Abstract
Molecular analyses of cochlear pathology rely on the acquisition of high-quality cochlear samples. For small rodents, isolating sensory cell-enriched samples with well-preserved RNA integrity for transcriptional analyses poses a significant challenge. Here, we report a microdissection technique for isolating sensory cell-enriched samples from the cochlea. We found that treating the tissue with RNAlater, a RNA preservation medium, alters the physical properties of the tissue and facilitates the dissection. Unlike previous samples that have been isolated from the sensory epithelium, our samples contain defined cell populations that have a consistent ratio of sensory cells to supporting cells. Importantly, the RNA components were well preserved. With this microdissection method, we collected three types of samples: sensory cell-enriched, outer hair cell-enriched, and inner hair cell-enriched. To demonstrate the feasibility of the method, we screened multiple reference genes in the sensory cell-enriched samples and identified stable genes in noise-traumatized cochleae. The method described here balances the need for both quality and purity of sensory cells and also circumvents many limitations of the currently-available techniques for collecting cochlear tissues. With our approach, the collected samples can be used in diverse downstream analyses, including qRT-PCR, microarray, and RNA sequencing.
Keywords: Hair cell, cochlea, method, dissection, mouse, RNA, RNAlater
1.1 Introduction
Cochlear hair cells are responsible for converting acoustic stimuli to neural impulses. They are also the major target of common pathological insults (Hu et al., 2002; Keithley and Feldman, 1982; Nakashima et al., 2000). Because hair cells in the mammalian cochlea are unable to regenerate, the loss of these sensory cells can lead to permanent hearing loss. Preventing such functional loss requires a better understanding of the molecular mechanisms that control hair cell pathogenesis.
The success of molecular analyses of cochlear pathology relies on the collection of high-quality cochlear samples. Due to the complexity of the cochlear structure, isolating sensory cell-enriched samples from mammalian cochleae has been a significant challenge for researchers in the field. This challenge is particularly striking for the mouse, which is a widely-used animal model for molecular studies of cochlear disorders.
To date, commonly-used methods for collecting sensory cell-specific samples involve disassociating the cells from the cochlea using mechanical and/or enzymatic approaches (Harter et al., 1999; He et al., 2000; Towers et al., 2011). The disassociated hair cells are then either manually picked up by a micropipette or automatically sorted via flow cytometry (Hertzano et al., 2011). Although these methods can result in a highly purified population of sensory cells, their ability to preserve RNA integrity has been a concern because without RNA protection, any disturbance of the cellular environment causes rapid RNA degradation and/or changes in RNA expression. Laser capture microdissection, another method that has been used to collect hair cell-specific samples in the cochlea (Anderson and Zheng, 2007), requires extensive tissue preparation, which can also be problematic in maintaining RNA integrity.
Traditionally, the most effective way to preserve the RNA integrity of cochlear tissues has been to extract the tissues in the shortest time possible in a cold medium (Christensen et al., 2009; Han et al., 2012; Szaumkessel et al., 2012). In recent years, RNA stabilization reagents have been developed to protect RNA (Mutter et al., 2004; Wang et al., 2006). These reagents allow high-quality RNA samples to be obtained from defined cochlear partitions, including the apical and basal segments of the cochlea containing the sensory epithelium and the modiolus (Sato et al., 2009), the lateral wall of the cochlea (Jin et al., 2008), and the sensory epithelium (Hu et al., 2009). Our recent study has further improved the spatial resolution of this partitioning by separating the apical and basal sections of the sensory epithelium (Cai et al., 2012). While the sensory epithelium sample contains a relatively higher percentage of hair cells than the samples from other preparations, the purity of sensory cells in this sample is still unsatisfactory because of the presence of a large quantity of non-sensory cell structures, such as supporting cells, mesothelial cells, blood vessels-derived cells, and extracellular matrix structures including the basilar membrane. Moreover, the precise cell composition of each individual sample is difficult to define. Inconsistent tissue composition can lead to a significant variation in the results of expression analyses conducted across several samples (Hertzano and Elkon, 2012). Because many new technologies, such as whole-transcriptome analysis using RNA-sequencing technology and miniature sample analysis using droplet PCR, are now available for cochlear tissues, there is an urgent need to develop a method to collect high-quality, sensory cell-enriched samples.
Here, we report a microdissection technique that permits the isolation of sensory cell-enriched samples with significantly improved sensory cell purity. Unlike previously obtained sensory epithelium samples, our samples contain defined cell populations with a relatively consistent ratio of sensory cells to supporting cells. Importantly, the RNA of these samples is well preserved. Using this microdissection method, we were able to collect three types of samples from the mouse cochlea: sensory cell-enriched, outer hair cell (OHC)-enriched, and inner hair cell (IHC)-enriched. To demonstrate the feasibility of this method, we screened 12 reference genes in the sensory cell-enriched samples and identified stable genes that are expressed in noise-traumatized cochleae. This method balances the need for both quality and purity of sensory cells and circumvents many of the limitations of the currently available techniques for collecting cochlear tissue.
2.1 Materials and methods
2.1.1 Animals
Both mice (C57BL/6J, 2–4 months old, male and female, the Jackson Laboratory, Bar Harbor, ME) and rats (Sprague Dawley, 2–3 months old, male and female, Charles River Laboratories, Wilmington, MA) were used in this study. All animals received a baseline hearing evaluation using auditory brainstem response testing. Only the subjects that exhibited a normal hearing sensitivity were included in this study. The procedures involving the use and care of the animals were approved by the Institutional Animal Care and Use Committee of the State University of New York at Buffalo.
2.1.2 Sample collection
To better describe the procedures used in this study, we used the following three anatomic distinctions: the cochlear sensory epithelium, the sensory organ partition, and the organ of Corti. The sensory epithelium was defined as the tissue located between the lateral wall and the modiolus, containing all components of the tissue, including cellular components, blood vessels, nerve fibers, the basilar membrane, and the osseous spiral lamina. The sensory organ partition consists of tissue that is similar to the sensory epithelium but lacks the structures medial to the inner border cell. The organ of Corti contains the cells between the Claudius cell and the inner sulcus cell on the top of the basilar membrane. It should be noted that the term “organ of Corti” has been used in some previous investigations to denote the tissue defined as the sensory epithelium in the current investigation. In the current investigation, we adopted the conventional meaning of the term that refers to a much more limited cell population (Iurato, 1961; Santi and Mancini, 1998).
The mouse cochlea has two turns. The first turn, also referred as the basal turn, is a major site of pathogenesis in many disease conditions; therefore, we selected this cochlear location for tissue collection. This region has a relatively wide distance between the inner and outer hair cells than does the basal end of the first cochlear turn, which mitigates the technical difficulty of separating the OHC-enriched and IHC-enriched tissues.
Initial preparation of the cochlea
The animal was decapitated under deep anesthesia with CO2. The cochleae were quickly removed from the skull and placed in an ice cold Dulbecco’s phosphate buffer saline solution (DPBS, GIBCO). Under a dissection microscope, the bony shell of the cochlea that faces the middle ear cavity was quickly removed. The modiolus was also removed along with the tissues of the lateral wall and the sensory epithelium; however, the section that was designated for sample collection in the apical portion of the first cochlear turn was left behind. Then, the cochlea was transferred into a PCR tube that contained 0.6 ml of an RNA-stabilizing reagent (RNAlater; Qiagen, Valencia, CA) and stored at 4°C until further dissection. The initial processing of the cochlea, which took only a few minutes, facilitated the rapid entry of the RNAlater reagent into the tissues. Moreover, the separation of the modiolus from the cochlea generated a smooth cut at the medial edge of the sensory organ partition (Fig. 1), which facilitated the subsequent microdissection.
Figure 1.
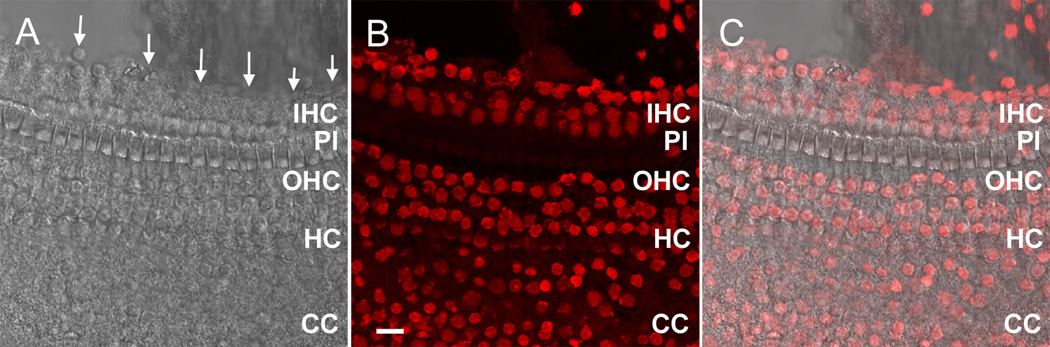
A microscopic view of the medial edge of the sensory organ partition following the initial cochlear preparation. A) A surface view of the tissue with arrows pointing to the medial edge. The osseous spiral lamina has been removed. B) The nuclei of the same tissue are stained with propidium iodide. The nuclei along the medial edge of the tissue (in the top section of the image) belong to IHCs, inner pillar cells, inner border cells and inner phalangeal cells. C) The merged image of (A) and (B). IHC: inner hair cell. PI: pillar cell. OHC: outer hair cell. HC: Hensen cell. CC: Claudius cell. Scale bar =15 µm.
