Abstract
Hypoxia-inducible factor (HIF) is a crucial regulator of cellular and systemic responses to low oxygen levels. Here we firstly cloned three HIF-α isoforms from the basal branches of Osteichthyes and used computational tools to characterise the molecular change underlying the functional divergence of HIF-α isoforms in different lineages. Only the HIF-1α and HIF-2α in African lungfish and amphibians were found under positive selection. HIF-1α and -2α were less functionally divergent in basal ray-finned fish than in teleosts, and showed conserved but different transcriptional activity towards specific target genes.
Keywords: HIF-α, Osteichthyes, Functional divergence, Positive selection
Abbreviations: N-ODD, N-terminal oxygen dependent degradation domain; C-ODD, C-terminal oxygen dependent degradation domain; N-TAD, N-terminal trans-activation domain; C-TAD, C-terminal trans-activation domain
Highlights
-
•
All three HIF-α isoforms are well preserved in basal ray-finned fish.
-
•
The HIF-1α and -2α in amphibians and lungfish are positively selected.
-
•
The HIF-1α and -2α are more functionally diverged in teleosts.
-
•
The HIF-1α and -2α in different lineages exhibit different levels of activity.
1. Introduction
Oxygen, which serves as the terminal electron acceptor in the production of aerobic energy, is a vital requirement for all metazoans at the systemic level. Maintaining oxygen homoeostasis is a crucial challenge for all aerobic organisms. An evolutionarily conserved pathway mediated by oxygen-dependent posttranslational hydroxylation of a transcription factor called hypoxia-inducible factor (HIF) plays a pivotal role in this process [1,2]. HIF is composed of an oxygen-dependent α-subunit and a constitutively expressed β-subunit, the HIF-α/β dimer binds to the hypoxia-response elements (HREs), which are associated with a wide range of transcriptional targets [3]. The transcriptional activity of HIF relies primarily on the stability of the HIF-α subunit. HIF-α is functionally inhibited and degraded by prolyl hydroxylase enzymes under normal oxygen levels (normoxia) but is stabilised in hypoxia. Three isoforms of HIF-α are expressed in higher metazoans, and each of these isoforms is encoded by a distinct gene locus. HIF-1α and HIF-2α, the best characterised of these isoforms, are similar in structure and function: each possesses two independent, oxygen-dependent degradation (ODD) domains, the N-ODD and C-ODD domains, and two trans-activation domains, N-TAD and C-TAD [4–6].
Fishes live in various types of aquatic environments in which the oxygen concentration differs both temporally and spatially; therefore, fishes encounter hypoxia more frequently than do air-breathing animals. Furthermore, large fluctuations in oxygen concentrations played a significant role in the evolution and distribution of fishes. Thus, the study of the evolutionary patterns of HIF-α in fishes should advance our understanding toward their biodiversity, evolution and adaptation to the environments. Members of Osteichthyes can be phylogenetically divided into Actinopterygii and Sarcopterygii. The former group contains the Chondrostei (including Polypteriformes and Acipenseriformes) and Neopterygii (including Lepisosteiformes, Amiiformes and teleosts); the latter includes the lungfish, coelacanths and tetrapods. To date, the evolution of HIF-α has been extensively investigated in teleosts and mammals, though no study has identified key links between these taxa. Bichir, sturgeon, paddlefish, gar (the four of which are generally referred to as “basal ray-finned fish” or “ancient fish”) and lungfish, which have been placed in the basal branches of the Osteichthyes, represent particularly important positions in the evolution of jawed vertebrate. Therefore, investigation of the evolutionary patterns of the HIF-α genes in these species is of great importance.
