Abstract
The last step in eukaryotic translational initiation involves the joining of the large and small subunit of the ribosome, with initiator tRNA (Met-tRNAiMet) positioned over the start codon of mRNA in the P site. This step is catalyzed by initiation factor eIF5B. We have used recent advances in electron cryo-microscopy (cryo-EM) to determine a structure of the eIF5B initiation complex to 6.6 Å resolution from <3% of the population comprising just 5,143 particles. The structure reveals conformational changes in eIF5B, initiator tRNA and the ribosome that provide insights into the role of eIF5B in translational initiation. The relatively high resolution obtained from such a small fraction of a heterogeneous sample suggests a general approach for characterizing the structure of other dynamic or transient biological complexes.
Introduction
Initiation of protein synthesis is a key step in the control of gene expression in all kingdoms of life (1). Specific exogenous factors, called initiator factors (IFs), assist the ribosome in positioning the initiation codon of the messenger RNA (mRNA) and a charged initiator transfer RNA in the P site of the ribosome. Bacteria employ just three initiation factors, but eukaryotes use almost a dozen initiation factors, many of which are large multisubunit complexes themselves (reviewed in refs.2, 3, 4).
Despite substantial differences in the mechanism of initiation, all kingdoms of life share a subset of universally conserved initiator factors (5). In particular, the three bacterial factors IF1, IF2 and IF3 are functionally related to the eukaryotic factors eIF1A, eIF5B and eIF1 respectively. Of these, IF2 and eIF5B (and the archaeal aIF5B) are universally conserved GTPases that promote recruitment of the large ribosomal subunit at the final step of initiation. Recruitment of the large subunit and recognition of the initiator tRNA results in GTP hydrolysis and dissociation of the factor, leaving the ribosome in a competent state for elongation (6). Recent reports also relate eIF5B to the last step of pre-40S subunit maturation (7). The GTPase activity of eIF5B is essential for a pseudo-translational initiation cycle where eIF5B helps to position the 60S subunit for the last cleavage of 20S pre-rRNA (8).
Crystal structures of a starvation-stalled 80S ribosome (9), the 40S subunit in complex with eIF1 (10), and the 60S subunit in complex with eIF6 (11) have provided the first high-resolution snapshots of the eukaryotic translational machinery. However, the difficulty of obtaining large amounts of homogeneous complexes trapped in a specific state of translation presents a major barrier to crystallography. On the other hand, using several orders of magnitude less material, cryo-EM makes it possible to determine the structure of a minor population in a heterogeneous sample (12). Moreover, recent developments in electron detectors and statistical image processing now make it feasible to rapidly determine cryo-EM structures for the ribosome to near-atomic resolutions from relatively small data sets (13). These advances make possible interplay between EM and biochemistry that guide the preparation of appropriate samples to characterize a specific functional state. In the present work, we exploited these developments to determine the structure at 6.6 Å resolution of the complex of 80S ribosome, Met-tRNAiMet, mRNA and eIF5B with the non-hydrolyzable GTP analogue β-γ-methyleneguanosine 5′-triphosphate (GDPCP), which provides a snapshot of the last step in eukaryotic initiation.
Results
Interplay between electron cryo-microscopy and sample preparation
We used rapid but high-quality single-particle reconstructions by cryo-EM to characterize ribosomal samples and provide feedback for optimizing preparation of the initiation complex of eIF5B (Fig. 1).
Fig.1. Sample optimization strategy.
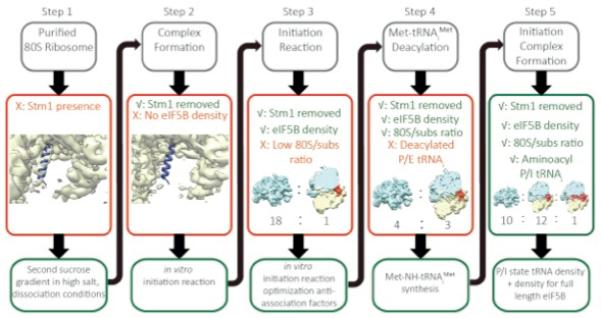
Our sample preparation procedure was based on structural feedback provided by cryo-EM reconstructions, and consisted of five steps that are described in detail in the main text. Cryo-EM reconstructed density for 60S subunits is indicated in cyan; density for the 40S in yellow; density for eIF5B/tRNA in red and Stm1 protein is depicted as blue ribbon. A similar colour scheme is used throughout all figures.
