Abstract
Mammalian neural development has been traditionally studied in the context of evolutionarily conserved signaling pathways and neurogenic transcription factors. Recent studies suggest that microRNAs, a group of highly conserved non-coding regulatory small RNAs also play essential roles in neural development and neuronal function. A part of their action in the developing nervous system is to regulate subunit compositions of BAF complexes (ATP-dependent chromatin remodeling complexes), which appear to have dedicated functions during neural development. Intriguingly, ectopic expression of a set of brain-enriched microRNAs, miR-9/9* and miR-124 that promote the assembly of neuron-specific BAF complexes, convert the nonneuronal fate of human dermal fibroblasts towards post-mitotic neurons, thereby revealing a previously unappreciated instructive role of these microRNAs. In addition to these global effects, accumulating evidence indicate that many microRNAs could also function locally, such as at the growth cone or at synapses modulating synaptic activity and neuronal connectivity. Here we discuss some of the recent findings about microRNAs’ activity in regulating various developmental stages of neurons.
Introduction
MicroRNAs (miRNAs) are endogenous 20~24 nucleotide RNAs that bind to target motifs in mRNAs of protein-coding genes to direct posttranscriptional silencing either through transcript degradation or translational repression [1,2]. Predominantly these binding motifs are found in the 3′ untranslated regions (UTRs) of target mRNAs, although examples of their presence in the coding regions of genes have been documented [3]. A single microRNA can have up to several hundreds of targets; and conversely, a gene can have target sites for different miRNAs synergizing the downregulation by multiple miRNAs [4-6]. So far more than one thousand microRNAs have been denoted in the human genome and the number is ever increasing, illustrating the potential of microRNAs as important players in gene regulation. In addition, aberrant expressions of miRNAs have been linked to various pathologies including tumors, highlighting their function in safeguarding normal cell growth and development [7]. This review mainly focuses on recent advances in our understanding of the role of miRNAs in neural development and miRNA-mediated neuronal conversion of mammalian cells.
Essential functions of microRNAs in neural development
MicroRNAs are abundantly expressed in the nervous system, with about half of known species detected in the human brain [8], implicating their significant contribution in neural development and function. Earlier studies utilizing genetic deletions of Dicer, a core component of the biogenesis of mature microRNAs demonstrated essential roles of miRNAs in diverse neural cell types at different stages of development. Conditional deletion of Dicer in neural progenitors in Emx1-Cre or Nestin-Cre mice resulted in markedly thinner cortices, increased apoptosis and disorganized cortical layering, indicating their essential function in neural progenitor expansion as well as neuronal differentiation [9-11]. Selective deletion of Dicer in specific neuronal types (including Purkinje cells (Pcp2-Cre), spinal cord neurons (VAChT-Cre), dopaminergic neurons (DAT-Cre) have consistently resulted in apoptosis of neurons [12-14]. Furthermore, in many cases, such as shown in spinal cord neurons, mutant mice exhibited marked decrease in activity and movement with an array of hallmarks reminiscent of spinal muscular atrophy (SMA), implicating miRNA deregulation in neurodegenerative disorders [14]. In addition, studies from deleting other components of microRNA biogenesis including Dgcr8 have largely yielded similar phenotypes including increased apoptosis and microcephaly, further illustrating the importance of miRNAs during neural development [15]. It should be noted that since the generation of several non-canonical miRNAs may bypass the requirement of Dgcr8 but not Dicer, comparison of the phenotypes of the two knockouts should reveal the contribution of noncanonical miRNAs, as recently shown in rodent neurons [15].
Specific MicroRNAs and their roles in neuronal differentiation, maturity and function
While interfering with miRNA machinery has demonstrated essential roles of miRNAs in brain development, recent development of various experimental techniques that allow selective overexpression or inhibition of individual miRNAs has been instrumental in delineating the role of specific miRNAs. We here review recently published literature of miRNAs with specific functions in different stages of neuronal development (Table 1).
Table 1.
