Abstract
Nuclear expression of CCAAT enhancer binding protein-α (C/EBPα), which supports tissue differentiation through several antiproliferative protein–protein interactions, augurs terminal differentiation of prostate epithelial cells. C/EBPα is also a tumor suppressor, but in many tumors its antiproliferative interactions may be attenuated by de-phosphorylation. C/EBPα acts as a corepressor of the classical androgen response element (ARE)-mediated gene activation by the androgen receptor (AR), but this is paradoxical as the genotropic actions of AR are crucial not only for the growth of the prostate but also for its maintenance and function. We show that DNA-bound C/EPBα recruits AR to activate transcription. C/EBPα-dependent trans-activation by AR also overrode suppression of AREs by C/EBPα elsewhere in a promoter. This mechanism was remarkable in that its androgen dependence was apparently for nuclear translocation of AR; it was otherwise androgen independent, flutamide insensitive and tolerant to disruption of AR dimerization. Gene response profiles and global chromatin associations in situ supported the direct bimodal regulation of AR transcriptional signaling by C/EBPα. This unique mechanism explains the functional coordination between AR and C/EPBα in the prostate and also shows that hormone-refractory AR signaling in prostate cancer could occur through receptor tethering.
Keywords: androgen receptor, C/EBPα, prostate, prostate cancer
Introduction
Testosterone and dihydrotestosterone, the two major natural androgens, exert their genotropic effects through the androgen receptor (AR) to have a primary role in reproduction and gender differentiation (Quigley et al., 1995) and also directly target non-reproductive tissues (Manolagas and Kousteni, 2001). AR has been grouped with class I nuclear receptors (steroid receptors) that, upon ligand binding, typically dissociate from a cytosolic complex containing heat shock proteins, homodimerize and translocate into the nucleus (Pratt and Toft, 1997) and bind to inverted repeat DNA response elements in their target genes (Tsai and O’Malley, 1994; Beato et al., 1995). The agonist bound receptors then recruit co-activators; in contrast, when bound to antagonists the receptors preferentially recruit corepressors (Glass and Rosenfeld, 2000; McKenna and O’Malley, 2002). Class I nuclear receptors typically share a domain structure that includes a ligandindependent activation function 1 in the N-terminal domain, a ligand-dependent activation function 2, a DNA-binding domain and a C-terminal ligand-binding domain (Bourguet et al., 2000). However, AR has several distinctive characteristics in its structural and functional organization compared with other steroid receptors (Jenster et al., 1995; Hong et al., 1996; Berrevoets et al., 1998; Gelmann, 2002; McEwan, 2004), including its ability to bind as a homodimer to both direct and inverted repeat androgen response elements (AREs) (Shaffer et al., 2004).
The action of AR is principally transcriptional (Gelmann, 2002; Xu et al., 2006) although some observations suggest a relatively minor contribution of cytosolic interactions of AR to cell survival (Kousteni et al., 2001; Sun et al., 2003; Baron et al., 2004). AR is required for the development, maintenance and function of the prostate (Roy et al., 1999). It is also responsible for prostatic hyperplasia and malignancy and is expressed in most androgen-independent prostate tumors (Ruizeveld de Winter et al., 1994) in which it is believed to have a significant role in tumor growth and refractoriness to androgen ablation (Zegarra-Moro et al., 2002). The exact mechanisms contributing to this androgen independence remain unclear, although AR gene amplification, AR mutations and an altered co-regulator complement as well as the phosphorylation or acetylation status of AR have been implicated (Miyamoto et al., 2004). Genetic lesions that support androgen-independent prostate cancer growth, including activation of mitogen-activated protein kinase, phosphatidylinositol 3 kinase /AKT and protein kinase C pathways, converge on activation of AR (Edwards and Bartlett, 2005; Shand and Gelmann, 2006). Cellular and molecular changes associated with androgen independence may also be expected to allow androgenindependent entry of AR into the nucleus. However, there is no clear evidence that, in truly hormone-refractory cells, hormone-independent transcriptional signaling by AR can occur through the classical DNA response elements (ARE) for AR; rather, in hormone refractory LNCaP prostate cancer cells, ARE-mediated gene activation was still ligand dependent, whereas AR supported hormone-independent growth by associating with target genes through other means (Jia and Coetzee, 2005; Gonit et al., publication pending); the putative tethered associations of AR with its target promoters have not been characterized.
In mammals, the CCAAT enhancer binding protein (C/EBP) family of homo- or hetero-dimeric basic/ leucine zipper transcription factors has at least six members designated α, β, γ, δ, ε and ζ (Ramji and Foka, 2002). The C/EBP element has a divergent dyad repeat sequence RTTGCGYAAY, in which R and Y represent A/G and C/T, respectively (Osada et al., 1996). C/EBP family proteins are functionally coordinated in inducing the differentiation and function of several tissues (Ramji and Foka, 2002); however, in this context, C/EBPα has the unique role of inhibiting cell proliferation (Umek et al., 1991; Hendricks-Taylor and Darlington, 1995; Watkins et al., 1996). C/EBPa also acts as a tumor suppressor (Watkins et al., 1996; Burel et al. 2001; Pabst et al., 2001a, b; Halmos et al., 2002; ; Gery et al., 2005; Schuster and Porse, 2006; Loomis et al., 2007). The antiproliferative action of C/EBPα can occur independent of its ability to bind to DNA (Harris et al., 2001) by protein–protein interactions that include stabilization of p21 (Timchenko et al., 1996, 1997), disruption of electro-acoustic 2 factor complexes (Timchenko et al., 1999a, b; Porse et al., 2001), inhibition/degradation of cyclin-dependent kinases 2 and 4 (Wang et al., 2001, 2002) and interaction with the switch/sucrose nonfermentable chromatin remodeling complex (Muller et al., 2004). In liver tumors, dephosphorylation of C/EBPα by activation of the phosphatidylinositol 3 kinase/AKT pathway abrogates its interactions with cyclin-dependent kinase 2 and electro-acoustic 2 factor complexes (Wang et al., 2004); dephosphorylated C/EBPα also promotes proliferation by sequestering retinoblastoma protein (Wang and Timchenko, 2005).
CCAAT enhancer binding protein-α is expressed in prostate epithelial cells, entering the nucleus at the onset of maturation (Zhang et al., 2008). Ectopic C/EBPα was antiproliferative in prostate cancer cells (Chattopadhyay et al., 2006). Remarkably, C/EBPα is commonly expressed in malignant prostate tumors in which its relative levels correlate significantly with those of AR, especially in metastatic tumors, which express the highest levels (Yu et al., 2004; Zhang et al., 2008), possibly reflecting functional modifications as noted above. In both normal and malignant prostate tissues, the function of C/EBPα also seems to be controlled through regulation of its nuclear-cytoplasmic distribution (Zhang et al., 2008) by mechanisms that are yet to be analysed.
When C/EBPα was ectopically expressed in cells that were C/EBPα negative, concomitant with growth inhibition, it bound to AR and inhibited ARE-mediated promoter activation by inhibiting co-activator recruitment (Chattopadhyay et al., 2006). This finding has obvious physiological significance as androgen-stimulated growth must be attenuated at the late stages of formation of the prostate epithelium. However, as the principal physiological action of AR is believed to be genotropic, attenuation of its classical transcriptional activity by C/EBPα also poses the question of how AR can then support differentiation, maintenance and function of the prostate when C/EBPα is co-expressed. The issue led to the exploration of alternate promoter models to elucidate the nature and functional consequence of interaction between AR and C/EBPα. The studies reveal an alternate mechanism of interaction between AR and C/EBPα at C/EBPα-binding sites that could override the suppression of classical AR signaling by C/EBPα elsewhere in a gene and also enable AR to activate different genes. The novel mechanism has several remarkable features including ligand-independent promoter association and trans-activation per se under conditions that permit nuclear localization of AR and the absence of a need for AR dimerization. The bimodal interaction of AR and C/EBPα was extended to global gene regulation and chromatin associations in situ. The findings enable a comprehensive mechanistic understanding of the coordinated actions of AR and C/ EBPα in the normal prostate and reveal a potentially significant mechanism of AR signaling in hormone-refractory and anti-androgen-resistant prostate tumors.
Results
In contrast to prostate epithelial cells and clinical prostate tumors, established prostate cell lines have lost C/EBPα expression. Therefore, in the following experiments the role of endogenous C/EBPα was examined in HeLa cells, which express relatively low levels of C/EBPα. In LNCaP cells as well as in HeLa cells, C/EBPα was introduced ectopically to cover a range of expression levels.
Androgen receptor (AR) can be functionally recruited by DNA-bound CCAAT enhancer binding protein-α (C/EBPα)
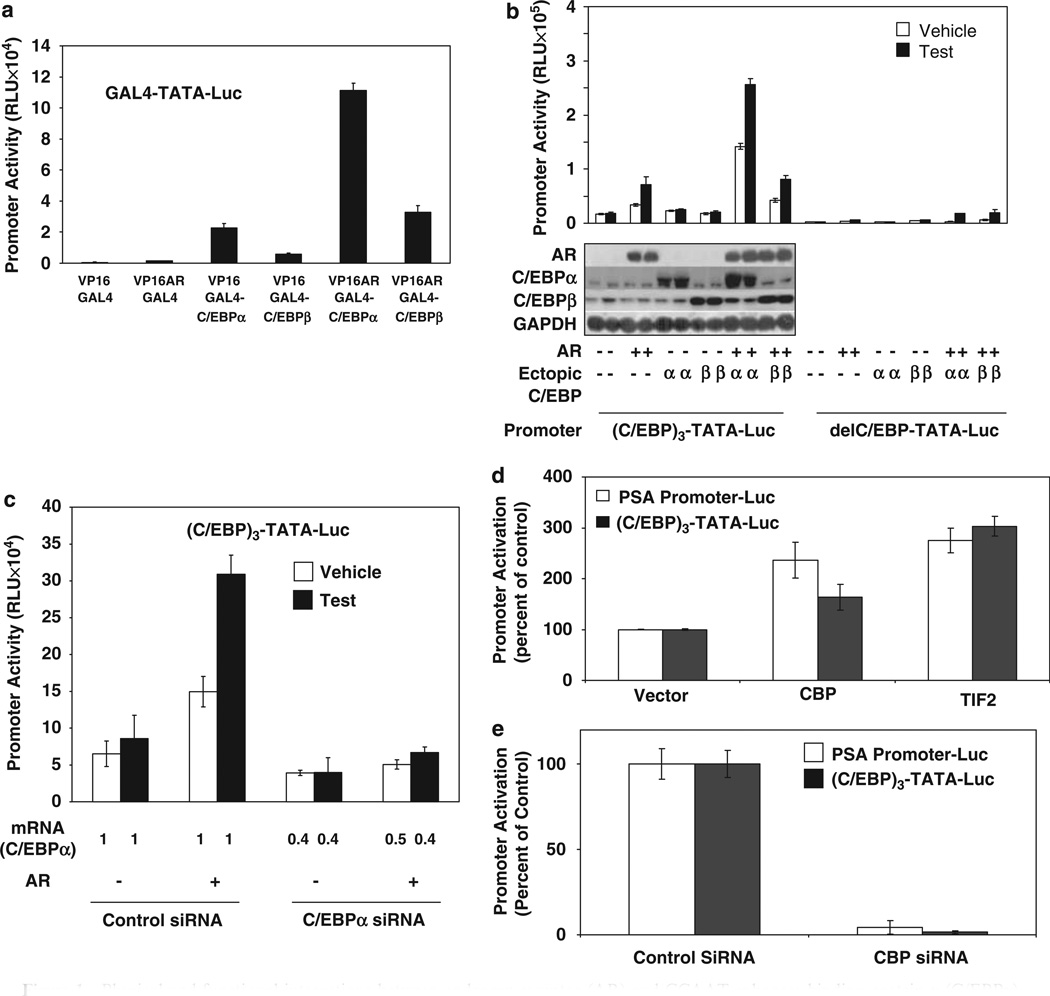
The possibility and functional consequence of recruitment of AR by DNA-bound C/EBPα on transcriptional activation of promoters was tested in two ways. First, a two-hybrid assay using Gal4 fusions of C/EBPα (amino acids 6–217) or C/EBPβ (amino acids 1–256) and VP16-AR was used with a minimal promoter containing GAL4 elements (Figure 1a). Both Gal4 fusion proteins produced basal trans-activation, but Gal4-C/EBPα mediated a striking increase in AR-specific transactivation by VP16-AR (Figure 1a); in comparison, Gal4-C/EBPβ only showed a modest effect (Figure 1a). Second, the effect of C/EBPα or C/EBPβ on testoster-one/AR-dependent activation of an artificial minimal promoter-luciferase reporter construct containing three tandem C/EBP elements upstream of a TATA box ((C/EBP)3-TATA-Luc) was determined in AR-transfected HeLa cells; an identical promoter that did not contain the C/EBP elements (delC/EBP) was used as negative control (Figure 1b). Testosterone/AR and C/EBP element-dependent activation of the promoter occurred in a manner that was further increased by ectopic C/EBPα but not C/EBPβ (Figure 1b). Thus, C/EBPα bound to its cognate element can recruit AR to positively regulate a minimal promoter.
