Abstract
Gene expression is an important phenotype that informs about genetic and environmental effects on cellular state. Many studies have previously identified genetic variants for gene expression phenotypes using custom and commercially available microarrays1-5. Second generation sequencing technologies are now providing unprecedented access to the fine structure of the transcriptome6-14. We have sequenced the mRNA fraction of the transcriptome in 60 CEU individuals and have combined these data with genetic variants from the HapMap3 project15. We have quantified exon abundance based on read depth and have also developed methods to quantify whole transcript abundance. We have found that approximately 10 million reads of sequencing can provide access to the same dynamic range as arrays with better quantification of alternative and highly abundant transcripts. Correlation with SNPs leads to a larger discovery of eQTLs than with arrays. We also detect a substantial number of variants that influence the structure of mature transcripts indicating variants responsible for alternative splicing. Finally, measures of allele-specific expression allowed the identification of rare eQTLs and allelic differences in transcript structure. This analysis shows that high throughput sequencing technologies reveal new properties of genetic effects on the transcriptome and allow the exploration of genetic effects in cellular processes.
Genetic variation in gene expression is an important determinant of human phenotypic variation; a number of studies have elucidated genome-wide patterns of heritability and population differentiation and are beginning to unravel the role of gene expression in the etiology of disease1-5. Interrogation of the transcriptome in these studies has been greatly facilitated by the use of microarrays, which quantify transcript abundance by hybridization. However, microarrays possess several limitations and recent advances in transcriptome sequencing in second generation sequencing platforms have now provided single nucleotide resolution of gene expression providing access to rare transcripts, more accurate quantification of abundant transcripts (above the signal saturation point of arrays), novel gene structure, alternative splicing and allele-specific expression6-14. Although RNA-Seq studies have addressed issues of transcript complexity, they have not yet addressed how genetic studies can benefit from this increased resolution to reveal novel effects of sequence variants on the transcriptome.
To understand the quantitative differences in gene expression within a human population as determined from second generation sequencing, we sequenced the mRNA fraction of the transcriptome of lymphoblastoid cell lines (LCLs) from 60 CEU individuals (from CEPH - Centre d’Etude du Polymorphisme Humain) using 37-bp paired-end Illumina sequencing. Each individual’s transcriptome was sequenced in one lane of an Illumina GAII analyzer and yielded 16.9±5.9 (mean ± SD) million reads which were then mapped to the NCBI36 assembly of the human genome (Supp. Fig 1) using MAQ16. We subsequently filtered reads which had low mapping quality, mapped sex chromosomes or mitochondrial DNA and were not correctly-paired which yielded 9.4±3.3 million reads. On average, 86% of the filtered reads mapped to known exons in EnsEMBL version 54)17 and 15% of read pairs spanned more than one exon. Evaluation of sequence and mapping quality measures was preformed in order to ensure that the data quality is acceptable for analysis (Supp. Fig 2 – see also methods).
We quantified reads for known exons, transcripts and whole genes. Read counts for each individual were scaled to a theoretical yield of 10 million reads and corrected for peak insert size across corresponding libraries. Each quantification was filtered to exclude those with missing data for >10% of the individuals. For exons, this resulted in data for 90,064 exons for 10,777 genes. Of these, 95% had on average more than 10 reads, 38% more than 50 reads and 20% had a mean quantification of ≥100 reads (Supp. Fig 3). For transcript quantification, new methods needed to be developed to map reads into specific isoforms18, 19. We developed a methodology, called the FluxCapacitor, to quantify abundances of annotated alternatively spliced transcripts (see Methods). Using this method, we obtain relative quantities for 15967 transcripts from 11674 genes. For each individual, we compared whole gene read counts to array intensities generated with Illumina HG-6 version 2 microarrays. Correlations coefficients between RNA-Seq and array quantities and among RNA-Seq samples were high and consistent with previous studies20 (Supp. Fig 4 and 5). Finally, we explored whether the correlation structure of abundance among exons could facilitate the development of a framework that will allow the imputation of abundance values for exons that are not screened, given a set of reference RNA-Seq samples. This is the same principle as using the correlation structure (Linkage Disequilibrium) of genetic variants to impute variants from a reference to any population sample of interest21. For each of the 10,777 genes, we assessed the pairwise correlation of all exons and on average, any two pairs of exons within a gene were moderately correlated (mean Pearson’s correlation R2 = 0.378 ± 0.261) (Supp. Fig. 6). This correlation increased with increase in total number of reads present in each exon. It is worth noting that the average correlation coefficient between SNPs within the same recombination hotspot interval in HapMap3 is r2 = 0.326±0.174, suggesting that the correlation structure within genes is stronger and likely more accessible by imputation methodologies than SNPs, however this needs to be assessed in a tissue-specific context.