Summary of specfic miRNAs discussed in the text, with their known functions and targets in neural development.
| miRNA | species | Function | Targets | Reference |
|---|---|---|---|---|
| miR-184 | mouse | Promotion of adult neural stem cell proliferation and inhibition of neuronal differentiation |
Numbl | 16 |
|
| ||||
| let-7b | mouse | Impairment of neural progenitor expansion and enhancement of cell cycle exit and neuronal |
Hmga2, TLX, and Cyclin D1 | 18,19 |
|
| ||||
| miR-137 | mouse | Inhibition of neural stem cell proliferation and promotion of neuronal differentiation |
LSD1, Ezh2 | 21,22 |
| mouse | Inhibition of dendritic development | Mib1 | 23 | |
|
| ||||
| miR-9 | mouse | Promotion of neuronal fate and inhibition of neural progenitor division |
TLX, Foxg1, Gsh2, SIRT1, REST/NRSF, Hes1 |
24-29, 31,33 |
| chick | Regulation of motor subtype determination | FoxP1 | 30 | |
| mouse | Regulation of axon development | Map1b | 32 | |
|
| ||||
| miR-124 | mouse | Promotion of neuronal fate and inhibition of neural progenitor division |
PTBP1, Sox9, SCP1, Ephrin-B1, JAG1, BAF53a |
4, 37, 42-44 |
| mouse | hippocampal axonogenesis and retinal cone survival | Lhx2 | 47 | |
| Aplysia | Regulation of synaptic activity | CREB | 48 | |
|
| ||||
| miR-134 | rat | Regulation of dendritic spine morphogenesis | Limk1 | 49 |
|
| ||||
| miR-133 | mouse | regulation of Dopaminergic neurons formation | Pitx3 | 13 |
|
| ||||
| miR-7a | mouse | regulation of Dopaminergic neurons formation in the olfactory bulb |
Pax6 | 55 |
A number of miRNAs have been demonstrated to regulate proliferation and differentiation of neural stem cells (NSCs). MiR-184 was found to be expressed in the adult neural stem cells in the subventricular zone and dentate gyrus, and promote their expansion. Its expression was negatively regulated by methyl-CpG binging protein 1 (MBD1), an epigenetic transcriptional repressor whose deficiency results in impaired adult neurogenesis [16]. In this study Numblike (Numbl), a molecule previously known to regulate stem cell asymmetric division, was identified as a target of miR-184, as its expression restored the imbalance between NSC proliferation and neuronal differentiation caused by either overexpressing miR-184 or abrogating MBD1 [16]. However, it is not clear exactly how Numbl functions in this context. Nevertheless, this study clearly demonstrated a cross-talk between epigenetic mechanisms and miRNAs in adult neurogenesis, and it would be interesting to examine whether similar genetic networks operate in embryonic brain development.
The evolutionarily conserved let-7 family miRNAs also regulate NSC self-renewal and differentiation. Overexpression of let-7a in neural stem cells induced neuron formation while blocking its activity retained them as proliferative Nestin-positive progenitors [17]. The activity of let-7a is enhanced along neuronal differentiation, as a result of binding of TRIM32, a mammalian homologue of Drosophila Brat and Argonaute 1 (Ago1) [17]. Another family member, let-7b was found to promote cell cycle exit and neuronal differentiation [18,19]. Several functional targets of let-7b have been identified, including high mobility group AT-hook 2 (Hmga2) [18], a known regulator of neural stem cell self-renewal, cyclin D1 [18], and an orphan nuclear receptor TLX [19], an neural stem cell fate determinant [20].
In proliferating neural stem cells, TLX forms a repressor complex with lysine specific demethylase 1 (LSD1) and transcriptionally repress miR-137 that is enriched in neurons [21]. Intriguingly, miR-137 targets LSD1, creating a double negative regulation involving TLX-miR-137-LSD1 as NSCs differentiate into post-mitotic neurons. miR-137 has also been shown to be regulated by SRY-box 2 (Sox2), a core transcription factor sustaining NSC self-renewal, and methyl CpG binding protein 2 (MeCP2) in adult mice [22]. The same group also reported that Ezh2 to be a target of miR-137 in adult NSCs, and suggested that reduced Ezh2 expression (resulting in the decrease in H3K27 level) may contribute to the Rett sydrome-like phenotype in MECP2 mutant mice. However, the in vivo relevance of this finding remains unclear. Additionally, miR-137 functions to inhibit dendritic morphogenesis and functional maturation of neurons through silencing of an ubiquitin ligase mindbomb homolog 1 (Mib1) [23].