Initially, we isolated 80S ribosomes directly from Saccharomyces cerevisiae strain YAS-2488 using cells that were collected in mid-log phase. Despite harvesting in mid-log phase, cryo-EM reconstructions showed that the majority of ribosomes contained Stm1 (Fig. 1, Step 1), a protein that previously was considered to be bound to the ribosome as a result of glucose starvation and required for its crystallization (9). The ribosomes from this sample also crystallized in the same form as previously reported. As seen in the crystal structure of the 80S ribosome (9), this protein partially overlaps with P-site tRNA on the 60S as well as with the mRNA channel in the 40S, thus precluding the formation of most functional complexes.
To obtain ribosomes free of Stm1, we purified 60S and 40S ribosomal subunits and reassociated them. The density corresponding to Stm1 was absent in the reassociated 80S ribosomes, but initial attempts of complex formation with Met-tRNAiMet, eIF5B and mRNA showed no density for the factor (Fig. 1, Step 2).
We then proceeded to form the eIF5B complex by using the method for reconstituting translational initiation in yeast (14). To stabilize eIF5B on the ribosome, a mutant defective in GTP hydrolysis (15) was used in conjunction with GDPCP. In addition, the eIF5B used lacks the N-terminal unstructured tail (15, 16) and consists of 4 domains: an N-terminal nucleotide binding domain (G-domain) followed by domains II to IV.
When we analyzed the resulting sample by cryo-EM, we observed a mixture of 80S ribosomes and free subunits, and a very noisy background, which we attributed to unbound initiation factors. However, an initial reconstruction of 2,000 selected 80S particles showed a prominent density near the GTPase center of the 80S, which was unambiguously assigned to the G-domain and domain II (Fig. 1, Step 3).
Reducing the excess concentration of eIF1 and eIF2/eIF5 from a 3× to 1× molar ratio relative to ribosomal subunits resulted in an increased fraction of 80S ribosomes in the sample. We observed reasonable amounts of 80S particles per micrograph (~100 particles / 0.5 μm2, Fig. 2A), and reconstructions showed good density for eIF5B. However, only the G-domain and domain II of the factor were clearly visible. Moreover, the tRNA was in a hybrid P/E state interacting with the L1-stalk. This suggested that the tRNA had become deacylated (Fig. 1, Step 4) and the eIF5B had become partially disordered.
Fig. 2. Micrograph example, maximum-likelihood classification scheme and final maps.
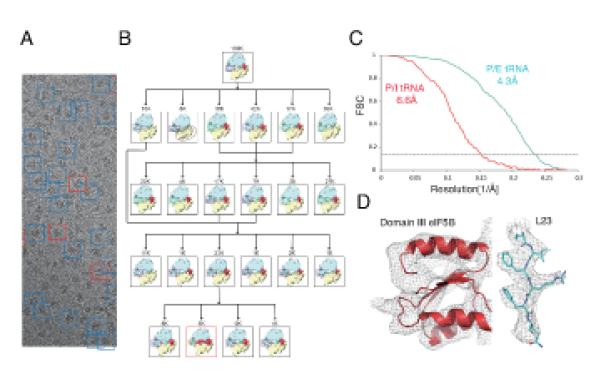
(A) Example of a micrograph obtained after the protocol optimization (B) Maximum-likelihood classification scheme used to localize the subpopulation of particles where density for all four domains of eIF5B could be identified. In (C), gold-standard Fourier shell correlation (FSC) curves for the small class (P/I-tRNA, red) and the larger class (P/E-tRNA, cyan) after refinement and statistical movie processing in RELION (34). Representative densities for the two maps are shown in (D).
To prevent deacylation, we synthesized a Met-NH-tRNAiMet version of the yeast initiator-tRNA, in which the ester bond between the methionine and the 3′ carbon of A76 of the tRNAiMet is replaced by an amide bond, thus exhibiting much slower deacylation under our experimental conditions (17). With this improved sample, we collected a cryo-EM dataset that comprised 130,030 particles. An elaborated classification protocol (Fig 2B) showed for the first time a class of 5,143 particles for which all four domains of eIF5B and the Met-NH-tRNAiMet were well defined in the reconstruction, with the tRNA in the P site (Fig. 1, Step 5). The resolution of this reconstruction was improved to 6.6 Å using statistical movie processing (13) (Fig. 2C). At this resolution, secondary structure elements were clearly resolved in the density (Fig. 2D, left) and could be assigned unambiguously to the factor and tRNA. A molecular model of the complex was obtained by rigid-body fitting of crystal structures of yeast 80S (9), and domains of eIF5B and tRNA, followed by molecular dynamics flexible fitting (MDFF) on the factor and tRNA (18). A second class of 40,729 80S particles was identified as the class described in Step 4 of figure 1. This class, which had an ordered G-domain and domain II of eIF5B, as well as a hybrid P/E-tRNA interacting with the L1 stalk, yielded a final resolution of 4.3 Å (Fig. 2C) at which amino acid side chains could be seen (Fig. 2D, right).