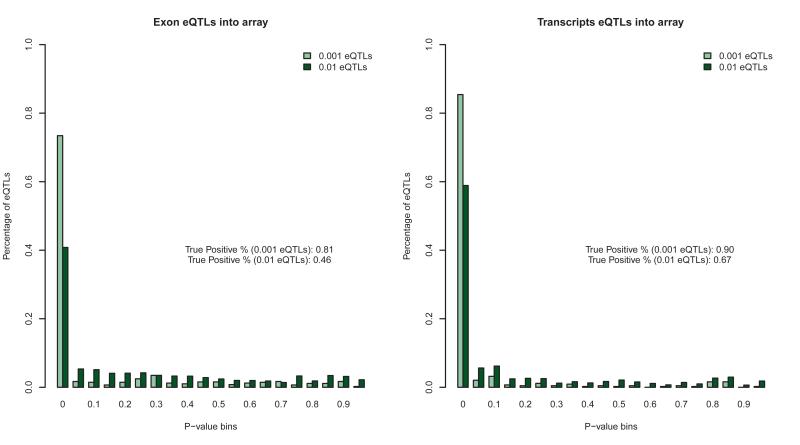
Association of gene expression measured by RNA-Seq with genetic variation was evaluated in cis with the use of 1.2 million HapMap3 SNPs (methods previously described22). We evaluated association in exons, transcripts and genes and determined the unique number of genes containing a significant association through permutation23 (Table 1). RNA-Seq eQTLs, significant at 0.001 and 0.01 permutation thresholds, replicate significantly (46% for 0.01 and 81% for 0.001) in the array data for the same SNP-gene combinations, as indicated by the enrichment in low p-values (Figure 1) and the effect sizes are of very similar magnitude (Supp. Fig. 7). Overall, the number of genes with eQTLs eQTL at the 0.01 permutation threshold using exon quantification was higher than the number of genes discovered by arrays for the same sample of individuals (836 genes vs. 539 genes at the 0.01 permutation threshold), even when normalized for the number of genes tested (Supp. Table 1), suggesting that increased resolution contributed to the identification of larger number of genetic regulatory effects. The RNA-Seq exon eQTLs were mainly enriched in the higher abundance classes relative to array eQTLs and whole gene eQTLs (Supp. Fig. 8-10). This is likely due to two reasons: (i) exons capture genetic effects in splicing complexity, which is higher in higher abundance genes (Spearman Rank Correlation between abundance and number of transcipts in EnsEmbl, p < 2.2 × 10−16); (ii) saturation of intensity signal above a certain abundance level in arrays but not in RNA-Seq data. RNA-Seq exon eQTLs have lower representation in low abundance genes suggesting that rare transcripts are not well quantified at this level of coverage. Finally, we performed eQTL analysis of 102 well-quantified long noncoding RNAs (not overlapping any known protein-coding gene, see Methods), and found 6 with significant eQTLs (Table 1), highlighting that regulatory variation extends beyond well characterized protein coding genes.
Table 1.
eQTL Discoveries. eQTL discoveries for genes, transcripts, exons, splicing events and long non-coding RNAs for each of the 2 sequencing based quantifications (by-transcript and by-exon) and matching array samples are shown using Spearman Rank correlation.
| ASSOCIATIONS | # OF TRAITS | # OF SNPS | PERMUTATION THRESHOLDS* | ||
|---|---|---|---|---|---|
|
| |||||
| 0.05 | 0.01** | 0.001 | |||
| Exon quantification | 90064 exons / 10777 genes | 1171085 | 3258 | 836 (0.13) | 103 |
| Transcript quantification | 15967 transcripts / 11674 genes | 1171085 | 1129 | 293 (0.40) | 66 |
| Whole Gene quantification | 11210 genes | 1171085 | 875 | 256 (0.43) | 62 |
| Long non-coding RNAs | 232 exons / 102 genes | 1171085 | 14 | 6 (0.17) | 1 |
| Transcript events | 6468 events | 1171085 | 416 | 110 (0.59) | 21 |
| Array-based quantification | 21800 probes / 17420 genes | 1171397 | 1682 | 539 (0.32) | 194 |
Thresholds at the gene level
False Discovery Rate (FDR) in parentheses
Figure 1. Array association p-values for RNA-Seq significant eQTLs.