miR-9, a highly expressed miRNA in the brain, promotes neuronal fate determination in the developing CNS as well as influencing neuronal subtype specification and regulating axonal growth, branching and targeting [24-33]. Reported targets of miR-9 that are important for neuronal differentiation include NEFH, TLX, Foxg1, Gsh2, SIRT1, and REST/NRSF [24-29]. miR-9 was recently demonstrated to promote neural stem cell differentiation by targeting TLX whereas in neural stem cells, TLX represses miR-9 expression [33]. In addition, miR-9 was also found to form a double negative feedback loop with Hes1, an essential molecule for NSC homeostasis [31]. Moreover, Otaegi et al. recently reported a role of the interplay between miR-9 and its target Foxp1 in establishing motor neuron identity and columnar architecture of developing chick spinal cord [30]. Furthermore, a role of miR-9 in regulating axon length and branching through Map1b, an important protein for microtubule stability, in mouse cortical neurons has been described [32]. Taken together, mounting evidence supports a multifaceted role of miR-9 during neuronal differentiation.
miR-124 is another highly abundant brain-enriched miRNA that has been studied extensively. While most loss-of-function and overexpression studies in vertebrates supports miR-124 as a promoter of neuronal differentiation and an inhibitor of progenitor self-renewal (discussed further below), independent reports that studied miR-124 knockout in Drosophila showed that it is grossly dispensable for neuronal differentiation, but rather required for proliferation of neural stem cells [34-36]. Several possibilities could underlie the apparent divergence in the function of miR-124. Notably in flies miR-124 is expressed in proliferating neuroblasts, whereas in vertebrates, its expression starts when double-cortin positive neurons appear [34,35,37,38]. Similarly, in human embryonic stem cell to neuron differentiation system, miR-124 expression was largely restricted to differentiated neurons [39]. Thus the different expression pattern could underlie the distinct activities of miR-124 in different species. Second, the targets of miR-124 could have evolved differentially in different organisms. Consistent with this, Anachronism, a target of miR-124 in neural stem cells in flies has no mammalian homologue [35]. In this review, we focus on studies using vertebrate systems where miR-124 has repeatedly been shown to promote cell cycle exit and neuronal differentiation.
Earlier studies showed that overexpression of miR-124 in HeLa cells shifted their transcriptome towards that of neurons and suggested the pro-neuronal activity of miR-124 [40]. To gain insight of miR-124’s function in vivo, two recent studies examined the role of miR-124 in the subventricular zone in mice [37,38]. In both studies ectopic expression of miR-124 in SVZ cells resulted in early exhaustion of neural progenitors and premature neuronal differentiation while knockdown of miR-124 rendered the opposite effects, thus establishing miR-124 as a neuronal fate determinant in the adult SVZ. Whether miR-124 has a role in neuron/glia lineage specification remains unclear, as the two reports arrived at different conclusions. This discrepancy may be due to the different knockdown efficacy and duration achieved owing to different techniques employed; a genetic loss-of-functional approach thus would be much needed to clarify this issue (which has been hindered so far due to the presence of 3 loci encoding miR-124). In one study Sox9, an important regulator of NSCs [41], and Jag1, a Notch ligand, were identified as targets of miR-124 [37]. In addition to Sox9 and Jag1, many other targets of miR-124 have been documented, whose repression are essential for establishment of neuronal programs, including PTBP1, SCP1, Ephrin-B1, and BAF53a (in conjunction with miR-9*) [4,37,42-44]. In particular, BAF53a, a subunit of a neural progenitor-specific BAF (npBAF) complexes [45]), is repressed and replaced by a homologous BAF53b as a part of neuron-specific BAF (nBAF) complexes. This subunit switching is important for dendritic arborization [46]. Yoo et al. demonstrated that miR-9* and miR-124 synergistically target BAF53a in neural progenitors during neuronal differentiation allowing BAF53b to be expressed in post-mitotic neurons [4]. The expression of miR-124 and miR-9* has been shown to be regulated by REST [24], thus miR-124 and miR-9/9* appear to lie in the center of a triple negative genetic circuit regulating mitotic exit of neural progenitors and the onset of neuronal differentiation [38].