Microdissection
The microdissection was performed within a period of 1 to 7 d after the initial cochlear processing. Each cochlea was transferred to a shallow, glass-bottom dish (35-mm Fluorodish with 10-mm glass, FD3510-100, WPI) that had been filled with 200–300 µl of the RNAlater reagent. Under a dissection microscope, the cochlea was oriented to visualize the stripe of the organ of Corti. Because of the differential light transmittance of cells in the fresh tissue that was stored in the RNAlater reagent, the three rows of outer hair cells and one row of inner hair cells were clearly visible under the dissection microscope (Figs. 2A and 2B).
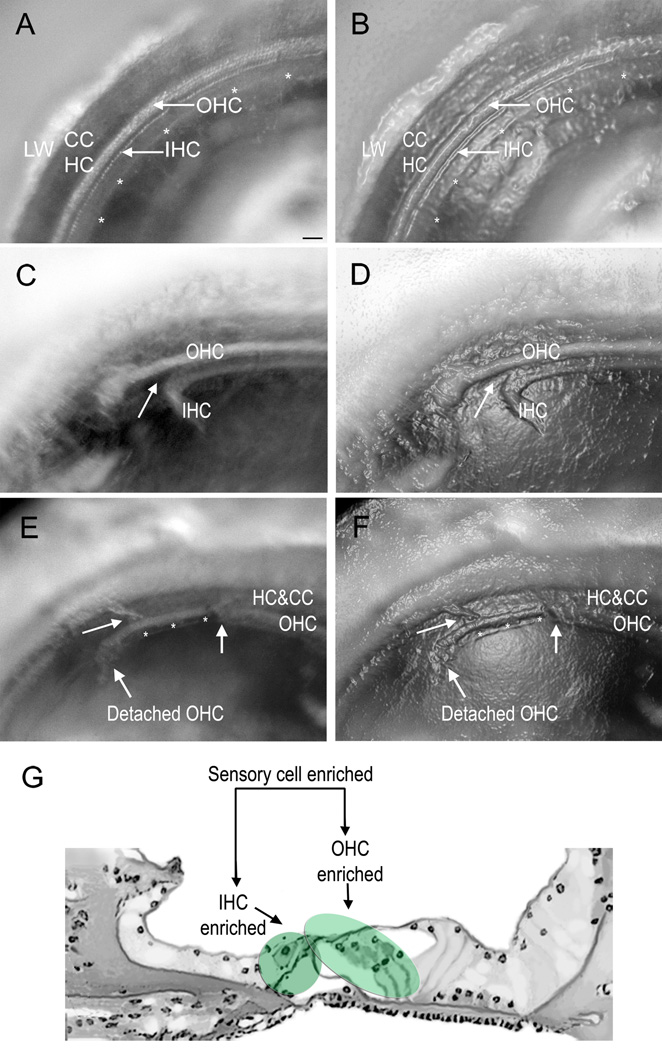
Figure 2.
Microphotographs of the sensory organ partition were taken at different stages of microdissection. To aid in the identification of the structures, the micrographs in (A), (C) and (E) have been enhanced with the “plastic wrap” function of Photoshop (Adobe Photoshop, CS3 Extended) to generate (B), (D) and (F), respectively. A) and B) A surface view of the tissue after the initial preparation but prior to microdissection. The asterisks mark the medial edge of the tissue. The white dots marked by arrows are hair cells. C) and D) A surface view of a cochlea after a portion of the IHC-containing region has been detached. On the left side, the IHC-containing region has been separated from the OHC-containing region. In contrast, the tissue remains intact on the right side. The arrow points to the site of separation in the pillar cell region. E) and F) A surface view of a cochlea where the OHC-enriched region has been partially separated from the Hensen cells. The IHC-containing region in this section has been removed (asterisks). On the right-hand side of the image marked by the arrow, both the IHC- and OHC-enriched tissues remain intact. G) A schematic drawing of the side view of the sensory organ partition. The shaded areas denote the regions for each type of enriched sample. LW: lateral wall. CC:claudius cell. HC: Hensen cell. OHC: outer hair cell. IHC: inner hair cell. Scale bar = 35 µm.
The medial edge of the sensory organ partition had already been disassociated from the osseous spiral lamina after the bony shelves of the osseous spiral lamina were removed during the initial tissue preparation phase. We therefore focused on separating the lateral edge of the tissue using a custom-made micro-knife. We found that the RNAlater treatment not only hardened the tissue but also weakened the cell-cell attachments, which facilitated the microdissection.
To collect sensory cell-enriched samples, we gently scraped the reticular laminar at the junction between the Deiters cells and the Hensen cells and pushed the tissue away from the basilar membrane. To collect OHC- and IHC-enriched samples, we first separated OHCs and IHCs at the site of pillar cells to collect the IHC-enriched tissues (Figs. 2C and 2D), and then we isolated the OHC-enriched tissue (Figs. 2E and 2F) using an identical method to the sensory cell-enriched sample collection. Fig. 2G shows the cochlear sites for the separation of the sensory cell-, OHC- and IHC-enriched samples.
2.1.3 Microscopic assessment of the size and cellular composition of the collected tissues
Surface view of collected tissues using an inverted microscope
The isolated tissue samples were transferred into a culture dish containing the RNAlater solution and were first examined using an inverted microscope (Eclipse TE300, Nikon, Inc.) to determine the general integrity and cellular composition of the tissue. The tissue was photographed using a camera attached to the microscope (SPOT RT, Diagnostic Instruments, Inc.), and the length and the area of the tissue stripe was measured using image processing software (ImagePro, Plus).
Nuclear inspection of the tissues using confocal microscopy
To verify its cellular composition, we stained each tissue with propidium iodide, a nuclear dye, and examined it under a confocal microscope (Carl Zeiss LSM510 multichannel laser scanning confocal imaging system) using a method modified from our previous study (Hu et al., 2002). Briefly, the collected tissue was fixed with 10% buffered formalin solution for 2 h. Then, the tissue was transferred to a propidium iodide solution (5 µg/ml in 10 mM PBS), and incubated for 10 min. After rinsing with PBS, the tissue was mounted on a slide containing an antifade reagent (Prolong® Gold antifade reagent, Invitrogen) and examined using the confocal microscope. A series of confocal images from the top to the bottom of the tissue were collected. The images were analyzed with imaging software (LSM Image Examiner, Version 40091) to identify the various cell types.
2.1.4 Quantification of cell numbers
To determine the number of cells in the tissue stripe, we counted the number of nuclei in the tissue. The nuclei at different locations of the tissue were distinguished using confocal microcopy. We also quantified the cells in the cochlear sensory epithelium, which we have defined as the region between the lateral wall and the modiolus. To arrive at this number, five cochleae were processed using a conventional surface preparation method (Hu et al., 2002). Briefly, mice were sacrificed, and their cochleae were collected and then fixed with a 10% buffered formalin solution. The cochleae were then dissected in 10 mM PBS to collect the sensory organ partition, and the tissues were stained with propidium iodide. The number of cells was determined in the apical portion of the first cochlear turn, which corresponds to the cochlear site where sensory cell-enriched samples were collected. While the collected tissues also contained cells from the lateral wall and the osseous spiral lamina, we only counted the cells in the sensory epithelium.
2.1.5 RNA extraction and quality assessment
Total RNA was extracted from the collected tissues using the RNeasy Micro Kit or RNeasy Mini Kit (Qiagen) as previously described (Hu et al., 2012). To analyze the expression pattern of reference genes, one sample was generated from each cochlea. To assess the quantity and quality of the total RNA, tissue from three cochleae was pooled to generate one sample, which was then analyzed using the Agilent RNA 6000 Pico Kit and the Agilent Bioanalyzer 2100 (Agilent Technologies) to obtain the RNA Integrity Number (RIN) and the RNA concentration.
2.1.6. Reference gene expression based on qRT-PCR arrays
To demonstrate the feasibility of using these RNA samples for transcriptional analysis, we examined the expression pattern of 12 reference genes with a previously reported qRT-PCR array technique (Hu et al., 2012). Briefly, the animals were exposed to an intense noise (see the "Noise Exposure" section for details) and were then sacrificed 2 h, 1 d, or 7 d later (n = 4 cochleae from 4 individual animals for each time point). Four additional animals that did not experience the noise exposure were used as controls. A sensory cell-enriched sample was collected from one cochlea (either the right or the left) of each animal. The length of the tissue stripe was measured, and this was used to normalize the tissue for expression analyses. Total RNA was also extracted from each sample.
We used a mouse reference array (PAMM-0002, Qiagen) to determine the expression pattern of 12 reference genes (18SrRNA, Actb, B2m, Gusb, Hprt, Hsp90ab1, Ldhal6b, Nono, Ppia, Rpl13a, Tbp, and Tfrc). Total RNA was isolated from each sample using the RNeasy Micro kit (Qiagen) according to the manufacturer's instructions. The isolated total RNA was used to generate cDNA, which was preamplified using the RT2 PreAMP cDNA Synthesis Kit and RT2 PreAMP Primer Mix (Qiagen). The PCR product was mixed with RT SYBR Green/Fluorescein qPCR Master Mix (Qiagen) and transferred to the PCR array plate. qRT-PCR was performed using a Bio-Rad MyiQ Single-Color Real-Time PCR System. The expression level of each reference gene was calculated as the averaged raw Ct value.
