, nerve regeneration, and vibratory perception in patients with chronic diabetic neuropathy: an analysis of two randomized placebo-controlled trials. Diabetes Care 2005;28:89–94 [DOI] [PubMed] [Google Scholar]
- 7.Wiggin TD, Sullivan KA, Pop-Busui R, Amato A, Sima AA, Feldman EL. Elevated triglycerides correlate with progression of diabetic neuropathy. Diabetes 2009;58:1634–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hur J, Sullivan KA, Pande M, et al. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain 2011;134:3222–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy JM, Zochodne DW. Impaired peripheral nerve regeneration in diabetes mellitus. J Peripher Nerv Syst 2005;10:144–157 [DOI] [PubMed] [Google Scholar]
- 10.Sullivan KA, Brown MS, Harmon L, Greene DA. Digital electron microscopic examination of human sural nerve biopsies. J Peripher Nerv Syst 2003;8:260–270 [DOI] [PubMed] [Google Scholar]
- 11.Curk T, Demsar J, Xu Q, et al. Microarray data mining with visual programming. Bioinformatics 2005;21:396–398 [DOI] [PubMed] [Google Scholar]
- 12.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264 [DOI] [PubMed] [Google Scholar]
- 13.Benito M, Parker J, Du Q, et al. Adjustment of systematic microarray data biases. Bioinformatics 2004;20:105–114 [DOI] [PubMed] [Google Scholar]
- 14.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 2006;7:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 16.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 2009;25:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyck PJ, O’Brien PC, Litchy WJ, Harper CM, Klein CJ, Dyck PJ. Monotonicity of nerve tests in diabetes: subclinical nerve dysfunction precedes diagnosis of polyneuropathy. Diabetes Care 2005;28:2192–2200 [DOI] [PubMed] [Google Scholar]
- 19.Sachs UJ, Andrei-Selmer CL, Maniar A, et al. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31). J Biol Chem 2007;282:23603–23612 [DOI] [PubMed] [Google Scholar]
- 20.Lee EY, Lee ZH, Song YW. CXCL10 and autoimmune diseases. Autoimmun Rev 2009;8:379–383 [DOI] [PubMed] [Google Scholar]
- 21.Coert JH, Dellon AL. Clinical implications of the surgical anatomy of the sural nerve. Plast Reconstr Surg 1994;94:850–855 [DOI] [PubMed] [Google Scholar]
- 22.Dyck PJ, Albers JW, Andersen H, et al. Toronto Expert Panel on Diabetic Neuropathy Diabetic polyneuropathies: update on research definition, diagnostic criteria and estimation of severity. Diabetes Metab Res Rev. 21 June 2011 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Kennedy JM, Zochodne DW. The regenerative deficit of peripheral nerves in experimental diabetes: its extent, timing and possible mechanisms. Brain 2000;123:2118–2129 [DOI] [PubMed] [Google Scholar]
- 24.Cui Q, Yin Y, Benowitz LI. The role of macrophages in optic nerve regeneration. Neuroscience 2009;158:1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patodia S, Raivich G. Downstream effector molecules in successful peripheral nerve regeneration. Cell Tissue Res 2012;349:15–26 [DOI] [PubMed] [Google Scholar]
- 26.Ydens E, Cauwels A, Asselbergh B, et al. Acute injury in the peripheral nervous system triggers an alternative macrophage response. J Neuroinflammation 2012;9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]