Besides its role in promoting neuronal fate acquisition, miR-124a is also required for hippocampal axonogenesis and retinal cone survival through Lhx2 suppression in mice [47] and constraining synaptic plasticity through CREB in Aplysia [48]. A similar role of miR-124 for regulating synaptic function in mammals will need to be confirmed.
In addition to neuronal fate acquisition, an increasing number of miRNAs, some of which have been described above, are implicated in dendritic morphogenesis and synaptic development in neurons. Interestingly, many of them including miR-134 displayed localization in dendrites or synapses, consistent with their purported role in generating rapid and local responses in an activity dependent manner [49]. Physiological functions, in vivo targets and regulators of such miRNAs are extremely fascinating topics, and have recently been reviewed elsewhere [50].
MicroRNAs and neuronal reprogramming
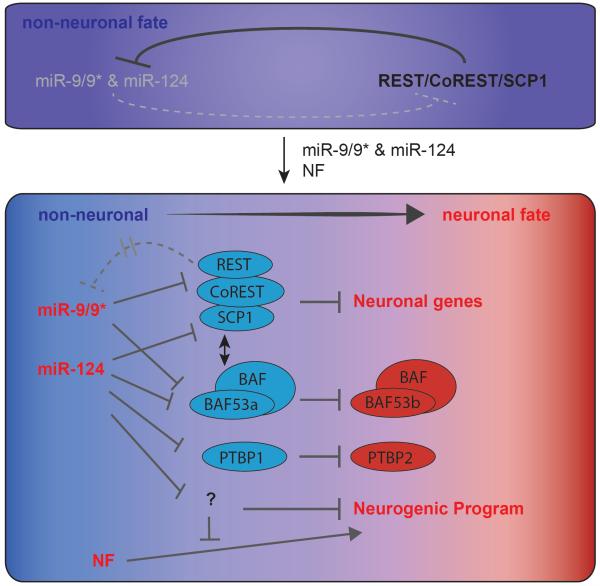
Because miRNAs typically mediate gene silencing, they are traditionally thought as fine-tuners of gene expression. Consistent with this view, most published papers on cell fate switches used transcription factor based cocktails, rather than miRNAs [51]. Recently, however, several studies reported the potency of microRNAs in influencing neuronal cell fates [52,53]. In one study, miR-9/9* and miR-124 alone were shown to able to convert human fibroblasts towards neurons and more so with as few as one transcription factor [53]. Synergism between mi-9/9* and miR-124 is crucial for this conversion, since expression of individual miRNAs was insufficient to induce neurons. Further addition of neurogenic factors enhanced both the efficiency and the maturity of the reprogrammed neurons, while expression of these factors alone was far less ineffective at generating neurons. These results clearly demonstrated an instructive role of these miRNAs in induction of neuronal fates, and highlighted the cooperation between miRNAs and transcription factors in neuronal reprogramming. What are the crucial targets of the miRNAs that mediate this transdifferentiation? Yoo et al. attempted to address this question by persistently expressing some of the known targets of miR-9/9* and miR-124, including SCP1, PTBP1 and BAF53a during reprogramming. However, none of these targets could completely block neuronal conversion when overexpressed singly. These data suggested that miR-9/9* and miR-124 could act synergistically and programmatically on multiple targets to exert their neurogenic activities. Fig. 1 depicts a model of the neurogenic activity of these miRNAs, illustrating multiple negative pathways that eventually lead to repression of factors that oppose neuronal fate acquisition.
Figure 1.