2.1.7 Individual qRT-PCR analysis of reference genes
We used a conventional qRT-PCR approach to analyze the expression levels of three reference genes (Hprt1, Actb and Rplp1) in sensory cell-enriched samples that had been collected from rat cochleae. The tissue from one cochlea was used to generate one sample, and four biological replications were performed. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and was then made into cDNA using a High Capacity cDNA reverse transcription kit (Life Technologies). The cDNA underwent an additional amplification stage using the TaqMan PreAmp Master Mix (Life Technologies) for 10 cycles. The resulting PCR product was used for qRT-PCR on a MyIQ two-color real time PCR detection system (BioRad). We also used this procedure to examine the expression level of solute carrier family 26, member 5 (Slc26a5), a gene that codes for prestin that is expressed exclusively in OHCs (Zheng et al., 2000).
2.1.8 Auditory brainstem response (ABR) measurement
To assess the hearing sensitivity of the subjects used for enriched sample preparation and to determine the threshold shifts of the animals used for reference gene analysis after noise exposure, ABR was measured in both the normal condition and after the noise exposure (2 h, 1 d and 7 d post-exposure) as previously described (Hu et al., 2012). Briefly, each animal was anesthetized with an intraperitoneal injection of ketamine (87 mg/kg) and xylazine (3 mg/kg). The ABRs were elicited with tone bursts at 5, 10, 20, 30, and 40 kHz (0.5 ms rise/fall Blackman ramp, 1 ms duration, alternating phase) at a rate of 21 bursts/s. The electrodes were placed subdermally over the vertex (non-inverting input) and posterior to the stimulated (inverting input) and nonstimulated ears (ground) of the animal. The outputs of the electrodes were delivered to a preamplifier/base station (RA4LI and RA4PA/RA16B; TDT). The responses were filtered (100–3000 Hz), amplified, averaged using TDT hardware and software, and stored and displayed on a computer. The ABR threshold was defined as the lowest intensity that reliably elicited detectable responses. During the ABR measurement, the animal was placed on a heating pad (Homeothermic Blanket Control Unit; Harvard Apparatus) to keep its body temperature at 37.5 °C.
2.1.9 Noise exposure
A continuous noise (1–7 kHz) at 120 dB (sound pressure level, re 20 µPa) for 1 h was used to induce acoustic trauma to the cochlea. This noise signal was generated by a real-time signal processor (RP2.1; TDT), and was subsequently routed through an attenuator (PA5; TDT) and a power amplifier (Crown XLS 202; Harman International Company) to a loudspeaker (NSD2005-8; Eminence). The noise level was calibrated with a condenser microphone (LDL 2559; Larson Davis) that was placed in the animal's holding cage at head level. The microphone output was sent to a preamplifier (model 825; Larson Davis) and was read through a sound level meter (model 800 B; Larson Davis).
3.1 Results
3.1.1 The cellular composition of the sensory cell-enriched sample
To determine the cellular composition of the collected tissues, we examined the surface view of the tissues under an inverted microscope. We also examined the position of cell nuclei that had been stained with propidium iodide to confirm the cellular populations. Because hair cells, supporting cells, and non- sensory epithelium cells are spatially organized in an orderly pattern within the sensory epithelium, inspecting their nuclear positions using confocal microscopy enabled us to identify specific cell populations.
The surface view of the sensory cell-enriched samples revealed the cellular boundary of the collected tissue stripe (Figs. 3A, 3B and 3C). The lateral edge of the tissue stripe consisted of the third row of OHCs/Deiters cells. The medial edge appeared to be defined by inner border cells. Based on the surface view, we estimated that the sensory cell-enriched sample contained sensory cells (IHCs and OHCs), pillar cells (both inner and outer), Deiters cells, inner phalangeal cells and inner border cells. All these cells are found in the organ of Corti (Iurato, 1961; Santi and Mancini, 1998).
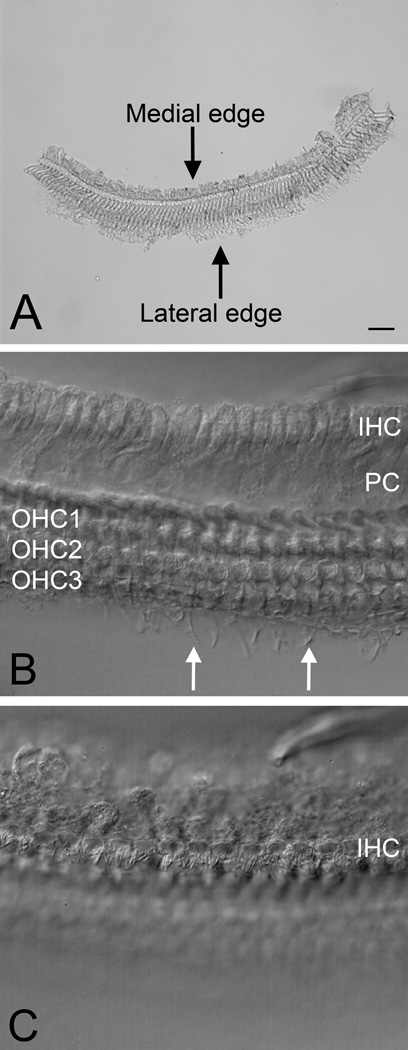
Figure 3.
A surface view of the sensory cell-enriched samples. A) A low magnification view of a sensory cell-enriched sample. B) A high magnification view of a sensory cell-enriched sample with the optical focus of the microscope placed at the level of the OHC region. The fiberlike structures outside the third row of OHCs are the phalangeal progress of Deiters cells (arrows). C) A high magnification view of a sensory cell-enriched sample with the optical focus placed at the level of the IHC region. The cuticular plate and the stereocilia of IHCs are visible. Scale bar = 10 µm.
To confirm the cellular composition that we had determined from the surface view, we inspected the nuclei of the cells in the collected tissues. This analysis identified IHCs/OHCs, Deiters cells, and inner/outer pillar cells (Fig. 4). We also observed numerous nuclei surrounding the IHC nuclei, which likely belong to inner border cells and inner phalangeal cells. However, the identification of these cells was less certain because the positions of the nuclei were intermingled.
Figure 4.
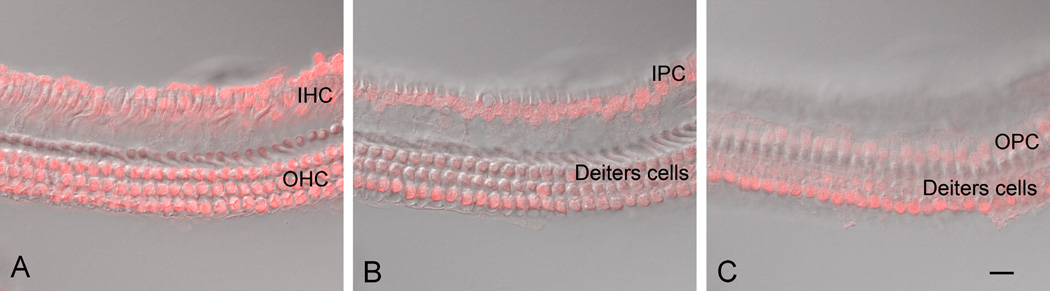
A confocal microscopic view of sensory cell-enriched nuclei. To better visualize the individual structures, images with nuclear staining were superimposed on the DIC view of the same tissues. A) This confocal image is focused at the level of OHCs and IHCs. Both OHC and IHC nuclei are present in this sample. The nuclei surrounding the IHCs belong to inner border cells or inner phalangeal cells. B) This confocal image is focused at the level of Deiters cell nuclei and inner pillar cell nuclei, all of which are visible in this sample. C) This confocal image is focused at the level of outer pillar cell nuclei, all of which are visible in this image. IHC: inner hair cell. OHC: outer hair cell. IPC: inner pillar cell. OPC: outer pillar cell. Scale bar = 15 µm.
To further elucidate the cellular composition of the tissues that we collected, we examined the remaining cochlear tissue from which the sensory cell-enriched sample had been removed (Fig. 5). We observed Hensen cell and Claudius cell nuclei on the lateral side of the region where the enriched tissue had been removed. We also found nuclei on the medial side of this region. While the identification of these cells was less certain, their anatomic location suggests that they are inner sulcus cells. The basilar membrane was intact, and mesothelial cell nuclei were visible in that region. Collectively, nuclear inspection provided additional evidence to support the characterization of cell composition that had been made via the surface view of the tissues.
