Abstract
Purpose
The G-protein coupled receptor (GPCR) CLR and its ligand peptide adrenomedullin (AM, encoded by ADM gene) are implicated in tumor angiogenesis in mouse models but poorly defined in human cancers. We therefore investigated the diagnostic/prognostic utility for CLR in human tumor types that may rely on AM signaling and in clear cell renal cell carcinoma (RCC), a highly vascular tumor, in particular.
Experimental design
In silico gene expression messenger RNA profiling microarray study (n=168 tumors) and cancer profiling cDNA array hybridization (n=241 pairs of patient-matched tumor/normal tissue samples) were performed to analyze ADM mRNA expression in 13 tumor types. Immunohistochemistry on tissue microarrays containing patient-matched renal tumor/normal tissues (n=87 pairs) was performed to study CLR expression and its association with clinico-pathological parameters and disease outcome.
Results
ADM expression was significantly up-regulated only in RCC and endometrial adenocarcinoma compared with normal tissue counterparts (P<0.01). CLR was localized in tumor cells and vessels in RCC and up-regulated compared to patient-matched normal control kidney (P<0.001). Higher CLR expression was found in advanced stages (P<0.05), correlated with high tumor grade (P<0.01) and conferred shorter overall survival (P<0.01).
Conclusions
In human tissues ADM expression is up-regulated in cancer-type specific manner, implicating potential role for AM signaling in particular in RCC, where CLR localization suggests autocrine/paracrine mode for AM action within the tumor microenvironment. Our findings reveal previously unrecognised CLR up-regulation in an autocrine loop with AM in RCC with potential application for this GPCR as a target for future functional studies and drug development.
Keywords: angiogenesis, CLR, G-protein coupled receptor, renal cancer
Introduction
Adrenomedullin (AM; encoded by an ADM gene) is a multifunctional peptide involved in cellular proliferation, survival and angiogenesis (1, 2). Whilst the role of AM in tumor biology has been established from studies using in vitro and mouse models and although ADM gene expression is known to be up-regulated by hypoxia (a typical feature of solid tumors) (3-5), there is a general lack of information about the expression of AM receptors in human cancers and its correlation with the disease outcome in particular (reviewed in (4)).
In vitro, AM mediates its activities through binding to G-protein-coupled receptor (GPCR) calcitonin receptor like-receptor (CLR, encoded by the CALCRL gene) (6, 7). The GPCRs are the largest family of cell-surface molecules involved in signal transduction and they control key physiological functions, including regulation of blood pressure, immune system activity and inflammation. These receptors are involved in some of the most prevalent human diseases, as reflected by the fact that GPCRs directly or indirectly represent the targets of 50-60% of all current therapeutic agents (8-10). For example, the analgesic drugs codeine and morphine target opioid receptors in the nervous system and the GPCR antagonist zibotentan (ZD4054) targets the endothelin A receptor in solid tumors (9, 11). Recently some GPCRs, such as chemokine and prostaglandin receptors, have emerged as crucial players in tumor growth and metastasis, suggesting their importance in initiation and progression of cancer in humans (4, 11, 12).
In the present study we tested the hypothesis that GPCR CLR may be important in tumor biology in man and could play a role in some human cancers. Our aims were to determine those tumor types that may rely on AM, examine the pattern and level of expression of AM receptor CLR in a prototypical example and perform a clinico-pathological analysis to assess its diagnostic and/or prognostic utility. We therefore performed a large-scale in silico gene expression (messenger RNA profiling) microarray study coupled to cancer profiling cDNA array hybridization and identified tumors that had up-regulated ADM expression compared with patient-matched normal tissue counterparts. Our data showed that ADM expression was significantly up-regulated only in clear cell renal cell carcinoma (RCC) and endometrial adenocarcinoma (P<0.01) but not other tumor types, suggesting that in human tissues/organs it is up-regulated in cancer-type specific manner. We then focused on RCC as potentially an AM signaling-driven tumor and, by using immunohistochemistry and a tissue microarray approach, analyzed CLR expression in a large cohort of patient-matched paired carcinoma and normal kidney tissue samples. Our data showed that CLR was localized in tumor cells and tumor vessels and that its expression was up-regulated in RCC, when compared with patient-matched controls. Moreover, CLR expression was higher in advanced stages and correlated with tumor grade and overall patient survival. Our findings suggest that GPCR CLR has clinical significance in RCC as a promising immunohistochemical biomarker of disease outcome and as a potential molecular target for therapeutic agents.
Materials and Methods
Please see details of the following methods in the Supplementary Data: Antibody, Cell culture, SDS–PAGE and Immunoblotting, Cancer Profiling Microarray Hybridization and Northern blotting, RNA Isolation, cDNA Synthesis and Quantitative Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR).
GeneChip® Human Genome Array analysis
Public Affymetrix GeneChip® HG-U133plus2 arrays data for 11 carcinoma types (n=168 samples) were obtained from the Expression Project for Oncology expO (http://www.intgen.org/expo.cfm) via the NCBI Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/projects/geo; accession GSE2109). Raw data were pre-processed and normalized using Bioconductor software for R (13), and the robust-multiarray algorithm (rma) as previously described (14-16).
The Cancer Profiling Microarray Hybridization and Northern blotting
The Cancer Profiling Microarray 7841 (cDNA microarray), consisting of normalized paired cDNA samples (n=241) generated from the total RNA from 13 organs (breast, uterus, colon, stomach, ovary, lung, kidney, rectum, thyroid, prostate, cervix, small intestine and pancreas), was obtained from Clontech (http://www.clontech.com). Each cDNA pair consisted of carcinoma and corresponding organ-matched normal tissue samples obtained from the same patient, represented the entire mRNA message expressed in given tissues and was used to provide comprehensive ADM gene-disease correlation data.