A model of neuronal reprogramming mediated by miR-9/9*-124 and neurogenic factors. MiR-9/9* and miR-124 in non-neuronal cells are repressed synergistically by neurogenic repressors such as REST complex, non-neuronal BAF complexes and others. The top panel is an example of REST repressing neuronal genes such as miR-9/9* and miR-124. Forced expression of miR-9/9* and miR-124 results in the break of genetic network involving repression of multiple factors including REST complexes, BAF53a, and PTBP1, which normally suppress neurogenesis. The synergism of miRNAs suggests that these miRNAs work programmatically on multiple targets during the process of neuronal conversion. In addition, it is possible that miRNAs may target factors whose silencing potentiates the activity of neurogenic transcription factors (NFs) (bottom panel).
MicroRNAs in specific neuron subtype identity
A hallmark of mammalian nervous system is its extraordinary diversity of neuronal subtypes. Do miRNAs play any important roles in specification of neuronal subtypes? In a study using in vitro differentiation of mouse ES cells to dopaminergic neurons (DNs), miR-133b has been suggested to be play important role in DN formation and function by targeting Pitx3 [13]. However, genetic deletion of miR-133b in mouse does not affect DN formation nor function [54], a discrepancy that may be possibly explained by compensatory actions by other miR-133 family miRNAs. In addition, miR-7a has recently been shown to regulate Pax6 in adult neurogenesis and affect dopaminergic fate in the olfactory bulb [55].
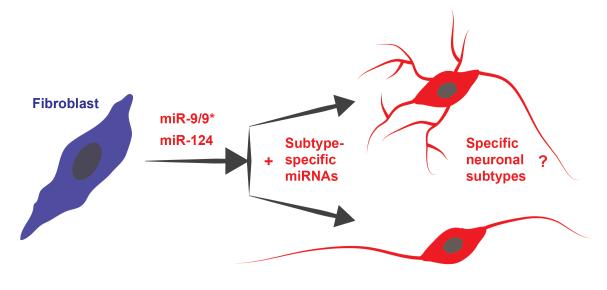
A significant advance in our understanding of neuron cell type specific miRNA expression came from a recent study using an ingenious miRNA tagging and affinity-purification (miRAP) method that is targeted to specific neuronal subtypes through the Cre-loxP technique in mice [56]. Using miRAP, the authors were able to identify a large number of miRNAs with distinct profiles in glutamatergic, gabaergic neurons and subtypes of gabaergic neurons, providing experimental support that the identity of neuronal subtypes could be related to differentially expressed microRNAs. Indeed, even between closely related interneurons types such as parvalumin (PV) neurons versus somatostatin expresssing (SST) neurons, more than half of the miRNAs profiled are differentially expressed. Interestingly, miR-124 and miR-9/9* were found to be highly expressed at relatively constant levels among all subtypes, consistent with their pan-neuronal neurogenic activities. Based on the potency of microRNAs in influencing cell fates, combining the neurogenic activity of miR-9/9* and miR-124 with subtype-specific microRNAs to reprogram into specific neuronal types remains an intriguing possibility to be tested (Fig. 2).
Figure 2.
A hypothetical scheme of production of subtype-specific neurons by reprogramming. Leveraging on the known pan-neuronal activity of miR-9/9* and miR-124, additional miRNAs specific to neuronal subtypes may lead to neuronal reprogramming into their respective neuronal subclasses.
Conclusions and Future Perspectives
MicroRNAs are emerging pivotal regulators of neurogenesis. In addition to being involved in neuronal differentiation, as reviewed here, miRNAs have also been shown to be play significant roles in other neural lineages such as oligodentrocytes [57,58]. Even within the neurons, increasing numbers of miRNAs are being identified to regulate various stages of neuronal development and function. Expression of miRNAs as well as the targets of miRNAs are dynamically regulated, both spatially and temporally, contributing to the diversity and plasticity of our brain. MiRNA profiling in distinctive neuronal populations have just started, and it would be exciting to see even more refined classification of miRNAs expression pattern with better cell isolation techniques. In addition, the link between miRNAs deregulation and neurological diseases are just beginning to be revealed and certainly merit further studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ebert M, Sharp P. Roles for microRNAs in conferring robustness to biological processes. Cell. 149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasquinelli A. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature reviews. Genetics. 13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 3.Tay Y, Zhang J, Thomson A, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 4.Yoo A, Staahl B, Chen L, Crabtree G. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. ** This paper described a series of elegantly designed BAC-transgenic reporter mouse embryos that demonstrated BAF53a to be simultaneously targeted by both miR-124 and miR-9*. Further experiments delineated a negative genetic circuit centered around these miRNAs that regulates neural progenitor mitotic exit and dendritic morphogenesis.