Overview of the structure
Relative to its conformation in the canonical ribosome, the 40S subunit in the complex is rotated anticlockwise by approximately 3.4 degrees with respect to the 60S subunit (Fig. 3A, movie S1). There is no additional swiveling of the head of the 40S subunit.
Fig.3. The eukaryotic 80S/ Met-tRNAiMet /eIF5B initiation complex.
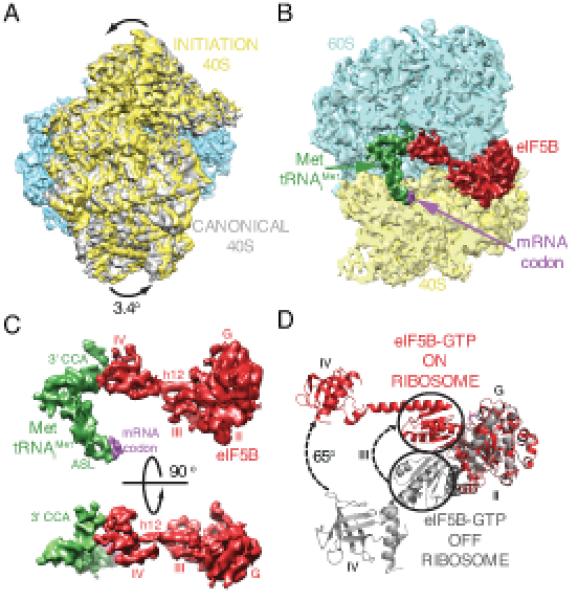
(A) Ratcheted overall conformation of the eukaryotic initiation complex. The 40S in the initiation complex (yellow) is rotated anticlockwise approximately 3.4 degrees from its position in the canonical 80S (gray (39)). (B) eIF5B (red), Met-tRNAiMet (green) and mRNA start codon (purple) bound to the 80S ribosome (60S in cyan, 40S in yellow). (C) Cryo-EM density showing the interaction between Met-tRNAiMet (green) and eIF5B (red) in two orientations. Domain IV of eIF5B contacts the 3′ CCA aminoacyl end of Met-tRNAiMet. (D) Conformational changes in eIF5B induced by during formation of the initiation complex on the ribosome. A coordinated displacement of domains III (circled) and IV, together with the linker α-helix 12 of eIF5B, is observed compared to the crystal structure of eIF5B in isolation (19).
The conformation of eIF5B in the initiation complex is markedly different from its crystal structure in isolation, either in complex with GDP or with the GTP non-hydrolyzable analog GDPNP (Fig. 3B-D) (Movie S2) (19). Domains III and IV, including the long α-helix h12 that links the two, move in a coordinated manner to adopt a position deep inside the inter-subunit space of the 80S. In this position, the β-barrel of domain IV is in close contact with the 3′-CCA end of the Met-tRNAiMet and in the vicinity of the peptidyl transferase center (PTC). A similar conformational change was also observed for the bacterial ortholog of eIF5B (IF2) on the bacterial initiation complex (20). In this conformation, eIF5B stabilizes the initiator tRNA in a conformation different from the canonical P-site orientation (17), preventing the 3′-CCA of the tRNA from reaching the PTC (Fig. 4; Movie S3). In addition, the anticodon stem loop (ASL) of the Met-tRNAiMet appears base paired with the start codon of mRNA and is slightly displaced from the position adopted by a canonical P-site tRNA (Fig. 4A, Movie S3). A similar change in the conformation of the P-site tRNA was also described in the bacterial initiation complex (20).
Fig.4. Met-tRNAiMet conformation on the initiation complex.
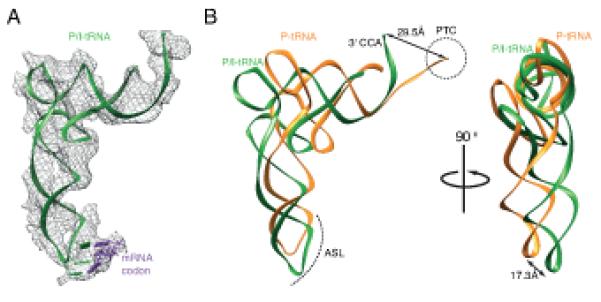
(A) Cryo-EM density for the Met-tRNAiMet allowed unambiguous docking of the tRNA (green) and the start codon of mRNA (purple). (B) Met-tRNAiMet (green) in the initiation complex compared to canonical P-site tRNA conformation (orange, PDB ID 2WDK/2WDL)(17). A displacement in the anticodon stem loop (ASL) is indicated. The 3′ CCA of the Met-tRNAiMet is stabilized in a conformation that prevents its entering the peptidyl transferase center (PTC), circled.