The p-value distribution using array data for RNA-Seq eQTLs significant at the 0.01 and 0.001 permutation threshold from both the exon and transcript quantification data is plotted. In each of the plots, the significant tail of the p-value distribution is substantially enriched indicating that eQTLs discovered through transcriptome sequencing are also significant in arrays. For each plot this excess is quantified using the q-value statistic 1-pi0 to estimate the proportion of true positives. Enrichment in the p-value distribution is higher for eQTLs discovered via transcript quantification and for eQTLs that are more significant at a more stringent permutation threshold.
To replicate our eQTL discoveries, we compared associations between our study and those obtained from sequencing the transcriptomes of an African population (Pickrell, submitted for the same issue)24. We assessed the p-value distribution of matching CEU associations given the top associated SNP for 500 genes from the African population (Supp. Fig. 11). We estimated that ~33% of these signals were shared (p < 0.0001 assessed by permutations). This result indicates the robustness of the eQTL discovery of the two transcriptome sequencing based studies and, given the degree of differentiation of the two populations, the magnitude of replication is consistent with past array studies for the same samples22
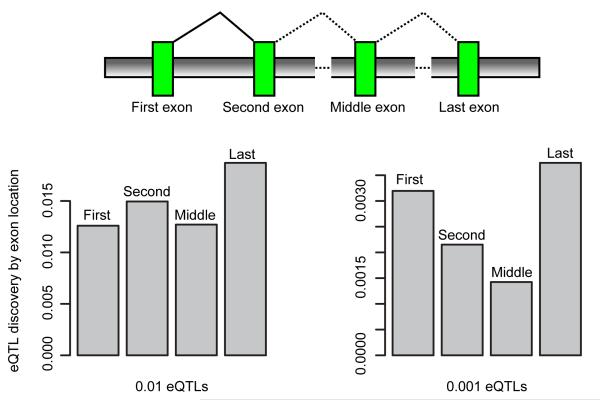
As previously observed22, we have detected enrichment of eQTLs around the transcription start site (TSS) (Supp. Fig 12). We have further investigated the discovery rate and distribution of eQTLs given an exon’s location in multi-exonic genes. We identified increased number of discoveries for the first, second and last exon compared to any middle exons (Figure 2). We find that we make more discoveries for the last exon than for the first exon. When we assess the distribution of significant eQTLs around the 5′ end of the exon of interest, we find that significant eQTLs when found associated with the last exon are closer to the last exon than any other exon followed by first exons, second exons and middle exons (Supp. Fig. 13). This is consistent with our understanding of expression modulating effects within the 3′ UTR and upstream region of genes25.
Figure 2. Exon eQTLs by exon relative location.
We investigated the proportion of discovered exon eQTLs relative to exon location in multi-exonic genes (normalized by number of exons tested within class). For 0.001 eQTLs we see that the proportion of discoveries is increased relative to middle exons in both first, second and last exons. We also see that we make proportionally more discoveries than any other class in the last exon of the gene.
Transcriptome sequencing allows the quantification of allele-specific expression (ASE)26-28. We found an average of 4000 heterozygote confirmed HapMap3 SNP positions per individual, which could be used to assess ASE. Of these we assessed the proportion where both alleles were detected in the sequencing of that individual as a function of mapping quality using Samtools (Supp. Fig. 14)29. At MAQ mapping quality 10, we find that 72% of heterozygote sites have from both alleles detectable at least once. As expected, this fraction slightly reduces with increasing mapping quality. Furthermore, we find 41% of the heterozygotes have more than 6 reads. We tested for ASE after correcting for reference to non-reference differential mapping for each library (Supp. Fig. 15). We tested the relationship between eQTLs and ASE by first phasing double heterozygotes for both. We found that as the number of reads increased, the correlation between the eQTL effect size and the strength of ASE increased (Supp. Fig 16). Reads were then summed across individuals to assess the 1-sided ASE binomial p-value distribution with respect to eQTL phasing. We found that for 0.01 and 0.001 significant eQTLs, the tail of the ASE p-value distribution was enriched. For exons without eQTLs, both tails of this distribution were enriched (Supp Fig 17), which highlights the presence of other non-genetic or rare genetic factors that affect ASE.