- 5.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 7.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola M, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, et al. MicroRNA profiling in human medulloblastoma. International journal of cancer. Journal international du cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 8.Shao N-Y, Hu H, Yan Z, Xu Y, Hu H, Menzel C, Li N, Chen W, Khaitovich P. Comprehensive survey of human brain microRNA by deep sequencing. BMC genomics. 11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Pietri Tonelli D, Pulvers J, Haffner C, Murchison E, Hannon G, Huttner W. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development (Cambridge, England) 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. Journal of cell science. 123:586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer A, O’Carroll Dn, Tan C, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. The Journal of experimental medicine. 2007;204:1553–1558. doi: 10.1084/jem.20070823. * This study examined the role of miRNAs in mature neurons. Genetic deletion of miRNAs in Purkinje cells resulted in locomotive impairment of animals before ataxia, as a result of Purkinje cell degeneration in the absence of miRNAs.
- 13.Kim J, Inoue K, Ishii J, Vanti W, Voronov S, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science (New York, N.Y.) 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser P, Wills A-M, Wirguin I, et al. miRNA malfunction causes spinal motor neuron disease. Proceedings of the National Academy of Sciences of the United States of America. 107:13111–13116. doi: 10.1073/pnas.1006151107. * To define the function of miRNAs in motor neurons, Haramati et al. generated a conditional KO mouse of miRNAs in spinal cord motor neurons. Mutant mice displayed many defining features of SMA, including sclerosis of the spinal cord ventral horns and myofiber atrophy. Notably, the authors were able to identify one important neural specific miRNA, miR-9, in motor neurons survival and physiology, which regulates intermediate neurofilament composition in neurons.
- 15.Babiarz J, Hsu R, Melton C, Thomas M, Ullian E, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA (New York, N.Y.) 17:1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Teng Z-Q, Santistevan N, Szulwach K, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell stem cell. 6:433–444. doi: 10.1016/j.stem.2010.02.017. * In MBD1 mutant mice miR-184 expression is elevated. Using both overexpression and knockdown techniques, the authors were able to show miR-184 as a functional mediator of MBD1 in regulating adult NSCs proliferation and differentiation in vitro and in vivo.
- 17.Schwamborn J, Berezikov E, Knoblich J. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. * This study demonstrated polarized distribution of TRIM32 in asymmetrically dividing neural progenitors and how this process influenced the cell fate of the daughter cells. The authors showed that through its NHL domain, TRIM32 binds to Ago1, and this binding enhanced the activity of selective miRNAs, including let-7a, which promotes neuronal differentiation.
- 18.Nishino J, Kim I, Chada K, Morrison S. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proceedings of the National Academy of Sciences. 107 doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, Yu R, Gage F, Evans R, Shi Y. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nature cell biology. 12:31. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun G, Ye P, Murai K, Lang M-F, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nature communications. 2:529. doi: 10.1038/ncomms1532. * This study defined a regulatory cascade involving TLX-miR-137-LSD1 which provides a critical control of neural stem cell proliferation, differentiation and migration.
