Abstract
Ectopic expression of mutant forms of PTEN lacking lipid (G129E) or lipid and protein (C124S) phosphatase activity decreased sensitivity of MCF-7 breast cancer cells, which have wild-type PTEN, to doxorubicin and increased sensitivity to the mTOR inhibitor rapamycin. Cells transfected with a mutant PTEN gene lacking both lipid and protein phosphatase activity were more resistant to doxorubicin than cells transfected with the PTEN mutant lacking lipid phosphatase activity indicating that the protein phosphatase activity of PTEN was also important in controlling the sensitivity to doxorubicin, while no difference was observed between the lipid (G129E) and lipid and protein (C124S) phosphatase PTEN mutants in terms of sensitivity to rapamycin. A synergistic inhibitory interaction was observed when doxorubicin was combined with rapamycin in the phosphatase-deficient PTEN-transfected cells. Interference with the lipid phosphatase activity of PTEN was sufficient to activate Akt/mTOR/p70S6K signaling. These studies indicate that disruption of the normal activity of the PTEN phosphatase can have dramatic effects on the therapeutic sensitivity of breast cancer cells. Mutations in the key residues which control PTEN lipid and protein phosphatase may act as dominant negative mutants to suppress endogenous PTEN and alter the sensitivity of breast cancer patients to chemo- and targeted therapies.
Keywords: PTEN, Akt, mTOR, Rapamycin, Chemotherapeutic Drugs
Introduction
Signal transduction cascades downstream of epidermal growth factor (EGF) receptor (EGFR) isoforms have been associated with breast cancer development and resistance to anticancer agents (Shelton et al, 2005). Among the signaling pathways downstream of the EGFR, the phosphatidylinositol 3 kinase (PI3K)/Akt pathway has been shown to regulate apoptosis and its deregulation is often implicated in malignant transformation (Luo et al, 2005; Martelli et al, 2006).
Phosphatidylinositol (PI) (3,4)P2 and PI(3,4,5)P3 produced by class 1A PI3Ks recruit phosphoinositide dependent kinase-1 (PDK1) as well as Akt isoforms to the plasma membrane by interacting with their pleckstrin homology (PH) domains (Franke et al, 1997; Frech et al, 1997; Martelli et al, 2007). Colocalization of PDK1 with Akts at the plasma membrane causes PDK1 to phosphorylate Akts at a threonine residue (T308) (Alessi et al, 1997; Stokoe et al, 1997) and a serine residue (S473). Activation of PDK1 and Akt by class 1A PI3Ks is negatively regulated by phosphatase and tensin homologue deleted on chromosome ten (PTEN) (Martelli et al, 2007; Steelman et al, 2004). PTEN removes phosphate groups from PI(3,4)P2 and PI(3,4,5)P3 added by PI3K as well as from tyrosine phosphorylated proteins including focal adhesion kinase (FAK) and Shc (Tamura et al, 1998; Machama et al, 1998; Nimwegen et al, 2006).
Amino acid substitutions within the phosphatase domain have been identified that affect PTEN activity. PTEN(C124S) lacks both lipid and protein phosphatase activities (Leslie et al, 2000). PTEN(G129E) lacks lipid phosphatase activity yet retains protein phosphatase activity. PTEN(399stop) lacks the PDZ domain binding motif. Cowden’s syndrome (CS) patients have germline PTEN mutations (Lynch et al, 1997; Steelman et al, 2004). These patients often have a high incidence (25–50%) of breast cancer. PTEN(C124S) and (G129E) mutations were originally identified in CS patients and have been postulated to function as naturally occurring dominant negative (DN) mutations (Marsh et al, 1998).
Diverse mechanisms regulate PTEN expression (Abdel-Rahman et al, 2006; Lee et al, 2004; Tsutsui et al, 2005; Agrawal et al, 2006). These range from gene deletion, alterations in mRNA splicing, subcellular localization or epigenetic mechanisms which prevent PTEN transcription. Mutations have been reported to occur at PTEN in breast cancer at varying frequencies (5–21%). While PTEN is deleted in certain cancers, loss of heterozygosity (LOH) is probably a more common genetic event (30%) leading to changes in PTEN expression (Singh et al, 1998; Eng, 2003). PTEN promoter methylation leads to low PTEN expression (Garcia et al, 2004). In one study, 26% of primary breast cancers had low PTEN levels which correlated with lymph node metastases and poor prognoses (Tsutsui et al, 2005). PTEN has both plasma membrane and nuclear localized activities. Disruption of PTEN activity by various mechanisms could have vast effects on different processes affecting breast cancer drug resistance.
A consequence of impaired PTEN expression is elevated activation of Akt. One downstream molecule of mTOR is ribosomal S6 kinase (p70S6K). This kinase regulates the efficiency of translation of certain mRNAs and also functions in a negative feedback loop to control Akt activity (Chiang et al, 2005; Inoki et al, 2005; Martelli et al, 2007). Akt, mTOR and p70S6K activation have been associated with a more severe prognosis in breast and other cancers (Bose et al, 2002; Clark et al., 2002; Tsutsui et al, 2005) (Martelli et al, 2007; Tokunaga et al, 2006; DeGraffenried et al, 2004; Nagata et al, 2004; Lin et al, 2005; Kios et al, 2006; Tokunaga et al., 2006). Targeting the PI3K/Akt/mTOR pathway may prove effective in various cancer therapies (Martelli et al, 2007). High levels of activated Akt expression have been associated with both chemo- and hormonal resistance in breast cancer (Clark et al, 2002; Tokunaga et al, 2006). Indeed some studies have evaluated the effectiveness of targeting mTOR in PTEN negative cells (DeGraffenried et al, 2004). Cells which express high levels of activated Akt may be more sensitive to mTOR inhibitors and inhibition of mTOR activity by rapamycin may restore their sensitivity to chemo- and hormonal based therapies (DeGraffenried et al, 2004).
In the following studies, the effects of ectopic expression of PTEN mutants lacking either lipid phosphatase or lipid and protein phosphatase activity, on the sensitivity of breast cancer cells to doxorubicin and the mTOR inhibitor rapamycin were determined. Our studies demonstrate for the first time that the protein as well as the lipid phosphatase activity of PTEN is important in determining the sensitivity of breast cancer cells to chemotherapeutic drugs such as doxorubicin and document a synergistic interaction between doxorubicin and mTOR inhibitors in inducing death of breast cancer cells harboring mutant PTEN genes.