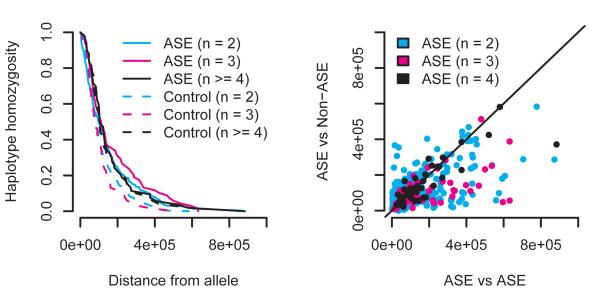
To investigate if ASE signals could be marking recent rare eQTLs, undetected through standard genotypic association, we selected SNPs heterozygous in 6 or more individuals in exons with no evidence for an eQTL (exons not significant at permutation threshold of 0.05), and examined patterns of haplotype homozygosity between individuals that shared a significant ASE signal (at p<0.05) with those that did not. Haplotype homozygosity assesses the length of perfectly shared alleles on a haplotype as a proxy for the age of a haplotype30. We calculated haplotype homozygosity comparing those haplotypes that had an ASE signal to each other and then separately with those that did not have an ASE signal and found greater haplotype homozygosity for haplotypes sharing a common ASE signal (Figure 3). This differentiation was highly significant when only 2-3 individuals had significant ASE (Wilcoxon Paired test, p = 0.00039) and disappeared when 4 or more individuals had significant ASE (Wilcoxon Paired test, p = 0.55), consistent with the idea that these rare ASE effects are a result of recent and rare eQTL variants. We also assessed the direction of effect for these potential rare eQTL haplotypes and found no significant bias in the direction of effect for new mutations (48.5% increased expression for 2-3 individuals compared to 47.1% for haplotypes shared in 4 or more individuals). These results highlight the potential of using second generation sequencing to identify rare regulatory haplotypes.
Figure 3. Haplotype homozygosity for shared ASE haplotypes versus shared and unshared ASE haplotypes.
(A) We assessed the degree to which shared ASE indicated a rare regulatory haplotype. We selected heterozygotes which were present in 6 or more individuals and assessed haplotype homozygosity between haplotypes which shared a significant ASE effect (p<0.05) (labelled as ASE in plot) versus those which shared and did not share an ASE effect (labelled as Control in plot) for all exons for which we did not have evidence for an eQTL (not significant at 0.05 permutation threshold). We see that when comparing the among significant (ASE) extent of haplotype homozygosity to that of significant vs. non-significant haplotypes (Control) where 2 or 3 individuals share the ASE significant signal there is greater haplotype homozygosity for the haplotypes which share the ASE signal indicating that these are on a more recent and rarer haplotype. This signal decreases when the ASE signal is shared in 4 or more individuals. Here the derived allele was selected as the one with the longest haplotype homozygosity without reference to the ASE signal. (B) For each heterozygote we plotted the extent of haplotype homozygosity for significant ASE haplotypes (X-axis) versus significant by not significant ASE haplotypes (Y-axis). We observed that the length of homozygosity is greater in the significant haplotypes compared to each other than when compared to non-significant corresponding haplotypes. Here the derived allele was selected as the one with the longest haplotype homozygosity without reference to the ASE signal.
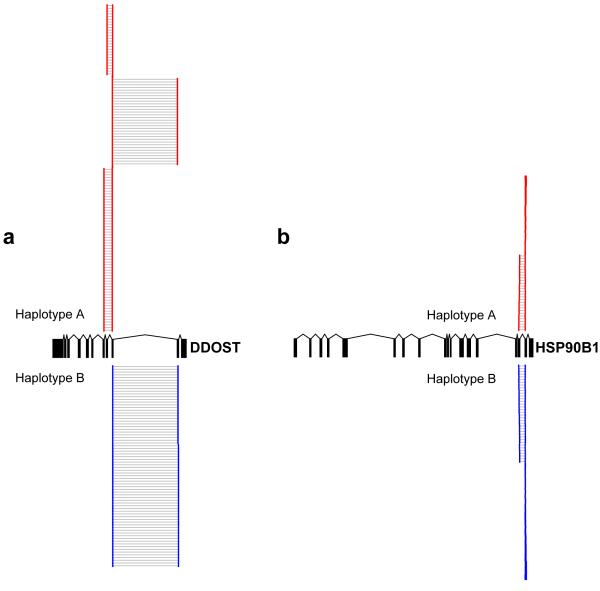
We have further investigated features of the genetic basis of alternative splicing. First, we performed association between known variants affecting splicing signals with their respective genes and exons; in total, we tested 963 variants for 788 genes. We compared associations for gene RNA-Seq quantification and arrays and found similar enrichment (8.30% versus 8.51% true positives). We stratified splice variants in donor and acceptor variants and tested against abundance of exons 5′ and 3′ to the intron they are residing. For donor variants we found a large enrichment (3.17 fold) of associations with the 5′ exon relative to the 3′ exon, while for acceptor variants we found large enrichment of associations with the 3′ exon relative to the 5′ exon (7.02 fold), consistent with them affecting the inclusion/exclusion of the associated exon in the mature transcript. We further hypothesize that if genetic variants are effecting transcript-specific expression, we should be able to detected heterogeneity in the transcript distribution found between chromosomes within an individual. To verify this hypothesis we tested for heterogeneity in paired-end insert sizes, used as proxy to heterogeneity in the transcript