Figure 1.
Physical and functional interactions between androgen receptor (AR) and CCAAT enhancer binding protein-α (C/EBPα). (a) The minimal promoter-luciferase reporter containing a GAL4 element (GAL4-TATA-Luc) was transfected into HeLa cells that were co-transfected with an expression plasmid for either VP16 or a VP16 fusion protein with AR (VP16-AR). The cells were additionally co-transfected with an expression plasmid for either Gal4 or the Gal4 fusion proteins, GAL4-C/EBPα(2–217) or GAL4-C/EBPβ(1–256). The cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h) after which they were subjected to luciferase assay. (b) The minimal promoter-luciferase reporter containing 3-tandem C/EBP elements ((C/EBP)3-TATA-Luc) or the same construct, in which the C/EBP elements were absent (delC/EBP- TATA-Luc), was transfected into HeLa cells with the co-transfection of AR plasmid or empty vector and C/EBPα expression plasmid, C/EBPβ expression plasmid or empty vector. The cells were treated with testosterone (10 nM) or vehicle as described for panel a and harvested either to measure luciferase activity or for western blot analysis using antibody to AR, C/EBPα, C/EBPβ or the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control. (c) HeLa cells were transfected with C/EBPα small interfering RNA (siRNA) or negative control siRNA. The cells were co-transfected with combinations of AR expression plasmid (or vector) and (C/EBP)3-TATA-Luc reporter as indicated. Cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h) and harvested either to measure luciferase activity or C/EBPα mRNA level. The mRNA level for C/EBPα is shown on the x axis as a ratio to the value for the control siRNA transfected samples. The expression of AR and treatment with testosterone had no significant effect on the C/EBPα mRNA level; conversely, the siRNA did not affect expression of AR (data not shown). The standard error in determining the C/EBPα mRNA level was –10%. (d) HeLa cells were separately transfected with two different promoter-luciferase constructs (prostate-specific antigen (PSA) Promoter-Luc or (C/EBP)3-TATA-Luc) and were co-transfected with an expression plasmid for AR together with the co-activator CREB-binding protein (CBP) or transcriptional intermediary factor 2 (TIF2) or with the vector control. The cells were treated with testosterone (10 nM) for the duration of the transfection (48h) and harvested for luciferase assays. (e) HeLa cells were separately transfected with the two promoter-luciferase constructs used in (d) and were co-transfected with an expression plasmid for AR and either control siRNA or siRNA for CBP. The cells were treated with testosterone (10 nM) for the duration of the transfection (48 h) and harvested for luciferase assays. For (a–d), the P-values for the differences noted in the text were <0.001.
Trans-activation by androgen receptor (AR) is supported by endogenous CCAAT enhancer binding protein-α (C/EBPα) and by AR co-activators
The role of endogenous C/EBPα expressed in HeLa cells in mediating trans-activation by AR was tested by examining the effect of knocking down C/EBPα on AR-dependent activation of the (C/EBP)3-TATA-Luc promoter (Figure 1c). Expression of AR was unaffected by knocking down C/EBPα, and AR did not alter the expression of endogenous C/EBPα (data not shown). The negative effect of knocking down C/EBPα on the testosterone/AR response of the promoter (Figure 1c) shows the role of endogenous C/EBPα in this regulation.
The effect of representative AR co-activators on the novel C/EBPα-dependent trans-activation by AR was examined (Figure 1d). The prostate-specific antigen (PSA) promoter (ARE-driven control) and the (C/EBP)3-TATA promoter were tested; in both cases, CREB-binding protein and transcriptional intermediary factor 2 acted as activators of the testosterone/AR response. Furthermore, knocking down CREB-binding protein expression virtually completely abrogated the activation of both promoters (Figure 1e). These results indicate co-activator dependence of the transcriptional activity of AR mediated through recruitment by C/EBPα.
C/EBPα has opposite but independent effects on C/EBP element versus ARE-targeted promoter activation by AR
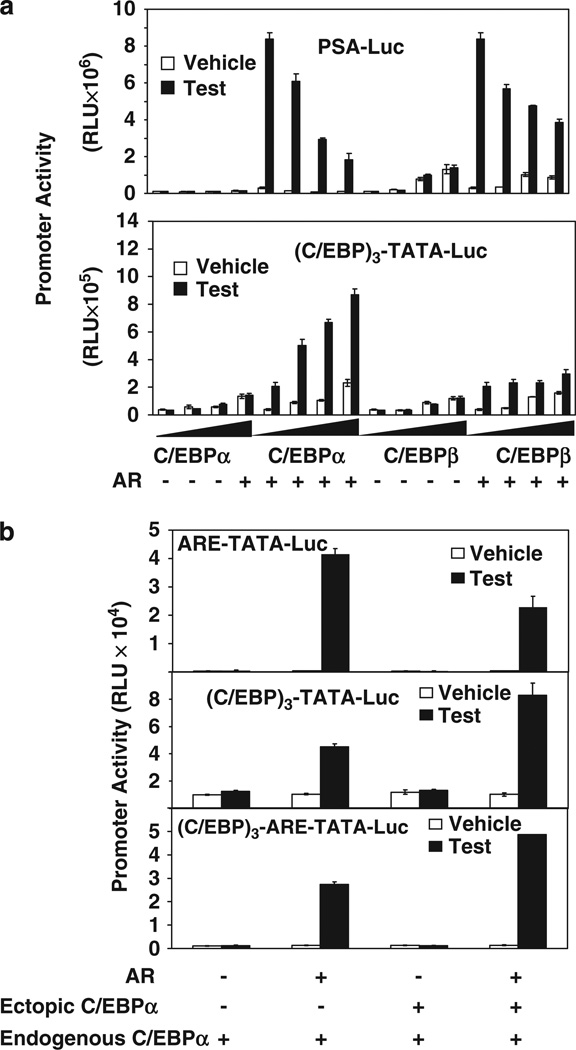
Physical association with C/EBPα has been reported to inhibit promoter activation by AR in the context of the DNA (ARE)-bound receptor, that is, C/EBPα is a corepressor of AR in this context. The observation was confirmed for the PSA promoter in transfected HeLa cells in which ectopic C/EBPα and, to a lesser extent, ectopic C/EBPβ inhibited the promoter activation by testosterone/AR in a dose-dependent manner (Figure 2a, top panel). Under identical conditions, C/EBPα caused a striking dose-dependent increase in the activation of the (C/EBP)3-TATA promoter by AR; C/EBPβ produced a similar effect but to a relatively modest extent (Figure 2a, bottom panel). When, in a minimal promoter containing an ARE, a canonical C/EBP element was inserted upstream of the ARE, ectopic C/EBPα caused an increase in promoter activation by testosterone/AR (Figure 2b). The results show that the interaction of C/EBPα and AR at a canonical C/EBP element, which result in trans-activation, overrides the inhibitory interaction of the two proteins at AREs elsewhere in the promoter.
Figure 2.
The effect of different promoter configurations of androgen response element (ARE) and CCAAT enhancer binding protein (C/EBP) elements on the transcriptional activities of androgen receptor (AR) and C/EBPα. (a) HeLa cells (5 × 105) were transfected with (top panel) prostate-specific antigen (PSA) promoter luciferase reporter (PSA-Luc) or (bottom panel) (C/EBP)3-TATA-Luc in the indicated combinations with AR expression plasmid (50 ng) or vector control and different amounts of C/EBPα expression plasmid (50, 100 and 200 ng), C/EBPβ expression plasmid (50, 100 and 200 ng) or vector control. The cells were treated with testosterone (10nM) for the duration of the transfection (48 h) and harvested for luciferase assays. (b) HeLa cells were transfected with the minimal promoter-luciferase reporter containing an ARE (ARE-TATA-Luc) (top panel), (C/EBP)3-TATA-Luc (middle panel), the same promoter construct containing three-tandem C/EBP elements upstream of the ARE ((C/EBP)3-ARE-TATA-Luc) (bottom panel). The cells were co-transfected with AR expression plasmid or vector control, and C/EBPα expression plasmid or vector control. The cells were treated with testosterone and harvested for luciferase assays as described for panel a. For panels a and b, the P-values for the differences noted in the text were <0.001.
