To the Editor: Efforts to contain drug-resistant tuberculosis depend on the rapid detection and effective treatment of cases, together with public health interventions to prevent and investigate ongoing transmission. The necessary laboratory support for these activities includes the identification of the Mycobacterium tuberculosis complex, antimicrobial susceptibility testing, and bacterial genotyping. However, even in well-resourced countries, it typically takes 1 to 2 months to achieve all these goals because of the slow growth rate of the M. tuberculosis complex.1,2 Moreover, phenotypic susceptibility testing can be unreliable and is not performed for some agents. Molecular techniques have accelerated some of these diagnostic functions, but they only interrogate a small part of the microbial genome and do not provide all the clinically relevant information.1-3 Whole-genome sequencing has not been used as a diagnostic tool for tuberculosis, in part because of the need to culture M. tubercu- complex for several weeks, until sufficient DNA can be extracted.2,4
Here we report the use of rapid whole-genome sequencing to investigate the case of a patient with extensively drug-resistant (XDR) tuberculosis (the case history is provided in the Supplementary Appendix, available with the full text of this letter at NEJM.org). His first sputum sample became culture-positive after 3 days in the mycobacterial growth indicator tube (MGIT) culture system. DNA was extracted directly from the MGIT tube and sequenced with the use of the Illumina MiSeq platform. Two distantly related Beijing strains of M. tuberculosis were identified (in a ratio of 7:3) (Fig. 1B). Mixed infection was not apparent when standard genotyping was performed on three additional samples from this patient by means of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat assay, which probably identified the majority strain. These findings have important implications for distinguishing relapse from reinfection and identifying secondary cases of infection.
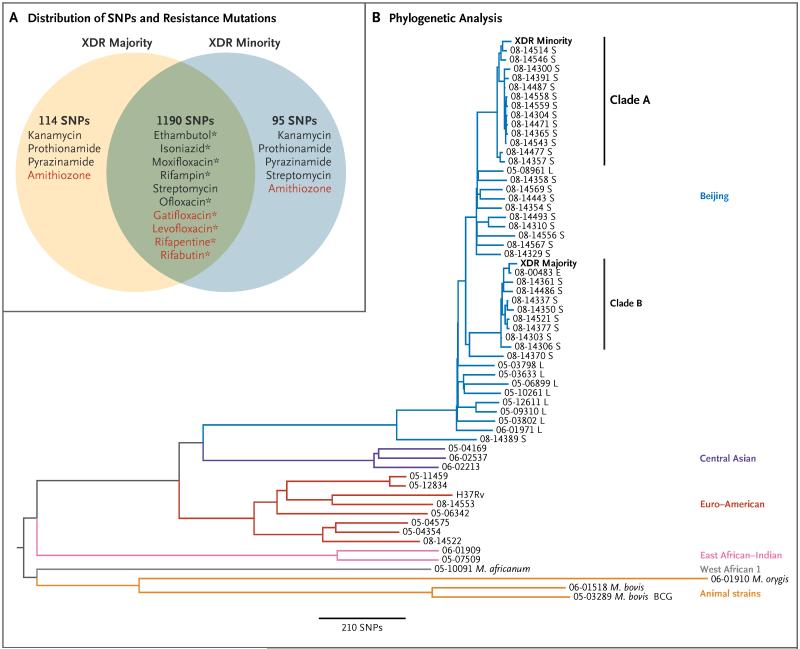
Figure 1. Phylogenetic Analysis and the Distribution of Drug-Resistance Mutations.
In Panel A, the numbers refer to the number of single-nucleotide polymorphisms (SNPs) that were shared by the two strains of extensively drug-resistant (XDR) Mycobacterium tuberculosis isolated from a single patient (termed majority and minority strains on the basis of numerical predominance) or were unique to one strain as compared with the M. tuberculosis H37Rv reference genome. The reference laboratory had reported resistance to nine antibiotics (black type). Genotypically, we found that both strains had mutations that were consistent with resistance to these nine drugs. The same mutation was present in both strains for six drugs (green overlap), but different mutations in each strain accounted for resistance to the remaining three drugs. Streptomycin is shown twice because both strains had a common resistance mutation, but the minority strain had a second resistance mutation. In addition, we found mutations that were associated with resistance to five antibiotics, for which no phenotypic results were available (red type). (Amithiozone is also known as thiacetazone.) Current genotypic tests could have detected only the nine antibiotic resistances marked by an asterisk (see Table S1 in the Supplementary Appendix). In Panel B, a phylogenetic tree based on whole-genome sequencing data shows the majority and minority strains from our patient as compared with data from a published study of XDR tuberculosis, which included representatives of the main lineages of M. tuberculosis and strains of M. africanum, M. orygis, M. bovis, and M. bovis bacillus Calmette-Guérin (BCG).4 For isolates in the Beijing lineage, the country of origin is shown after the name of the strain (London [L]; Samara Oblast, Russia [S], or Estonia [E]). The minority and majority XDR strains from our patient fell into clades A and B, respectively, of the Beijing lineage, which together account for 36% of Beijing cases in Samara. The closest relative of the majority strain was isolated in Estonia and is a representative of the dominant clone in this country.4 The results of mycobacterial inter spersed repetitive-unit-variable-number tandem-repeat genotyping of three samples from this patient were consistent with the majority strain (Table S2 in the Supplementary Appendix).