distribution. We compared reads over one allele relative to the other in significant ASE SNPs vs. non-significant ASE SNPs for positions with at least 50 reads to have adequate comparable transcript distributions, which resulted in 901 heterozygote positions. We found a significant enrichment in transcript distribution (insert size) heterogeneity (KS p-value < 0.05) over significant ASE SNPs relative to non-significant ASE SNPs (Supp Fig 18 and example in Fig 4). Of the heterozygotes, 235 were significant for ASE and of those 105 had significant transcript distribution heterogeneity; this corresponded to 72 of 105 genes that contained an ASE significant heterozygote. Visual inspection suggested that such heterogeneity is driven both by differential structure in internal exons as well as alternative 3′ends of genes. Genotypic associations with mean insert size and 3′ ends of genes showed enrichment in low p-values suggesting the presence of genetic variants affecting such processes (Suppl Figures 19 and 20). Finally, we assessed the effect of genetic variants on events that contribute to alternative isoforms (e.g. inclusion/exclusion of exons) derived from the FluxCapacitor quantification. We found that of 6600 quantified events 110 are significant at the 0.01 permutation threshold (Table 1). Of these 41% were exon skipping, 17% were due to an alternative acceptor, 13% were double or triple exon skipping, 6% were alternative donors, 5% were mutually exclusive exons and 5% were retained introns. This analysis suggests extensive genetic variation in the determination of isoform diversity and transcript structure, which is expected to have direct consequences in protein sequence diversity.
Figure 4. Allelic alternative splicing effects.
The two panels show examples of alternative use of exons centered on two significant ASE SNPs. In panel (A) there is a very significant signal of alternative use of exons (KS, P<10−14) between alleles where larger abundance coincides with larger diversity in transcript structure. In panel (B) there is significant ASE signal but not significant diversity in transcript structure between alleles.
Our study and ref 24 have described the first attempts of interrogation of genetic effects on the transcriptome using 2nd generation sequencing technologies. The increasing accessibility of sequencing has increased our ability to resolve new features of regulatory complexity. We have confirmed the feasibility of interrogating eQTLs in population transcriptome sequencing and have discovered more eQTLs than with array data for the same population sample. Furthermore,, despite relatively low sequencing depth, the association signals were well replicated across populations. We have also identified the potential and power of such studies in resolving rare regulatory haplotypes. Finally, we have uncovered a variety of genetic effects influencing isoform abundance and transcript structure. As sequencing technologies continue to increase the depth and breadth of the interrogation of the genome and the transcriptome, it is anticipated that our understanding of finer scale cellular processes will become more detailed and robust.
Methods
RNA preparation, library construction and sequencing
Total RNA was extracted from lymphoblastoid cell lines in 63 individuals of CEPH origin from the HapMap Consortium. Poly-A containing mRNA were purified using poly-T oligo-attached magnetic beads and subsequently fragmented using divalent cations under elevated temperature. Single strand cDNA was made from RNA fragments using reverse transcriptase followed by second strand synthesis with DNA Polymerase I and RNaseH. We sequenced each individual with 37-bp paired end sequenced in one sequencing lane in an Illumina GAII. Lanes were assessed for multiple quality metrics including number of reads, read quality and percentage of reads mapping known exons (Supplemental Figure 2). 2 individuals failed sequencing QC in three rounds of sequencing and were excluded from further analysis.
Read mapping
Reads were mapped to the reference human genome (NCBI36) using MAQ (using a theoretical insert-size upper limit of 2Mb). Reads were subsequently filtered to include only those which were called as properly paired and had a mapping quality score greater than or equal to 10. This yielded between 3.5 and 17.1 million reads per individual (9.4 mean +/− 3.3 SD million reads).