Trans-activation per se by androgen receptor-CCAAT enhancer binding protein-α (AR-C/EBPα) occurs without androgen-dependence or sensitivity to flutamide
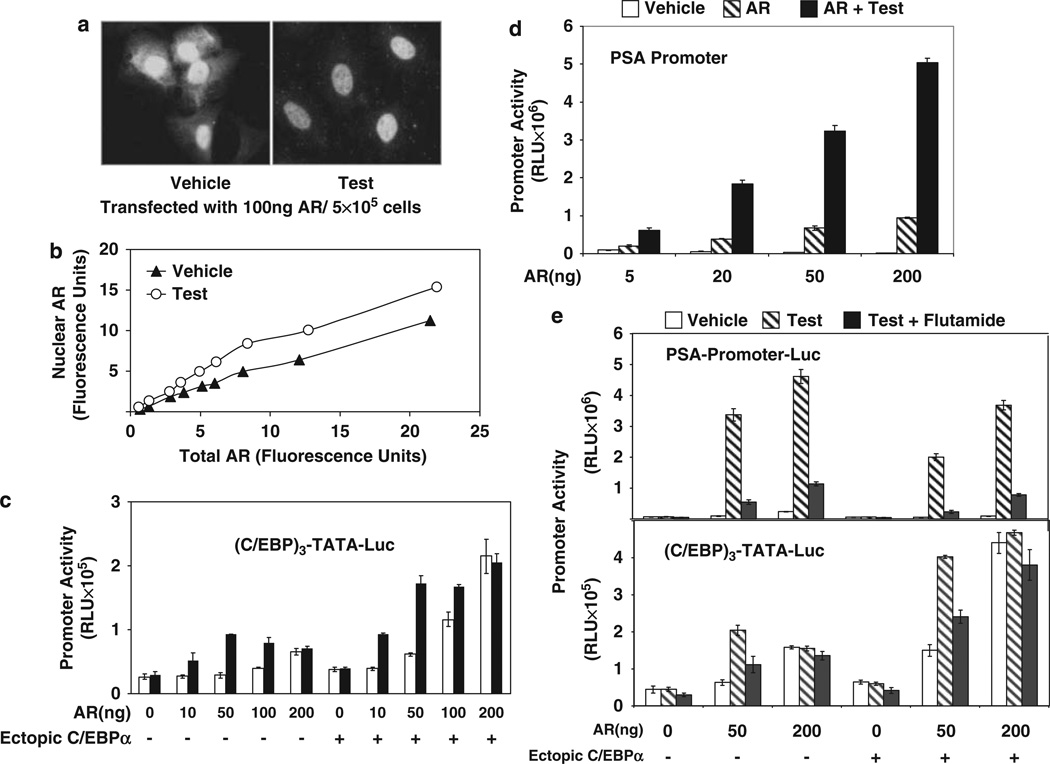
As observed in Figure 1, AR recruitment by C/EBPα and the associated trans-activation was only partially androgen dependent for wild-type AR. This could be explained by reported observations (Jenster et al., 1993) that in HeLa cells a substantial amount of ectopic AR can enter the nucleus independent of ligand and also hints that the primary role of androgen may be to further enhance nuclear translocation of AR in the transfected HeLa cells. To analyse this possibility, the intracellular localization of AR was examined using quantitative confocal microscopy in HeLa cells expressing different amounts of ectopic AR. In the absence of androgen treatment, the ectopic AR was localized in the nucleus as well as in the cytosol (Figure 3a). Treatment with testosterone further mobilized the cytosolic AR to the nucleus (Figure 3a). Under both conditions, the amount of AR in the nucleus was a function of the total AR expressed (Figure 3b).
Figure 3.
The effect of forced nuclear localization of androgen receptor (AR) on ligand sensitivity of promoter activation by AR-CCAAT enhancer binding protein-a (C/EBPα). (a, b) Sub-cellular localization of ectopic AR in HeLa cells was examined using immunofluorescence. HeLa cells grown in chamber slides were transfected with different doses of AR expression plasmid (10–200 ng) or vector control and treated with either testosterone(10 nM) or vehicle for the duration of the transfection (48 h). Immunofluorescence staining for AR was performed using a primary rabbit antibody to AR and a bovine anti-rabbit immunoglobulin G (IgG)-fluorescein isothiocyanate (FITC) as the secondary antibody. The nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; not shown) and fluorescence images were captured using confocal microscopy. In (a), the representative images show a mixed distribution of AR (green fluorescence) between nuclear and cytosolic compartments in the absence of hormone but a predominantly nuclear distribution after testosterone treatment in the cells transfected with 100 ng AR expression plasmid per 5 × 105 cells. In panel b, the total amount of AR fluorescence in the cell and the amount of AR fluorescence localized in the nucleus were quantified using LASAF software. The values are plotted as arbitrary fluorescence units. (c) HeLa cells were co-transfected with (C/EBP)3-TATA-Luc and different amounts of AR expression plasmid (10–200 ng per 5 × 105 cells) or vector control together with C/EBPa expression plasmid or vector control. The cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h) and harvested for luciferase assays. (d) HeLa cells were transfected with the prostate-specific antigen (PSA)-promoter luciferase reporter construct together with different amounts of AR expression plasmid (5–200 ng per 5 × 105 cells). The cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h) and harvested for luciferase assays. (e) HeLa cells were transfected with either PSA-promoter-Luc or (C/EBP)3-TATA-Luc and co-transfected with different amounts of AR expression plasmid (50–200 ng per 5 × 105 cells) or vector control and C/EBPa expression plasmid or vector. The cells were treated with testosterone (10 nM), vehicle or the combination of testosterone (10 nM) and flutamide (25 µM) for the duration of the transfection (48 h) and then harvested for luciferase assays. For panels b-e, the P-values for the differences noted in the text were <0.001. A full colour version of this figure is available at the Oncogene journal online.
In HeLa cells, increasing expression levels of ectopic AR caused activation of the (C/EBP)3-TATA promoter by the receptor to become progressively androgen independent, both in the absence and in the presence of ectopic C/EBPα, reaching complete androgen independence at the highest level of AR (Figure 3c). In contrast, under the same conditions, the AR activation of a promoter dependent entirely on AREs (the PSA promoter, Figure 3d) was predominantly androgen dependent. In addition, the ARE-mediated promoter activation was sensitive to the androgen antagonist flutamide (Figure 3e), whereas for C/EBP element-mediated activation insensitivity to flutamide accompanied androgen independence (Figure 3e). As shown in Figure 3b, as increasing the level of ectopic AR results in increased hormone-independent entry of AR into the nucleus, these results indicate that for C/EBP element-targeted promoter activation by AR, recruitment of AR to the promoter and trans-activation per se are independent of ligand. This conclusion is strongly supported by the contrasting observation that regardless of ectopic AR levels, ARE-mediated trans-activation required treatment with androgen. The principal role of androgen in C/EBP element-targeted promoter activation must thus be to enhance the localization of AR in the nucleus in the normal cell context, which limits androgen-independent entry of AR into the nucleus.
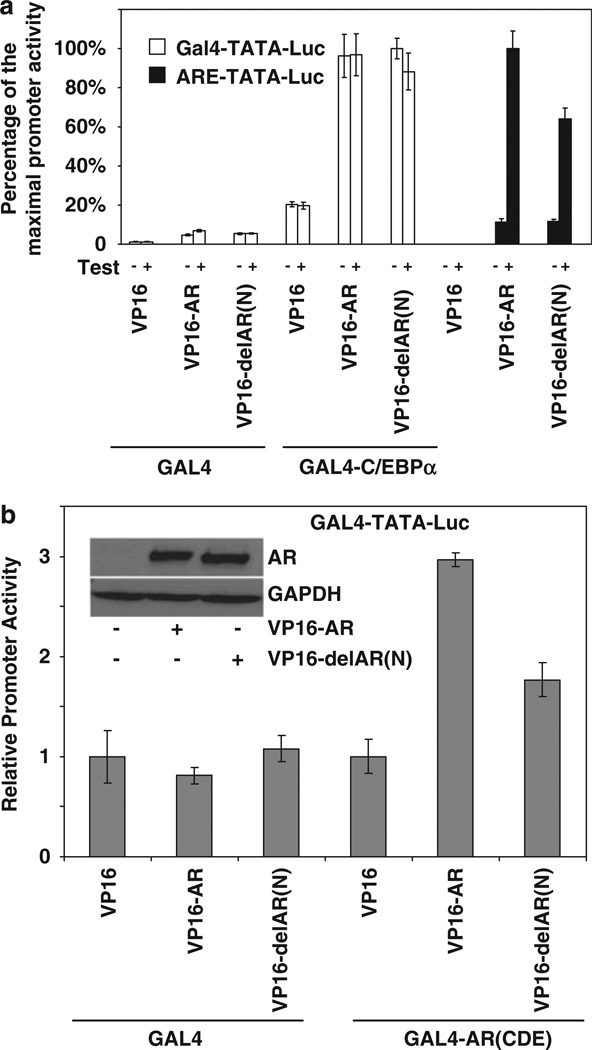
Multiple androgen receptor (AR) domains and a CCAAT enhancer binding protein-α (C/EBPα) peptide are involved in the association of AR with DNA-bound C/EBPα
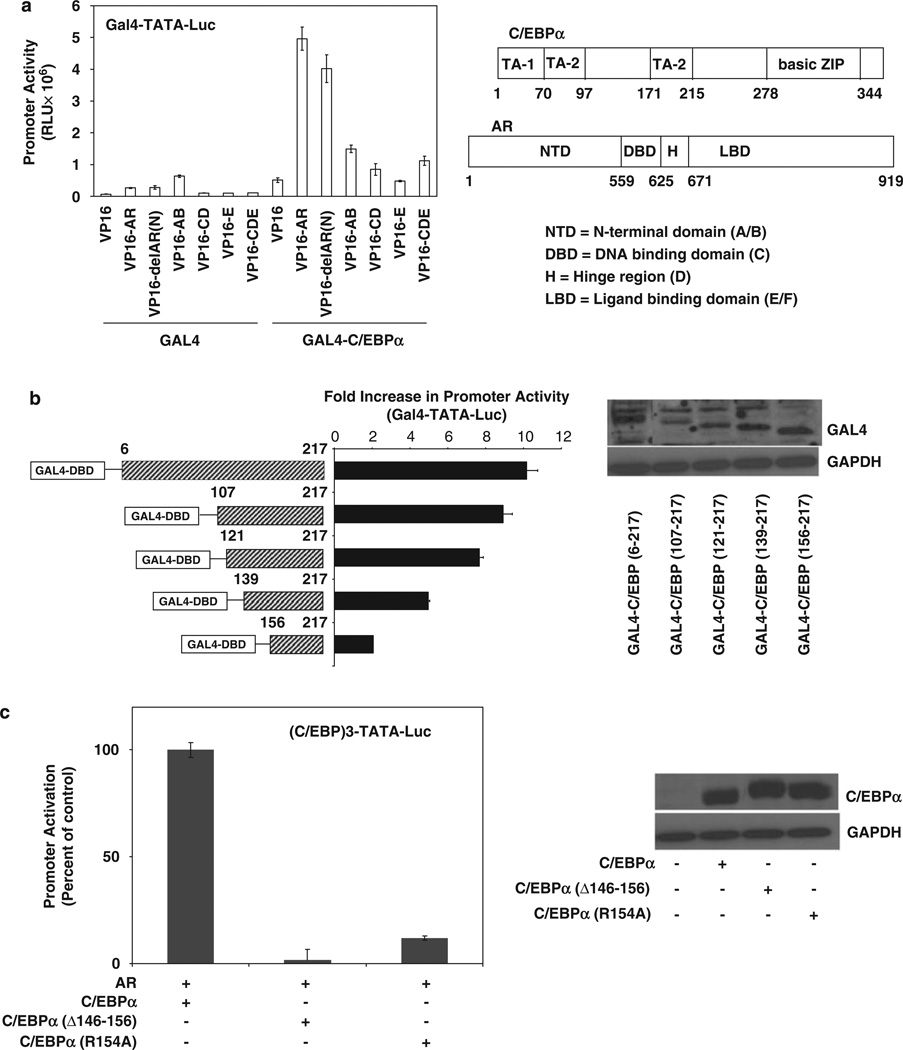
The specific AR domains required for interaction with C/EBPα in the context of AR recruitment by DNA-bound C/EBPα was determined by a two-hybrid assay using Gal4-C/EBPα and N-terminal VP16 fusions of various AR domains. AR domains AB, CD, E and CDE showed variable but modest association with Gal4-C/ EBPα in comparison with the full-length VP16-AR (Figure 4a). The results indicate that in the context of DNA-bound C/EBPα, more than one AR domain must collectively contribute to the ability of the two proteins to associate.