Initiator-tRNA recognition is coupled to GTPase activation by the simultaneous interaction of eIF5B with the initiator-tRNA via domain IV at the C-terminal end and the G-domain with the sarcin-ricin loop (SRL) of 28S RNA in the 60S subunit at the N-terminal end (Fig. 5A). This coupling is made possible by a shift of domain III relative to domain II and the G-domain. In the ribosome-bound conformation, the shifted domain III repositions the base of the connecting α-helix 12 (residues 835-840) to directly contact the SRL of the 28S-rRNA (Fig. 5B). This interaction aligns the SRL with the GTP-binding site on the G-domain in a conformation similar to what has been seen for the activated form of EF-Tu (21). A sequence alignment with the bacterial and archaeal orthologues of eIF5B (Fig. 5C) reveals a universally conserved sequence for these residues, providing further evidence for their importance in eIF5B function.
Fig.5. Role of domains III and the connecting helix 12 of eIF5B in GTPase activation.
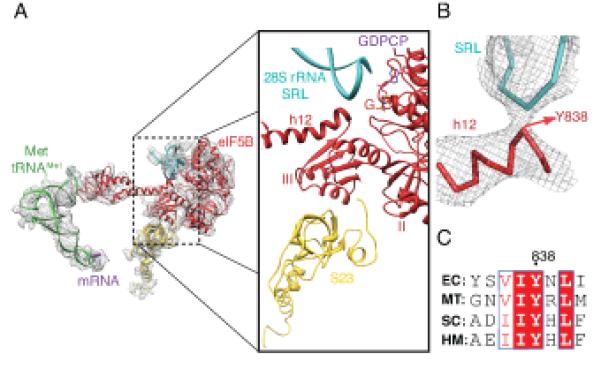
(A) Details showing the interaction of domain III of eIF5B with ribosomal protein S23 from the 40S (yellow) and the base of helix 12 with the sarcin ricin loop (SRL) of the 28S rRNA (B) Zoomed-in view of the interaction between helix 12 and the SRL around residue Y838. (C) A sequence alignment of IF2 from the bacteria Escherichia coli (EC), aIF5B from the archaea Methanothermobacter thermautotrophicus (MT) and eIF5B from the eukaryotes Saccharomyces cerevisiae (SC) and humans (HM) highlights the conservation of the Y838 and surrounding residues suggesting the importance of this interaction.
As expected, the G-domain is closely packed against the GTPase center of the 80S (Fig. 6). The GTP analog GDPCP is directly adjacent to the sarcin-ricin loop (SRL) of the 28S-rRNA (nucleotides 3014-3039). Ribosomal protein L9 (the eukaryotic orthologue of the bacterial protein L6) is within interaction distance of helix 6 of eIF5B, and orients the SRL for optimal contact with the G-domain. Moreover, the eukaryote-specific ribosomal protein L40 is in the vicinity of the GTPase center, and its N-terminal end is in close contact with helix 6 (Fig 6, top left). This protein is expressed as an N-terminally tagged ubiquitin-fusion protein, and the cleavage of the ubiquitin moiety is required for 40S subunit maturation (22). This close contact between the G-domain and the N-terminus of the cleaved L40 protein suggests that binding of translational GTPases is only possible after cleavage of the ubiquitin domain (11).
Fig.6. Two views of the eukaryotic ribosomal GTPase centre.
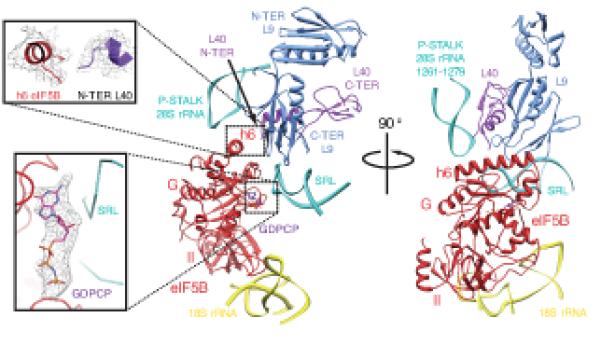
The G-domain and domain II of eIF5B (red) contact the GTPase center of the 80S formed by the ribosomal proteins L9 (blue), L40 (purple) and regions of the 28S rRNA (cyan) and the 18S rRNA (yellow). Helix 6 of eIF5B packs tightly against the C-terminal domain of L9 and the N-terminal domain of the eukaryote-specific protein L40 (inset, top left). Density for the GTP analog GDPCP was clearly seen in the G-domain of eIF5B facing the 28S-rRNA SRL (inset, bottom left).