ADM cDNA was cloned into TOPO vector (Promega) and ubiquitin cDNA probe was from BD Biosciences. Inserts were excised with restriction enzymes and labelled with 32P-dCTP using MegaPrime labeling Kit (GE Healthcare). The specificity of the probes was confirmed by Northern blotting, which was performed as previously described (17). Cancer Profiling Microarray was consequentially hybridised with each individual probe according to the manufacturer’s instructions. The microarray was exposed to Hyperfilm (GE Healthcare) and subsequently to Phosphoscreen. The hybridization signals were further analyzed using ImageQuant software. Ratio ADM/Ubiquitin was calculated to determine the relative mRNA expression levels and scatter plot graphs were generated using GraphPrism software.
Patients
A total of 131 patients with histologically confirmed renal carcinoma from two centers (John Radcliffe Hospital, Oxford, U.K. (cohort 1; n=113 patients) and Canterbury District Health Board, Christchurch, New Zealand (cohort 2; n=18 patients) were included in this study.
Baseline histological characteristics on all analyzed renal tissue samples from cohort 1 (used for immunohistochemical analysis as described below) and the information on age and tumor characteristics (e.g. stage, grade, size etc.) for RCC patients are provided in the Tables 1 and 2 respectively. The cause of death was obtained from the Oxford Cancer Registry from the Oxford Cancer Intelligence Unit (http://www.ociu.nhs.uk/index.html) (18). The median follow-up time after nephrectomy was 43 months (ranging from 1 to 296 months) and a mean overall survival of 3.3 years. None of the patients received novel experimental therapies, such as tumor vaccines or molecular targeted medicine, nor had received any conventional treatment for at least six months before the surgery. Overall survival was defined as the time from nephrectomy and cancer diagnosis until death, the last time that the patient was known to be alive, or until the end of follow-up. All H&E-stained slides were reviewed by experienced genitourinary pathologists and histologically classified according to the WHO classification of tumors (19). Tumors were classified according to the Fuhrman grade (20) and TNM staging system (21) and data deposited in the Cancer Registry. T stage data was available for all patients, whilst N and M staging data was not and therefore not included in analysis. The samples from the cohort 2 patients were collected for routine histological evaluation and also for reverse transcriptase PCR (RT-PCR) and quantitative real-time polymerase chain reaction (qRT-PCR) analysis as described below.
Table 1.
Histological characteristics of analyzed renal tumor tissue samples
| Histology | Number of tumor (paired) samples* | |
|---|---|---|
| Clear cell RCC | (RCC) | 87 (69) |
| Papillary RCC | (PRCC) | 3(3) |
| Transitional cell carcinoma | (TCC) | 6(0) |
| Oncocytoma | 3(3) | |
| Sarcomatoid | 8(8) | |
| Other (including chromophobe) | 4(4) | |
| Total | 113(87) | |
The number of patient-matched normal kidney tissue samples is indicated in brackets.
Table 2.
Clinicopathological characteristics of RCC patients
| Age, years (median±S.D.) | 62.6±10.7 |
|---|---|
| Gender (male/female), n | 31/51 |
| T stage (TNM system), n | |
| T1 | 28 |
| 12 | 19 |
| T3 | 40 |
| T4 | 0 |
| Grade (Fuhrman), n | |
| G1 | 14 |
| G2 | 49 |
| G3 | 15 |
| G4 | 8 |
| Tumor size, cm (mean±S.D.) | 8.0 ±3.39 |
| Follow up, median (range), days |
43.4 (0.1-296.2) |
| Mortality information renal cancer-related death, n (%) |
43 (50%) |
All patients were confirmed to have sporadic disease based on their medical records taking (obtained from Oxford Cancer Registry), according to which no family history of RCC was elucidated. The Central Oxfordshire Research Ethics Committee (C00.147; C02.216) and Canterbury Ethics Committee (V2-4 02.06.98-01.05.2002) approved the use of all human tissues used in this study.
Tissue Microarray and Immunohistochemistry
Tissue microarray (TMA) of 1mm cores (from formalin-fixed, paraffin-embedded specimens of renal carcinoma and normal tissues) with two-fold redundancy from 113 patients (including 87 pairs of patient-matched samples) from the cohort 1 was constructed (18) and 4 μm sections cut. The presence of all analyzed patient-matched normal and tumor tissue samples on the same TMA section enabled the performance of immunostaining under standardized conditions and its quantitative comparison. Immunohistochemical analysis of CLR expression was done using previously characterized rabbit polyclonal anti-human CLR antibody LN1436 (22) and pre-immune serum from the same rabbit, in which the antibody was raised, as a control. Additional control was performed using primary antibody pre-incubated with 10μg/ml peptide, against which it was raised, for 90 minutes at room temperature prior to immunostaining. Scoring of the intensity and proportion of tumor or normal epithelial cells was performed by two independent pathologists and on two independently immunostained TMA using a semi-quantitative analysis as described elsewhere (23). Briefly, the intensity of the staining (“no staining” (0), “weak staining” (1), “moderate staining” (2), or “strong staining” (3) and the percentage of stained cells (0-10% (1), 10-50% (2), 51-80% (3) or 81-100% (4)) were determined. Intensity was multiplied by percentage to obtain “CLR intensity and percentage score” for each tissue sample on TMA. The scoring of the intensity (but not the percentage) of the CLR immunoreactivity in tumor vessels or normal vessels was performed following the same approach.