Figure 4.
Mapping the protein domains of androgen receptor (AR) and CCAAT enhancer binding protein-α (C/EBPα) required for their interaction in the context of DNA-bound C/EBPα. (a) HeLa cells were transfected with GAL4-C/EBPα or Gal4 expression plasmid together with an expression plasmid for either VP16 or VP16 fusion proteins with the full-length AR (VP16-AR), AR with its N-terminal amino acids 1–37 deleted (VP16-delAR(N)), the AB domains of AR (VP16-AB), the CD domains of AR (VP16-CD), the E domain of AR (VP16-E) or the CDE domains of AR (VP16-CDE). The cells were co-transfected with the Gal4-TATA-Luc reporter. At 48h after transfection, the cells were harvested to measure luciferase activity. (b) HeLa cells were transfected with an expression plasmid for either Gal4, the Gal4 fusion protein GAL4-C/EBPα(2–217) or GAL4 fusion proteins with various portions of C/EBPα polypeptide, as illustrated in the figure. The cells were co-transfected with the AR expression plasmid or vector control together with the Gal4-TATA-Luc promoter construct. The cells were treated with testosterone (10 nM) for the duration of the transfection (48 h) after which they were harvested for luciferase assays. The values are plotted as the fold increase in promoter activity relative to the values in the absence of AR. The western blot shows the relative expression levels of the fusion proteins by probing with antibody to GAL4. (c) HeLa cells were transfected with expression plasmids for C/EBPα or its mutant forms containing either an internal deletion of amino acids 146–156 or mutation of R154 to A. The cells were co-transfected with the AR expression plasmid or vector control together with the (C/EBP)3-TATA-Luc promoter construct. The cells were treated with testosterone (10 nM) for the duration of the transfection (48h) after which they were harvested for luciferase assays. The western blot shows the relative expression levels by probing with antibody to C/EBPα. For panels a–c, the P-values for the differences noted in the text were <0.001.
In similar two-hybrid assays, deletional analysis of GAL4-C/EBPα(6–217), which encompasses all of the activation domains of C/EBPα upstream of its conserved basic leucine zipper domain (Nerlov and Ziff, 1994), showed that the C-terminal half of this polypeptide (amino acids 107–217) contains within it, the motif(s) required for binding to AR with most of the binding activity being dependent on the peptide 121–156 (Figure 4b). This peptide sequence spans one of three trans-activation elements of C/EBPα. Moreover, an internal deletion of peptide 146–156, as well as mutation of arginine at position 154 to alanine, strikingly decreased the ability of C/EBPα to support AR-dependent promoter activation through the C/EBP element (Figure 4c), consistent with the importance of this region for binding to AR.
A dimerization mutant of androgen receptor (AR) interacts with CCAAT enhancer binding protein-α (C/EBPα)
Deletion of amino acids 1 to 37 of AR (delAR(N)) did not significantly diminish association of the fusion protein with Gal4-C/EBPα (Figure 4a). As amino acids 3 to 36 are required for amino- to carboxyl-terminal (N- to C-) interaction of AR, which is believed to be responsible for the homodimerization of the receptor (Berrevoets et al., 1998), it was of interest to examine the effect of the deletion on the association of AR with C/EBPα versus DNA sequence elements that make direct contact with AR. Accordingly, the effect of testosterone was tested in the two-hybrid assay using VP16-delAR(N) and Gal4-C/EBPα and was compared with the agonist effect in one-hybrid assays using the minimal Gal4 promoter in which the Gal4 element was replaced by a canonical ARE (Figure 5a). Fusion with VP16 may be expected to allow AR to localize in the nucleus independent of ligand binding. Both VP16-AR and VP16-delAR(N) bound to Gal4-C/EBPα to the same extent with or without ligand (Figure 4c). In contrast, the binding of both VP16-AR and VP16-delAR(N) to ARE required agonist and further, the amino-terminal (1–37) deletion in AR partially reduced binding of the fusion protein to the ARE (Figure 5a); as observed in Figure 5b, this partial decrease is consistent with the partial loss of dimerization of VP16-AR with the GAL4 fusion of the CDE domains of AR upon deletion of amino acids 1–37 (Figure 5b).
Figure 5.
The effect of disrupting androgen receptor (AR) dimerization on the ability of CCAAT enhancer binding protein-a (C/EBPα) to recruit AR. (a) HeLa cells were transfected with GAL4-C/EBPα or Gal4 expression plasmid together with an expression plasmid for VP16, VP16-AR or VP16-delAR(N) (deletion of amino acids 1–37). The cells were co-transfected with either Gal4-TATA-Luc or ARE-TATA-Luc. The cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48h) after which they were harvested for luciferase assays. The luciferase activities are plotted as the percentage of the maximal activity for each promoter construct. (b) HeLa cells were transfected with expression plasmid for Gal4 or GAL4 fused to AR containing only its CDE domains (GAL4-AR(CDE)) together with an expression plasmid for VP16, VP16-AR or VP16-delAR(N). The cells were co-transfected with either Gal4-TATA-Luc. The cells were treated with testosterone (10 nM) for the duration of the transfection (48h) after which they were harvested for luciferase assays. The western blot shows the relative levels of VP16-AR and VP16-delAR(N) probed with antibody to AR. For panels a and b, the P-values for the differences noted in the text were <0.001.
The results show that whereas ligand-dependent dimerization of AR is essential for its interaction with a canonical ARE, recruitment of AR by DNA-bound C/EBPα can occur without AR dimerization. This observation is also consistent with the ability of DNA-bound C/EBPα to recruit and mediate trans-activation by AR in the absence of androgen, which is required for AR dimerization.
CCAAT enhancer binding protein-α (C/EBPα) orchestrates global gene regulation by androgen receptor (AR)
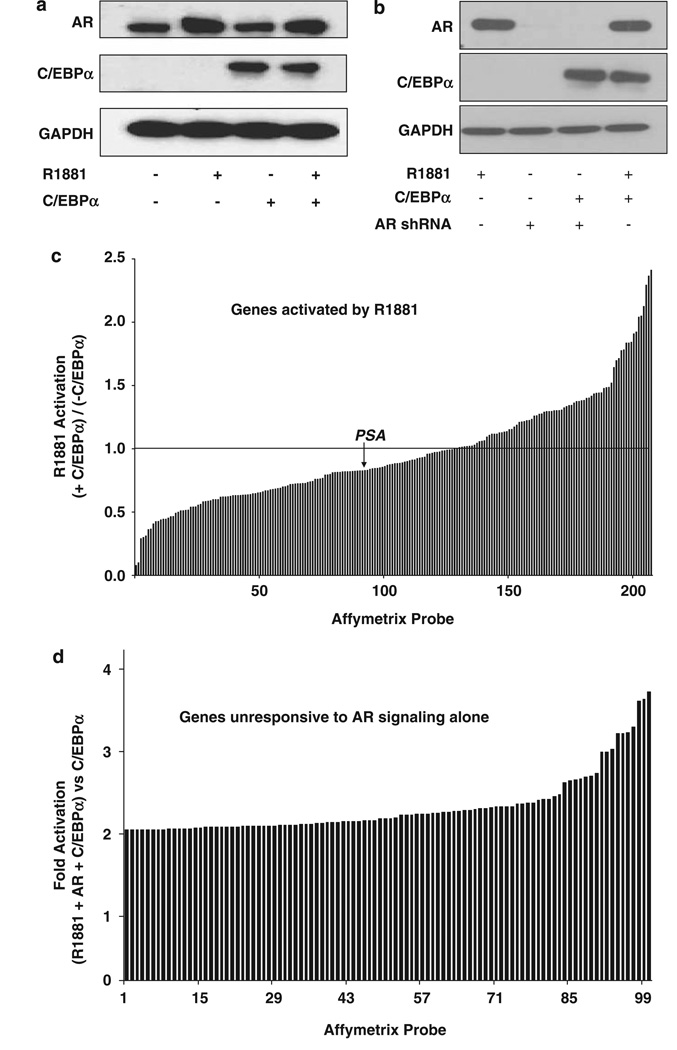
Affymetrix (Santa Clara, CA, USA) DNA microarray analysis was used to examine the genome-wide effect of C/EBPα on androgen signaling in LNCaP cells, which are deficient in C/EBPα. First, C/EBPα was introduced by transient transfection of an expression plasmid to minimize possible phenotypic alterations that might result from selection pressures in long-term cell culture that would occur in the production of stable recombinant cells. The transfection efficiency for C/EBPα was ~60%, as determined using flow cytometric analysis of co-expressed green fluorescent protein (data not shown). Ectopic expression of C/EBPα did not significantly alter the expression of endogenous AR and a 6-h treatment with the synthetic AR agonist R1881 (Dupont Inc., Wilmington, MA, USA) did not alter the expression of C/EBPα (Figure 6a).
Figure 6.
The combined effect of androgen receptor (AR) and CCAAT enhancer binding protein-α (C/EBPα) on global gene regulation. (a, c) LNCaP cells were transfected by nucleofection with either C/EBPα or vector. After 48 h, the cells were treated with either R1881(1 nM) or vehicle for 6 h. The cells were then harvested either for western blot analysis using antibody to AR or C/EBPα (a) or to obtain total RNA (c). In panel c, the mRNA profile was determined using replicate samples by Affymetrix microarray analysis and genes that were induced ≥twofold by R1881 in 6 h were selected; the data are plotted to show the effect of ectopic C/EBPα on the fold activation by R1881 of each gene; a value of 1 on the y axis indicates that R1881 stimulation was unaffected by C/EBPα. (b, d) LNCaP cells were transfected by nucleofection with C/EBPα or vector. After 24h, the cells were infected with AR short hairpin RNA (shRNA) lentivirus or non-target control lentivirus; the cells were then grown in fresh hormone-free media. The cells were then treated for 48h with either vehicle or R1881 (1 nM) as shown in (b). The cells were harvested at 72 h after nucleofection for either western blot analysis (b) or to obtain total RNA (d). In panel d, the mRNA profile was determined using replicate samples by Affymetrix microarray analysis; genes whose expression was unaffected by R1881/AR were selected; the data are plotted to show the increase in expression in the presence of R1881+AR+C/EBPα compared with C/EBPα alone.