Domains II and III of eIF5B make interactions with the 40S subunit (Fig. 6). An interaction between domain II and the 40S subunit was suggested by previous genetic and biochemical data on rRNA suppressor mutations in the body of the 40S that are able to restore the wild-type phenotype of a GTPase-deficient eIF5B mutant (16). Our structure also identifies a second major interaction through domain III of eIF5B and ribosomal protein S23, which is the yeast orthologue of bacterial protein S12 (Fig. 5A).
The P stalk of the 60S is the equivalent of the L12 stalk in bacteria that is implicated in interaction with translational GTPases (23). Upon interaction with eIF5B, the P stalk as a whole moves downwards compared with its position in the empty 80S, to closely contact the G-domain of eIF5B. In particular, the element of secondary structure formed by the P-stalk rRNA residues 1265-1275 is projected towards eIF5B, contacting the G-domain of the initiation factor (Fig 6, center). The result is a further stabilization of eIF5B in the active conformation by the P stalk.
Discussion
In this work, we show how recent developments in cryo-EM that allow reconstructions to high resolution from an order of magnitude fewer particles than previously (13) can accelerate the determination of structures by guiding the biochemistry of sample preparation. The rapidity of cryo-EM reconstructions from a relatively small number of particles provided iterative feedback to efficiently steer us towards an optimized strategy for increasing the fraction of the sample that had the initiation complex of eIF5B (Fig.1). Even with this optimization, the fraction of this complex in the population was very small. However, when combined with new, powerful image classification algorithms (12), we were able to determine a 6.6 Å resolution structure of the eIF5B initiation complex on the ribosome from only ~5,000 particles comprising less than 3% of the particles in our sample.
The structure both allows for a comparison with a previous, lower-resolution structure of the corresponding bacterial complex (20), as well as sheds insights into the mechanism of this step in both kingdoms. The rotation of the small subunit relative to the large subunit, the change in the orientation of the initiator tRNA from the canonical P-site conformation and the change in the orientation of the long connecting helix in the factor that allows it to make contact with the 3′ end of tRNA in the ribosome are all features that appear to be common to both eukaryotes and bacteria. They therefore represent essential features of subunit joining and eIF5B/IF2 function. In addition, the molecular model derived from the substantially higher resolution of the current structure provides further mechanistic insights into eIF5B function.
The large conformational change in eIF5B compared to the isolated structure couples initiator tRNA recognition to GTP hydrolysis. It is known that the A1-U72 base pair is an essential discriminating feature of initiator tRNA (24). The C-terminal domain of eIF5B makes extensive contacts around this base pair, thus possibly specifically recognizing initiator tRNA. The Met-tRNAiMet is also bent at the anticodon stem exactly in the region characterized by three consecutive GC pairs that are the hallmark of all initiator tRNAs. It is possible that this sequence allows the stabilization of the bent conformation by eIF5B. The result is that the tRNA is stabilized by eIF5B in a conformation that places the aminoacyl 3′ end out of the PTC, thus preventing the formation of an elongation-competent complex until GTP hydrolysis and dissociation of eIF5B from the ribosome takes place.
Simultaneously, the altered conformation places the base of the long connecting helix 12 of eIF5B in contact with the SRL. This in turn, places the catalytic site of eIF5B in a conformation seen in the activated forms of EF-Tu and EF-G (21, 25). Thus the conformational change in eIF5B, by simultaneously stabilizing the GTPase activated catalytic site and the unusual conformation of the initiator tRNA, couples GTP hydrolysis to specific recognition of the initiator tRNA in the 40S subunit.
The large number of contacts made by eIF5B with both the 40S and 60S subunits is consistent with its role in facilitating subunit joining. However, it is known that eIF5B also accelerates the rate of 60S recruitment (26, 27). It is likely that the conformational changes in eIF5B occur when it binds the 40S subunit, where they are stabilized by its specific interaction with initiator tRNA. These result in additional contacts with both the 60S and 40S subunits. Moreover, the P-stalk of the ribosome is also stabilized as a result of factor binding. Acceleration of forward rates has often been suggested to be the result of induced conformational changes (e.g. ref. (28), and it is likely that the induced changes in the ribosome, tRNA and eIF5B during the formation of this initiation complex lead to an acceleration of the forward rate here. Finally, the close contact of eIF5B with the P stalk RNA and proteins L9 and L40 at its base (Fig. 6) would be sterically impossible with the immature ubiquitinated form of L40, thus precluding the recruitment of immature 60S subunits, which is another essential feature of eIF5B.