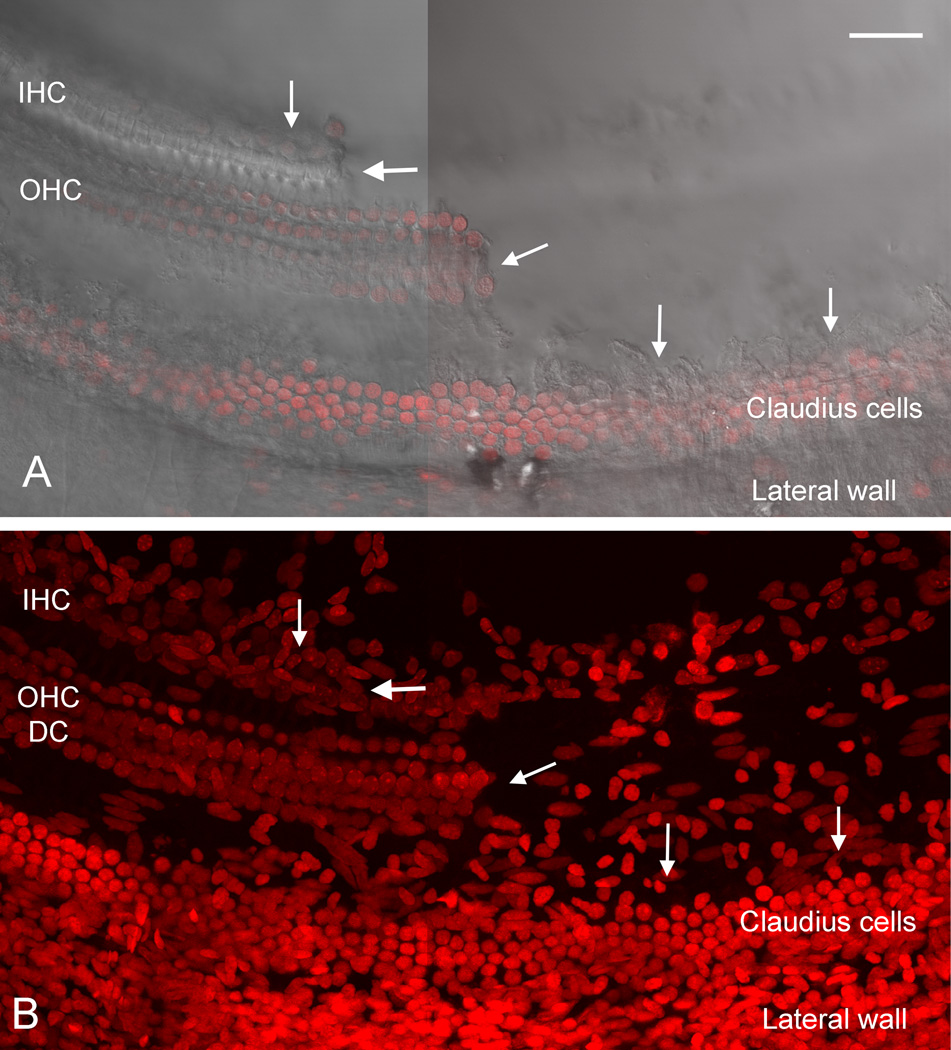
Figure 5.
Images showing a region of the sensory cell partition where the sensory cell-enriched tissue has been removed. A) A surface view of the reticular lamina of the tissue. On the right side of the tissue, the sensory cell-enriched region has been removed, and arrows mark the edge where the separation was made. On the left side of the image, the sensory cells remain in place. B) Nuclei in the same tissue have been stained with propidium iodide. On the left side of the tissue, all of the sensory and supporting cell nuclei are still present. In the right side of the image where the sensory cell-enriched tissue has been removed, Claudius cell nuclei, cells from the scala tympani side of the basilar membrane and cells in the lateral wall are present. IHC: inner hair cell. OHC: outer hair cell. DC: Deiters cell. Scale bar = 45 µm
To demonstrate that we had enriched the sensory cells, we examined the expression levels of the Slc26a5 gene, which encodes the protein prestin and is expressed exclusively in OHCs (Zheng et al., 2000). qRT-PCR was performed to determine the difference in mRNA abundance between the sensory-cell-enriched samples and the remaining cochlear tissue, from which the sensory-cell-enriched sample had been removed (n = 3 experimental repetitions for each sample type). We first examined the expression levels of 3 reference genes (Hsp90, Rpl13 and Hprt1). Because the two types of the tissues to be compared had different cellular compositions, their reference gene expression levels (the average of the three genes) were different (average Ct values: 20.2 ±1.9 for the remaining tissues vs. 22.5 ± 2.7 for the enriched tissues). Given that the aim of the analysis was to compare the Slc26a5 mRNA abundances between two types of tissues, but not to define an expression change after an experimental treatment, we decided not to use the reference genes to normalize the measurement of Slc26a5 abundance. Instead, we calculated the fold difference in the Slc26a5 mRNA abundance between the two types of the tissues by comparing the Ct values of the Slc26a5 measurement. This analysis revealed an 1145-fold difference between the two sample types (Student’s t-test, p < 0.001), suggesting that our sensory cell samples were highly enriched.
3.1.2 Consistency of sample collection
To determine their consistency, we inspected the tissues that had been obtained from rodent cochleae and stored in RNAlater for various amounts of time (1 to 7 d). The surface view of these tissues revealed that the length of the tissue stripe in samples collected from different cochleae was variable (mean ± SD: 580.75 ± 270.11 µm). The shape of the tissue was also variable due to tissue twisting. However, the cellular composition of the tissues was fairly consistent (Figs. 6A–F). Occasionally, an attachment of Claudius cells was seen in the lateral edge of the tissue stripe (Fig. 6G). This attachment could be removed upon further dissection. Some samples also suffered cellular loss (Fig. 6H). This defect often occurred towards the end of the samples and these defect sections could be removed with a micro-knife. We noticed that as our dissection skill improved over time, the defects were less commonly observed.
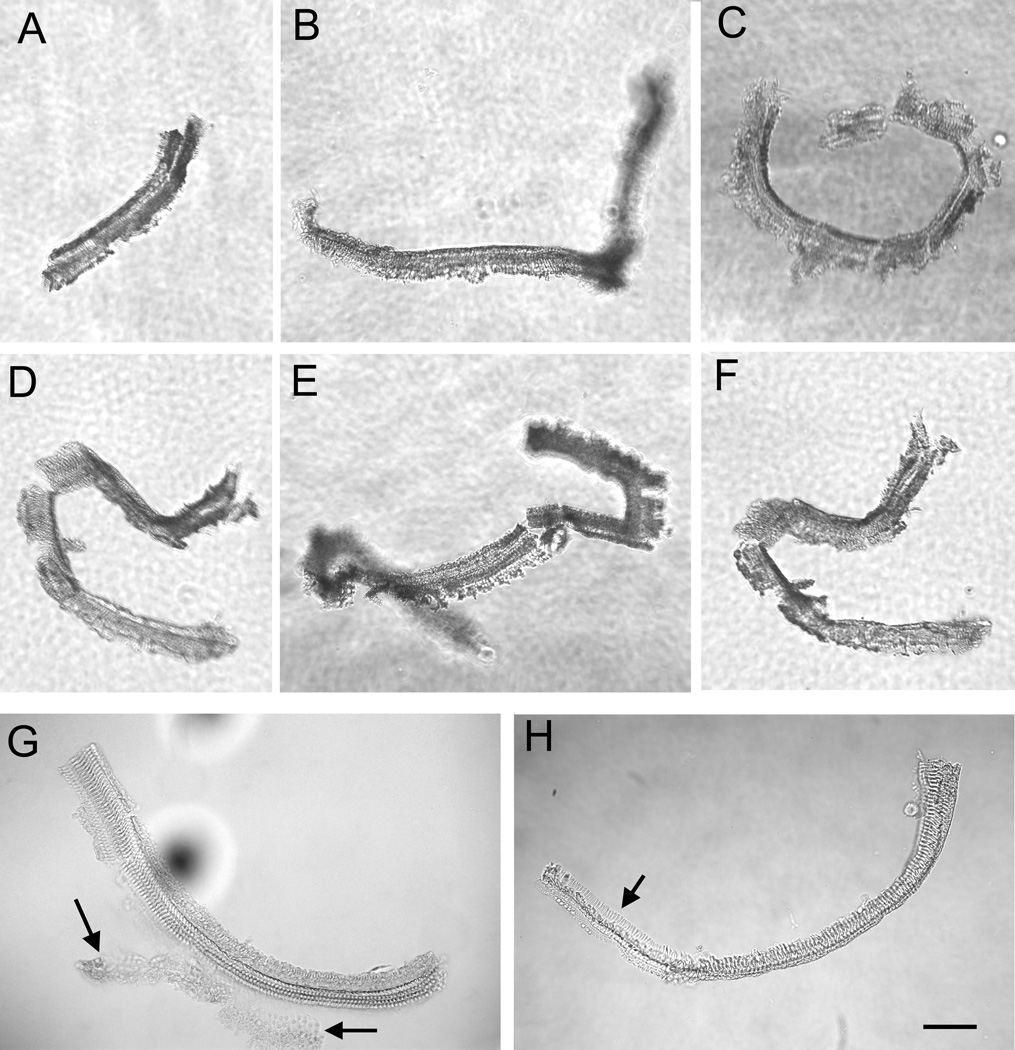
Figure 6.
Multiple sensory cell-enriched samples were photographed using an inverted microscope to illustrate the consistency among samples collected from different cochleae. The arrows in G point to a region of Claudius cells that remained attached to the sensory cell-enriched sample. The arrow in H points to the region of the sensory cell-enriched sample where cells are missing. Scale bar = 80 µm.