RNA Isolation, cDNA Synthesis, RT-PCR and qRT-PCR were performed as previously described (17, 24) using tissue samples collected into the liquid nitrogen and stored at −80°C. For qRT-PCR, the following primer/probe kits were used: Hs00181605_m1 (ADM) and Hs99999903_m1 (ACTB) (both from Applied Biosystems). Relative quantification of gene expression was performed using previously described method (25), based on the mean value of qRT-PCR reactions performed in triplicate. Human ACTB was used as reference gene to normalize for differences in the amount of total RNA in each sample. The comparator for the clinical samples was the median from normal kidney samples.
Statistical analysis
All statistical analyses were done using SPSS Statistics software (version 16.0), GraphPrism or Microsoft Excel computer programs. Messenger RNA expression levels in patient-matched normal and cancer tissues were analyzed using Wilcoxon signed-rank t test. Analysis between the level of CLR immunostaining in tumor cells on TMA and various clinicopathological parameters (patient’s age, tumor size, histology, T stage and Fuhrman grade) was performed using Spearman rank correlation or paired t-tests depending on the data set as previously performed in other studies (26, 27). For all tests, two-sided analysis was performed and actual P values are shown. P values of <0.05 were considered statistically significant.
For the survival analysis, all immunostained on the TMA RCC samples were divided into two categories according to CLR immunostaining in tumor cells - “CLR positive” (1-12) or “CLR negative” (0) based on intensity and percentage score; or divided at the median into two categories according to the staining in tumor/normal vessels - “CLR high” and “CLR low”. Kaplan-Meier curves and log-rank Mantel-Cox test was used to estimate the association of CLR expression levels in tumor cells or tumor vessels with an overall survival in order to obtain protein-disease outcome data based on available data for RCC patients (total n=87; Table 2). All survival analyses refer to overall survival times, where time to death from disease represents an event. Patients were censored in survival according to the date last seen by a doctor as indicated above. The median survival times and hazard ratios within each subgroup were estimated from Kaplan-Meier curves.
Results
ADM expression in human tissues is up-regulated in a cancer type-specific manner
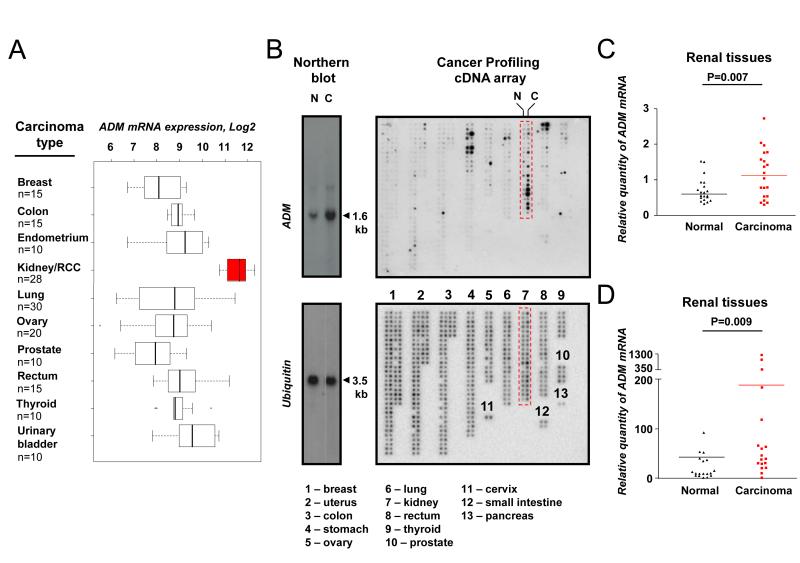
There was a significant heterogeneity of ADM expression across 11 human tumor types studied here (Figure 1A). The highest (mean log2 value 11.5) levels were in RCC and the lowest (mean log2 value 8) - in breast carcinoma, i.e. approximately ten-fold variation. Furthermore, ADM expression was significantly altered only in three tumor types out of 13 analyzed carcinomas (up-regulated in kidney and uterine tumors, whilst down-regulated in breast carcinoma; Figure 1B; Supplementary Figure S1). The highest up-regulation of ADM mRNA expression was found in RCC when compared with paired normal kidney (Figure 1C and Supplementary Figure S1) and this was confirmed by qRT-PCR using an independent cohort of 18 pairs of patient-matched renal carcinoma and normal control tissue samples (Figure 1D).
Figure 1. ADM mRNA expression in patient-matched renal carcinoma and normal tissues.
GeneChip® Human Genome Array data was obtained from the Expression Project for Oncology (expO) via the NCBI GEO repository (accession GSE2109, n=168 covering 11 most common types of human carcinomas). A, ADM mRNA expression levels were compared and presented on the box (interquartiles) and whisker (95th percentiles) plots, showing Log2 expression of ADM probe 202912_at. B, ADM expression was studied using the Cancer Profiling cDNA Microarray (Clonetics), which consists of normalized paired normal and cancer cDNA samples (n=241 pairs) generated from the total RNA from 13 organs (as labelled and specified below on the bottom cDNA Microarray image). The microarray was subsequently hybridized with ADM and ubiquitin probes labeled with 32P-dCTP, the specificity of which was confirmed on Northern blot (Northern blot), using RNA obtained from paired normal (lane N) and carcinoma (lane C) patient-matched tissue samples. The highest ADM mRNA expression was found in renal tissues (boxed on top cDNA microarray image), where it was up-regulated in carcinoma (right column, C), when compared to normal tissue (left column, N) samples. C, The microarray was exposed to Phosphoscreen and hybridization signals were analyzed using ImageQuant software. Ratio ADM/ubiquitin was calculated to determine the relative ADM mRNA expression levels and to generate jittered dot-plot plot graph for patient-matched normal and carcinoma renal cDNA samples (n=20 pairs). Statistical analysis was performed using Wilcoxon signed-rank test. Note that the quantitative data and graphs for all other carcinoma types from the cDNA microarray are presented in the Supplementary Figure S1. D, ADM mRNA up-regulation in renal cancers was confirmed by qRT-PCR using an independent cohort of paired patient-matched normal and carcinoma RNA samples (n=18; RCC n=14, papillary n=2, oncocytoma n=2). Ratio ADM/ACTB was calculated to determine relative quantity of ADM mRNA and shown on jittered dot-plot graph for normal and carcinoma samples. Wilcoxon signed-rank test was used. C-D, Actual P values are shown.