The preceding mechanistic studies predict that the androgen responsiveness of genes may be differentially regulated by C/EBPα depending on the promoter contexts of the target genes; ARE-mediated gene activation would be inhibited by C/EBPα in one group of genes but in others C/EBPα may interact at other promoter sites to override this inhibition or even further increase transcriptional activation by AR. Consistent with this prediction, among genes that were activated (≥twofold) by a 6-h treatment with R1881, the effect of ectopic C/EBPa on the androgen-stimulated mRNA expression ranged from a significant (~70%) inhibition to a further ~2.5-fold activation (Figure 6c; annotated data provided in Supplementary Table 1).
Next, to identify genes whose induction was dependent on the presence of both AR signaling and C/EBPα, the mRNA profiles were examined in AR + cells treated with R1881 or cells in which AR was completely knocked down (Figure 6b); in both cases, the mRNA profiles were determined either in the absence or in the presence of ectopic C/EBPα. As predicted from the mechanistic studies, among genes that were unresponsive to AR signaling alone, many genes were synergistically activated by AR signaling and C/EBPα. The fold-change for AR and C/EBPα versus C/EBPα was adjusted to compensate for the fact that only ~50% of the cells were transfected by the C/EBPα expression plasmid, as determined using co-transfected green fluorescent protein (data not shown). The list was filtered to remove all probe sets with activation by AR + R1881/AR control >1.2, those with average expression levels o64 and those with P-values for increase >0.01. The adjusted increases for the 100 genes with the greatest increases are plotted in Figure 6d (annotated data provided in Supplementary Table 2).
To examine their potential mechanisms of regulation, the top 100 genes showing C/EBPα-dependent AR activation (from Supplementary Table 2) were submitted to the promoter analysis pipeline program (http://bioinformatics.wustl.edu/webTools/PromoterSearch.do). This program reports enrichment for predicted binding sites for transcription factors in the promoter regions (−10 kb to +5 kb) of sets of genes. The submitted list of 100 genes matched 87 genes in the promoter analysis pipeline database. C/EBP sites were the top three highest scoring sites, with the lowest P-value of 0.2156, which from our experience indicates highly significant enrichment. For comparison, the top 100 genes whose expression increased in C/EBPα-transfected cells lacking AR, as compared with controls lacking AR, showed enrichment for C/EBP sites with a lowest P-value of 0.1852. Genes with adjusted values of C/EBPa-dependent AR activation <1.8-fold showed only marginal enrichment for C/EBP sites (data not shown).
Whereas regulation of any individual gene by AR and C/EBPa may be influenced or determined by additional factors related to their unique and complex promoter contexts, gene expression profiling presents a scenario for the physiological relevance of the bimodal regulation of AR signaling by C/EBPa elucidated in the preceding sections.
CCAAT enhancer binding protein-a (C/EBPα) induces functional chromatin associations of androgen receptor (AR) in situ
Direct evidence for C/EBPa-dependent recruitment of AR at chromatin sites in situ was sought using chromatin immunoprecipitation-DNA microarray chip (ChIP-chip) analysis. Functional AREs are believed to generally map to chromatin sites distal (>10 kb) from their target promoters (Wang et al., 2007), although association of those sites with the relevant target genes has only been made in a few cases. However, this study sought a more limited analysis of the genome to explore a significant presence of chromatin sites of C/EBPα-dependent AR recruitment. Accordingly, the Roche-Nimblegen (Madison, WI, USA) promoter tiling arrays covering promoter regions −3500 to +1000nt were chosen for the analysis. Co-expression of C/EBPa was associated with 169 unique peaks of AR recruitment with a false discovery rate of ≤0.05 and 33 unique peaks of AR recruitment with a false discovery rate of ≤0.01 (annotated data provided in Supplementary Table 3). The 169 peak sequences were submitted for analysis to the Trawler program, which identifies families of sequence motifs enriched in peak sequences compared with comparable background sequences (Ettwiller et al., 2007). The family with the highest z-score (12.42) had several clusters that were identified as having significant matches to C/EBPα-binding motifs, as defined using the TRANSFAC and JASPAR databases. No enrichment for AREs was observed.
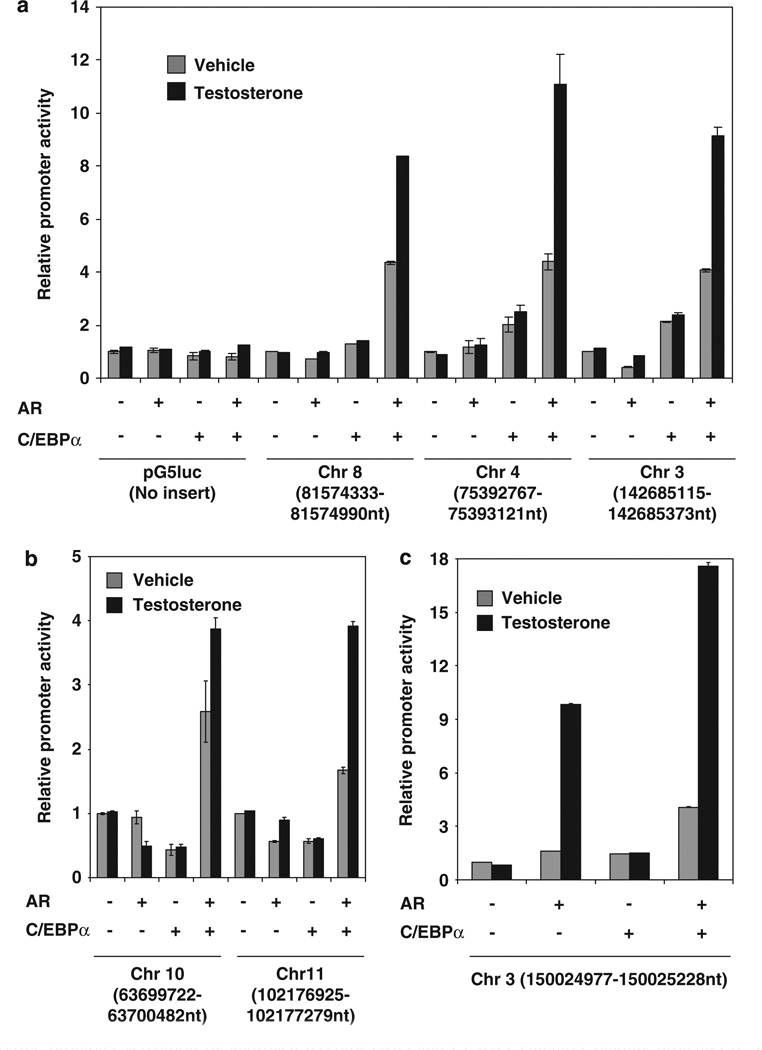
To test whether C/EBPα recruited AR to chromatin sites in a functional manner, several AR-associated peak genomic DNA sequences identified above were inserted upstream of the minimal promoter within the GAL4-TATA-Luc (pG5luc) reporter plasmid and tested in the AR-negative HeLa cells for their ability to enhance promoter activity in response to ectopic AR. Among the top 25 peaks, 10 peak sequences were selected, based on the ease of PCR amplification from genomic DNA and subcloning into the pG5luc reporter plasmid. In all, six of the sequences supported C/EBPα-dependent promoter activation by AR (Figure 7a – c); one of the sequences also showed C/EBPα-independent activation by AR (Figure 7c), suggesting the presence of an additional tethering mechanism for AR within this sequence. Thus, the ChIP-chip peaks identified above include functional target sites of C/EBPα-dependent transcriptional activity of AR. It is possible that the peak sequences that did not give a functional response were limited by the promoter context.
Figure 7.
Synergistic promoter activation by androgen receptor (AR) and CCAAT enhancer binding protein-α (C/EBPα) through chromatin sites identified using chromatin immunoprecipitation-DNA microarray chip (ChIP-chip). HeLa cells (3 × 105) were transfected with either a minimal promoter-luciferase reporter (pG5luc) or pG5luc in which the indicated genomic DNA fragments were inserted upstream of the minimal promoter. The chromosomal locations of the insert sequences are indicated on the x axis. The cells were co-transfected with an expression plasmid for AR (200 ng) or vector control and/or C/EBPa (50 ng) expression plasmid or vector control for 48 h. The cells were treated with testosterone (10 nM) or vehicle for the last 12 h of the transfection and harvested for luciferase assay. The relative promoter activities are plotted as the ratio to that of the corresponding promoter in the absence of AR and ectopic C/EBPα. The data for the different promoter constructs are plotted in a-c. For a-c, the P-values for the differences noted in the text were <0.001.
Androgen receptor (AR) upregulates the peroxisome proliferator-activated receptor-γ (PPARγ) gene from a CCAAT enhancer binding protein (C/EBP)-dependent AR recruitment site in the first intron
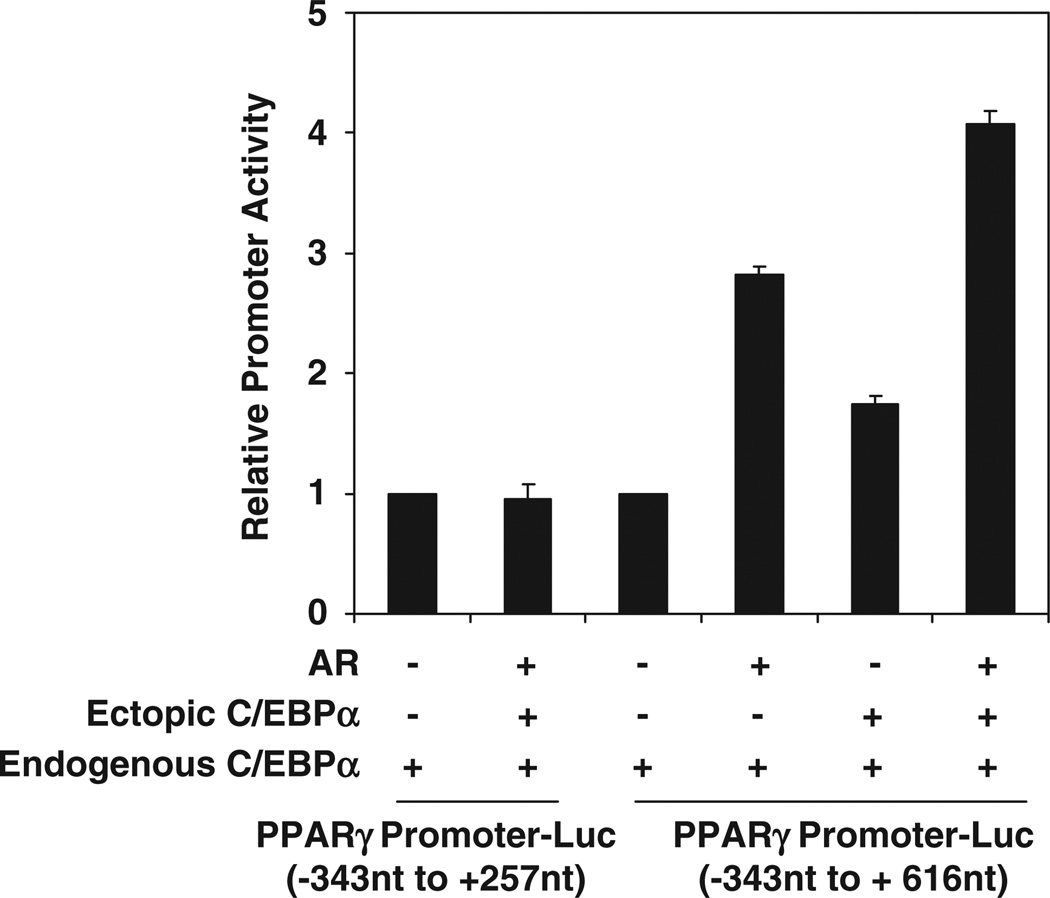
By inspection of genes that showed both C/EBPα-dependent AR recruitment and C/EBPα-dependent gene activation by AR, the PPARγ gene was identified as a direct target gene with an obvious physiological significance in prostate tissue. A site of C/EBPa-dependent recruitment of AR was identified in the first intron of the PPARγ gene, at +257 to +616nt. This region contained two C/EBP elements. A genomic fragment spanning the PPARγ promoter and including this intronic region (−343 to +616nt) was inserted upstream of a luciferase reporter gene in the pGL3 basic plasmid and tested in the AR-negative HeLa cells. As observed in Figure 8, the promoter was activated by ectopic AR in a manner that was enhanced by ectopic C/EBPα; AR and C/EBPa could not activate the promoter construct in which the region of AR recruitment mapped by ChIP-chip (−343 to +616nt) was deleted (Figure 8).