3.1.3 Quality of total RNA extracted from the sensory cell-enriched samples
Maintaining RNA integrity is a key prerequisite for high-quality downstream analysis. To demonstrate the capacity of the current method to preserve RNA integrity, we examined the RIN, an RNA integrity parameter, using an Agilent's 2100 Bioanalyzer. Initially, we found that even with the Pico assay (the most sensitive kit that can be used with Agilent’s Bioanalyzer), the amount of total RNA extracted from a single cochlea was insufficient for the analysis. We therefore pooled three sensory cell-enriched samples that had been collected from three different cochleae. The average RIN from four experimental repetitions was 7.6 ± 0.9 (mean ± SD). Among these four experimental repetitions, the two in which the tissues had been stored in RNAlater for 5 d at 4 °C had relatively low RIN values (6.6 and 7). In contrast, the remaining two samples, which had been processed within 1 d after cochlear collection, had relatively high RIN values (8.2 and 8.4).
To further improve the quality of the RNA samples, we reduced the sample storage time from 5 d to 10–12 h. We also reduced the buffer temperature from approximately 20–24 °C (room-temperature) to ice-cold during the initial sample preparation. With these changes, we collected additional 8 samples. The average RIN was increased to 9.2 ± 0.2 (Mean ± SD). Together, these observations indicate that our sample preparation method is able to yield a high-quality RNA sample.
3.1.4 Estimating the amount of total RNA from the sensory cell-enriched samples
The amount of total RNA extracted from the sensory cell-enriched samples was measured using the Agilent RNA 6000 Pico Kit. Using samples pooled from three cochleae, we found that the average total RNA concentration in our preparation was 165.2 ±37.7 pg/µl (n=4 experimental repetitions). Because the total volume of the RNA solution was 10 µl and each sample was pooled from three cochleae, we estimated that the average amount of total RNA extracted from the sensory cell-enriched tissue of one cochlea (average length = 580.75 ± 270.11 µm) was approximately 550 pg. It should be noted that the RNA Pico kit is not intended as a quantitative assay. Therefore, the concentration reported here should be regarded only as an estimate.
3.1.5 Normalizing the starting materials for future expression analyses
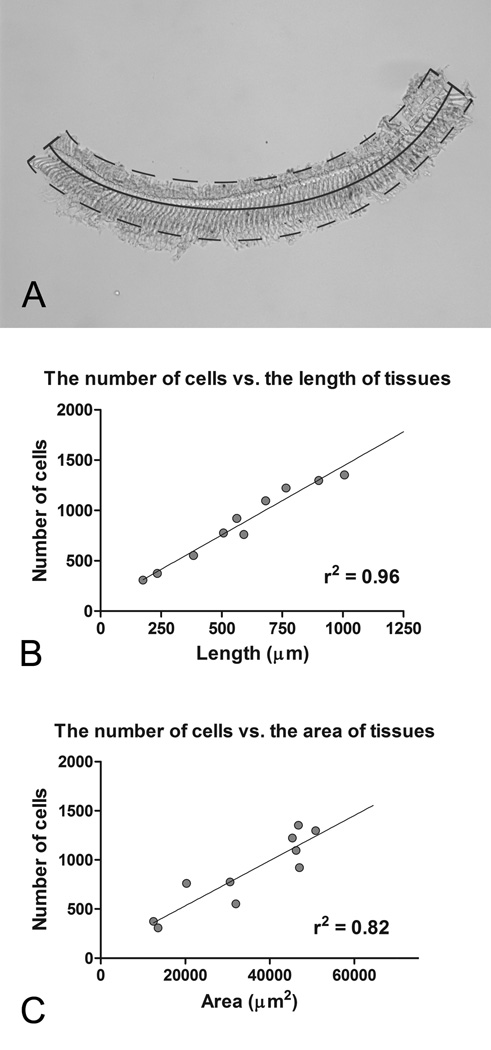
To compare the RNA expression levels across multiple samples or experimental conditions, the starting material must be normalized. While the most common method of accomplishing this is to measure total RNA concentration, this is not feasible with our sample preparation because of the limited amount of total RNA. To solve this problem, we instead used tissue size to normalize the amount total RNA extracted. To accomplish this, we photographed each tissue and then measured both the length and the area of the tissue stripe using image analysis software (ImageJ, NIH; Fig. 7A). We also counted the number of nuclei, which represents the number of cells in the tissue. Pearson’s correlation analysis revealed that both the area and the length of the tissue are correlated with the number of cells in the tissue (Figs. 7B and 7C). Contrary to our expectation, tissue area was less stringently correlated to the number of cells than tissue length (r2 = 0.82 for the area correlation vs. r2 = 0.96 for the length correlation). This observation likely resulted from the difficulty in accurately measuring the area of the tissue because it naturally twisted when placed in the RNAlater medium. Because length measurement is simple and provides a stringent correlation with the number of cells in the tissue, we used this parameter to normalize the samples for the subsequent expression analyses.
Figure 7.
The number of cells in each tissue stripe was correlated with the length/area of the tissue. A) The length and area of this sensory cell-enriched sample was measured. The dotted line denotes the area of the tissue, and the solid line indicates the length of the tissue. B) The correlation between the number of cells and the length of the tissue stripes is shown. C) The correlation between the number of cells and the area of the tissue stripes is shown. The number of cells in each tissue stripe is more stringently correlated to the length to the tissues as opposed to the area.
3.1.6 Measurement of the purity of sensory cells in the sensory cell-enriched samples
Previous investigations from our lab and others have used sensory epithelium tissue for expression analyses. Although this sample preparation generates higher sensory cell purity than using the entire cochlea, it still contains a large quantity of non-sensory cells. To demonstrate that our sensory-cell enriched preparation improves the purity of sensory cells, we calculated the percentage of sensory cells among the total number of cells in each sample (n = 10 cochleae) and found that the average proportion of sensory cells was 46.1 ± 5.8 %. This value is much higher than the percentage of sensory cells in sensory epithelium tissues (n= 5 cochleae, 21.6 ±1.3 %, Student’s t test, t = 9.2, p <0.0001, Fig. 8). It should be noted that for conventional sensory cell epithelium samples, the actual cell composition in individual samples varies considerably due to the variation in tissue collection. It is likely that cells from both the lateral wall and the bony shelves of the osseous spiral lamina are included in the sensory epithelium samples. Because the value presented here (21.6 ±1.3 %) includes only the cells in the sensory epithelium region located between the lateral wall and the medial edge of the sensory organ partition, this number is not typical for most sensory epithelium preparations. Even with this overestimation of sensory cell purity in the sensory epithelium sample, we are able to achieve a higher level of purity with the new enriched preparation described here.
Figure 8.
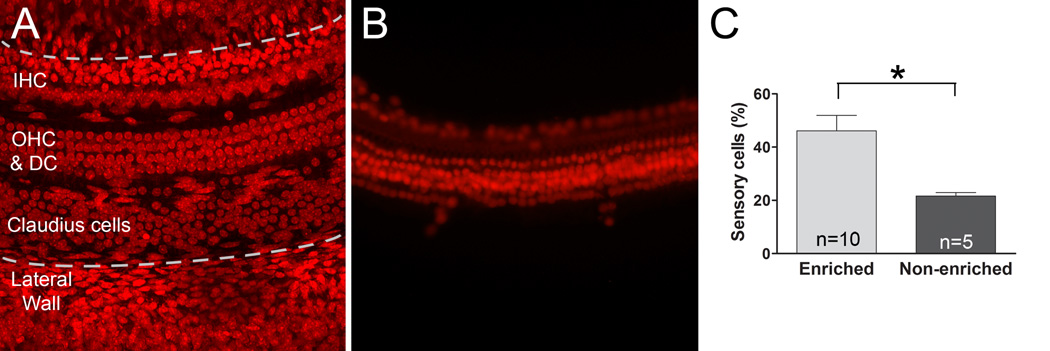
The purity of sensory cells is higher in the sensory cell-enriched samples than the sensory epithelium samples. A) A typical cochlear sensory organ partition was stained with propidium iodide. The area between the two dotted lines is the region of the sensory epithelium. B) A typical sensory cell enriched sample is shown here. C) A comparison of the percentage of sensory cells in relation to the total number of cells was made between the sensory cell-enriched samples and the sensory epithelium samples. The asterisk indicates a significant difference (Student’ s t test, p < 0.0001). n= the number of cochleae.