Results
Ectopic PTEN Expression Suppresses Colony Formation of MCF-7 Cells
The activity of the PI3K/PTEN/Akt/mTOR cascade was manipulated in MCF-7 cells in order to determine how signals transduced by this pathway may control the sensitivity of breast cancer cells to various therapies. MCF-7 cells were transfected with plasmids encoding PTEN(WT), PTEN(C124S), PTEN(G129E), PTEN(399stop) or the empty vector pEGFP-C2 (Leslie et al, 2000).
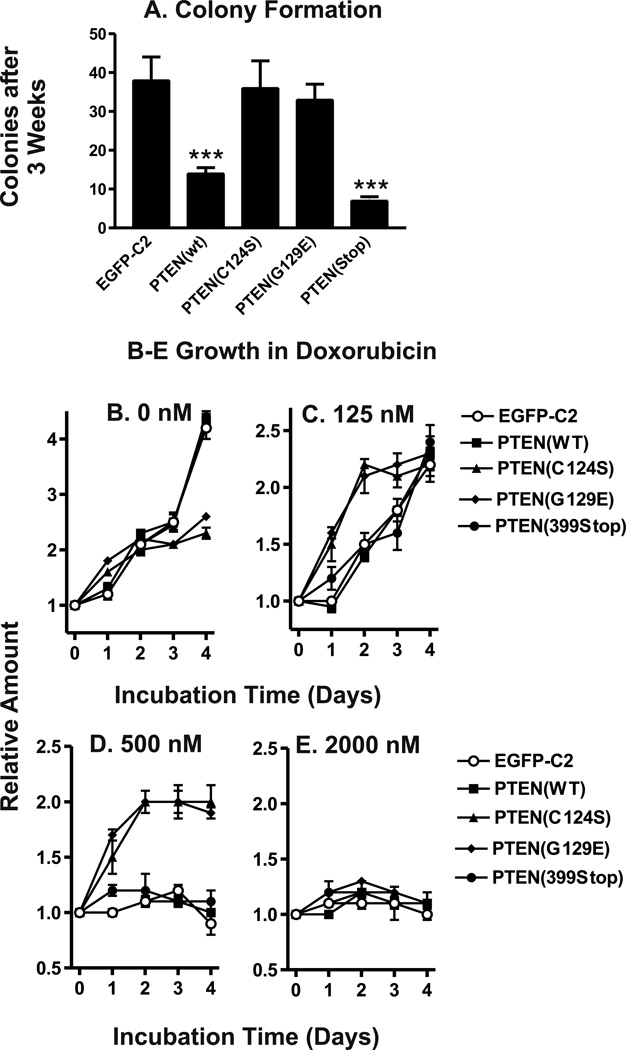
The mean number of colonies which arose after transfection of MCF-7 cells with the various PTEN constructs followed by G418 selection was calculated from nine replicate wells (Figure 1, Panel A). Transfection with pEGFP-C2 yielded 38 ± 12 colonies per well. Transfection with PTEN(C124S) yielded 36 ± 14 colonies per well and transfection with PTEN(G129E) yielded 33 ± 8 colonies per well. Differences between the number of colonies present after EGFP-C2 transfection and the number of colonies present after PTEN(C124S) or PTEN(G129E) transfection were not statistically significant (P>0.10). However, statistically significant decreases in the number of colonies present were evident after transfection with PTEN(WT) or PTEN(399stop) in comparison to transfection with pEGFP-C2 (P<0.001). Transfection with PTEN(WT) yielded 14 ± 3 colonies per well and transfection with pPTEN(399stop) yielded 7 ± 2 colonies per well. These results indicated that inheritance of an additional WT PTEN gene or the PTEN(399stop) genes decreased the number of colonies obtained compared to cells transfected with an empty vector or either a PTEN mutant lacking lipid and protein or just lipid phosphatase activity.
Figure 1. Effects of WT and Mutant PTEN Genes on Colony Formation and Proliferation.
Panel A) MCF-7 cells were transfected with plasmid DNA encoding PTEN(WT), PTEN(C124S), PTEN(G129E), PTEN(399stop) or the empty vector pEGFP-C2. The mean number colonies present after selection in 2 mg/ml G418 for approximately 3 weeks and corresponding standard deviation was calculated from 9 replicate wells for each plasmid. Asterisks indicate statistically significant decreases in the number of colonies present in comparison to pEGFP-C2 transfected cells (***, P<0.001). Panels B–E) Proliferation of: MCF/EGFP-C2 cells (open circles, ○) MCF/PTEN(WT) (solid squares, ■), MCF/PTEN(C124S) (solid triangles, ▲), MCF/PTEN(G129E) (solid diamonds, ◆), and MCF/PTEN(399stop) (solid circles, ●) in the presence of 0 nM (Panel B) 125 nM doxorubicin (Panel C), 500 nM doxorubicin (Panel D) or 2,000 nM doxorubicin. The amount of cells present after incubation in these conditions for 1, 2, 3, or 4 days was normalized to the mean amount of cells present at the time of treatment.
Proliferation Analysis of PTEN-Transfected MCF-7 cells
Proliferation rates of MCF-7 cells which inherited the PTEN mutants that lacked lipid phosphatase activity [PTEN(C124S) or PTEN(G129E)] were different than the MCF-7 cells which inherited the PTEN(WT), PTEN(399stop) or the empty vector (pEGFP-C2). However, differences were not observed in these proliferation assays between the lipid phosphatase-deficient (G129E) and lipid and protein phosphatase-deficient (C124S) transfected cells. In the absence of doxorubicin, the levels of proliferation of the MCF-7 cells transfected with the phosphatase-deficient PTEN(C124S) or PTEN(G129E) did not reach as high levels as the cells transfected with PTEN(WT), PTEN(399stop) or the empty vector (Panel B). While inheritance of PTEN(WT) or PTEN(399Stop) suppressed initial colony formation, they did not appear to affect the growth rates of stably transfected cells.