CLR is expressed in tumor cells and tumor vessels in RCC and terminally glycosylated in tumor tissues and cell lines
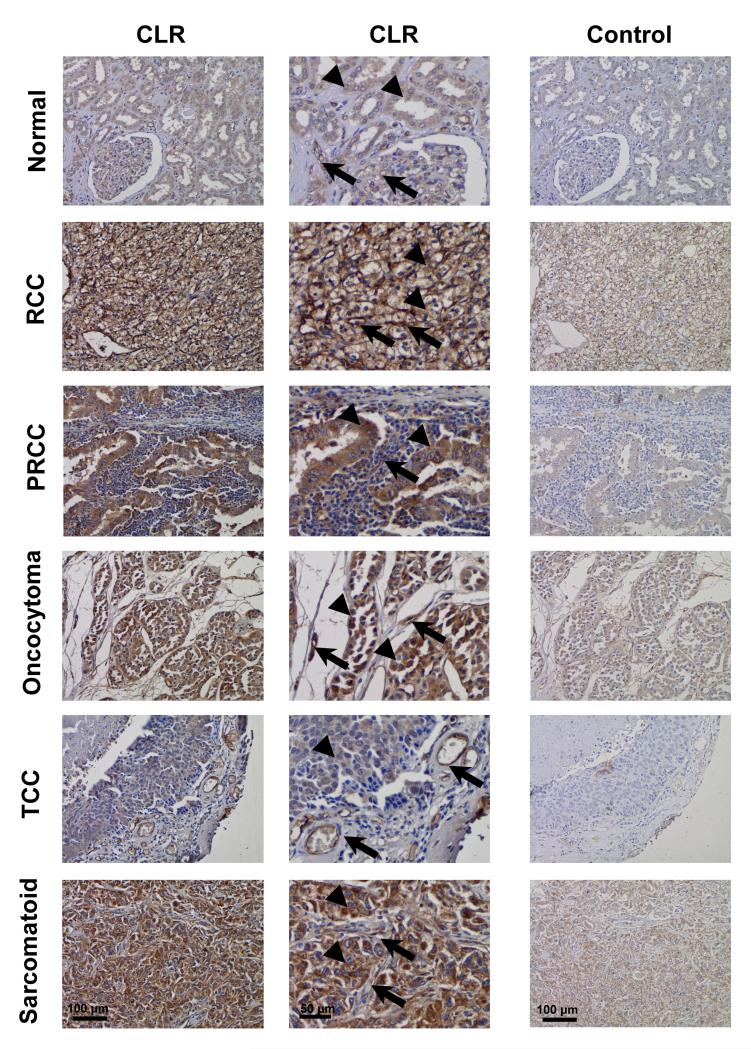
CLR was variably expressed in tumor cells and tumor vessels in RCC and in normal patient-matched tissue samples (n=87 pairs, including 69 RCC cases) and in RCC cell lines, as shown by immunohistochemistry and/or immunoblotting. CLR was localized in both tumor cells and tumor vessels (Figure 2; Supplementary Figure S2 A and B) and its expression was up-regulated in renal tumors in general and in individual types (Supplementary Figure S3 A and B), including RCC (Figure 3A), when compared with control patient-matched normal tissues.
Figure 2. CLR expression in tumor cells and tumor vessels in renal carcinoma.
The renal cancer profiling TMA included paired carcinoma and corresponding normal tissue samples from individual patients (n=87; including 69 RCC cases) as described in the Materials and Methods and Table 1. CLR localization (CLR; left column of images – at low magnification and central column of images - at higher magnification) was assessed by immunohistochemistry using anti-CLR antibody LN1436. Secondary goat anti-rabbit antibody conjugated to horseradish peroxidase was used and further detected with DAB (brown color). Cell nuclei were counter stained with haematoxylin (blue color). Immunostaining using primary antibody pre-adsorbed with peptide antigen, against which it was raised, was used as a control (Control; right image). Note that the receptor is localized predominantly in the vessels (arrows) in glomeruli and between tubules in normal tissue (upper row of images) and in carcinomas (RCC – clear cell and PRCC – papillary carcinomas and TCC – transitional cell carcinoma of the renal pelvis) as well as oncocytoma and sarcomatoid renal cancers; and also in normal epithelial cells and in renal neoplastic cells (both - arrowheads). Note that one example is given here for RCC, although CLR expression levels vary in tumor cells in this type of renal cancer; to demonstrate this, six RCC cases are presented in the Supplementary Figure S2.