In conclusion, this work shows that the structure of a macromolecular complex can be determined by cryo-EM to high enough resolution to resolve secondary structures with relatively small numbers of particles that represent a very small fraction of the sample population. Since the preparation of significant amounts of homogeneous sample is a major barrier in the structural biology of large complexes, this approach is likely to be useful in the study of a variety of important processes. We have used these advances to shed light on the role of eIF5B in the final step of eukaryotic initiation.
Materials and methods
Preparation of ribosomes and initiation factors
Ribosomes were purified from the Saccharomyces cerevisiae strain YAS-2488. Cells were harvested in mid-log phase (OD600 = 2-4) and resuspended in 20 mM Hepes-KOH pH 7.45, 150 mM KCl, 150 mM K-acetate, 10 mM Mg+2-acetate, 1 mg/ml heparin, 0.1 mM PMSF, 0.1 mM benzamidine, 2 mM DTT. Cell pellets frozen in liquid nitrogen were mechanically disrupted by a blender. The lysate was thawed at 4°C, clarified by centrifugation for 20 mins at 14,500 g. Ribosomes in the supernatant were pelleted through a sucrose cushion for 4 hrs at 45,000 rpm in a Ti45 rotor (Beckman Coulter) in the same buffer supplemented with 1M sucrose. The pellets were resuspended in sucrose gradient buffer without sucrose (20 mM Hepes-KOH pH 7.45, 50 mM KCl, 5 mM Mg+2-acetate, 0.1 mM PMSF, 0.1 mM benzamidine, 2 mM DTT) and incubated for 15 minutes with 1 mM puromycin on ice. The sample was loaded on a 10-40% sucrose gradient and centrifuged for 16 hrs at 28,000 rpm in a Ti25 zonal rotor (Beckman Coulter). The single 80S peak was concentrated. For subunit purification, 80S ribosomes were exchanged into dissociation buffer (20 mM Hepes-KOH pH 7.45, 600 mM KCl, 8 mM Mg+2-acetate, 1 mg/ml heparin, 0.1 mM PMSF, 0.1 mM benzamidine, 2 mM DTT) before loading onto a sucrose gradient in the same buffer and centrifuged for 19 hours at 28,500 rpm in the Ti25 rotor. For re-association, the 60S and 40S subunits were exchanged separately to re-association buffer (3mM Hepes-KOH pH 7.45, 6.6mM Tris-acetate pH 7.2, 3mM NH4Cl, 6.6mM NH4-acetate, 48mM K-acetate, 4mM Mg+2-acetate, 2.4mM DTT), concentrated to 6 μM and stored at −80 C after being flash frozen in liquid nitrogen.
Initiation factors eIF1, eIF1A, eIF2, eIF5 were produced as previously described (29). An N-terminally truncated eIF5B version (residues 396-1002, with a molecular weight of 68 kDa) was cloned in a T7 based expression vector that added a 6xHis-tag followed by a TEV protease cleavage site at the N-terminal. A point mutation eIF5B-T439A was introduced using QuickChange Mutagenesis Kit (Invitrogen) and was confirmed by DNA sequencing. Over-expression in E.coli strain BL21-AI (Invitrogen) was followed by chromatography using HisTrap, Q-HP and Sephacryl S-200 (GE Healthcare). The resulting highly pure protein that was concentrated to 100 μM and snap frozen in liquid nitrogen.
tRNA and mRNA production
Yeast tRNAiMet purification was purified as previously described with minor modifications (30): following phenol extraction of cell lysate, the aqueous phase was ethanol precipitated and the re-suspended pellet was applied to a Q sepharose column in buffer A (20 mM Tris/HCl pH 7.5, 5 mM MgCl, 300 mM NaCl). tRNA was eluted with a linear gradient with buffer B (20 mM Tris/HCl pH 7.5, 5 mM MgCl, 1 M NaCl). The eluted peak was further purified using a TSK phenyl 5PW hydrophobic column in buffer A (10 mM ammonium acetate pH 6.3, 1.7 M ammonium sulphate) and eluted with an inverse ammonium sulphate gradient in the same buffer. A final purification step was performed in a C18 reverse phase HPLC column using as solvent A: 20 mM Tris-acetate pH 6, 20 mM Mg2+-acetate, 400 mM NaCl; and solvent B: 20 mM Tris-acetate pH 6, 20 mM Mg2+-acetate, 400 mM NaCl, 60% v/v methanol. The acylated version of tRNAiMet as well as the Met-NH-tRNAiMet were produced following previously established protocols (17). The mRNA with sequence 5′ GGAA[UC]4UAUG[CU]4C 3′ was chemically synthesised by IDT.