The proliferation rates of PTEN(WT), PTEN(C124S), PTEN(G129E), and PTEN(399stop) cells were compared with that of pEGFP-C2 transfected cells, in the presence of 125 nM doxorubicin (Panel C). The relative growth rates of PTEN(WT) and PTEN(399stop) transfected cells were also similar to that of pEGFP-C2 cells in presence of 125 nM doxorubicin. In contrast, the proliferation rates of PTEN(C124S) and PTEN(G129E) transfected cells differed from that of pEGFP-C2 cells both in the absence of doxorubicin or in the presence of 125 nM doxorubicin. PTEN(C124S) and PTEN(G129E) transfected cells proliferated at a slower rate than pEGFP-C2 cells in the absence of doxorubicin. However, PTEN(C124S) (Panel D) and PTEN(G129E) cells proliferated at a faster rate than pEGFP-C2 cells in the presence of 125 nM doxorubicin. Interesting, all cell lines reached similar levels after 4 days of culture in 125 nM doxorubicin. The difference in proliferation rates in the presence of 125 nM doxorubicin was most evident during the first 3 days of treatment. The observation that PTEN(C124S) and PTEN(G129E) transfected cells proliferated slower than pEGFP-C2 cells in the absence of doxorubicin yet proliferated faster than these cells in the presence of 125 nM doxorubicin demonstrated some of the effects of inheritance of the phosphatase-deficient PTEN genes on proliferation and will be discussed below.
The most significant differences in proliferation were observed when the different transfected cells were cultured in medium containing 500 nM doxorubicin (Panel D). Cells transfected with PTEN(WT) or PTEN(399Stop) did not proliferate in medium containing 500 nM doxorubicin. In contrast, the cells transfected with the PTEN phosphatase-deficient mutants proliferated in the presence of 500 nM doxorubicin. None of the PTEN or empty vector transfected cells proliferated in the presence of 2000 nM doxorubicin (Panel E).
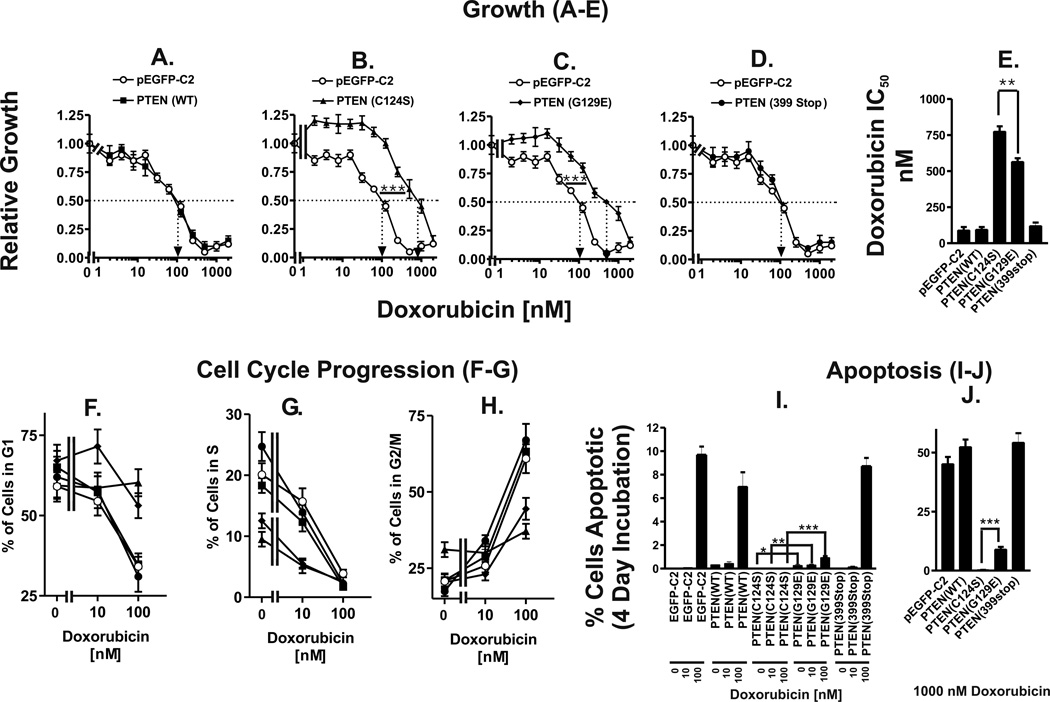
The relative growth rates of PTEN(WT), PTEN(C124S), PTEN(G129E), and PTEN(399stop) transfected cells at each doxorubicin concentration were compared with that of pEGFP-C2 transfected cells (Figure 2). The growth of PTEN(WT) (Panel A) and PTEN(399stop) (Panel D) cells were similar to that of pEGFP-C2 cells for all doxorubicin concentrations tested. In contrast, the relative growth rates of PTEN(C124S) (Panel B) and PTEN(G129E) (Panel C) were higher than that of pEGFP-C2 cells for all doxorubicin concentrations tested with the exception of 2000 nM.
Figure 2. Effects of Mutant PTEN Genes on Sensitivity to Doxorubicin.
MTT was utilized to monitor proliferation of: Panel A) MCF/PTEN(WT) (solid squares, ■), Panel B) MCF/PTEN(C124S) (solid triangles, ▲), Panel C) MCF/PTEN(G129E) (solid diamonds, ◆), Panel D) MCF/PTEN(399stop) (solid circles, ●). The sensitivity of MCF/EGFP-C2 cells to doxorubicin (open circles, ○) is presented in each panel for comparison purposes. Relative growth rates were calculated by dividing the increase in the amount of cells after 4 days of treatment with each doxorubicin concentration by the increase in the amount of cells after 4 days of proliferation in the absence of doxorubicin. The mean relative growth rate and corresponding standard deviation for each condition was calculated from absorbance values of 8 replicate wells. The dotted arrow represents where 50% inhibition of growth intercepts with the X axis and is used to estimate the IC50. The statistical significance was determined by the unpaired t test (***, P<0.001). Panel E) doxorubicin IC50 values of PTEN transfected MCF-7 cells. The statistical significance was determined by the unpaired t test (**, P<0.005). Panels F, G & H) Cell cycle analysis of doxorubicin treated PTEN-transfected cells. Panels I & J) effects of PTEN mutants on sensitivity of MCF-7 cells to induction of apoptosis induced by doxorubicin. MCF-7 cells transfected with the different PTEN constructs were exposed to the indicated concentrations of doxorubicin for 4 days. The cells were then collected and the extent of apoptosis estimated by determining the percentage of subgenomic cells after propidium iodine staining following by flow cytometric analysis. The percentage of subgenomic cells was quantified by the ModFit program. The percentage of apoptotic MCF/PTEN(C124S) and MCF/PTEN(G129E) cells, treated with the same concentrations of doxorubicin, were statistical analyzed by the unpaired t test (***, P<0.001, **, P<0.005, *, P<0.05).