We interrogated the known genes associated with resistance to 39 antibiotics. The reference laboratory reported phenotypic resistance 9 drugs, the genetic basis for which was detected in all cases (Fig. 1A). Phenotypic susceptibility to amikacin, capreomycin, clofazimine, and linezolid was reported, and these findings coincided with the genotypic results. We found mutations that were consistent with resistance to amithiozone (also known as thiacetazone), gatifloxacin, levofloxacin, rifapentine, and rifabutin, which were not tested at the reference laboratory. Phenotypic susceptibility was reported for para-aminosalicylic acid, which was consistent for the majority strain, whereas in minority strain, one of the genes that is involved in the activation of this drug was mutated, consequence of which was unknown.
This retrospective study revealed the potential of rapid whole-genome sequencing to reduce the time taken to diagnose XDR tuberculosis weeks to days, depending on the time to culture positivity and the turnaround time for sequencing and analysis.2 In well-resourced countries, rapid whole-genome sequencing may replace current methods of identifying and typing M. tuberculosis complex, since it offers the ultimate molecular resolution for outbreak investigations.2,5 Whole-genome sequencing cannot replace phenotypic susceptibility testing for all antibiotics, given the incomplete understanding of the genetic basis of drug resistance. Nevertheless, it can be used to rapidly identify resistance when mutations known to confer resistance are detected, a finding that has the potential to guide clinicians and reference laboratories.
Supplementary Material
Acknowledgments
Supported by grants from the U.K. Clinical Research Collaboration Translational Infection Research Initiative and the Medical Research Council, with contributions from the Biotechnology and Biological Sciences Research Council, the National Institute for Health Research (NIHR) on behalf of the U.K. Department of Health, and the Chief Scientist Office of the Scottish Government Health Directorate (G1000803, to Dr. Peacock); Public Health England (to Dr. Peacock); the NIHR Cambridge Biomedical Research Centre (to Drs. Peacock, Török, and Carmichael); Medical Research Council (to Ms. Bryant); the Wellcome Trust Sanger Institute (WT098051, to Dr. Parkhill and Ms. Bryant); and the Ministry of Economy and Competitiveness, Spain (BFU2010-19310 and PIM2010EPA-00719, to Dr. Marti-Renom).
Footnotes
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
Contributor Information
Claudio U. Köser, Public Health England, Cambridge, United Kingdom
Josephine M. Bryant, Wellcome Trust Sanger Institute Hinxton, United Kingdom
Jennifer Becq, Illumina (Cambridge), Little Chesterford, United Kingdom
M. Estée Török, University of Cambridge, Cambridge, United Kingdom
Matthew J. Ellington, Public Health England, Cambridge, United Kingdom
Marc A. Marti-Renom, Centre Nacional d’Anàlisi Genòmica, Barcelona, Spain
Andrew J. Carmichael, Cambridge University Hospitals National Health Service Foundation Trust, Cambridge, United Kingdom
Julian Parkhill, Wellcome Trust Sanger Institute, Hinxton, United Kingdom
Geoffrey P. Smith, Illumina (Cambridge), Little Chesterford, United Kingdom
Sharon J. Peacock, University of Cambridge, Cambridge, United Kingdom, sjp97@medschl.cam.ac.uka
References
- 1.Parrish N, Carrol K. Importance of improved TB diagnostics in addressing the extensively drug-resistant TB crisis. Future Microbiol. 2008;3:405–13. doi: 10.2217/17460913.3.4.405. [DOI] [PubMed] [Google Scholar]
- 2.Köser CU, Ellington MJ, Cartwright EJ, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8(8):e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heysell SK, Houpt ER. The future of molecular diagnostics for drug-resistant tuberculosis. Expert Rev Mol Diagn. 2012;12:395–405. doi: 10.1586/erm.12.25. [DOI] [PubMed] [Google Scholar]
- 4.Casali N, Nikolayevskyy V, Balabanova Y, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–45. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker TM, Ip CL, Harrell RH, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.