Read quantification of known exon and genes
Each paired end read was assessed for overlap with known transcripts from EnsEMBL (version 54). Reads were only considered if the overlap for each mate pair was constrained to be first in the same transcript or in two separate genes. Reads spanning multiple exons were independently quantified given the same conditions in addition to the restriction that exons were located more than 200bp away from one another. Furthermore, in the case of multiple overlapping exons for one mate, we considered only the exon with the largest number of supporting reads. These conditions were to prevent spurious relationships being quantified when multiple overlapping exon annotations existed.
Raw exon read counts were subsequently normalized by scaling read count to a total of 10 million reads per sample. For gene counts, the same procedure was applied to the summed raw reads determined for exons of the respective gene. It has been observed by other investigators that the Poisson nature of the data can create a correlation between read depth and abundance estimation as well as GC content of the reads analyzed. We tested this hypothesis and found that in our data no such correlation was observed. As a further normalization check, after eQTL analysis, we assessed the degree to which the Poisson nature of RNA-seq data as reported by Marioni et al.31 affected exon quantification and eQTL discovery by examining the correlation p-values of expression abundance of all genes with read depth (Supplemental Figure 22). We observed no difference in the effect of read depth between genes with and without an eQTL.
Read quantification of known transcripts
We have developed a method we call the FluxCapacitor, to recontruct abundances of known transcript forms from RNAseq data. Our algorithm works by distributing the reads mapping to a given exonic region (or splice junction) among the transcripts including the exon (or splice junction). For each locus, that is, a set of overlapping transcripts {ti}, the FluxCapacitor adopts an underlying graph structure G = (V.E) similar to splicing graphs32. The nodes v∈V in the graph are sites (i.e., transcription start or termination sites, splice donor or acceptor sites). These are ordered by their genomic position pv in directionality < of the annotated transcript, from 5′ to 3′. Edges e=(v→w),e∈E are correspondingly non-overlapping (parts of) exons and introns. The support of each edge is the set of transcripts which include the edge: support(e)={t1,…,tn}, (Supplemental Figure 23).
Then, assuming a strict uniform read distribution along transcripts, the observed number of reads mapping to edge Xi (the flux of Xi) is equal to the sum of number or reads (the fluxes) produced by each transcript that includes the edge tj∈ support(Xi), normalized by the edge length:
| (1) |
where flux(tj) is the a priori unknown number of reads that were produced by transcipt tj normalized to the edge length.
Each edge recasts the sequence of a unique genomic region to which a given read is mapped iff it aligns to a substring of the edge sequence (sv,…sw). Reads that align to a consecutive suffix/prefix pairs of exonic edges that are adjacent in ordering <, i.e., reads that align to splice junctions or span alternative exon boundaries, are correspondingly aligned to k-super edge tuples (e1,…,ek), where k depends on the length ∣pw–pv∣ spanned by each of the edges compared to the length of the read. During the alignment of reads we require minimality on the size k of a tuple a read aligns to, i.e., a read that aligns to (e1,…,ek) will not get assigned to tuple (e1,…,ek+1). The minimality criterion assigns each region along the genomic sequence uniquely to one tuple (e1,…,ek),k≥1, and we denote the sum of reads aligned to that tuple as the frequency of the observation freq((e1,…,ek)).
In the practice, the assumption of strict uniformity in read distribution along the transcript is too strong, since RNA-Seq experiments suffer from systematic biases in read coverage (Supplemental Figure 23)33. In our approach we estimate the biases characteristic of each experiment by collecting read distribution profiles in non-overlapping transcripts, binned by several transcript lengths and expression levels. From these profiles, we estimate for each edge and transcript a flux correction factor , that following the language of hydro-dynamic flow networks34 we denote as the capacity of the edge, as the area under the transcript profile between the edge boundaries (Supplemental Figure 23).
Additionally we allow at each edge a certain deviation that accounts for statistical fluctuations resulting from the limited depth of the sequencing process and deviation from the estimated flux capacity corrections. Equation 1 is thus extended in the following way:
| (2) |
The application of equation (2) to each edge in the splicing graph for a given locus results in a system of linear equations. The FluxCapacitor solves the linear system as an optimization problem with the objective of minimizing the deviation from all observations Σi∣Δi∣. To find a solution we apply a standard linear program solver35. As a result all transcripts ti corresponding to a locus are quantified by the value flux(tj).