Doxorubicin IC50 values were calculated for pEGFP-C2, PTEN(WT), PTEN(C124S), PTEN(G129E), and PTEN(399stop) transfected cells to serve as benchmarks of doxorubicin sensitivity (Panel E). pEGFP-C2 transfected cells had a doxorubicin IC50 value of approximately 90 nM. Doxorubicin IC50 values of PTEN(WT) and PTEN(399stop) cells were similar to that of pEGFP-C2 cells. In contrast, doxorubicin IC50 values of PTEN(C124S) and PTEN(G129E) were several times higher (8.6- and 6.3-fold respectively) than that of pEGFP-C2 cells. PTEN(C124S) and PTEN(G129E) cells had doxorubicin IC50 values of approximately 775 nM and 565 nM, respectively. MCF-7 cells transfected with the PTEN mutant which lacked both lipid and protein phosphatase activity [PTEN(C124S)] had a 1.4-fold higher doxorubicin IC50 than MCF-7 cells transfected with the PTEN mutant which lacked only the lipid phosphatase activity [PTEN(G129E)]. This was determined to be statistically significant by the unpaired t test.
The effects of the PTEN mutants on cell cycle progression were also examined (Panels F-H). The effects of the phosphatase mutants on S phase were very evident, as less cells were in S phase in the PTEN(C124S) and PTEN(G129E) cells than in the pEGFP-c2, PTEN(WT) or PTEN(399stop) transfected cells (Panel G). After incubation with 100 nM doxorubicin for 4 days, pEGFP-c2, PTEN(WT) or PTEN(399stop) transfected cells were blocked in G2/M (Panel H), whereas PTEN(C124S) and PTEN(G129E) transfected cells still had some cells present in G1 documenting the effects that the PTEN phosphatase mutants had on cell cycle progression in the presence of doxorubicin (Panel F).
To determine whether the mutant PTEN genes affected the chemosensitivity of the cells by altering the induction of apoptosis, the effects of the various PTEN mutants on the suppression of apoptosis were determined (Panels I & J). MCF-7 cells transfected with WT PTEN, the various PTEN mutants or the empty pEGFP-c2 vector were treated with 0, 10 and 100 nM (Panel I) and 1000 nM (Panel J) doxorubicin and then the extent of apoptosis measured by determining the percentage of sub-G1 cells after PI staining followed by flow cytometric analysis. 100 nM doxorubicin induced between 7 to 12 % apoptotic cells in the empty vector, PTEN(WT) and PTEN(399Stop) transfected cells. In contrast, less apoptotic cells were observed in MCF-7 cells transfected with the PTEN mutants lacking lipid or lipid [PTEN(G129E)] and protein phosphatase [PTEN(C124S)] activity. The PTEN mutant (C124S) which lacked both lipid and protein phosphatase activity suppressed more apoptosis than the PTEN construct (G129E) which lacked only lipid phosphatase activity. Namely, there was approximately 0, 0, 0 and 0.3% apoptosis in the PTEN(C124S) transfected cells in response to 0, 10, 100 & 1000 nM doxorubicin while there was approximately 0.27, 0.32, 0.95 and 9 % apoptosis in the PTEN(G129E) transfected cells at the respective doxorubicin concentrations. This corresponded to at least 2.7, 3.2, 9.5 and 30-fold differences in the induction of apoptosis between these two different PTEN deficient mutants at the various doxorubicin concentrations. Thus MCF-7 cells transfected with the PTEN mutants which altered its phosphatase activity were more resistant to the induction of apoptosis induced by doxorubicin.
Effects of PTEN Mutants on Colony Formation and Induction of Cellular Senescence
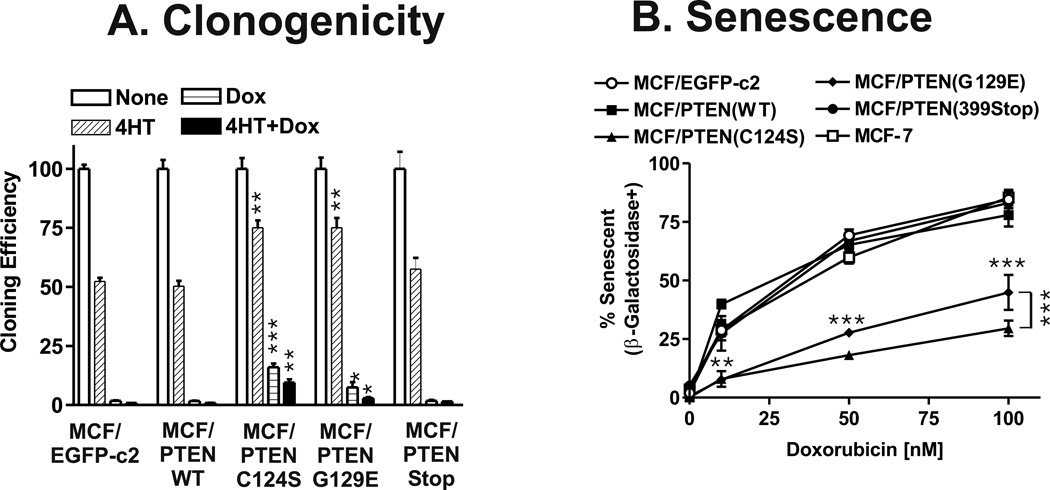
The number of colonies obtained with the different stably selected pools, after approximately one month in culture, in medium containing 4HT, doxorubicin, or 4HT + doxorubicin was normalized to the number of colonies observed in the absence of 4HT and doxorubicin for each type of PTEN-transfected cells (Figure 3, Panel A). The phosphatase-deficient PTEN mutants had higher cloning efficiencies in medium containing either 4HT (1.5-fold) or doxorubicin (approximately 8.8- and 4.2-fold for PTEN(C124S) and PTEN(G129E) than the PTEN (WT), PTEN(399Stop) or pEGFP-C2 transfected cells). Again more colonies (approximately 2.1-fold) in doxorubicin were obtained from cells transfected with PTEN mutants lacking the lipid and protein than the lipid phosphatase activities. Statistically significant higher cloning efficiencies were also observed when cells transfected with either the lipid or the lipid and protein phosphatase-deficient PTEN mutants when they were plated in medium containing 4HT and doxorubicin compared with the empty vector, PTEN(WT) or PTEN(399stop) transfected cells (approximately 3.5- and 10.5-fold respectively). Thus the presence of either the lipid or lipid and protein phosphatase-deficient mutant increased colony formation in medium containing doxorubicin, 4HT or the combination of 4HT + doxorubicin. Again the PTEN mutant (C124S) which eliminated both lipid and protein phosphatase activity yielded approximately 3.3-fold more clones in the presence of 4HT + doxorubicin than the PTEN mutant (G129E) which just eliminated lipid phosphatase activity.