2. Material and methods
2.1. Total RNA extraction and cDNA synthesis
Tissue samples were collected from bighead carp (Hypophthalmichthys nobilis), silver carp (Hypophthalmichthys molitrix), Prenant's schizothoracin (Schizothorax prenanti), Namucuo naked carp (Gymnocypris namensis), Chinese sturgeon (Acipenser sinensis), North American paddlefish (Polyodon spathula), shortnose gar (Lepisosteus platostomus), Senegal bichir (Polypterus senegalus) and African lungfish (Protopterus annectens). Total RNA was isolated using TRIzol reagent (TaKaRa, Japan), suspended in RNase-free water and evaluated by electrophoresis and a BioPhotometer plus 6132 instrument (Eppendorf, Germany). The first-strand cDNA was synthesised using M-MLV reverse transcriptase (Promega, USA) according to the manufacturer's instructions. All the experimental procedures were approved by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (ETS No. 123), and by the institution of animal care and use committee of Institute of Hydrobiology, Chinese Academy of Sciences.
2.2. Gene amplification and rapid amplification of cDNA ends (RACE)
Partial cDNA fragments of HIF-1α, -2α and -3α were obtained by PCR amplification. The PCR reaction was performed as follows: one cycle of 94 °C for 5 min; 6 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 2 min; 27 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min and one cycle of 72 °C for 10 min. The PCR products were separated by 1% agarose gel electrophoresis, purified, ligated, cloned into the pMD18-T vector (TaKaRa, Japan) and sequenced.
To obtain the complete full-length sequences of the HIF-α genes, we performed 5′ and 3′ RACE for all the HIF-α fragments obtained. The templates were prepared using the FirstChoice RLM-RACE Kit (Ambion, USA). Gene-specific primers were designed for the initial and nested PCRs (Table S2), which were performed using 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 90 s. The obtained fragments were purified, subcloned into the pMD18-T vector (TaKaRa, Japan) and sequenced.
2.3. Phylogenetic analysis
Known sequences were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) or obtained through tBLASTn searches of the various genome assemblies. Protein sequence alignments were performed using Clustal W [7]. The aligned sequences were concatenated and subsequently used for phylogenetic inferences. Neighbour-joining trees of the Osteichthyes were constructed in MEGA 4 [8].
2.4. Sequence divergence and molecular evolution
The synonymous and nonsynonymous substitution rate was calculated with the CODEML programme in PAML version 4 using the maximum likelihood method [9]. The comparison of relative fixation rates of nonsynonymous (dN) and synonymous (dS) substitutions is known to offer insight into the molecular evolution of protein-coding genes. Multiple evolutionary models were tested. The branch model, which utilises different ω ratio parameters for different branches on the phylogeny, can be used to detect positive selection acting on the lineages of interest. The site model treats the ω ratio for any site in the gene as a random variable selected from a statistical distribution and allows it to vary among sites.
Two pairs of nested models that have proved to be particularly effective, M1a (neutral)–M2a (selection) and M7 (beta)–M8 (beta and ω), were used in the likelihood ratio test (LRT) in this research. The branch-site model A aims to identify the positive selection that affects only a few sites on the pre-specified foreground branches. A Bayes empirical Bayes (BEB) approach was used to calculate the posterior probabilities for the site classes under the site models (M2a and M8) and branch-site model A.
2.5. Functional divergence analysis
We adopted Gu's methods [10,11] to analyse the functional divergence of HIF-1α and HIF-2α within and between the ancient fish and teleosts (using cartilaginous fish as an outgroup). In theory, the evolution of duplicated genes can be divided into two stages. In the early stage, the evolutionary rate may increase for functional divergence, resulting in shifted functional constraints between clusters. In the late stage, purifying selection plays a central role in maintaining the related but distinct functions of two duplicate genes. The evolutionary rate in the late stage may be different (higher or lower) than the original rate, resulting in altered functional constraints, i.e., type-I functional divergence [10]; alternatively, it may be the same as the original rate, resulting in a radical shift in amino acid properties, i.e., type-II functional divergence [11]. The methods used to analyse these two types of functional divergence are integrated in the DIVERGE software [12].