In order to account for reads that have been derived from different ends of the same cDNA molecule (so-called mates), we generalize our mapping to k-super edges so that the set of edges that form a k-super edge no longer need to be consecutive, but can be formed by two distant regions, each one corresponding to the minimal edge set covering the region to which one of the mate aligns. Mate-specific orientations are taken into account, when deciding on valid mate pairs, but no constraints on the size of the insert are applied. Flux capacity correction factors are estimated as before, as the sum of areas under the read distribution profile a certain k-super edge covers.
Read quantification of long non-coding RNAs
We used the Gencode annotation (data freeze 3b) which contains 9,937 long non-coding transcripts (attribute transcript_type “non-coding” or “processed”) with 32,979 exons in 6,333 non-protein coding loci (without attribute gene_type “protein_coding”). In order to exclude any influences in cis with protein-coding genes transcription, we additionally filtered out about half (3,031 of the loci with 4,875 transcripts) which are located close (i.e., 1kb upstream or downstream, regardless of the strand) of an annotated protein-coding gene. The majority (96%) of the remaining 5,062 transcripts have been annotated manually-some of them even including experimental confirmation-such that we further excluded 221 transcripts that stem from computational prediction pipelines (i.e., attribute annotation level “3”). Filtering for those lncRNAs with <= 10% missingness across individuals resulted in 232 quantifiable exons for association analysis.
Mixed insert size normalization
RNA-seq quantifications for each individual/lane were identified as having excess correlation with the maximum peak of their respective insert size distributions. We modelled this relationship as polynomials of order −1, 0.5, 1, 2 and 3 and found the best fit, where correlation was maximized, using a linear (order 1) model. This fit was similarly observed when excluding genes which contained a transcript less than 500bp, 1000bp and 2000bp in length. The residuals of the regression were used as input to the association analyses.
Hybridization protocol and quantification
Array-based gene expression data was collected for each of the individuals on the Illumina HG-6 version 2 array. Two technical replicates were performed for each. We quantile normalized within replicates and calculate a weighted mean at each rank given the average signal by the number of beads. Individuals are subsequently median-normalized and log2 transformed. We selected only probes which uniquely mapped to an EnsEMBL gene and did not contain a SNP; this resulted in 21,800 probes corresponding to 17,420 genes.
Array versus sequencing comparison
To compare array versus sequencing quantification we determined RPKM values for each gene for each individual and compared them to the mean probe intensity for the same gene. For association analyses (described below) 2 sequenced individuals did not have matching array data. For these individuals we replaced them with their fathers for whom we had expression data for. As such, NA10847 was replaced with father NA12146 and NA10851 with father NA12056.
Association analyses and multiple testing corrections
We conducted spearman rank correlation analyses of 1.2 million HapMap3 SNPs with MAF >5% to the exon, transcript and gene sequencing quantifications and the array-based data. For the exon quantification, we selected 90,064 exons corresponding to 10,777 genes where at least 10% of the individuals had data. We similarly selected from the transcript quantification 15,967 transcripts corresponding to 11,674 genes and from the gene quantification we obtained 11210 genes For each of the 3 datasets, exon, transcript, gene and array we performed SNP-by functional unit associations within 1Mb of the transcription start site. P-value significance was evaluated in each data set by permuting the expression phenotype 10,000 times and summarizing the extreme p-value distribution for each particular exon, transcript, gene or probe. To further control for multiple testing within each analysis given that several quantifications were available for a gene (such as for the exon, transcript and array-based quantifications) we set gene-level permutation thresholds by taking the most stringent p-value distribution.
Imputation
We imputed genotypes in four individuals (NA0851, NA12004, NA12414 and NA12717) with Beagle version 3.0.4 that were not present in the HapMap 3 (release 3) but had been typed on the Affymetrix 6.0. Imputation of Affymetrix 6.0 into the full HapMap 3 set, for SNPs greater than 5% minor allele frequency has a demonstrated true positive rate of 96.4% (A. Price, personal communication). We further used 410 CEU+TSI (Tuscans from Italy) phased chromosomes from HapMap 3 to conduct the imputation. Inclusion of the phased chromosomes from TSI has also demonstrated an increased true positive rate (J. Barrett, personal communication). In total, 595,716 SNPs were imputed. We assessed the mean difference in genotype probability between the imputed genotype and second best call as 0.95, 0.94, 0.94 and 0.94, across each of the imputed individuals respectively. Two of these individuals (NA12004 and NA12717) had been genotyped previously in HapMap 2 and we replaced all imputed genotypes with these genotypes where possible. We assessed genotype concordance between the imputed genotypes against the HapMap Phase 2 genotypes for both individuals as 3.4%. We also performed PCA analysis within CEU and across the eleven populations in HapMap 3 and observed no significant clustering of the imputed individuals outside the component 1 and 2 CEU group means (Supplemental Figure 21).