Figure 3. Effects of PTEN Mutants on Colony Formation and Cellular Senescence.
Panel A) Colony formation of PTEN transfected cells in 4HT and doxorubicin. MCF-7 cells transfected with the various PTEN plasmids were plated in triplicate in 6 well plates. The following day, the cells were exposed to 100 nM doxorubicin, 100 nM 4HT or 100 nM doxorubicin + 100 nM 4HT or no supplement. The medium was changed every three days. After 3 weeks the wells were stained with Giemsa and the number of colonies determined. The percentage colony formation was normalized to the number of colonies observed in cells not treated with 4HT, doxorubicin or 4HT + doxorubicin. The cloning efficiencies of MCF/PTEN(C124S) and MCF/PTEN(G129E) at the indicated culture conditions were statistically compared with the cloning efficiency of MCF/EGFP-c2 cells (***, P<0.001, **, P<0.005, *, P<0.05). Panel B) Effects of PTEN mutants on induction of senescence. Induction of cellular senescence was examined in the PTEN transfected cells as well as control MCF-7 and MCF/EGFP-c2 cells. The percent senescent vs. non senescent cells was determined in at least 6 fields per indicated culture condition and averaged together and the statistical significance determined by the student’s t test. The differences in induction of senescence between MCF/PTEN(C124S) and MCF/PTEN(G129E) and MCF/EGFP-c2 were statistically compared and are indicated by horizontal asterisks. The difference in induction of senescence between MCF/PTEN(C124S) and MCF/PTEN(G129E) in 100 nM doxorubicin was statistically compared and is indicated by vertical asterisks to the right of the 100 nM data points (***, P<0.001, **, P<0.005).
When certain cells are treated with chemotherapeutic drugs, they undergo cellular senescence and express senescence associated (SA) β-galactosidase (SA-β-gal) (Lehmann et al, 2007). The effects of the PTEN mutants on the induction of cellular senescence after doxorubicin treatment were examined (Figure 3, Panel B). In the absence of doxorubicin, very little senescence was observed after a four day incubation period and essentially no differences were observed between the different PTEN-transfected cells. In contrast in the MCF-7 cells transfected with the PTEN mutations resulting in loss of lipid (G129E) or lipid and protein (C124S) phosphatase activity, increased resistance to chemotherapeutic drug-induced cellular senescence was observed. MCF-7 cells transfected with PTEN(G129E) or PTEN(C124S) mutants yielded statistically significantly fewer senescent cells after exposure to doxorubicin than cells transfected with PTEN(WT), PTEN(399Stop) or the empty vector EGFP-c2 (Panel B). The C124S mutation deactivates both the lipid and protein phosphatase activity of PTEN, and conferred the strongest resistance to drug induced senescence.
Effects of Altered PTEN Expression on Akt, Ask-1 and p70S6K Phosphorylation
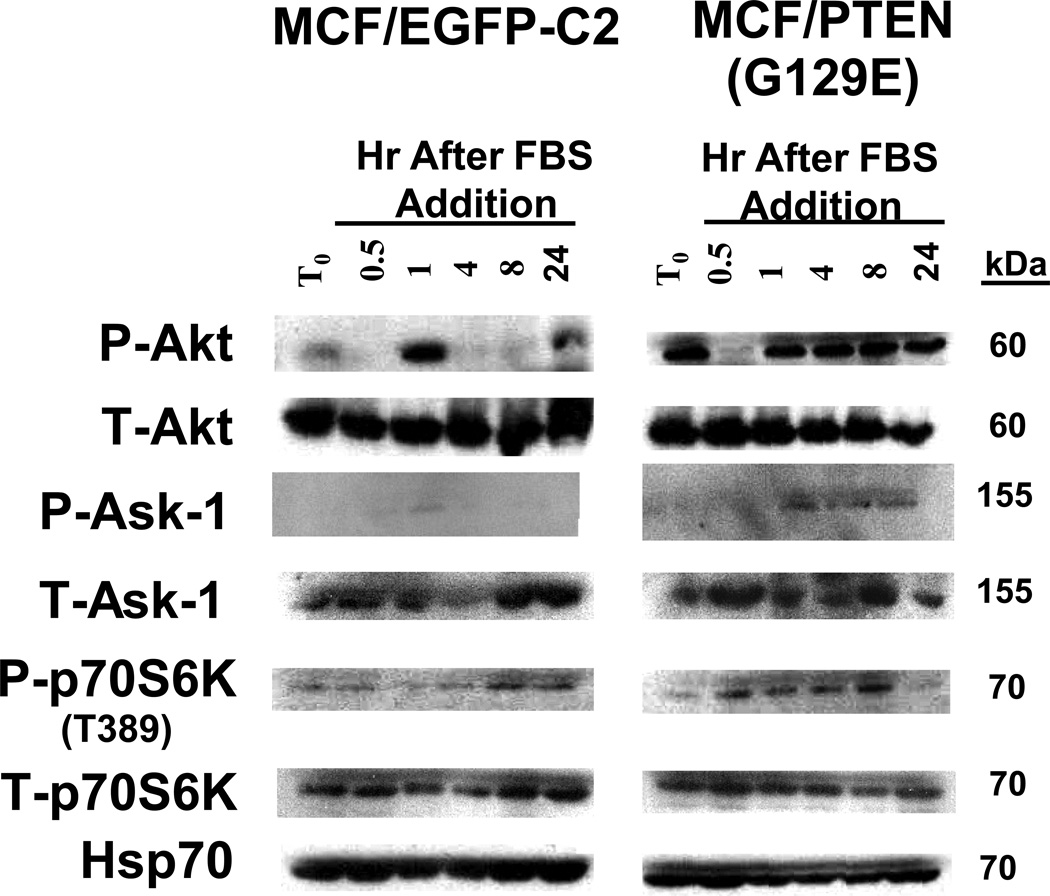
The effects of inheritance of the lipid phosphatase-deficient PTEN gene on phosphorylation of Akt, Ask-1, and p70S6K were compared in EGFP-C2 and PTEN(G129E) transfected cells by western blot analysis (Figure 4). Akt activation was examined as decreased PTEN expression could result in Akt activation. Apoptotic signal kinase-1 (Ask-1) and p70S6K phosphorylation were also examined as they lie downstream of Akt.