2.6. Plasmid construction and luciferase reporter assays
Full-length HIF-1α (from Senegal bichir, shortnose gar, bighead carp and Namucuo naked carp) and HIF-2α (from Senegal bichir and Namucuo naked carp) cDNAs were generated by reverse transcription PCR and inserted into a pCMV-Tag2C expression vector using primers that incorporated restriction sites. The HRE-luciferase reporter construct was provided by Professor Wuhan Xiao (w-xiao@ihb.ac.cn). Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% foetal bovine serum (HyClone). The cells were seeded in 24-well plates for 24 h prior to transfection and then co-transfected with pCMV-Tag2C-HIF-1α (or 2α) and the HRE-luciferase reporter construct. The pTK-Renilla luciferase reporter was used as an internal control.
The cells were separated into two groups and cultured under normoxic or 1% O2 hypoxic conditions for 18 h, respectively, after co-transfection. The luciferase activity was determined using the Dual-luciferase Reporter Assay System (Promega). The data were normalised to the Renilla luciferase enzyme activity and were reported as the mean ± SEM of three independent experiments performed in triplicate. The statistical analysis (paired t-test) was performed using GraphPad Prism 5.
3. Results and discussion
We cloned 25 HIF-α sequences from 9 species (GenBank accession numbers JQ031035–JQ031059, Table S1). Sequence comparisons of HIF-1α between representatives of Actinopterygians (zebrafish and bichir) and Sarcopterygians (human and lungfish) showed high conservation in the crucial domains (Fig. S1), indicating the maintenance of significant functions throughout the bony fish evolutionary history. A deletion of approximately ten codons, which alters a highly conserved Asp-Pro-Ala/Val-Leu-Asn signature, has been noticed in the region between the CODD and NODD/N-TAD domains in the teleosts HIF-1α (Fig. S2). The deletion is not within any of the most important, currently known, functional domains, thus the functional consequences of this mutation remain unclear.
Lungfish HIF-3α was not detected in any tissues we examined, by using several pairs of primers and different PCR procedures, or in the lungfish EST and transcriptome databases we searched. We excluded the possibility of gene silencing or special-temporal expression by trial of amplifying single exons. Previous studies demonstrated that all three HIF-αs, corresponding to those in teleosts and tetrapods, have been found in sharks and share high similarity with their counterparts [13], indicating that HIF-α had already diverged into three classes before the split of bony fish and cartilaginous fish (Fig. 1). Therefore, considering that the HIF-3α was also absent from the amphibian genome, we speculated that HIF-3α might have been lost in the ancestors of basal Sarcopterygians after the divergence of Osteichthyes, which is probably due to the sharp decrement of oxygen concentration in the late Middle and Late Devonian [14,15]. As is known to us, HIF-3α can competitively targets and forms abortive transcriptional complexes with HIF-1α or HIF-2α, preventing them from binding to the HREs of their target genes [16–18]. Thus, the loss of HIF-3α would enable organisms to a more robust response to hypoxia.
Fig. 1.
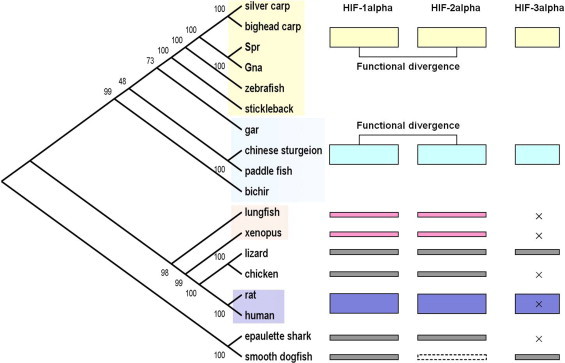
An overview of the HIF-α proteins in each lineage of Osteichthyes and their functional divergence in ray-finned fish. The phylogeny on the left was constructed using concatenated HIF-α sequences, with cartilaginous fish as an outgroup. Spr, Schizothorax prenanti; Gna, Gymnocypris namensis. The species marked with light yellow rectangles represent the teleosts, the species marked with light cyan rectangles represent the basal ray-finned fish, and the species marked with blue rectangles represent the mammals. All three HIF-α isoforms are present in each ray-finned fish species, but HIF-3α was absent (marked by ×) in the lungfish, amphibians, Aves and specific mammalian species (opossums); it is also absent from the epaulette shark, according to previous study. The dotted box represents the HIF-α sequences that should exist but have not yet been sequenced or predicted. The functional divergence between HIF-1α and HIF-2α in the ray-finned fish are marked in the upper right corner. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Phylogenetic relationship of bony fish was reconstructed using the concatenated HIF-1α, HIF-2α and HIF-3α sequences. Bichir was basal to all other ray-finned fish, while lungfish was placed at the bottom of the lobe-finned fish branch and formed a monophyletic group with tetrapods (Fig. 1). The relationship is supported by up-to-date opinions [19,20]. Based on this phylogeny, we investigated the selective pressure acting on each HIF-α isoform among the different lineages. The results in the the basal ray-finned fish were coordinated with that in teleost [13], each HIF-α isoform evolved under different negative selection pressure, indicating that the basal ray-finned fish HIF-α did not experience much genetic alteration that was then preserved by natural selection. The HIF-α exhibited a faster evolutionary rate in the Senegal bichir and shortnose gar than in the sturgeon and paddlefish (P < 0.05), most likely due to the slowed evolutionary rate of the Acipenseriformes genome [21]. In Sarcopterygii branch, positively selected sites were detected both in the lungfish and amphibian HIF-1α and HIF-2α (Table 1), some were located in the bHLH domain and ODD domain, suggesting a possible change in the regulatory mechanism. This may have some relationship with the loss of HIF-3α in these two lineages, because when a new genetic trait is beneficial and heritable, it tends to undergo positive selection.
Table 1.
Positively selected sites detected in lungfish and clawed frog HIF-1α & 2α genes.
| Gene | dN/dS value | Positively selected sitesa |
|---|---|---|
| Lungfish HIF-1α | 4.26723 | 74S 362P 434V 716T |
| Lungfish HIF-2α | 2.19682 | 383R 623Q |
| Clawed frog HIF-1α | 16.34256 | 491S 510S |
| Clawed frog HIF-2α | 30.54111 | 583T 816S |
Posterior probability >95%.
Computational methods [10,11] were used to obtain a rough estimate of the functional divergence of HIF-1α and HIF-2α in the basal ray-finned fish or teleosts. The results of two types of functional divergence reflected that HIF-1α and HIF-2α were more diverged in the teleosts than in the basal ray-finned fish (Figs. S3 and S4). Gene duplication is thought to be a major mechanism that provides raw materials for functional innovation. One possible fate of gene copies after duplication is that one copy maintains its original function, whereas the other is free to accumulate amino acid changes that lead to functional divergence [22]. As a result, many genes are represented by several paralogues in the genome that have related but distinct functions. Since the discovery of HIF, it has become increasingly clear that HIF-1 and HIF-2 perform similar functions, i.e., regulating target genes. Some target genes can be regulated by both of these proteins, whereas others can be regulated by only HIF-1 or HIF-2 [23–25]. Therefore, these proteins have related but distinct functions [26]. Thus, the results suggested that the responsibilities of HIF-1 and HIF-2 have become more defined during the evolution from basal ray-finned fish to teleosts and that the number of their common targets has decreased.
To investigate whether the functional divergence affect the transcriptional activities of HIF-1α and HIF-2α toward their common targets, we constructed HIF-1α expression plasmids using the HIF-1α sequences from Senegal bichir, shortnose gar, bighead carp and Namucuo naked carp and HIF-2α expression plasmids using HIF-2α from Senegal bichir and Namucuo naked carp. Each HIF-1α can significantly upregulate the luciferase activity (P < 0.001, Fig. 2A), suggesting that the HIF-1α function is conserved in different evolutionary lineages. However, it appears that the HIF-1α of basal ray-finned fish is more efficient and powerful. Additionally, it should be noted that even under normoxic conditions, the luciferase activity was very high in the cells that were transfected with shortnose gar hif-1α, indicating that the shortnose gar HIF-1α protein might be more stable than the others when exposed to oxygen. The transcriptional activity of HIF-2α is as robust in Senegal bichir as it is in Namucuo naked carp (Fig. 2B), revealing no difference between basal fish and teleost. Additionally, we found that the transcriptional activity of HIF-1α was much more robust than that of HIF-2α in Senegal bichir (Fig. 2C), whereas the opposite was true in Namucuo naked carp (Fig. 2D), suggesting that the HIF-1α and HIF-2α exhibit different levels of activity in different lineages.