- 22.Szulwach K, Li X, Smrt R, Li Y, Luo Y, Lin L, Santistevan N, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. The Journal of cell biology. 189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smrt R, Szulwach K, Pfeiffer R, Li X, Guo W, Pathania M, Teng Z-Q, Luo Y, Peng J, Bordey A, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem cells (Dayton, Ohio) 28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Packer A, Xing Y, Harper S, Jones L, Davidson B. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders L, Sharma A, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging. 2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 38:6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. The Journal of neuroscience : the official journal of the Society for Neuroscience. 31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonev B, Stanley P, Papalopulu N. MicroRNA-9 Modulates Hes1 Ultradian Oscillations by Forming a Double-Negative Feedback Loop. Cell reports. 2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nature neuroscience. doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature structural & molecular biology. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun K, Westholm J, Tsurudome K, Hagen J, Lu Y, Kohwi M, Betel D, Gao F-B, Haghighi A, Doe C, et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS genetics. 8 doi: 10.1371/journal.pgen.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng R, Cohen S. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development (Cambridge, England) 139:1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 36.Clark A, Goldstein L, Tevlin M, Tavaré S, Shaham S, Miska E. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic acids research. 38:3780–3793. doi: 10.1093/nar/gkq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature neuroscience. 2009;12:399–408. doi: 10.1038/nn.2294. * This report provided in vivo study of miR-124 function in adult SVZ. Careful analysis were done using both FACS-purified aSVZ cells or in vivo cells after either overexpression or knockdown via 2O’ME anstisense RNA, demonstrating the role of miR-124 in adult neurogensis.
- 38.Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. * Using a transgenic miR-124 sensor, the authors provided an additional means of evaluating its activity in vivo, and confirmed its neuronal expression pattern. Lentiviral manipulations of miR-124 levels allowed the authors to confirm the essential function of miR-124 in adult SVZ.
- 39.Liu J, Githinji J, McLaughlin B, Wilczek K, Nolta J. Role of miRNAs in Neuronal Differentiation from Human Embryonic Stem Cell-Derived Neural Stem Cells. Stem Cell Rev. doi: 10.1007/s12015-012-9411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim L, Lau N, Garrett-Engele P, Grimson A, Schelter J, Castle J, Bartel D, Linsley P, Johnson J. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 41.Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, et al. SOX9 induces and maintains neural stem cells. Nat Neurosci. 13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- 42.Arvanitis D, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Molecular and cellular biology. 30:2508–2517. doi: 10.1128/MCB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visvanathan J, Lee S, Lee B, Lee J, Lee S-K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes & development. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lessard J, Wu J, Ranish J, Wan M, Winslow M, Staahl B, Wu H, Aebersold R, Graef I, Crabtree G. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Lessard J, Olave I, Qiu Z, Ghosh A, Graef I, Crabtree G. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 47.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nature neuroscience. 14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 48.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil S, Russo J, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schratt G, Tuebing F, Nigh E, Kane C, Sabatini M, Kiebler M, Greenberg M. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 50.McNeill E, Van Vactor D. MicroRNAs Shape the Neuronal Landscape. Neuron. 75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nature biotechnology. 29:892–907. doi: 10.1038/nbt.1946. * This review summarized recent advances in reprogramming of cellular fates and provides a quick overview of the current status of this field.
- 52.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo A, Sun A, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch R, Tsien R, Crabtree G. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 476:228–231. doi: 10.1038/nature10323. ** Continuing with previous work involving miR-9* and miR-124, this study took a step further and showed that miR-9/9* and miR-124 were capable of inducing neuronal fates in non-neural cells. Highlighting the synergism between miRNAs, this study is the first to demonstrate the instructive role of miR-9/9*-124 in neuronal fate acquisition and implies miRNAs as important regulators of cellular identity.
- 54.Heyer M, Pani A, Smeyne R, Kenny P, Feng G. Normal Midbrain Dopaminergic Neuron Development and Function in miR-133b Mutant Mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 32:10887–10894. doi: 10.1523/JNEUROSCI.1732-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Chevigny A, Coré N, Follert P, Gaudin M, Barbry P, Béclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nature neuroscience. 15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- 56.He M, Liu Y, Wang X, Zhang M, Hannon G, Huang Z. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 73:35–48. doi: 10.1016/j.neuron.2011.11.010. ** Using a novel miRAP and Cre-LoxP system, this paper profiled miRNAs in different classes of neurons, and demonstrated that different neuronal subtypes expressed different miRNAs. This pioneering work suggests that miRNAs could be part of the signature defining the extraordinary diversity of neurons.
- 57.Dugas J, Cuellar T, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian J, Foo L, McManus M, Barres B. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi Q-S, Xin M, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]