Figure 8.
Synergistic activation of a genomic peroxisome proliferator-activated receptor-γ (PPARγ) promoter fragment by androgen receptor (AR) and CCAAT enhancer binding protein-α (C/EBPα) through a chromatin site in intron 1 identified using chromatin immunoprecipitation-DNA microarray chip (ChIP-chip). HeLa cells were transfected with the pGL3-basic vector in which genomic DNA fragments corresponding to the indicated PPARγ gene sequences were inserted upstream of the luciferase reporter. The cells were co-transfected with an expression plasmid for AR or vector control and/or C/EBPα expression plasmid or vector control for 48h. The relative promoter activities are plotted as the ratio to that of the corresponding promoter in the absence of AR and ectopic C/EBPα. The P-values for the differences noted in the text were <0.001.
Discussion
The positive regulation of the transcriptional activity of AR by C/EBPα elucidated in this study, together with the negative regulation of AR activity by C/EBPα reported previously (Chattopadhyay et al., 2006), reveal a unique bimodal association between two DNA-binding transcription factors that also interact reciprocally as co-regulators of each other. This type of interaction enables a switch in the gene regulatory pattern of the one transcription factor coincident with co-expression of the second. The exact nature of this interaction depends on the target promoter context, specifically the occurrence and configuration of DNA elements that bind either AR or C/EBPα. The association of AR with an ARE is ligand dependent and in this context, C/EBPα binds to AR as a corepressor; in contrast, the binding of C/EBPα to a C/EBP element and its recruitment of AR is entirely independent of ligand (barring ligand dependence for nuclear translocation of AR), resulting in promoter activation through further recruitment of AR co-activators. When target genes contain non-interacting ARE and C/EBP elements at distinct sites in their promoter regions, recruitment of AR at the C/EBP element leads to gene activation in a manner that is not influenced by events at the ARE site; in such promoters, androgen-dependent gene activation can occur through the ARE in the absence of C/EBPα. Inhibition of ARE-mediated gene activation has been attributed to competition between C/EBPα and co-activators for binding to AR (Chattopadhyay et al., 2006); however, it may be noted here that recruitment of AR at C/EBP elements, as shown in this study, would also predict inhibition of classical androgen target genes through sequestering of AR at non-classical target promoters by C/EBPα. In the normal prostate, both ligand-dependent and ligand-independent mechanisms of target gene interactions of AR must depend on androgen for translocation of the receptor to the nucleus. On the other hand, in hormone refractory prostate tumors, in which AR is localized in the nucleus independent of hormone, C/EBPα-mediated gene activation by AR would be entirely independent of hormone.
Although androgens are necessary for the growth and maintenance of the normal prostate and for prostatic hyperplasia, none of these processes have been linked with an alteration in the hormonal status. Therefore, androgens have a permissive role in prostate physiology in a manner that is regulated by other factors. In this context, the actions of C/EBPα and its interactions with AR are particularly significant. The diversity of gene regulatory patterns allowed by the AR-C/EBPα interactions adds to the independent activities of the two proteins, offering a significant physiological advantage in the development and function of the prostate. AR is necessary for supporting virtually all aspects of prostate physiology, but C/EBPα, a strongly antiproliferative protein, is only expressed in the nucleus of prostate epithelial cells beginning at the time of prostate maturation when it may be expected to inhibit the classical mechanism of gene activation by androgen. Whereas negative regulation of androgen signaling by C/EBPα may be needed to inhibit androgen-stimulated growth, alternate mechanisms of gene activation by androgen could be simultaneously facilitated by C/EBPα, appropriately redirecting androgen signaling to support its other functions. Consistent with this view, gene ontology analysis indicated a broad range of functions for the direct and indirect target genes of AR-C/EBPα. The physiological significance of gene co-regulation by AR and C/EBPα is also illustrated by the specific example shown above of the upregulation of the PPARγ gene; PPARγ is expressed in prostate tissues, is antiproliferative and is a prostate tumor suppressor (Paltoo et al., 2003; Han and Roman, 2007), in keeping with the known physiological effects of C/EBPα.
This study has elucidated in detail the unique characteristics of a novel molecular mechanism of interaction between AR and C/EBPα and established the prevalence of these interactions in regulating androgen-induced gene expression patterns. It must be noted that the profound effects of AR on the growth, differentiation and maintenance of the prostate are associated with a broad and complex (direct and indirect) transcriptional regulation, and it has been virtually impossible to assign specific functional groups of ultimate target genes of AR to the regulation of specific aspects of prostate physiology (Thomson and Marker, 2006). Such correlations are further confounded by the fact that even after terminal differentiation, the prostatic epithelium constantly undergoes apoptosis and androgen-dependent regeneration. Therefore, the extensive nature of the influence of C/EBPα on the AR-regulated gene expression profile in LNCaP cells should, of itself, be taken as an indicator of a profound physiological role of C/EBPα in androgen regulation of prostate physiology.
Direct target gene activation by tethering to DNA-bound transcription factors, such as Sp family proteins and activator protein-1, has been well established for the estrogen receptor (Jakacka et al., 2001; Safe and Kim, 2004). AR is known to repress gene activation by activator protein-1 and nuclear factor-κB (RelA) but this seems to occur principally through sequestration of limiting amounts of the co-activator, CREB-binding protein (Aarnisalo et al., 1998; Frønsdal et al., 1998). Tethering mechanisms have not been adequately elucidated for AR, notwithstanding a report that in myoblasts, AR activates the skeletal α-actin gene through its recruitment to the target promoter by serum response factor (Vlahopoulos et al., 2005). As elucidated in this study, C/EBPα-dependent direct gene activation by AR is a clear example of tethering of AR to its target genes. Interestingly, this mechanism of transcriptional activation by AR occurs independently of hormone and is insensitive to a classical androgen antagonist. The degree of promoter activation by AR through association with DNA-bound C/EBPα was not diminished in a dimerization mutant of the receptor, indicating that the ligand-independent gene activation by AR through tethering to C/EBPα does not involve AR dimerization. The results represent the first detailed characterization of a tethering mechanism that supports the transcriptional activity of AR and that further, is independent of ligand.
Androgen receptor has a predominantly nuclear localization in hormone-refractory prostate cancer cells that are ‘truly’ independent of androgen. However, hormone-independent localization of AR in the nucleus will not result in ligand-independent activation through classical AREs; therefore, in the absence of hormone, AR-dependent growth must be supported by alternate modes of association of the receptor with its target genes, rather than by binding to its classical response elements (Jia and Coetzee, 2005) (Gonit et al., publication pending). C/EBPα frequently manifests in clinical prostate cancer similar to a variety of other tumors (Sundfeldt et al., 1999; Zhang et al., 2008; Oncomine microarray data repository, http://www.oncomine.org/) in which the cellular contexts may be expected to attenuate or overcome its antiproliferative effects (Wang et al., 2004; Wang and Timchenko, 2005). C/EBPa-mediated trans-activation by AR in prostate cancer could therefore represent a significant aspect of AR function that is both independent of androgen and refractory to anti-androgen therapy.
Materials and methods
Chemicals and reagents
Dulbecco’s minimum essential medium, RPMI 1640, sodium pyruvate and penicillin/streptomycin/l-glutamine stock mix were purchased from Life Technologies, Inc. (Carlsbad, CA, USA); fetal bovine serum (FBS) and charcoal-stripped FBS (CS-FBS) were from Invitrogen (Carlsbad, CA, USA). FUGENE 6 was purchased from Roche Diagnostics (Indianapolis, IN, USA). Luciferase assay reagents were from Promega (Madison, WI, USA). Affinity purified rabbit anti-human AR (sc-816)), C/EBPα(sc-61), C/EBPβ(sc-150), mouse anti- glyceraldehyde-3-phosphate dehydrogenase (sc-47724) and normal rabbit immunoglobulin G control (sc-2027) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Peroxidase or fluorescein isothiocyanate-conjugated secondary antibodies were from Vector Laboratories (Burlin-game, CA, USA). Cell Line Nucleofector Kits (R) were purchased from Amaxa Biosystems (Cologne, Germany). Custom oligonucleotide primers were from Life Technologies. Lipofectamine 2000 reagent was from Invitrogen. C/EBPα small interfering RNA (siRNA; s2888) and control siRNA were purchased from Ambion (Branchburg, NJ, USA). Vent DNA polymerase was purchased from New England Biolabs (Beverly, MA, USA). Custom oligonucleotide primers were from Integrated DNA Technologies (Coralville, IA, USA). The reagents for reverse transcription-PCR and real-time PCR were purchased from Applied Biosystems (Branchburg, NJ, USA).