Complex formation
Initially ribosomes were reassembled by mixing equimolar amounts of 40S and 60S subunits, either by themselves or with an excess of Met-tRNAiMet, mRNA and eIF5B with GDPCP. Subsequently, the complex of eIF5B via the initiation pathway using 60S, 40S, Met-NH-tRNAiMet and initiation factors was assembled following procedures previously described (31) with modifications to the final concentrations of eIF1, eIF2 and eIF5 (1:1 molar ratio each with respect to ribosomal subunits). The 40S mix (40S, eIF1, eIF1A and mRNA), the ternary complex mix (eIF2, GTP and Met-tRNAiMet), and the 60S mix (60S, eIF5, eIF5B and GDPCP) were incubated separately for 5 minutes at 30 C. The ternary complex added to the 40S mix and incubated for an additional 5 minutes before addition of the 60S mix. After a further incubation of 5 minutes, the sample was cooled to 4 C and used immediately to make cryo-EM grids.
Electron microscopy
Aliquots of 3μl of the 80S initiation complex at a concentration of ~80 nM were incubated for 30 s on glow-discharged holey carbon grids (Quantifoil R2/2), on which a home-made continuous carbon film (estimated to be ~30 Å thick) had previously been deposited. Grids were blotted for 2.5s and flash cooled in liquid ethane using an FEI Vitrobot. Grids were transferred to an FEI Polara G2 microscope that was operated at 300 kV. Defocus values in the final data set ranged from 1.6-4.6 μm. Images were recorded manually on a back-thinned FEI Falcon II detector at a calibrated magnification of 79,096 (yielding a pixel size of 1.77 Å). A system built in house was used to intercept the videos from the detector at a speed of 16 frames for the 1 s exposures (13). All electron micrographs that showed signs of significant astigmatism or drift were discarded.
Image processing
All reconstructions described in Fig.1 were calculated using semi-automated image processing as outlined below. We used the swarm tool in the e2boxer.py program of EMAN2 (32) for semi-automated particle picking. For our final data set, we selected 193,297 particles from 1,012 micrographs. Contrast transfer function parameters were estimated using CTFFIND3 (33). All 2D and 3D refinements were performed using RELION (34).
We used reference-free 2D class averaging to discard 60S subunits and defective particles, resulting in 130,030 particles of the final data set for subsequent 3D refinement and classification. Refinement of all particles against a single model (a 60 Å low-pass filtered version of EMDB-2275 (13)) yielded a preliminary, consensus reconstruction with local fuzzy density for the 40S subunit, the L1 stalk and the factor. Subsequently, we employed a cascaded 3D classification scheme (Fig. 2,B) to identify a class of 5,143 particles, for which all four domains of the factor showed clear density. A second class of 40,729 80S ribosomes with tRNA in the P/E-site and only density for the G-domain and domain II of eIF5B were identified by a separate classification of all particles into 10 classes.
To further increase the resolution of both classes, we performed statistical movie processing as described previously (13). In this procedure, we used running averages of five movie frames; a standard deviation of 1° for the priors on the Euler angles; and a standard deviation of 1 pixel for the translations. Reported resolutions are based on the gold-standard FSC=0.143 criterion (35) and involved only soft masking. (Note that the gold-standard approach yields refinements that are free from overfitting, and that the previously common FSC=0.5 criterion would severely underestimate resolution in the absence of overfitting.) Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector, and then sharpened by applying a negative B-factor that was estimated using automated procedures (36) (−217 Å2 for the small class; −155 Å2 for the larger class).
Rigid-body and flexible fitting of 80S, tRNA and eIF5B crystal structures were performed using UCSF Chimera and MDFF (18, 37), respectively. We split the structure into multiple rigid-body sub-structures, comprising 40S and 60S subunits, eIF5B, tRNA, L1-stalk and P-stalk, and manually rebuilt those parts that were outside the cryo-EM density using COOT (38). MDFF refinements for the reported structures were performed at 300 K for 200,000 steps using both hydrogen bond and dihedral angle restraints and a weight on the EM density (GSCALE) of 0.3 kcal/mol. After MD, another 5,000 steps of energy minimization were performed using GSCALE = 5.0 kcal/mol. Compared to the rigid-body fit, numerous attempts at flexible fitting (with varying GSCALEs) of the entire 80S did not improve FSCs between the model and the map beyond the resolution used in MDFF. This lack of improvement suggests that at this resolution, the flexible fitting procedure generally results in overfitting. However, the eIF5B, the tRNA and the parts of the ribosome that are in contact with those (e.g. the L1-stalk and the P-stalk) showed large and obvious conformational changes with respect to available crystal structures. For these entities, rigid-body fitting was done for the various domains followed by minor adjustments of the interface between the domains using flexible fitting. A manual inspection using COOT was performed to check the accuracy of the final fit. For the tRNA, the anticodon stem loop and the rest of the molecule were fit as rigid bodies, followed by flexible fitting of the acceptor region which shows considerable deviation from canonical tRNA (17). Additional density corresponding to the mRNA start codon was clearly visible next to the anticodon in a conformation consistent with codon-anticodon base pairing. The rest of the mRNA appears disordered in the map. Since the fitted coordinates of the G-domain and domain II of eIF5B showed no obvious differences between the small and the larger class (with an r.m.s.d. of 1.2 Å between the two), we used the coordinates from the higher-resolution map of the larger class to describe the GTPase centre of the 80S interacting with the G-domain and domain II of eIF5B.