3.1.7 Identification of stable genes in the noise-traumatized samples
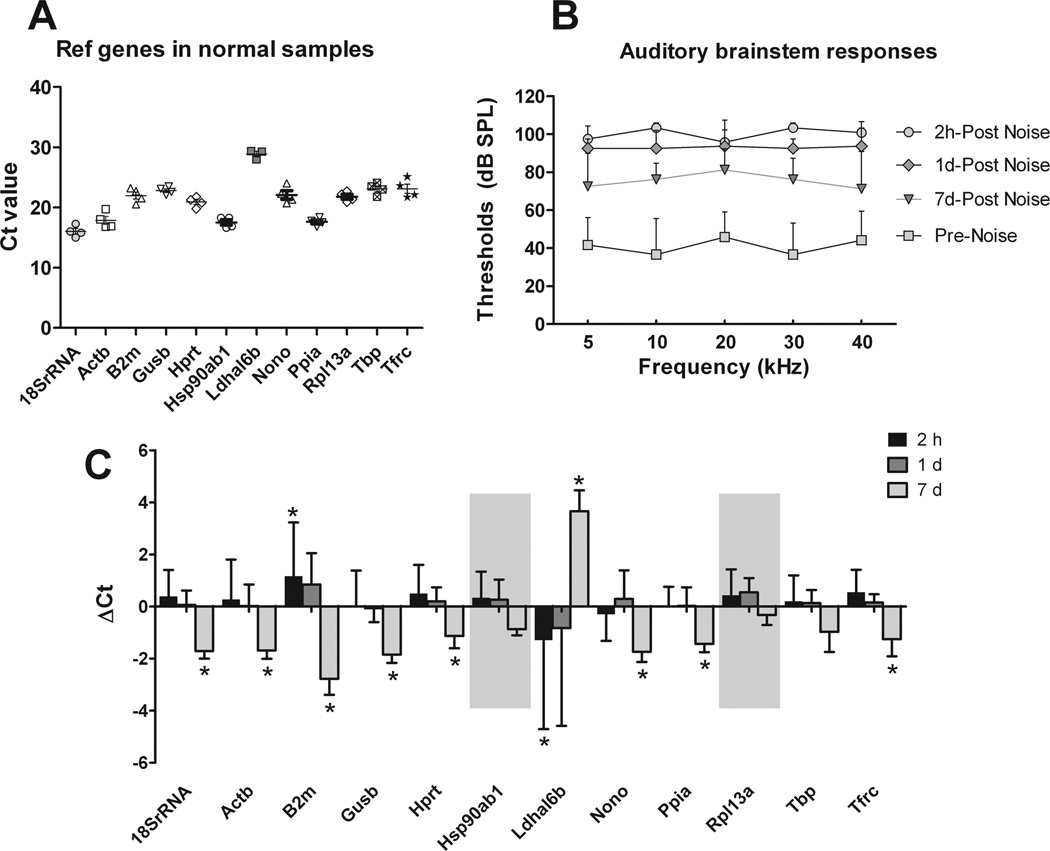
To demonstrate the feasibility of performing transcriptional analyses on the samples collected with our new microdissection technique, we examined the expression pattern of 12 reference genes (18SrRNA, Actb, B2m, Gusb, Hprt, Hsp90ab1, Ldhal6b, Nono, Ppia, Rpl13a, Tbp, Tfrc) in sensory cell-enriched samples. We found that all twelve genes were detectable (Fig. 9A), with average raw Ct values ranging from 16.0 ± 1.0 for 18SrRNA (the gene with the highest expression level) to 28.9 ± 0.7 for Ldhal6b (the gene with the lowest expression level).
Figure 9.
The expression of 12 reference genes among the sensory cell-enriched samples collected from normal and noise-traumatized mouse cochleae. A) Ct values of each reference gene were determined from four experimental repetitions. Each dot represents the Ct value of each sample, and the lines represent the mean. B) Auditory brainstem response thresholds were measured before and at various time points after the animals were exposed to a 120-dB noise for 1 h. C) The Ct values of the reference genes after the noise exposure were compared with those measured in the normal cochleae. The asterisks mark the genes where a change in Ct value > 1 was seen. The shaded areas denote the two genes for which less than a two-fold change in expression was seen at all the time points after the noise exposure. The lines above the bars represent one standard deviation above the mean.
We also examined the changes in the expression levels of these reference genes following acoustic overstimulation. This allowed us to identify genes that remain stable and could therefore be used in the future to normalize the transcriptional expression levels of various genes of interest. Mice were exposed to a broadband noise at 120 dB SPL for 1 h. This noise caused a significant hearing loss with average threshold shifts of 59.2 ± 15.2, 47.8 ± 12.4, and 30.3 ± 14.3 dB (mean ± SD) at 2 h, 1 d and 7 d after noise exposure, respectively (Fig. 9B). The expression analysis revealed that most genes exhibited stable expression levels at 2 h and 1 d post-noise exposure, as illustrated by an average Ct change of less than 1 (fold change less than 2). Only two genes (Ldhal6b and B2m) showed a large variation in their expression, as determined by an average Ct change greater than 1 (fold change greater than 2) (Fig. 9C). However, at 7 d post-noise exposure, 10 out of the 12 examined genes displayed an average Ct change > 1, which indicated a reduced expression (Fig. 9C). Only two genes (Hsp90ab1 and Rpl13a) were stable after 7 d. This pattern of changes in reference gene levels at the chronic phases of cochlear pathogenesis was previously described (Hu et al., 2012; Hu et al., 2009) and is likely related to the accumulation of sensory cell death over time after the noise exposure.
3.1.8 Collection of sensory cell-enriched samples from the rat cochlea
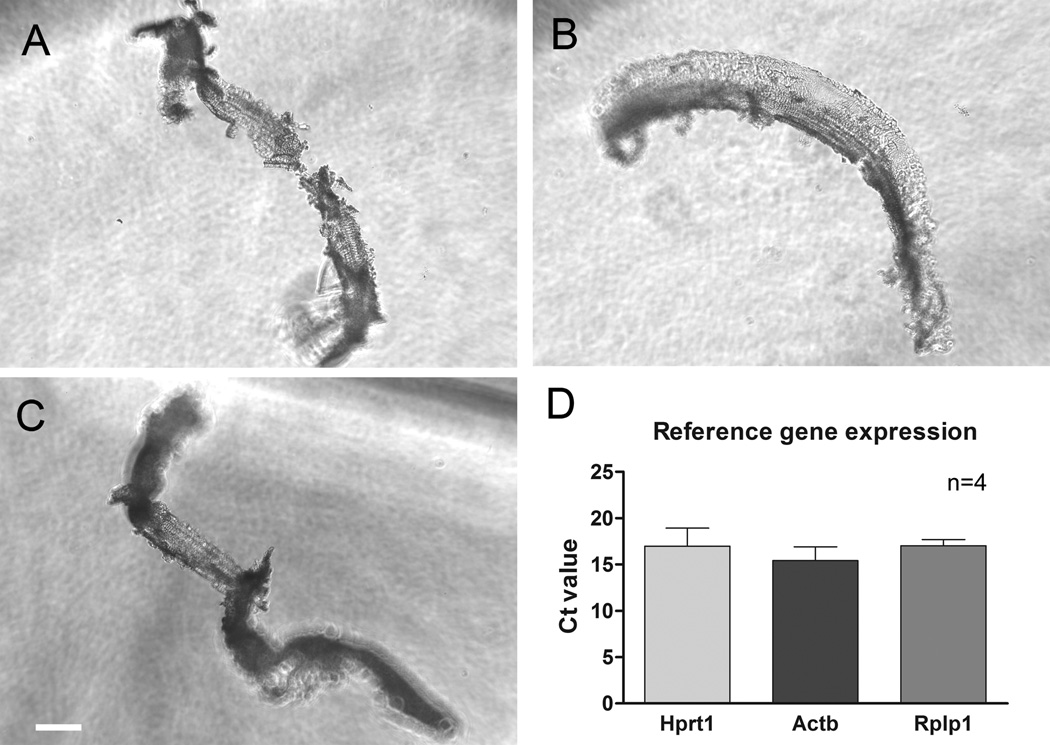
While the mouse is the most widely-used animal model for investigating the molecular mechanism of inner ear disorders, other rodents are also valuable for these types of studies. To determine whether the current microdissection method can be applied to additional animal models to collect sensory cell-enriched samples, we also dissected rat cochleae using a similar method. We found that the bony shell of the rat cochlea is thicker than that of the mouse cochlea, which blocks the light available to illuminate the cochlear tissue and therefore blurs the view of the organ of Corti. Even with this challenge, we were able to isolate sensory cell-enriched tissue from the rat cochlea (Figs. 10A, B and C). We also examined the expression level of three reference genes (Hprt1, Actb, and Rplp1), all of which were detectable and displayed consistent Ct values across the four experimental repetitions (Fig. 10D). This observation indicates that the application of the current microdissection can be expanded to other rodent models.
Figure 10.
Examples of sensory cell-enriched samples collected from rat cochleae. A), B) and C) A surface view of three typical sensory cell-enriched tissues is shown. The samples display a similar morphology to those collected from mouse cochleae. D) The average Ct values of three reference genes were calculated from the sensory cell-enriched samples. The lines above the bars represent one standard deviation above the mean. n= 4 experimental repetitions.