Figure 4. Effects of PTEN Expression on Akt, Ask-1 and p70S6K Phosphorylation.
Cells were collected after trypsinization and plated in RPMI 1640 containing 10% FBS. After allowing the cells to adhere for 24 hrs, the monolayers were washed twice with PBS and then phenol red free RPMI 1640 containing 0.5% CS FBS was added for 24 hrs. Cells were then stimulated with 10% FBS for the indicated time periods. Protein lysates were prepared and subjected to western blot analysis.
Upon serum stimulation of serum-deprived pEGFP-C2 cells, activated Akt was detected at higher levels after 1 hr than after ½ hr. Levels of activated Akt decreased after 1 hr of treatment and increased after 24 hr of serum addition. In contrast, the levels of total Akt protein remained constant. Thus serum treatment, led to a transient decrease then a transient increase in phosphorylated Akt levels in pEGFP-C2 cells. Phosphorylated Akt was detected in the serum deprived PTEN(G129C) cells at the T0 time point. Similar to the results observed with empty vector transfected MCF-7 cells, when PTEN(G129E) transfected cells were stimulated with serum for ½ hr, the level of phosphorylated Akt decreased. After 1 hr of serum stimulation, the levels of phosphorylated Akt increased and in contrast with the results observed in empty vector transfected cells, remained constant.
Higher levels of phosphorylated Ask-1 were detected in the PTEN(G129E) than in the EGFP-C2 cells, consistent with the higher levels of activated Akt in these cells. The phosphorylated and ubiquitinated Ask-1 may contribute to the apoptotic resistance of the PTEN(G129E) cells.
The levels of phosphorylated p70S6K at the mTOR-regulated site T389 increased in pEGFP-C2 cells 8 and 24 hrs after serum addition. In contrast, relatively equal levels of total p70S6K were detected in pEGFP-C2 cells. The levels of phosphorylated p70S6K were low after serum deprivation of PTEN(G129E) cells but increased after 0.5 hr and remained elevated when the cells were stimulated with FBS. A more rapid increase in activated p70S6K was observed in the cells transfected with PTEN(G129E) than in the cells transfected with the empty pEGFP-C2 vector. Elevated levels of activated Akt and p70S6K were also detected in similar experiments with PTEN(C124S) cells (data not presented).
As a control, the levels of Hsp70 were examined. Relatively constant levels of Hsp70 were detected in both pEGFP-C2 and PTEN(G129E) cells over these time course experiments. In summary, the cells transfected with the PTEN(G129E) mutant, which lacked lipid phosphatase activity, expressed higher and prolonged levels of activated Akt and p70S6K than the cells transfected with the empty vector. Disruption of the lipid phosphatase PTEN activity by the G129E mutant can lead to alterations in Akt, Ask-1 and p70S6K phosphorylation.
Sensitivity of Mutant PTEN Transfected Cells to the mTOR Inhibitor Rapamycin
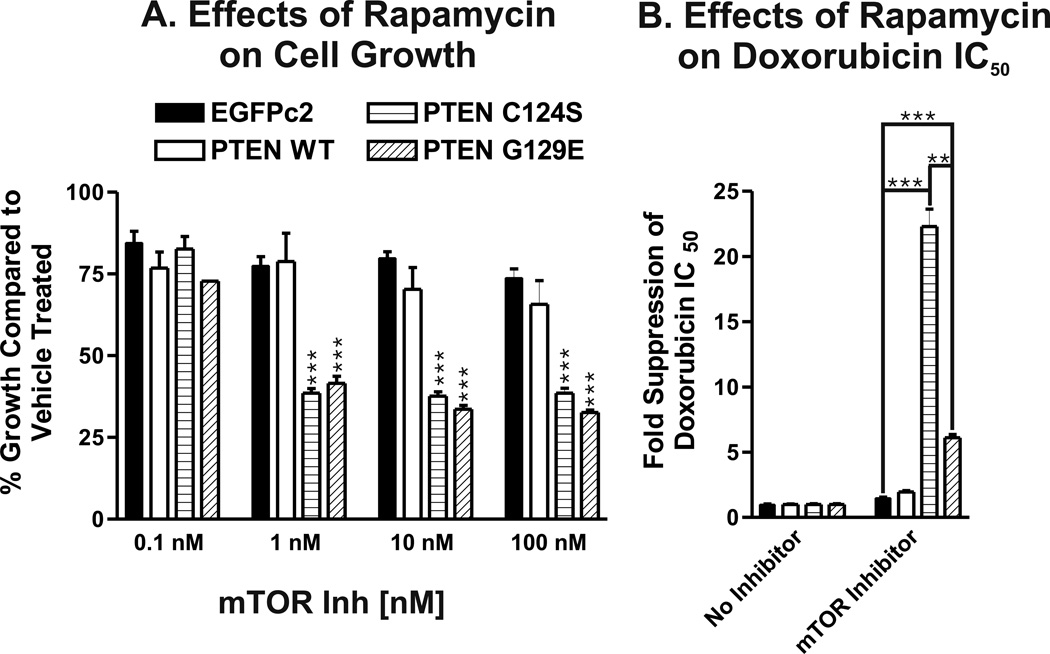
Altering Akt or PTEN levels changes the sensitivity of breast cancer cells to mTOR inhibitor rapamycin (DeGraffenried et al, 2004). The sensitivity of the different PTEN transfected MCF-7 cells to rapamycin was examined (Figure 5). The cells transfected with the lipid or lipid and protein phosphatase deficient PTEN (C124S and G129E) genes were more sensitive to rapamycin than the cells transfected with WT or the empty EGFPc2 vector (Panel A). The effects of rapamycin were observed over a broad dose range (1 to 100 nM).
Figure 5. Effects of the mTOR Inhibitor Rapamycin on the Growth of PTEN Transfected Cells.