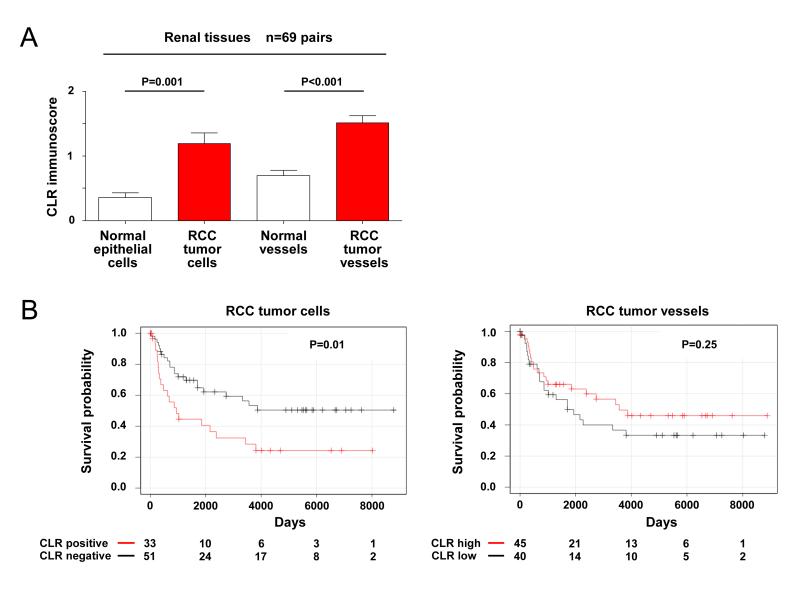
Figure 3. High CLR expression in tumor cells and tumor vessels in RCC compared to normal tissues and its association with patient survival.
The semi-quantitative analysis of the TMA data for CLR immunostaining (as shown on Figure 2) was used to provide comprehensive CLR protein-disease (RCC) outcome correlation data. A, CLR expression in tumor cells and tumor vessels in RCC was compared to that in normal epithelial cells of vessels in control histologically normal kidney tissue (n=69 patient-matched paired tumor and corresponding control/normal tissue samples) as shown on bar graphs (mean ± S.D.). Statistical analysis was performed using paired t test. B, Kaplan-Meier overall survival curve for renal cancer patients sub-grouped according to CLR expression in tumor cells or tumor vessels in RCC. All samples used for the analysis were divided/stratified into two categories/groups based on quantification of CLR immunostaining as described in Statistical Analysis section of the Materials and Methods: “CLR positive” and “CLR negative“ for tumor cells and “CLR high” and “CLR low“ for tumor vessels. The representative images, showing variations in CLR expression levels in tumor cells in individual RCC cases, are presented in the Supplementary Figure S2. Log-rank Mantel-Cox test was used for the statistical analysis. A-B, Actual P values and numbers of analysed pairs of patient-matched tissue samples or RCC cases are shown.
CLR was terminally- but not core-glycosylated when expressed in RCC tissues (Supplementary Figure S4A). Fully processed terminally glycosylated form of CLR is only produced in the presence of the RAMPs and only in this particular form this GPCR is expressed at the cell surface and can bind the ligand, i.e. to be “functional” (28). Therefore our findings suggest that CLR is likely to be in its cell surface-bound/functional form (6, 22, 28, 29), when expressed in RCC tumor cells and tumor vessels in situ (Figure 2; Supplementary Figure S2). In vitro, CLR was also expressed in some, but not all, renal tumor cell lines (Supplementary Figure S4B). We confirmed that the presence of terminally glycosylated CLR in renal tumor tissues or cultured renal tumor cell lines and primary microvascular endothelium was associated with the expression of RAMP1, 2 and 3 mRNAs (Supplementary Figure S5), which encode proteins that facilitate terminal glycosylation and transport of the CLR to the cell membrane (6).
CLR expression in tumor cells in RCC correlates with tumor grade and patient survival rates
CLR expression in tumor cells (but not in tumor vessels) was significantly higher (P<0.05) in advanced stages (2 and 3) (Supplementary Figure S6) and correlated with high tumor grade (P<0.01), but not with tumor size or patient age (Table 3). Furthermore, RCC patients with CLR expressed in tumor cells had significantly shorter overall survival when compared with those with no CLR (with median survival times of 543 and 784 days respectively, and hazard ratio 0.4182; P=0.01; Figure 3B).
Table 3.
Spearman-rank correlation analysis of CLR immunostaining in RCC tumor cells and tumor vessels with some important clinico-pathological or prognostic parameters
| Variables | Tumor cells |
Tumor vessels |
||||
|---|---|---|---|---|---|---|
| Spearman R | 95% CI | P | Spearman R | 95% CI | P | |
| Age | 0.026 | (−0.19)-0.24 | 0.815 | −0.053 | (−0.27)-0.17 | 0.625 |
| Grade (G1-4) | 0.31 | 0.098 - 0.50 | 0.004 | −0.16 | (−0.37) - 0.057 | 0.133 |
| Tumor size | 0.12 | (−0.10)-0.34 | 0.275 | −0.017 | (−0.24) - 0.21 | 0.881 |
Discussion
Renal cancers account for 2-4% of cancers worldwide (http://www.cancer.org). The five year survival of renal cancer patients is 40.5%, largely due to metastasis which accompany 75% of the cases (30, 31). In these patients systemic or adjuvant treatments, such as conventional cytotoxic chemotherapy or cytokine immunotherapy, give a sustained response rate of only 10% and they are associated with considerable toxic effects (30-32). Recently the unravelling of the hypoxic response in RCC through von Hippel-Lindau (VHL)/hypoxia-inducible factor (HIF)-mediated signaling has demonstrated how its downstream pathways and processes, such as angiogenesis, are integral to the pathogenesis/progression of renal cancer (5). Indeed tumor responsiveness to the newer anti-cancer and anti-angiogenic drugs such as sorafenib, sunitinib and bevacizumab, targeting vascular endothelial and/or platelet-derived growth factor (VEGF and PDGF respectively; both - products of the genes which are transcriptional targets of HIF) pathways, demonstrated the benefit of exploiting them for targeted therapy in human cancers, including RCC (33-36). Nevertheless, a significant number of RCC patients do not completely respond (or become resistant) to these anti-angiogenic or current combination therapies, which often require long-term administration for continued disease control and have several adverse effects. This suggests that other molecular pathways may be activated in this type of cancer and play a significant role in its progression and also supports the need for the continued exploration of these pathways and prognostic or predictive markers for this disease as well as for the development of novel agents and new treatment combinations (32).