DNA constructs and expression plasmids
Construct design used either natural restriction sites or restriction sites created by PCR using Vent DNA polymerase (New England Biolabs) and custom oligonucleotides (Integrated DNA Technologies). GAL4-TATA-Luc plasmid (pG5luc) and expression plasmid for VP16 and Gal4 were purchased from Promega (CheckMate Mammalian Two-hybrid System). ARE-TATA-Luc, (C/EBP)3_TATA-luc and (C/EBP)3-ARE-TATA-luc were made by cloning appropriate annealed oligos with the addition of KpnI(5′) and NheI(3′) terminal restriction sites into the large segment of GAL4-TATA-Luc digested by KpnI and NheI. A high-affinity ARE, 5′-AGTACGTGATGTTCT-3′ (Schoenmakers et al., 2000) was inserted upstream of the TATA box. (C/EBP)3 is a three-tandem repeated C/EBP consensus element, (TGCAGA TTGCGCCAATCTGCA)3; the sequence underlined is the central binding motif. delC/EBP-TATA-Luc was made by deleting the GAL4 element from GAL4-TATA-Luc with KpnI and NheI digestion and then re-ligating the reporter after blunting of both restriction ends. The Renilla luciferase transfection control was the pRL-null plasmid from Promega. Gal4 fusions of various deleted versions of C/EBPα activation domain, that is, C/EBPα(107–217), C/EBPα(121–217), C/ EBPα(139–217) and C/EBPα(156–217), were all constructed by PCR using Gal4-C/EBPα(6–217) plasmid as the template and the appropriate primers and subcloned at EcoRI (upstream) and XbaI (downstream) sites in a vector for expressing Gal4 fusions (pSG424) (Sadowski and Ptashne, 1989). To test the functionality of chromatin sites mapped by ChIP-chip analyses, genomic DNA fragments were amplified using PCR, digested with the appropriate restriction enzymes and cloned upstream of the minimal promoter in the pG5luc plasmid in place of the GAL4 element. The PPARγ promoter plasmids were constructed by amplifying genomic DNA fragments by PCR using Pfu Turbo Polymerase followed by insertion upstream of the luciferase reporter in the pGL3-basic vector (Promega). The recombinant plasmids were amplified in Escherichia coli strain XL1 Blue and purified using the Qiagen plasmid kit (Qiagen, Chatsworth, CA, USA). The entire cloned DNA sequence in each construct was verified using automated DNA sequence analysis. The AR-specific short hairpin RNA (shRNA) and non-targeting shRNA control in the lentiviral expression vector, pLKO.1 puro, were purchased from Sigma-Aldrich (St Louis, MO, USA). The shRNA sequence for AR is: 5′-CCGGCACCAATGTCAACTCCAGGATCTCGAGCTC CTGGAGTTGACATTGGTGTTTTT-3′ (TRCN0000003718, MISSION TRC shRNA Target Set, Sigma). The control non-targeting shRNA sequence is: 5′-CCGGCAACAAGATGAA GAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG TTTTT-3′ (MISSION Non-Target shRNA Control Vector, Sigma). The co-regulator expression plasmids were provided by Dr Brian Rowan at Tulane University. The C/EBP expression plasmids were provided by Dr Steven L McKnight at University of Texas Southwestern Medical Center. The Gal4-C/EBP plasmids were from Dr William J Roesler at University of Saskatchewan, Canada.
Cell culture and transfection
HeLa (American Type Culture Collection) cells were cultured in Dulbecco’s minimum essential medium supplemented with FBS (10%), penicillin (100 unit/ml), streptomycin (100 µg/ml) and l-glutamine (2mm). LNCaP cells (American Type Culture Collection) were maintained in RPMI 1640 medium supplemented with FBS (10%), sodium pyruvate (1mm), penicillin (100 unit/ml), streptomycin (100 µg/ml) and L-glutamine (2 mM). To obtain hormone depletion, in all the experiment, HeLa cells were grown in phenol red-free media supplemented with charcoal-stripped FBS (5% v/v), L-glutamine (2mM), insulin (2µg/ml) and transferrin (40 µg/ml) for 72h. HeLa cells were transfected with DNA constructs in six-well plates (Corning, New York, NY, USA) using FuGENE 6 (Roche Diagnostics), according to the manufacturer’s protocol. Normally, 500 ng reporter and 25–100 ng of each expression plasmid were used where not specially indicated. LNCaP cells were nucleofected with Amaxa nucleofector Kit R (Amaxa), according to the manufacturer’s protocol. A total of 2µg of expression plasmid was used in the nucleofection. Immediately after the nucleofection, the cells were transferred to phenol red-free RPMI 1640 supplemented with CS-FBS (5% v/v), sodium pyruvate (1mM), L-glutamine (2 mM), insulin (2µg/ml) and transferrin (40 µg/ml). Transfection efficiency and promoter specificity were controlled using the pRL-null plasmid expressing Renilla luciferase and measurement of Renilla luciferase activity in the cell lysates. In the gene expression profiling experiments, transfection efficiency was determined using flow cytometry to determine the proportion of cells that expressed co-transfected green fluorescent protein expression plasmid.
Luciferase assay
The procedure for the preparation of cell lysates of transfected cells and the use of the Firefly luciferase assay system were as described (Shatnawi et al., 2007). The activity of co-transfected Renilla luciferase (transfectyion control) was also measured as described (Shatnawi et al., 2007). All luciferase assays were performed at least in triplicate.
Western blots
Cell lysates were prepared and analysed using western blot as described (Shatnawi et al., 2007).
RNA isolation, reverse transcription-PCR and real-time PCR
Total RNA from HeLa cells or LNCaP cells was prepared using RNeasy Mini kit (Qiagen). Reverse transcription-PCR followed by real-time PCR was used to measure mRNAs for luciferase or C/EBPα as well as glyceraldehyde-3-phosphate dehydrogenase. For the reverse transcription reaction, 200 ng of total RNA was reverse transcribed with random primers by using the high-capacity complementary DNA Archive kit (Applied Biosystems). The resulting complementary DNA was measured by quantitative real-time PCR using the real-time PCR master mix (Applied Biosystems) in the 7500 StepOne Plus Real Time PCR System (Applied Biosystems). The primers and TaqMan probe for Luciferase and glyceralde-hyde-3-phosphate dehydrogenase were obtained from Integrated DNA Technologies, Inc. Primers and probes for C/ EBPα were obtained from Applied Biosystems. All samples were assayed in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase values in the same samples.
Gene silencing with small interfering RNA (siRNA)
HeLa cells were plated at 40% confluency six-well plates and transfected with C/EBPα siRNA (5 nmol per well) or negative control siRNA together with 50 ng AR expression plasmid and 500 ng luciferase reporter using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s protocol. Cells were treated with testosterone (10 nM) or vehicle during the transfection (48 h) and then harvested for luciferase assay, RNA isolation or western blot analysis. Knocking down of C/ EBPα was confirmed at the mRNA level by real-time reverse transcription–PCR. The siRNA sequence for C/EBPα is: Sense, 5′-CCGCUCCAAUGCCU ACUGAtt-3′; Antisense, 5′-UCAGUAGGCAUUGGAGCGGtg-3′ (Ambion). The control siRNA is the Silencer Negative Control no. 1 siRNA from Ambion.
Gene silencing by infection with short hairpin RNA (shRNA) lentivirus
The AR shRNA vector or the non-targeting shRNA control vector was packaged into lentivirus. The virus particles were generated by transfecting 293FT cells using Lipofectamine 2000 reagent. Lentivirus was harvested from the supernatant at 48 h and 72 h after transfection. LNCaP cells were plated in 12-well plates the day before infection (1.5 × 105 cells per well); 0.5 ml virus supernatant combined with polybrene (800 µg/ml) was used for the infection of each well together with 0.5 ml media (RPMI 1640 media containing 10% heat-inactivated FBS, 2mML-glutamine and 1mM sodium pyruvate); after 4–5 h, the infection procedure was repeated. At approximately 4–5 h after the second infection, the cells were placed in fresh media.
Immunofluorescence and confocal microscopy
HeLa cells (2 × 105) were plated in chamber slides and transfected at 50% confluence with different doses of AR expression plasmid (10 ng, 50 ng, 100 ng and 200 ng) or vector and treated with either testosterone (10nM) or vehicle for the duration of the transfection (48 h). The cells were washed twice with phosphate-buffered saline PBS (2mM potassium dihydrogen phosphate, 10mM disodium phosphate, 2.7mM potassium chloride and 137 mM sodium chloride) and were fixed with freshly prepared 3.7% paraformaldehyde in PBS for 10min at room temperature. The cells were then permeabilized in PBS containing 0.1% bovine serum albumin and 0.3% Triton X-100 for 5min at room temperature. The cells were then washed and blocked with PBS containing 5% serum and 0.2% Triton X-100 at room temperature for 1 h. The cells were incubated overnight at 4°C with the antibody to AR in a dilution of 1:50. After washing thrice with PBS containing 5% serum and 0.2% Triton X-100, the cells were incubated with bovine anti-rabbit immunoglobulin G conjugated to fluorescein isothiocyanate for 1 h in the dark at room temperature in a dilution of 1:100. After the final wash, the cells were incubated for 2min in 4,6-diamidino-2-phenylindole, washed thrice and mounted using Vectashield (Burlingame, CA, USA) mounting medium. Images were acquired using the Leica TCS SP5 Broad band confocal microscope system (Leica, Wetzlar, Germany). The acquisition settings were kept constant between specimens and a negative control sample incubated with normal immunoglobulin G primary antibody was used to adjust for background. Sub-cellular localization of AR was examined by acquiring 10 optical sections using × 40 oil immersion with additional × 3.5 optical zoom. The images were compiled as projections using the Leica LAS software package. The total amount of fluorescence in the cell and the amount of fluorescence localized in the nucleus were quantified using the LASAF software (Wetzlar, Germany).
mRNA profiling
The Affymetrix DNA microarray analysis was performed as a full service global gene expression study at the transcriptional profiling core facility of the Cancer Institute of New Jersey. Total RNA samples were used to generate labeled cRNAs, which were hybridized to human U133 Plus2.0 Affymetrix microarrays. The expression data were analysed initially using Affymetrix GeneChip Operating Software to create CEL files. The CEL files were imported into the Bioconductor program affylmGUI (Wettenhall et al., 2006). The probe set level intensities were quantified and normalized using robust multi-array averaging and quantile normalization. Differential expression between treatments was determined using the limma linear modeling method, and the significance of differences were ranked by the moderated t-statistic.
Chromatin immunoprecipitation-DNA microarray chip (ChIP-chip) analyses
LNCaP cells were nucleofected with either 2 µg of C/EBPα or the corresponding empty vector for 48 h, and then the cells were treated for 2h with either R1881 (1 nM) or vehicle and subjected to ChIP as described (Shatnawi et al., 2007) using anti-AR antibody. The recruitment of AR to the major ARE enhancer region of the PSA promoter (−4366 to − 3874 nt) was measured using real-time PCR. An irrelevant target sequence within the downstream coding region was used as a negative control. The primers and TaqMan probe used to target the PSA ARE enhancer region were: 5′-GCCTGGATCTGAG AGAGATATCATC-3′ (forward primer), 5′-ACACCTTTT TTTTTCTGGATTGTTG-3′ (reverse primer) and 56FAM-TGCAAGGCCTGCTTTACAAACTTCC-36TAM (probe). The primers and TaqMan probe used to target the irrelevant coding sequence of PSA were: 5′-CACACCCGCTCTACG ATATGA (forward primer), 5′-GAGCTCGGCAGGCTCT GA-3′ (reverse primer) and 56-FAM-CTCCAGC CCG ACCTCATGCTGCT-36TAM (probe). The immunoprecipi-tated DNA samples (10 ng) that were validated for PSA as well as corresponding input DNA were amplified using the Sigma GenomePlex WGA2 kit (Sigma-Aldrich) to generate at least 4µg of DNA for chip analysis using the Nimblegen 385K H18 tiling array platform. The labeling and hybridization of DNA samples were performed by NimbleGen Systems, Inc (Madison, WI, USA). As described in the Nimblegen website, the data were analysed using the Nimblescan software to identify peaks by searching for four or more probes whose signals were above the specified cutoff values, ranging from 90 to 15%, using a 500-bp sliding window. The cutoff values are a percentage of a hypothetical maximum, which is the mean + 6 s.d. The ratio data were then randomized 20 times to assign a false discovery rate score. A false discovery rate of ≤0.05 was considered to be highly indicative of binding sites for AR.