Supplementary Material
Movie S1 Movie showing an overview of the 6.6 Å cryo-EM density map of the initiation complex with eIF5B. Shown are the 60S subunit (cyan), the 40S subunit (yellow), the tRNA(green), mRNA start codon (purple) and eIF5B (red). The canonical conformation of the 80S is shown in gray(39).
Movie S2 Movie illustrating the conformational changes that occur in eIF5B (red) upon binding to the ribosome (40S subunit in yellow) during the formation of the initiation complex.
Movie S3 Movie illustrating the quality of the tRNA density and the fitted atomic model in the P/I state. The P/I-tRNA is shown in green, highlighting conformational differences with the canonical P-tRNA state (orange). The mRNA start codon is shown in purple.
Acknowledgements
We are grateful to Shaoxia Chen for technical support with cryo-EM, Greg McMullan for help in movie data acquisition, Toby Darling and Jake Grimmett for help with computing, and David Tourigny for discussions. We are also grateful to one of the reviewers for suggestions on properly modeling the P/I tRNA. XCB is supported by a EU FP7 Marie Curie postdoctoral fellowship. This work was funded by grants from the UK Medical Research Council (MC_U105184332 to VR and MC_UP_A025_1013 to SHWS); a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute and the Jeantet Foundation (VR); and by the Human Frontiers Science Programme (grant RGP 28/2009 to VR and JRL). The cryo-EM density maps have been deposited in the EMDB with accession numbers EMD-2421 (for the large class with P/E-tRNA) and EMD-2422 (for the small class with P/I-tRNA). Fitted atomic coordinates have been deposited in the PDB, with entry codes 4byl, 4byn, 4byo, 4byp, 4byq, 4byr and 4bys for the large class, and entry codes 4byt, 4byu, 4byv, 4byw, and 4byx for the small class.
References
- 1.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 5.Roll-Mecak A, Shin BS, Dever TE, Burley SK. Engaging the ribosome: universal IFs of translation. Trends Biochem Sci. 2001;26:705. doi: 10.1016/s0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- 6.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebaron S, et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19:744. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Shem A, et al. The Structure of the Eukaryotic Ribosome at 3.0 A Resolution. Science. 2011 doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 10.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2011;331:730. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 11.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 12.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 15.Shin BS, et al. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell. 2002;111:1015. doi: 10.1016/s0092-8674(02)01171-6. [DOI] [PubMed] [Google Scholar]
- 16.Shin BS, et al. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol Cell Biol. 2009;29:808. doi: 10.1128/MCB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabuco LG, et al. Applications of the molecular dynamics flexible fitting method. J Struct Biol. 2011;173:420. doi: 10.1016/j.jsb.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 20.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacombe T, et al. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol. 2009;72:69. doi: 10.1111/j.1365-2958.2009.06622.x. [DOI] [PubMed] [Google Scholar]
- 23.Ballesta JP, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol. 1996;55:157. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 24.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova TV, et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 27.Acker MG, et al. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol. 2009;385:491. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Algire MA, et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 31.Acker MG, Shin BS, Dever TE, Lorsch JR. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem. 2006;281:8469. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 32.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 34.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armache JP, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci U S A. 2010;107:19748. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 Movie showing an overview of the 6.6 Å cryo-EM density map of the initiation complex with eIF5B. Shown are the 60S subunit (cyan), the 40S subunit (yellow), the tRNA(green), mRNA start codon (purple) and eIF5B (red). The canonical conformation of the 80S is shown in gray(39).
Movie S2 Movie illustrating the conformational changes that occur in eIF5B (red) upon binding to the ribosome (40S subunit in yellow) during the formation of the initiation complex.
Movie S3 Movie illustrating the quality of the tRNA density and the fitted atomic model in the P/I state. The P/I-tRNA is shown in green, highlighting conformational differences with the canonical P-tRNA state (orange). The mRNA start codon is shown in purple.