AM signaling pathway(s) mediated by GPCR CLR could be one such potentially significant axis for therapeutic intervention in RCC due to the up-regulation of ADM expression by hypoxia and HIF (similar to VEGF and PDGF genes), and also because the role for AM in tumor growth and angiogenesis has been already established by studies using mouse xenografted tumor models and in vitro assays (reviewed in (3, 4)). A recent study suggested that signaling via CLR has potential role in bypassing the normal requirement for vascular endothelial growth factor (VEGF) signaling in endothelial cells during embryogenesis (37). However, while these observations pointed to a potential role for AM in tumor biology and angiogenesis, the clinical significance or functional contribution of AM signaling to tumorigenesis in human tissues is as yet poorly defined (reviewed in (4)). To date studies in man have been limited to measurements of ADM mRNA levels (38, 39) or AM peptide concentration in blood and tissues (40) and to the use of few tumor cell lines or a limited number of neoplastic tissues; all only occasionally done with the comparison between the patient-matched normal and cancer samples. The data about CLR expression, including distribution/localization and presentation at the cell surface in vivo, in human tumor tissues is also very limited and has not been systematically studied (22, 38, 41). In particular, there is a general lack of information about the association of AM and/or CLR expression with disease outcome and patient survival in various cancer types, including RCC. As a result, limited data is available not only for the evaluation of a possible role for AM and CLR during initiation and progression of cancer in humans but also for an overall assessment of their value and potential in clinical oncology, e.g. for diagnostics, prognosis and/or targeted therapy (4, 29).
In the present study, by using large-scale in silico gene expression data screening coupled to the analysis of patient-matched carcinoma and normal tissue samples, we found that ADM mRNA expression in human organs and tissues is up-regulated only in a small number of tumors and in RCC in particular; in keeping with two previous studies of ADM expression in renal cancer with combined numbers of 62 cases (38, 39). Furthermore, our TMA and immunoblotting data showed that AM receptor CLR is localized both in tumor cells and tumor vessels and expressed in its functional (i.e. terminally glycosylated/cell surface-bound) form in renal carcinoma tissues. These findings suggest a potent autocrine loop or paracrine mode for AM action and possibly biologically active in vivo roles for this CLR-mediated signaling in both angiogenesis and/or tumor cell biology within RCC microenvironment, especially upon up-regulation of ADM expression. We therefore hypothesized (and tested) that CLR might be important in RCC progression.
In support of this hypothesis, we found that CLR expression in tumor cells is significantly up-regulated compared to patient-matched normal epithelial cells and that its high levels strongly associate with advanced stages and correlate with tumor grade as well as with shorter survival of RCC patients. To our knowledge, this is the first report demonstrating that in human cancers the AM receptor CLR has a prognostic value as the immunohistochemical biomarker of patient survival. Another important site for AM action within the RCC microenvironment is tumor vessels, where CLR is also significantly up-regulated when compared to patient-matched normal kidney vessels. We have previously shown that AM interacts with endogenous CLR in cultured primary human microvascular endothelial cells, which express this GPCR in tissues, and induces their proliferation and migration in vitro (22), suggesting the role for AM during angiogenesis and in vascular biology in general in human organs. In the present study, despite the lack of correlation of up-regulated CLR expression in tumor vessels with various clinico-pathological parameters and patients survival, the increased levels of this GPCR might nevertheless have a direct impact on tumor angiogenesis – the hallmark of cancer progression in the kidney (5, 32, 35, 36, 42) - due to simultaneous up-regulation in the expression of both the ligand and its receptor within the tumor microenvironment, as revealed in this study. Alternatively, since some of the RCC vessels may be lymphatics and because AM may play a role in lymphatic endothelial cell biology (43-46), CLR can affect tumor lymphangiogenesis (and possibly tumor spread via lymphatic vessels) and therefore further vessel subtyping may provide additional prognostic information in the future.
It is widely acknowledged that the differential expression of a specific molecule in tumor compared to normal tissues is often crucial for the development of targeted therapies and strategies in cancer (47). Therefore, up-regulated (in an autocrine loop with AM) by RCC tumor cells and tumor vessels CLR has a potential as a cell surface-presented molecular target for future functional studies in renal cancer using already developed range of modulators of AM-induced effects (and hence downstream signaling pathways mediated by this GPCR within human tumor tissue), including antibodies and peptide antagonists of AM receptors (4, 48, 49).
In summary, the present study reveals previously unrecognised cancer-type specific up-regulation of ADM expression in human tissues/organs and prognostic value for AM receptor CLR in RCC. Our findings suggest the potential use for CLR as a novel target for future clinical studies and drug development for the therapy of RCC, and possibly other hypoxia-driven cancers. In particular, drug-resistant RCC could be an ideal model for further evaluating the prognostic utility of CLR as a biomarker in first-line as well as refractory to anti-angiogenic or combination treatments patients and for investigating the potential role for AM and CLR-mediated signaling in tumor cells and tumor vessels resistance to conventional chemotherapy and current targeted therapies.
Supplementary Material
Translational relevance.
Renal cancer is often resistant to conventional systemic treatments or current targeted therapies. This study reveals that G-protein coupled receptor CLR expression in RCC is associated with poor outcome for patients’ survival. These findings suggest application of CLR as a molecular target for therapeutic agents/drug development, whilst avoiding adverse effects on normal tissues.