Panel A) The effects of rapamycin on the growth of PTEN transfected cells were examined by MTT analysis. Panel B). The effects of a constant amount of rapamycin (1 nM) on the doxorubicin IC50s were examined. Upon treatment with rapamycin, the combined doxorubicin IC50s were approximately 60, 47.5, 34.8 and 92.6 nM for the EGFP-c2, PTEN(WT), PTEN(C124S) and PTEN(G129E) cells, whereas in the absence of rapamycin they were approximately 90, 95, 775 and 565 nM respectively. Again, the doxorubicin IC50 in the PTEN(C124S), which lacks both lipid and protein phosphatase activity was affected to the greatest level. The differences in doxorubicin IC50 after treatment with 1 nM between MCF/PTEN(C124S), MCF/PTEN(G129E) and MCF/EGFP-c2 were statistically compared and are indicated by asterisks (***, P<0.001). The differences in doxorubicin IC50 after treatment with 1 nM between MCF/PTEN(C124S) and MCF/PTEN(G129E) were statistically compared and are indicated by asterisks (**, P<0.005). Rapamycin was determined to synergize with doxorubicin by the CalcuSyn program with combination indexes less than one. Rapamycin suppressed the detection of activated p70S6K (T389) in these cells by western blot analysis (data not presented).
The effects of combining chemo- and targeted therapies were examined (Panel B). Rapamycin also suppressed (decreased) the IC50 for doxorubicin in the cells transfected with the PTEN mutant which lacked both lipid and protein phosphatase activity (C124S) more than the PTEN transfected cells (G129E) which lacked just lipid phosphatase activity (Panel B). These and other experiments indicated a synergistic interaction occurred between doxorubicin and rapamycin in suppressing the doxorubicin IC50. While the lipid or lipid and phosphatase deficient PTEN transfected cells have a similar sensitivity to treatment with rapamycin by itself, they have a differential sensitivity to the combination of doxorubicin and rapamycin. Thus the protein phosphatase activity of PTEN appears to be more important in determining the sensitivity of the cells to doxorubicin than to rapamycin.
Discussion
In this manuscript, the effects of introduction of mutant and WT PTEN genes on the clonogenicity and sensitivity of MCF-7 breast cancer cells to doxorubicin, 4HT and rapamycin were examined. Introduction of additional copies of WT PTEN or the PDZ-defective PTEN(399Stop) mutant genes into PTEN-positive MCF-7 cells, inhibited initial colony formation in medium lacking doxorubicin when compared to MCF-7 cells transfected with PTEN(C124S), PTEN(G129E) or the control pEGFPc2 plasmid. While the PTEN(WT) or the PTEN mutant with a deleted PDZ domain [PTEN(399Stop)], reduced the initial colony forming ability, they did not alter the growth rate or sensitivity to either doxorubicin or rapamycin when stably transfected pools of cells were expanded and examined. The two different assays differed in the time frames of the experiments; the growth assays were performed over a 5 day time period on pools of stably transfected cells while the initial colony formation assays were performed over a 3 to 4 week period. In these studies, the PTEN(399Stop) mutant behaved similarly to PTEN(WT).
Interfering with the lipid phosphatase activity of PTEN also resulted in higher levels of activated Akt detected in the T0 sample and prolonged detection of activated Akt after FBS stimulation. Akt has been reported to slow the growth rate of certain cells as well as increase the cell size (Jin et al, 2005). We have shown that these phosphatase deficient PTEN mutants also decreased the sensitivity of MCF-7 cells to doxorubicin and increased their sensitivity to rapamycin. The elevated activated Akt present in the PTEN(C124S) and PTEN(G129E) cells may effect cyclin proteolysis by decreasing GSK-3β activity. This may alter cell cycle progression and growth both in the presence and absence of doxorubicin. This may be important clinically as cells with mutant PTEN/Akt will respond differently to various therapeutic approaches.
Importantly inheritance of a PTEN mutant lacking both lipid and protein phosphatase activity conferred more resistance to doxorubicin than inheritance of the PTEN mutant lacking lipid phosphatase activity, indicating that interfering with both lipid and protein phosphatase activities of PTEN were important in terms of chemotherapeutic resistance. While both lipid (G129E) and lipid and protein (C124S) phosphatase-deficient PTEN mutants were more sensitive to rapamycin than WT PTEN transfected cells, there did not appear to be a significant difference in the sensitivity of the lipid (G129E) and lipid and protein (C124S) phosphatase-deficient mutant transfected cells to rapamycin. However, rapamycin exerted a greater degree of inhibition of the doxorubicin IC50 in cells transfected with the lipid and protein phosphatase deficient PTEN gene than in the cells transfected with the lipid phosphatase PTEN mutant. The protein phosphatase activity of PTEN may be important in the regulation of PTEN itself which alters its sensitivity to doxorubicin.
MCF-7 cells transfected with a mutant PTEN gene lacking lipid phosphatase resulted in an increased level of activated Akt and earlier p70S6K activation compared to empty vector transfected MCF-7 cells indicating that interfering with the lipid phosphatase activity was sufficient to alter the induction of Akt and downstream p70S6K. It is likely that other proteins besides Akt and p70S6K are important in terms of sensitivity to doxorubicin, as PTEN(C124S) conferred more chemoresistance than PTEN(G129E). Identification of these PTEN substrates may provide additional targets to enhance current therapeutic approaches to treat breast cancer.
PTEN activity is critically involved in regulation of downstream Akt activity which is important in chemo- and hormonal drug sensitivity. Altering normal PTEN activity by some PTEN mutations may lead to drug resistance. Augmented Akt activity may increase resistance to doxorubicin. In fact, published reports (Clark et al, 2002) and our own preliminary data indicate that ectopic Akt expression increases the resistance of MCF-7 breast cancer cells to doxorubicin and tamoxifen.
The mechanisms by which doxorubicin and rapamycin synergize to inhibit the growth of breast cancer cells are under further investigation. Doxorubicin can induce reactive oxygen species (ROS) which have many effects on cell growth and the prevention of apoptosis. A consequence of doxorubicin treatment may be the induction of the PI3K/Akt/mTOR pathway which may have some anti-apoptotic effects. Indeed we have observed activation of Akt after doxorubicin treatment of MCF-7 cells. Treatment with rapamycin may inhibit the anti-apoptotic consequences of doxorubicin treatment. Hence combining doxorubicin and mTOR inhibitors such as rapamycin may be an effective means to treat certain breast cancers. This synergy may be elevated in MCF/PTEN(C124S) and MCF/PTEN(G129E) cells as in these cells with mutant PTEN phosphatase activity there is increased activated Akt present.