Fig. 2.
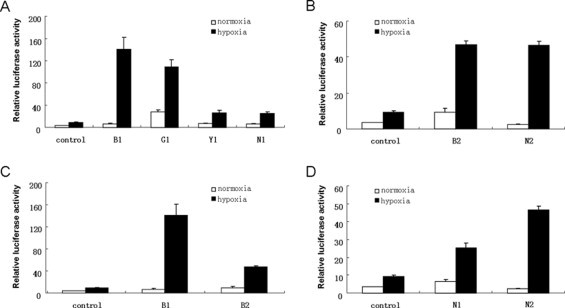
HEK-293T cells were co-transfected with pTK-Renilla luciferase control along with an HRE-luciferase reporter construct and HIF-1α (or HIF-2α) expression plasmid. The cells were divided into two groups and exposed to 21% O2 or 1% O2 for 18 h. The data are reported as the relative luciferase activities. (A) The increase in luciferase activity stimulated by HIF-1α of fish from different lineages due to hypoxia. (B) The increase in luciferase activity stimulated by HIF-2α of fish from different lineages due to hypoxia. (C) Comparison of the activity toward common targets between the HIF-1α and HIF-2α genes of Senegal bichir. (D) Comparison of the activity toward common targets between the HIF-1α and HIF-2α genes of Namucuo naked carp. B1, Senegal bichir HIF-1α; G1, shortnose gar HIF-1α; Y1, bighead carp HIF-1α; N1, Namucuo naked carp HIF-1α; B2, Senegal bichir HIF-2α; and N2, Namucuo naked carp HIF-2α.
HIF showed great significance in human adaptation to high altitude. Genome wide scans and exomes sequencing analysis of highland population in Tibet revealed the strongest signal of adaptive evolution in HIF-2α and PHD2, suggesting that during the long-term occupation of high-altitude areas, the functional sequence variations for acquiring biological adaptation to high-altitude hypoxia have been enriched in the HIF–PHD–VHL system in Tibetan populations [27–29]. In our study we adopted two high-altitude living fish, Prenant's schizothoracin and Namucuo naked carp, however, no clear signal of adaptive evolution of HIF-α was detected through sequence divergence analysis, selective pressure test or transcriptional activity examination, suggesting that natural selection did not directly force any crucial changes to the HIF-α sequences for the high-altitude environment adaptation.
Our studies answer several open questions about the evolution of HIF in the basal branches of Osteichthyes and serve as a foundation for future investigations of the speciation and migration of aquatic vertebrates due to fluctuations in oxygen content. It is common knowledge that the transcriptional activity of HIF relies primarily on the stability of HIF-α, which is sensitive to the oxygen availability. However, recent studies revealed a novel mechanism of HIF regulation. Patrick and his colleagues demonstrated that NF-κB can directly regulate HIF-β mRNA and protein, resulting in modulation of HIF-2α protein [30]. In another case, specific microRNA (miR-107) can also decreases hypoxia signalling by suppressing expression of HIF-β [31]. These studies suggesting an innegligible role HIF-β plays in the HIF regulation. Therefore, combined study of HIF-α and HIF-β in the future should benefit more to understanding animal diversity, their evolution and adaptation to environment.
Acknowledgements
We thank Doctor Yuanyuan Li for the helpful suggestions. Funding support was provided by the Grants 31090254 from the Natural Science Foundation of China (NSFC), the grants from Chinese Academy of Sciences (KSCX2-YW-Z-0807) and the National Basic Research Program of China (No. 2007CB411601).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Wei Chi, Email: bluetorry@163.com.
Xiaoni Gan, Email: ganxn@ihb.ac.cnl.