Acknowledgements
We thank Dr Rajeev Gupta (UCL Cancer Institute, London) for helpful discussions and comments.
Grant support: This study was supported in part by the Cancer Research UK (grants C575/A6125 and C575/A13100; LLN, DG, SH, ALH, SBF), The Wellcome Trust (grant 063353/Z/00Z; LLN, MCPR), The Cancer Society, Canterbury West Coast Division, NZ (HRM), Medical Research Fund, University of Oxford, UK (LLN), The Royal Society UK, Cancer Research UK and The Maurice & Phyllis Paykel Trust travel awards (LLN).
Footnotes
Disclosure of Potential Conflicts of Interest: L.L. Nikitenko is a scientific consultant for the Scientific Centre of the Family Health and Human Reproduction Problems, Siberian Branch of Russian Academy of Medical Sciences, Irkutsk, Russia. No potential conflicts of interest are disclosed by other co-authors.
References
- 1.Kitamura K, Kangawa K, Kawamoto M, Ichiki Y, Nakamura S, Matsuo H, et al. Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem Biophys Res Commun. 1993;192:553–60. doi: 10.1006/bbrc.1993.1451. [DOI] [PubMed] [Google Scholar]
- 2.Hinson JP, Kapas S, Smith DM. Adrenomedullin, a multifunctional regulatory peptide. Endocr Rev. 2000;21:138–67. doi: 10.1210/edrv.21.2.0396. [DOI] [PubMed] [Google Scholar]
- 3.Zudaire E, Martinez A, Cuttitta F. Adrenomedullin and cancer. Regul Pept. 2003;112:175–83. doi: 10.1016/s0167-0115(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 4.Nikitenko LL, Fox SB, Kehoe S, Rees MC, Bicknell R. Adrenomedullin and tumour angiogenesis. Br J Cancer. 2006;94:1–7. doi: 10.1038/sj.bjc.6602832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris AL. Hypoxia - a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 6.McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–9. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 7.Poyner DR, Sexton PM, Marshall I, Smith DM, Quirion R, Born W, et al. International Union of Pharmacology. XXXII. The mammalian calcitonin gene-related peptides, adrenomedullin, amylin, and calcitonin receptors. Pharmacol Rev. 2002;54:233–46. doi: 10.1124/pr.54.2.233. [DOI] [PubMed] [Google Scholar]
- 8.Kroeze WK, Sheffler DJ, Roth BL. G-protein-coupled receptors at a glance. J Cell Sci. 2003;116:4867–9. doi: 10.1242/jcs.00902. [DOI] [PubMed] [Google Scholar]
- 9.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 10.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends in Molecular Medicine. 2011;17:126–39. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 12.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 13.Gentleman R. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; New York ; Cambridge: 2005. [Google Scholar]
- 14.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 15.Henderson SR, Guiliano D, Presneau N, McLean S, Frow R, Vujovic S, et al. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–93. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 17.Nikitenko LL, Brown NS, Smith DM, MacKenzie IZ, Bicknell R, Rees MC. Differential and cell-specific expression of calcitonin receptor-like receptor and receptor activity modifying proteins in the human uterus. Mol Hum Reprod. 2001;7:655–64. doi: 10.1093/molehr/7.7.655. [DOI] [PubMed] [Google Scholar]
- 18.Charlesworth PJ, Kilbey N, Taylor M, Leek R, Cranston D, Turner G, et al. Automated uro-oncology data collection: the Cancer Research Uro-Oncology Database. BJU international. 2010;105:1663–6. doi: 10.1111/j.1464-410X.2009.09058.x. [DOI] [PubMed] [Google Scholar]
- 19.Eble JN. Pathology and genetics of tumours of the urinary system and male genital organs. IARC Press; Oxford University Press [distributor]; Lyon: Oxford: 2004. [Google Scholar]
- 20.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–63. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Hermanek P, Scheibe O, Spiessl B, Wagner G. [TNM classification of malignant tumors: the new 1987 edition] Radiobiol Radiother (Berl) 1987;28:845–6. [PubMed] [Google Scholar]
- 22.Nikitenko LL, Blucher N, Fox SB, Bicknell R, Smith DM, Rees MC. Adrenomedullin and CGRP interact with endogenous calcitonin-receptor-like receptor in endothelial cells and induce its desensitisation by different mechanisms. J Cell Sci. 2006;119:910–22. doi: 10.1242/jcs.02783. [DOI] [PubMed] [Google Scholar]
- 23.Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer. 2009;100:405–11. doi: 10.1038/sj.bjc.6604844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Research. 2005;65:8690–7. doi: 10.1158/0008-5472.CAN-05-1208. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jubb AM, Soilleux EJ, Turley H, Steers G, Parker A, Low I, et al. Expression of vascular notch ligand delta-like 4 and inflammatory markers in breast cancer. Am J Pathol. 2010;176:2019–28. doi: 10.2353/ajpath.2010.090908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leek RD, Landers RJ, Harris AL, Lewis CE. Necrosis correlates with high vascular density and focal macrophage infiltration in invasive carcinoma of the breast. Br J Cancer. 1999;79:991–5. doi: 10.1038/sj.bjc.6690158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilairet S, Foord SM, Marshall FH, Bouvier M. Protein-protein interaction and not glycosylation determines the binding selectivity of heterodimers between the calcitonin receptor-like receptor and the receptor activity-modifying proteins. J Biol Chem. 2001;276:29575–81. doi: 10.1074/jbc.M102722200. [DOI] [PubMed] [Google Scholar]