3.1.9 Collection of OHC- and IHC-enriched samples
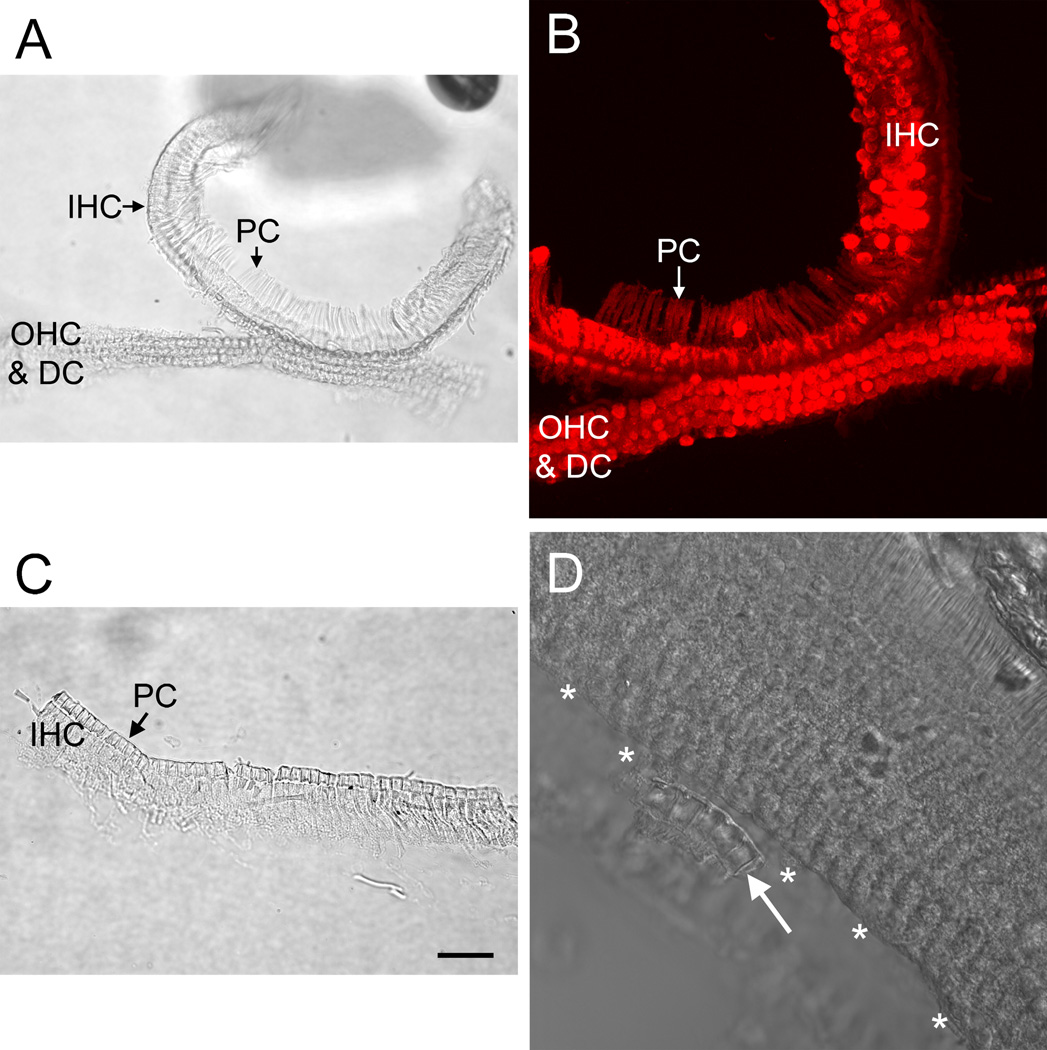
To further improve the specificity of sample cell population, we collected IHC- and OHC-enriched samples. The surface view of the IHC- and OHC-enriched tissues revealed that the pillar cells were the site of separation between these two regions (Figs. 11A–C). The pillar cells could be seen in both the IHC- and OHC-enriched tissues, but they appeared more frequently in the IHC-enriched samples. Examining propidium iodide-stained tissues under the confocal microscope confirmed that the OHC-enriched tissue contained OHCs, Deiters cells and pillar cells, while the IHC-enriched tissue contained IHCs, pillar cells and various cells surrounding the IHCs (inner border cells and phalangeal cells). This observation demonstrated that the current microdissection method could separate IHC- and OHC-enriched populations.
Figure 11.
Micrographs of OHC- and IHC-enriched samples. A) The surface view of both an OHC-enriched and an IHC-enriched tissue is shown. To illustrate the site of separation, the middle portion of the tissue stripe was intentionally kept intact. B) Nuclear staining of the same sample presented in (A). Notice that the OHC-enriched portion of the tissue contains the nuclei of OHCs and Deiters cells and that the IHC-enriched portion of the tissue contains IHCs and neighboring cells, including pillar cells, inner border cells and inner phalangeal cells. C) An example of an IHC-enriched sample is shown. D) The surface view of a sensory organ partition after the IHC-enriched portion was isolated illustrates that the IHC sample collection was incomplete. Asterisks mark the OHC edge where the IHC region has been removed. The arrow points to a small piece of the IHC-enriched tissue that was not removed during the microdissection. OHC: outer hair cell. IHC: inner hair cell. PC: pillar cell. DC: Deiters cell. Scale bar: 25 µm.
It should be noted that the OHC and IHC populations were incompletely separated in some samples (Fig. 11D). Therefore, it is essential to observe the collected tissues under a microscope to visually verify this separation.
4.1 Discussion
Acquiring high-quality, sensory cell-enriched samples from the mouse cochlea for use in transcriptional analysis has represented a significant challenge in the field of hearing research. Here, we report that treating the tissue with RNAlater alters tissue properties and facilitates tissue collection. We have developed a novel microdissection technique that allows us to isolate sensory cells and their immediate neighbors from the mouse cochlea. This method enables us to collect sensory cell-, IHC- and OHC-enriched samples with well-defined cell populations. Importantly, the RNA integrity of the samples is well preserved. Using this technique, we were able to collect sensory cell-enriched tissues that could be used in the transcriptional analysis of common reference genes, which allowed us to identify genes that remain stable following acoustic trauma. Finally, we have demonstrated that our method of collecting sensory cell-enriched samples from the mouse can also be applied to rat cochleae.
Isolating sensory cell-enriched samples, particularly IHC- and OHC-enriched samples, is hindered by the technical challenge of separating the tightly-bonded cells at defined cochlear sites. Moreover, the dissection should not compromise the gross structure of the tissue so that the collected tissue can be microscopically inspected to determine its cellular composition. We found that RNAlater treatment addresses these challenges because it alters the physical properties of the tissue in several ways. First, cell junctions are weakened, so cells can be easily separated from the sites where they are attached. While the precise mechanism behind this junction weakening is not clear, the presence of a divalent cation chelator in the RNAlater reagent (US patent No. 6204375B1) may affect the function of Ca++-dependent adhesion molecules. Moreover, high salt concentration in RNAlater can precipitate membrane proteins, which further weakens the cellular junctions. This effect of RNAlater enabled us to isolate cell populations of interest without compromising the gross tissue structure. Second, the RNAlater treatment improves the visualization of structural details of the tissue. This phenomenon is likely related to the high fluid viscosity of the RNA preservation medium, which alters the light-scattering property of the tissue. In addition, the decalcification of the bony shell of the cochlea by RNAlater improves light transmission. This effect enabled us to identify the sensory cells in the tissue stripe under a dissection microscope. Third, the tissue becomes hardened after treatment with RNAlater. This change enabled us to remove a longer piece of the tissue stripe than was previously possible. Finally, the pressure for a quick dissection is reduced because RNAlater preserves the RNA integrity at room temperature. Taken together, these observations suggest that the use of RNAlater as a preservation medium is required for the success of the current microdissection method.
Improving sensory cell purity is a significant advantage of our preparation. Previous studies in which transcriptional analyses of gene expression are performed often employ samples from the entire cochlea or tissues from a cochlear partition (Christensen et al., 2009; Han et al., 2012; Hu et al., 2009; Szaumkessel et al., 2012). These tissues contain a large quantity of non-sensory cells. As shown in a recent investigation using the flow cytometry to sort and capture cells, hair cells constitute only 2% of the total number of cells in the auditory tissue of the mouse inner ear (Hertzano et al., 2011). As demonstrated in this study, even if the sensory epithelium sample does not contain the lateral wall and the modulus tissues, sensory cells account for less than 20% of the total cells. Here, we demonstrate that using the current method for sample collection allows for a marked improvement in sensory cell purity (> 40% for the sensory cell-enriched sample). Although our samples still contain certain types of supporting cells, all of these cells have direct contact with sensory cells and are part of the organ of Corti (Iurato, 1961; Santi and Mancini, 1998). An investigation of these cells is expected to provide more relevant information on the sensory cell-related responses under various pathological conditions.
Knowing the cell composition in the collected tissues is another advantage of the current method. Our sample preparation includes a step of tissue assessment. This microscopic observation allows us to determine the cellular composition of the samples. Because the tissues are submersed in the RNA stabilization solution during this period of sample inspection, the RNA integrity is preserved. Knowing the precise cellular composition of the sample allows for greater accuracy in data analyses and interpretation. Moreover, inspecting the tissue provides another potential benefit in that the cellular pathology and gene expression in the tissue can be correlated. Although we did not assess this parameter in this study, we expect that this potential benefit will be important in future studies that aim to provide a better understanding of the molecular mechanism of inner ear disorders.