Wuhan Xiao, Email: w-xiao@ihb.ac.cn.
Wen Wang, Email: wwang@mail.kiz.ac.cn.
Shunping He, Email: clad@ihb.ac.cn.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.09.004.
Appendix. Supplementary materials
This document contains Supplementary Figs. 1–4 and Tables 1–3.
References
- 1.Schofield C.J., Ratcliffe P.J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin W.G., Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Semenza G.L. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 4.Semenza G.L. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 5.O’Rourke J.F., Tian Y.M., Ratcliffe P.J., Pugh C.W. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1alpha. J. Biol. Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- 6.Masson N., Willam C., Maxwell P.H., Pugh C.W., Ratcliffe P.J. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 10.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 11.Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol. Biol. Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 12.Gu X., Vander V.K. DIVERGE: phylogeny-based analysis for functional–structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- 13.Rytkonen K.T., Williams T.A., Renshaw G.M., Primmer C.R., Nikinmaa M. Molecular evolution of the metazoan PHD–HIF oxygen-sensing system. Mol. Biol. Evol. 2011;28:1913–1926. doi: 10.1093/molbev/msr012. [DOI] [PubMed] [Google Scholar]
- 14.Scott A.C., Glasspool I.J. The diversification of Paleozoic fire systems and fluctuations in atmospheric oxygen concentration. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10861–10865. doi: 10.1073/pnas.0604090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berner R. Phanerozoic atmospheric oxygen: new results using the geocarbsulf model. Am. J. Sci. 2009;309:603–606. [Google Scholar]
- 16.Hara S., Hamada J., Kobayashi C., Kondo Y., Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem. Biophys. Res. Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- 17.Maynard M.A., Evans A.J., Hosomi T., Hara S., Jewett M.A., Ohh M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–1406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- 18.Heikkila M., Pasanen A., Kivirikko K.I., Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell. Mol. Life Sci. 2011;68:3885–3901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broughton R.E., Betancur R.R., Li C., Arratia G., Ortí G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLOS Curr. 2013 doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betancur R.R., Broughton R.E., Wiley E.O., Carpenter K., López J.A., Li C., Holcroft N.I., Arcila D., Sanciangco M., Cureton I.I. JC. The tree of life and a new classification of Bony fishes. PLOS Curr. 2013 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieger J., Fuerst P.A. Evidence for a slowed rate of molecular evolution in the order acipenseriformes. Mol. Biol. Evol. 2002;19:891–897. doi: 10.1093/oxfordjournals.molbev.a004146. [DOI] [PubMed] [Google Scholar]
- 22.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;15:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C.J., Wang L.Y., Chodosh L.A., Keith B., Simon M.C. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covello K.L., Simon M.C., Keith B. Targeted replacement of hypoxia-inducible factor-1alpha by a hypoxia-inducible factor-2alpha knock-in allele promotes tumor growth. Cancer Res. 2005;65:2277–2286. doi: 10.1158/0008-5472.CAN-04-3246. [DOI] [PubMed] [Google Scholar]
- 25.Carroll V.A., Ashcroft M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66:6264–6270. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- 26.Rytkönen K.T., Akbarzadeh A., Miandare H.K., Kamei H., Duan C., Leder E.H., Williams T.A., Nikinmaa M. Subfunctionalization of cyprinid hypoxia-inducible factors for roles in development and oxygen sensing. Evolution. 2013;67:873–882. doi: 10.1111/j.1558-5646.2012.01820.x. [DOI] [PubMed] [Google Scholar]
- 27.Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 29.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Uden P., Kenneth N.S., Webster R., Muller H.A., Mudie S., Rocha S. Evolutionary conserved regulation of HIF-1beta by NF-kappaB. PLoS Genet. 2011;7:1–15. doi: 10.1371/journal.pgen.1001285. e1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamakuchi M., Lotterman C.D., Bao C., Hruban R.H., Karim B., Mendell J.T., Huso D., Lowenstein C.J. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains Supplementary Figs. 1–4 and Tables 1–3.