- 29.Nikitenko LL, Cross T, Campo L, Turley H, Leek R, Manek S, et al. Expression of terminally glycosylated calcitonin receptor-like receptor in uterine leiomyoma: endothelial phenotype and association with microvascular density. Clinical Cancer Research. 2006;12:5648–58. doi: 10.1158/1078-0432.CCR-06-0852. [DOI] [PubMed] [Google Scholar]
- 30.Dawson C, Whitfield H. ABC of urology - Urological malignancy .3. Renal and testicular carcinoma. British Medical Journal. 1996;312:1146–8. doi: 10.1136/bmj.312.7039.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godley PA, Stinchcombe TE. Renal cell carcinoma. Curr Opin Oncol. 1999;11:213–7. doi: 10.1097/00001622-199905000-00014. [DOI] [PubMed] [Google Scholar]
- 32.Atkins MB, Ernstoff MS, Figlin RA, Flaherty KT, George DJ, Kaelin WG, Jr., et al. Innovations and challenges in renal cell carcinoma: summary statement from the Second Cambridge Conference. Clinical Cancer Research. 2007;13:667s–70s. doi: 10.1158/1078-0432.CCR-06-2231. [DOI] [PubMed] [Google Scholar]
- 33.Jain RK. Lessons from multidisciplinary translational trials on anti-angiogenic therapy of cancer. Nat Rev Cancer. 2008;8:309–16. doi: 10.1038/nrc2346. [DOI] [PubMed] [Google Scholar]
- 34.Jain RK, Duda DG, Willett CG, Sahani DV, Zhu AX, Loeffler JS, et al. Biomarkers of response and resistance to antiangiogenic therapy. Nat Rev Clin Oncol. 2009;6:327–38. doi: 10.1038/nrclinonc.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Favaro JP, George DJ. Targeted therapy in renal cell carcinoma. Expert Opinion on Investigational Drugs. 2005;14:1251–8. doi: 10.1517/13543784.14.10.1251. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan R, Armstrong AJ, Dahut W, George DJ. Anti-angiogenic therapy in renal cell cancer. BJU International. 2007;99:1296–300. doi: 10.1111/j.1464-410X.2007.06834.x. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson RN, Koudijs MJ, Patient RK, Ingham PW, Schulte-Merker S, van Eeden FJ. Hedgehog signaling via a calcitonin receptor-like receptor can induce arterial differentiation independently of VEGF signaling in zebrafish. Blood. 2012;120:477–88. doi: 10.1182/blood-2011-10-383729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deville JL, Bartoli C, Berenguer C, Fernandez-Sauze S, Kaafarani I, Delfino C, et al. Expression and role of adrenomedullin in renal tumors and value of its mRNA levels as prognostic factor in clear-cell renal carcinoma. Int J Cancer. 2009;125:2307–15. doi: 10.1002/ijc.24568. [DOI] [PubMed] [Google Scholar]
- 39.Fujita Y, Mimata H, Nasu N, Nomura T, Nomura Y, Nakagawa M. Involvement of adrenomedullin induced by hypoxia in angiogenesis in human renal cell carcinoma. Int J Urol. 2002;9:285–95. doi: 10.1046/j.1442-2042.2002.00469.x. [DOI] [PubMed] [Google Scholar]
- 40.Michelsen J, Thiesson H, Walter S, Ottosen PD, Skott O, Jensen BL. Tissue expression and plasma levels of adrenomedullin in renal cancer patients. Clin Sci (Lond) 2006;111:61–70. doi: 10.1042/CS20060030. [DOI] [PubMed] [Google Scholar]
- 41.Nouguerede E, Berenguer C, Garcia S, Bennani B, Delfino C, Nanni I, et al. Expression of adrenomedullin in human colorectal tumors and its role in cell growth and invasion in vitro and in xenograft growth in vivo. Cancer Medicine. 2013;2:196–207. doi: 10.1002/cam4.51. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Fritz-Six KL, Dunworth WP, Li M, Caron KM. Adrenomedullin signaling is necessary for murine lymphatic vascular development. J Clin Invest. 2008;118:40–50. doi: 10.1172/JCI33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin D, Harada K, Ohnishi S, Yamahara K, Kangawa K, Nagaya N. Adrenomedullin induces lymphangiogenesis and ameliorates secondary lymphoedema. Cardiovasc Res. 2008;80:339–45. doi: 10.1093/cvr/cvn228. [DOI] [PubMed] [Google Scholar]
- 45.Maybin JA, Battersby S, Hirani N, Nikitenko LL, Critchley HO, Jabbour HN. The expression and regulation of adrenomedullin in the human endometrium: a candidate for endometrial repair. Endocrinology. 2011;152:2845–56. doi: 10.1210/en.2010-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikitenko LL, Shimosawa T, Henderson S, Makinen T, Shimosawa H, Qureshi U, et al. Adrenomedullin Haploinsufficiency Predisposes to Secondary Lymphedema. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–9. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 48.Kaafarani I, Fernandez-Sauze S, Berenguer C, Chinot O, Delfino C, Dussert C, et al. Targeting adrenomedullin receptors with systemic delivery of neutralizing antibodies inhibits tumor angiogenesis and suppresses growth of human tumor xenografts in mice. FASEB J. 2009;23:3424–35. doi: 10.1096/fj.08-127852. [DOI] [PubMed] [Google Scholar]
- 49.Archbold JK, Flanagan JU, Watkins HA, Gingell JJ, Hay DL. Structural insights into RAMP modification of secretin family G protein-coupled receptors: implications for drug development. Trends in Pharmacological Sciences. 2011;32:591–600. doi: 10.1016/j.tips.2011.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.