Preserving the RNA integrity of tissue samples is a fundamental prerequisite for high-quality downstream analyses. While previous techniques, including in vitro hair cell isolation, laser-capture microdissection, and flow cytometry are capable of providing a highly pure population of sensory cells, the ability of these methods to preserve RNA is a concern. Cochlear sensory cells are susceptible to mechanical disturbance and to changes in their microenvironments. Without proper RNA stabilization, a cascade of biological processes can occur in the cell while the tissue is being processed, which can alter cellular gene expression and/or cause rapid RNA degradation. Because our tissue was prepared in an RNAlater solution, we are able to reduce the amount of RNA degradation. The RNA quality assessment revealed a high RIN, indicating that our method of preparation allows for the preservation of RNA. It should be noted that, even with the protection of the RNA stabilizing reagent, prompt handling of the samples is still desirable. Moreover, we recommend that the initial sample preparation be performed in an ice-cold buffer.
The microdissection technique described here involves manual manipulation of the cochlea to obtain sensory cell-enriched samples. To ensure success, care must be taken in several steps of the process. First, a clear visualization of sensory cells in the stripe of the organ of Corti is essential for the success of the sample isolation. We prefer a high-quality dissection microscope with a focused illumination. We have noticed that the thick bony shell of the rat cochlea decreases light transmittance and blurs the anatomical details; therefore, the mouse cochlea, with its thinner bony shell, offers a better view of the tissue. Second, the medial edge of the organ of Corti stripe is difficult to separate with a micro-knife due to the lack of a clear anatomical mark. We found that pulling the osseous spiral lamina during the initial phase of tissue preparation can separate the medial edge at the region of inner sulcus cells with a smooth cut, which allows us to avoid the subsequent dissection with a micro-knife. We therefore recommend that this initial preparation is conducted before the RNAlater treatment. Third, transferring the isolated tissue from the dissection dish to a PCR tube for downstream processing requires extra caution. The collected tissues, particularly the IHC-enriched and OHC-enriched samples, are small and can be lost during this transfer. A micro-mesh used for transferring sliced tissue sections for scanning electron microscopy can be used to prevent tissue loss.
While the current method represents a valuable tool for tissue isolation, it also has certain limitations. First, the microdissection requires a high level of skill; however, this can be obtained with practice. Second, the abundance of total RNA extracted from our preparation is limited due to the small size of the cochlear tissue available for dissection. The limited abundance of total RNA may prove challenging for subsequent downstream analyses. By pre-amplifying RNAs, we have been able to conduct expression analyses of multiple reference genes as well as certain genes of interest (data not shown). As the sensitivity of transcriptional analysis techniques rapidly improves, we expect that this disadvantage will instead become an advantage of the method for a site-specific analysis of cochlear pathogenesis. Third, assessment of the amount of total RNA extracted from the tissue requires an assay with high sensitivity for the tiny samples generated from the current investigation. We found that even with an Agilent RNA 6000 Pico Kit, the measurement of individual samples is challenging. Therefore, we recommend measuring pre-amplified products as well as using reference genes for assessing the validity of the sample handing and for normalization of samples for RT-PCR.
In summary, the mouse is a valuable model for molecular studies of cochlear pathogenesis. The method described here offers a reliable tool for the collection of high-quality, sensory cell-enriched samples that are suitable for further transcriptional analyses. This method enables us to collect cochlear tissues that have a known cellular composition from a defined site within the cochlea. Importantly, the inner hair cell and outer hair cell populations can be collected separately. These samples can then be used in diverse downstream analyses, including qRT-PCR, microarray, and RNA sequencing.
Highlights.
A novel microdissection technique for cochlear tissue collection is described.
Collected samples contain defined sensory cell and supporting cell populations.
The RNA integrity of the samples is well preserved.
Stable reference genes have been identified for noise-traumatized samples.
The microdissection technique is applicable to both mouse and rat cochleae.
Acknowledgement
This work was supported by NIH grant R01DC010154 (BH). The authors thank Angela Moltrup for her assistance in preparation of the manuscript and figures.
Abbreviations
- OHC
outer hair cell
- IHC
inner hair cell
- RIN
RNA Integrity Number
- ABR
Auditory brainstem response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson CT, Zheng J. Isolation of outer hair cells from the cochlear sensory epithelium in whole-mount preparation using laser capture microdissection. J Neurosci Methods. 2007;162:229–236. doi: 10.1016/j.jneumeth.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Patel M, Coling D, Hu BH. Transcriptional changes in adhesion-related genes are site-specific during noise-induced cochlear pathogenesis. Neurobiology of disease. 2012;45:723–732. doi: 10.1016/j.nbd.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen N, D'Souza M, Zhu X, Frisina RD. Age-related hearing loss: aquaporin 4 gene expression changes in the mouse cochlea and auditory midbrain. Brain Res. 2009;1253:27–34. doi: 10.1016/j.brainres.2008.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Hong L, Zhong C, Chen Y, Wang Y, Mao X, Zhao D, Qiu J. Identification of new altered genes in rat cochleae with noise-induced hearing loss. Gene. 2012;499:318–322. doi: 10.1016/j.gene.2012.02.042. [DOI] [PubMed] [Google Scholar]
- Harter C, Ripoll C, Lenoir M, Hamel CP, Rebillard G. Expression pattern of mammalian cochlea outer hair cell (OHC) mRNA: screening of a rat OHC cDNA library. DNA and cell biology. 1999;18:1–10. doi: 10.1089/104454999315574. [DOI] [PubMed] [Google Scholar]
- He DZ, Zheng J, Edge R, Dallos P. Isolation of cochlear inner hair cells. Hear Res. 2000;145:156–160. doi: 10.1016/s0378-5955(00)00084-8. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R. High throughput gene expression analysis of the inner ear. Hear Res. 2012;288:77–88. doi: 10.1016/j.heares.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ, Eisenman DJ, Strome SE. Cell type-specific transcriptome analysis reveals a major role for Zebl and miR-200b in mouse inner ear morphogenesis. PLoS genetics. 2011;7:el002309. doi: 10.1371/journal.pgen.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BH, Cai Q, Hu Z, Patel M, Bard J, Jamison J, Coling D. Metalloproteinases and their associated genes contribute to the functional integrity and noise-induced damage in the cochlear sensory epithelium. J Neurosci. 2012;32:14927–14941. doi: 10.1523/JNEUROSCI.1588-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BH, Cai Q, Manohar S, Jiang H, Ding D, Coling DE, Zheng G, Salvi R. Differential expression of apoptosis-related genes in the cochlea of noise-exposed rats. Neuroscience. 2009;161:915–925. doi: 10.1016/j.neuroscience.2009.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BH, Henderson D, Nicotera TM. Involvement of apoptosis in progression of cochlear lesion following exposure to intense noise. Hear Res. 2002;166:62–71. doi: 10.1016/s0378-5955(02)00286-1. [DOI] [PubMed] [Google Scholar]
- Iurato S. Submicroscopic structure of the membranous labyrinth. 2. The epithelium of Corti's organ. Z Zellforsch Mikrosk Anat. 1961;53:259–298. doi: 10.1007/BF00339444. [DOI] [PubMed] [Google Scholar]
- Jin Z, Ulfendahl M, Jarlebark L. Spatiotemporal loss of K+ transport proteins in the developing cochlear lateral wall of guinea pigs with hereditary deafness. Eur J Neurosci. 2008;27:145–154. doi: 10.1111/j.1460-9568.2007.05994.x. [DOI] [PubMed] [Google Scholar]
- Keithley EM, Feldman ML. Hair cell counts in an age-graded series of rat cochleas. Hear Res. 1982;8:249–262. doi: 10.1016/0378-5955(82)90017-x. [DOI] [PubMed] [Google Scholar]
- Mutter GL, Zahrieh D, Liu C, Neuberg D, Finkelstein D, Baker HE, Warrington JA. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC genomics. 2004;5:88. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T, Teranishi M, Hibi T, Kobayashi M, Umemura M. Vestibular and cochlear toxicity of aminoglycosides—a review. Acta Otolaryngol. 2000;120:904–911. doi: 10.1080/00016480050218627. [DOI] [PubMed] [Google Scholar]
- Santi PA, Mancini P. Cummings CW, Fredrickson JM, Harker LA, Krause CJ, Richardson MA, Schuller DE, editors. Cochlear anatomy and central auditory pathways. Otolaryngology head and neck surgery. 1998:2803–3830. [Google Scholar]
- Sato T, Doi K, Hibino H, Kubo T. Analysis of gene expression profiles along the tonotopic map of mouse cochlea by cDNA microarrays. Acta Otolaryngol Suppl. 2009:12–17. doi: 10.1080/00016480902926464. [DOI] [PubMed] [Google Scholar]
- Szaumkessel M, Brauze D, Rydzanicz M, Karlik M, Szyfter K, Szyfter W, Maciej W. Simple technique for RNA purification from mouse inner ear hair cells. Molecular biology reports. 2012;39:6467–6469. doi: 10.1007/s11033-012-1465-7. [DOI] [PubMed] [Google Scholar]
- Towers ER, Kelly JJ, Sud R, Gale JE, Dawson SJ. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J Cell Sci. 2011;124:1145–1155. doi: 10.1242/jcs.076141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Sherman ME, Rader JS, Carreon J, Schiffman M, Baker CC. Cervical tissue collection methods for RNA preservation: comparison of snap-frozen, ethanol-fixed, and RNAlater-fixation. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2006;15:144–148. doi: 10.1097/01.pdm.0000213460.53021.cd. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shen W, He DZ, Long KB, Madison LD, Dallos P. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]