Statistical analyses
All experimental values in Figure 1, 2, 3, 4 and 6 are presented as mean±s.d. The statistical significance of differences (P-value) between values being compared was determined using analysis of variance. In all cases, the differences noted in the text are reflected by a P-value of <0.001.
Supplementary Material
Acknowledgements
Supported by NIH R01 grants CA 103964 and CA 80183 to MR.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Aarnisalo P, Palvimo JJ, Janne OA. CREB-binding protein in androgen receptor-mediated signaling. Proc Natl Acad Sci. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S, Manin M, Beaudoin C, Leotoing L, Communal Y, Veyssiere G, et al. Androgen receptor mediates non-genomic activation of phosphatidylinositol 3-OH kinase in androgen-sensitive epithelial cells. J Biol Chem. 2004;279:14579–14586. doi: 10.1074/jbc.M306143200. [DOI] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Berrevoets CA, Doesburg P, Steketee K, Trapman J, Brinkmann AO. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- Burel SA, Harakawa N, Zhou L, Pabst T, Tenen DG, Zhang DE. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol Cell Biol. 2001;21:5577–5590. doi: 10.1128/MCB.21.16.5577-5590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Gong EY, Hwang M, Park E, Lee HJ, Hong CY, et al. The CCAAT enhancer-binding protein-alpha negatively regulates the transactivation of androgen receptor in prostate cancer cells. Mol Endocrinol. 2006;20:984–995. doi: 10.1210/me.2005-0240. [DOI] [PubMed] [Google Scholar]
- Edwards J, Bartlett JM. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 1: modifications to the androgen receptor. BJU Int. 2005;95:1320–1326. doi: 10.1111/j.1464-410X.2005.05526.x. [DOI] [PubMed] [Google Scholar]
- Ettwiller L, Paten B, Ramialison M, Birney E, Wittbrodt J. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat Methods. 2007;4:563–565. doi: 10.1038/nmeth1061. [DOI] [PubMed] [Google Scholar]
- Frønsdal K, Engedal N, Slagsvold T, Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- Gery S, Tanosaki S, Bose S, Bose N, Vadgama J, Koeffler HP. Down-regulation and growth inhibitory role of C/EBPalpha in breast cancer. Clin Cancer Res. 2005;11:3184–3190. doi: 10.1158/1078-0432.CCR-04-2625. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Halmos B, Huettner CS, Kocher O, Ferenczi K, Karp DD, Tenen DG. Down-regulation and antiproliferative role of C/EBPalpha in lung cancer. Cancer Res. 2002;62:528–534. [PubMed] [Google Scholar]
- Han S, Roman J. Peroxisome proliferator-activated receptor gamma: a novel target for cancer theraputics? Anticancer Drugs. 2007;18:237–244. doi: 10.1097/CAD.0b013e328011e67d. [DOI] [PubMed] [Google Scholar]
- Harris TE, Albrecht JH, Nakanishi M, Darlington GJ. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J Biol Chem. 2001;276:29200–29209. doi: 10.1074/jbc.M011587200. [DOI] [PubMed] [Google Scholar]
- Hendricks-Taylor LR, Darlington GJ. Inhibition of cell proliferation by C/EBP alpha occurs in many cell types, does not require the presence of p53 or Rb, and is not affected by large T-antigen. Nucleic Acids Res. 1995;23:4726–4733. doi: 10.1093/nar/23.22.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Jenster G, Trapman J, Brinkmann AO. Nuclear import of the human androgen receptor. Biochem J. 1993;293(Pt 3):761–768. doi: 10.1042/bj2930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenster G, van der Korput HA, Trapman J, Brinkmann AO. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- Jia L, Coetzee GA. Androgen receptor-dependent PSA expression in androgen-independent prostate cancer cells does not involve androgen receptor occupancy of the PSA locus. Cancer Res. 2005;65:8003–8008. doi: 10.1158/0008-5472.CAN-04-3679. [DOI] [PubMed] [Google Scholar]
- Kousteni S, Bellido T, Plotkin LI, O’Brien CA, Bodenner DL, Han L, et al. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- Loomis KD, Zhu S, Yoon K, Johnson PF, Smart RC. Genetic ablation of CCAAT/enhancer binding protein alpha in epidermis reveals its role in suppression of epithelial tumorigenesis. Cancer Res. 2007;67:6768–6776. doi: 10.1158/0008-5472.CAN-07-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolagas SC, Kousteni S. Perspective: nonreproductive sites of action of reproductive hormones. Endocrinology. 2001;142:2200–2204. doi: 10.1210/endo.142.6.8221. [DOI] [PubMed] [Google Scholar]
- McEwan IJ. Molecular mechanisms of androgen receptormediated gene regulation: structure-function analysis of the AF-1 domain. Endocr Relat Cancer. 2004;11:281–293. doi: 10.1677/erc.0.0110281. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Messing EM, Chang C. Androgen deprivation therapy for prostate cancer: current status and future prospects. Prostate. 2004;61:332–353. doi: 10.1002/pros.20115. [DOI] [PubMed] [Google Scholar]
- Muller C, Calkhoven CF, Sha X, Leutz A. The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a SWI/SNF complex for proliferation arrest. J Biol Chem. 2004;279:7353–7358. doi: 10.1074/jbc.M312709200. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Ziff EB. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001a;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001b;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Paltoo D, Woodson K, Taylor P, Albanes D, Virtamo J, Tangerea J. Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma (PPAR-gamma) gene and risk of prostate cancer among men in a large cancer prevention study. Cancer Lett. 2003;191:67–74. doi: 10.1016/s0304-3835(02)00617-1. [DOI] [PubMed] [Google Scholar]
- Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, et al. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo . Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AK, Lavrovsky Y, Song CS, Chen S, Jung MH, Velu NK, et al. Regulation of androgen action. Vitam Horm. 1999;55:309–352. doi: 10.1016/s0083-6729(08)60938-3. [DOI] [PubMed] [Google Scholar]
- Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooij-man MC, Trapman J, Brinkmann AO, et al. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am J Pathol. 1994;144:735–746. [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog Nucleic Acid Res Mol Biol. 2004;77:1–36. doi: 10.1016/S0079-6603(04)77001-4. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem. 2000;275:12290–12297. doi: 10.1074/jbc.275.16.12290. [DOI] [PubMed] [Google Scholar]
- Schuster MB, Porse BT. C/EBPalpha: a tumour suppressor in multiple tissues? Biochim Biophys Acta. 2006;1766:88–103. doi: 10.1016/j.bbcan.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci USA. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand RL, Gelmann EP. Molecular biology of prostate-cancer pathogenesis. Curr Opin Urol. 2006;16:123–131. doi: 10.1097/01.mou.0000193384.39351.64. [DOI] [PubMed] [Google Scholar]
- Shatnawi A, Tran T, Ratnam M. R5020 and RU486 act as progesterone receptor agonists to enhance Sp1/Sp4-dependent gene transcription by an indirect mechanism. Mol Endocrinol. 2007;21:635–650. doi: 10.1210/me.2006-0274. [DOI] [PubMed] [Google Scholar]
- Sun M, Yang L, Feldman RI, Sun XM, Bhalla KN, Jove R, et al. Activation of phosphatidylinositol 3-kinase/Akt pathway by androgen through interaction of p85alpha, androgen receptor, and Src. J Biol Chem. 2003;278:42992–43000. doi: 10.1074/jbc.M306295200. [DOI] [PubMed] [Google Scholar]
- Sundfeldt K, Ivarsson K, Carlsson M, Enerback S, Janson PO, Brannstrom M, et al. The expression of CCAAT/enhancer binding protein (C/EBP) in the human ovary in vivo: specific increase in C/EBPbeta during epithelial tumour progression. Br J Cancer. 1999;79:1240–1248. doi: 10.1038/sj.bjc.6690199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AA, Marker PC. Branching morphogenesis in the prostate gland and seminal vesicles. Differentiation. 2006;74:382–392. doi: 10.1111/j.1432-0436.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Harris TE, Wilde M, Bilyeu TA, Burgess-Beusse BL, Finegold MJ, et al. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Darlington GJ. C/EBPalpha regulates formation of S-phase-specific E2F–p107 complexes in livers of newborn mice. Mol Cell Biol. 1999a;19:2936–2945. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Iakova P, Albrecht JH, Darlington GJ. E2F/p107 and E2F/p130 complexes are regulated by C/EBPalpha in 3T3-L1 adipocytes. Nucleic Acids Res. 1999b;27:3621–3630. doi: 10.1093/nar/27.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- Vlahopoulos S, Zimmer WE, Jenster G, Belaguli NS, Balk SP, Brinkmann AO, et al. Recruitment of the androgen receptor via serum response factor facilitates expression of a myogenic gene. J Biol Chem. 2005;280:7786–7792. doi: 10.1074/jbc.M413992200. [DOI] [PubMed] [Google Scholar]
- Wang GL, Iakova P, Wilde M, Awad S, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP alpha growth inhibitory activity. Genes Dev. 2004;18:912–925. doi: 10.1101/gad.1183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Timchenko NA. Dephosphorylated C/EBPalpha accelerates cell proliferation through sequestering retinoblastoma protein. Mol Cell Biol. 2005;25:1325–1338. doi: 10.1128/MCB.25.4.1325-1338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Goode T, Iakova P, Albrecht JH, Timchenko NA. C/EBPalpha triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 2002;21:930–941. doi: 10.1093/emboj/21.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PJ, Condreay JP, Huber BE, Jacobs SJ, Adams DJ. Impaired proliferation and tumorigenicity induced by CCAAT/ enhancer-binding protein. Cancer Res. 1996;56:1063–1067. [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- Xu Y, Chen SY, Ross KN, Balk SP. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66:7783–7792. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Zegarra-Moro OL, Schmidt LJ, Huang H, Tindall DJ. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 2002;62:1008–1013. [PubMed] [Google Scholar]
- Zhang J, Wilkinson JE, Gonit M, Keck R, Selman S, Ratnam M. Expression and sub-cellular localization of the CCAAT/ enhancer binding protein alpha in relation to postnatal development and malignancy of the prostate. Prostate. 2008;68:1206–1214. doi: 10.1002/pros.20779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.