Allele-specific expression detection
Allele-specific expression was determined on a per-heterozygote per-individual basis. Reads from heterozygote SNPs overlapping exons were assessed directly for their allelic state given SNP calls from HapMap3 (www.hapmap.org) and by using the Samtool pileup utility29. Reads were filtered to be above MAQ 10 mapping quality. For each individual’s sequencing lane a binomial probability of success was determined based on the probability that a reference allele maps to the genome compared to a non-reference allele. As such each individual had their own binomial probability of success given a heterozygote allele matching the reference sequence that accounted for potential biases in the sequencing reaction. We also computed a weighted metric of effect size using this probability where each occurrence of an allele was weighted by the probability of observing it. A weighted difference between both alleles was then computed by summing all observations.
When comparing to eQTLs, phasing data from HapMap3 was used to phase the eQTL with respect to heterozygote SNPs. Here, a one side binomial p-value was then utilized using the individual specific reference allele mapping probability to enforce the direction of effect in same direction.
For haplotype homozygosity based analyses, haplotype homozygosity was assessed for a heterozygote by comparing the extent of homozygosity for each allele and choosing the allele with the longer homozygous tract as the derived allele. This allele was then used to compare haplotype homozygosity within ASE-containing and ASE-containing versus no ASE haplotypes.
Alternative splicing analyses
Known splice variants were taken from EnsEMBL (version 54). We selected those variants which we had tested as part of our association analysis and for which at least one exon had been quantified within the relevant gene. We quantified the proportion of true positives (using q-value statistics) for associations of splicing variants to gene RNA-seq and array quantification. For arrays, we chose only genes which had be quantified once to correct for multiple testing. We further quantified and compared the proportion of true positives for donor SNP-donor exon, donor SNP-acceptor exon, acceptor SNP-donor exon and acceptor SNP-acceptor exon associations.
To perform genotypic associations of mean insert size we selected the best quantified exon per gene and determined the mean of all insert sizes over this exon. For genotypic associations of 3′ ends, we selected reads which only mapped to the 3′ exon and calculated the mean of distance from the exon’s start to the end of any given read. Both associations were corrected for insert size heterogeneity across individuals using the same method as in the quantification-based associations.
Acknowledgements
The authors would like to acknowledge Heng Li, Simon White, John O’Brien, Steve Searle, Michael Quail, Sri Vishnu Vardhan Deevi and the Sequencing Core facility at the Wellcome Trust Sanger Institute. We would also like to thank Claude Beazley, Alexandra Nica, Luke Jostins, Katherine Morley, Jeffrey Barrett and Verneri Anttila. Funding was provided by the Wellcome Trust and the Louis-Jeantet foundation to ETD.
Footnotes
DATA RELEASE All RNA-Seq data in raw and normalized form is available in ArrayExpress under accession numbers E-MTAB-197 and E-MTAB-198
References
- 1.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 2.Goring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 4.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 9.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 10.t Hoen PA, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 14.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–9. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 15.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng S, Chen L. A hierarchical Bayesian model for comparing transcriptomes at the individual transcript isoform level. Nucleic Acids Res. 2009;37:e75. doi: 10.1093/nar/gkp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiller D, Jiang H, Xu W, Wong WH. Identifiability of isoform deconvolution from junction arrays and RNA-Seq. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 22.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. 2009. submitted. [DOI] [PMC free article] [PubMed]
- 25.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–50. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 27.Verlaan DJ, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–27. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;6:613–8. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabatti C, Risch N. Homozygosity and linkage disequilibrium. Genetics. 2002;160:1707–19. doi: 10.1093/genetics/160.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sammeth M. Alternative Splicing events are bubbles in splicing graphs. Journal of Computational Biology. 2009;16:1117–1140. doi: 10.1089/cmb.2009.0108. [DOI] [PubMed] [Google Scholar]
- 33.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahuja RK, Magnanti TL, Orlin JB. Network Flows: Theory, Algorithms and Applications. Prentice Hall; 1993. ISBN 0-13-617549-X. [Google Scholar]
- 35.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. Second Edition. MIT Press and McGraw-Hill; 2001. Chapter 29: Linear Programming; pp. 770–821. ISBN 0-262-03293-7. [Google Scholar]