These results are relevant to potential cancer therapies as the PTEN gene is frequently mutated or silenced by various mechanisms in human cancer. Mutations occur which either delete the PTEN gene or alter its activity. Sometimes these mutations actually make the cells sensitive to Akt and mTOR inhibitors as the growth of the cells becomes dependent upon elevated Akt levels and downstream mTOR and p70S6K activities (DeGraffenried et al, 2004). Furthermore, epigenetic mechanisms may silence PTEN transcription and posttranscriptional mechanisms may alter the splicing of the primary PTEN mRNA transcript. Restoration of WT PTEN activity or inhibition of Akt activity are key strategies to prevent chemo- and hormonal resistance.
Some point mutations that hinder PTEN phosphatase activity such as the C124S and G129E may result in PTEN genes which serve as DN mutants suppressing the normal PTEN activity encoded by the remaining endogenous WT allele. These PTEN mutants may serve to alter chemo-, hormonal- and targeted therapy sensitivity. Identification of such mutations is becoming important in breast cancer therapy as it has been shown that the loss of PTEN activity is associated with Herceptin resistance (Nagata et al, 2004). Only one allele of PTEN may be mutated in many human cancers and they may serve as DN mutations.
Materials and Methods
Cell Culture
MCF-7 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Cell culture medium for MCF-7 cells consisted of Roswell Park Memorial Institute-1640 (RPMI 1640) medium (Invitrogen. Carlsbad CA) supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS) as described (Weinstein-Oppenheimer et al., 2001).
PTEN(WT), PTEN(C124S), PTEN(G129E), and PTEN(399stop) Plasmids
Mutated PTEN cDNAs were inserted into the multiple cloning site of pEGFP-C2 to generate plasmids encoding various PTEN mutants (Leslie et al, 2000). These PTEN mutants include: PTEN(C124S), PTEN(G129E), and PTEN(399Stop). PTEN(C124S) and PTEN(G129E) each differ from PTEN(WT) by substitution of a single amino acid. PTEN(399Stop) lacks the final five carboxyl terminal amino acids of PTEN(WT).
Transfection of MCF-7 cells with PTEN Constructs
5 × 105 MCF-7 cells were plated into 6-well cell culture plates (BD Biosciences, Mountainview, CA) one day prior to transfection and subsequently transfected with the various plasmid DNAs as described (Davis et al., 2003; Lehmann et al., 2007). The nomenclature of the transfected cells is MCF/ followed by the name of the introduced plasmid DNA.
Analysis of Sensitivity to Doxorubicin and Rapamycin
Cells were seeded in 96-well cell culture plates (BD Biosciences) at a density of 5,000 cells/well in 100 µl/well of phenol red free RPMI-1640 containing 5% charcoal stripped (CS) FBS as described (Weinstein-Oppenheimer et al., 2001). Cells were subsequently treated with serial 2-fold dilutions of either doxorubicin or rapamycin or a constant suboptimal concentration of rapamycin. Cells were incubated for 1 day to permit cells to adhere to the bottom of each well. Cells were incubated at 37 °C until extent of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma, St. Louis, MO) reduction in each well was quantified at 530 nm (Weinstein-Oppenheimer et al., 2001).
Cell Cycle and Apoptosis Analysis
The extent of cell cycle progression and apoptosis in the PTEN-transfected cells was estimated by flow cytometric analysis after propidium iodide (PI) (Roche Diagnostics, Indianapolis, IN) staining of the cells. Results were analyzed by the ModFit LT 3.1 software program (Verity Software, Topsham, ME).
Clonogenic Assays
MCF-7 cells were collected and seeded in 6-well cell culture plates at densities of 1000 and 2000 cells/well (3 wells at each density) as described (Lehmann et al., 2007).
Cellular Senescence Assay
Senescent cells were identified by a senescence associated (SA) β-galactosidase (SA-β-gal) assay as described (Lehmann et al, 2007).
Western blot analysis
Western blots were probed with antibodies specific for phospho and total Akt, Ask-1, p70S6K, and Hsp70 (as a loading control), as previously described (Shelton et al, 2005). Antibodies used in this study were purchased from Cell Signaling (Beverly, MA).
Acknowledgements
This work was supported in part by a grant from the US National Institutes of Health to JAM (R01CA098195). AMM has been supported in part by grants from the CARISBO Foundation and the Progetti Strategici Università di Bologna EF2006.
References
- Abdel-Rahman MH, Yang Y, Zhou XP, Craig EL, Davidorf FH, Eng C. J Clin Oncol. 2006;24:288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Eng C. Hu Mol Gen. 2006;5:777–787. doi: 10.1093/hmg/ddi492. [DOI] [PubMed] [Google Scholar]
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, et al. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. Develop Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Bose S, Crane A, Hibshoosh H, Mansukhani M, Sandweis L, Parsons R. Hu Pathol. 2002;33:405–409. doi: 10.1053/hupa.2002.124721. [DOI] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- Chung JH, Eng C. Cancer Res. 2006;65:8096–8100. doi: 10.1158/0008-5472.CAN-05-1888. [DOI] [PubMed] [Google Scholar]
- Clark AS, West K, Streicher S, Dennis PA. Mol Cancer Ther. 2002;1:707–717. [PubMed] [Google Scholar]
- Davis JM, Navolanic PM, Weinstein-Oppenheimer CR, Steelman LS, Hu W, Konopleva M, et al. Clin Cancer Res. 2003;9:1161–1170. [PubMed] [Google Scholar]
- DeGraffenried LA, Friedrichs WE, Russell DH, Donzis EJ, Middleton AK, Silva JM, et al. Clin Cancer Res. 2004;23:8059–8067. doi: 10.1158/1078-0432.CCR-04-0035. [DOI] [PubMed] [Google Scholar]
- Eng C. PTEN: One gene, many syndromes. Hu Mut. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR Hemmings. J Biol Chem. 1997;272:8474–8881. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- Garcia JM, Silva J, Pena C, Garcia V, Rodríguez R, Cruz MA, et al. Genes Chrom Can. 2004;41:117–124. doi: 10.1002/gcc.20062. [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Nature Gen. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Jin J, Woodgett JR. Oncogene. 2005;24:5459–5470. doi: 10.1038/sj.onc.1208704. [DOI] [PubMed] [Google Scholar]
- Kang S, Bader AG, Zhao L, Vogt PK. Cell Cycle. 2005;4:578–581. doi: 10.4161/cc.4.4.1586. [DOI] [PubMed] [Google Scholar]
- Kios KS, Wyszomierski SL, Sun M, Tan M, Zhou X, Li P, Yang W, et al. Cancer Res. 2006;66:2028–2037. doi: 10.1158/0008-5472.CAN-04-4559. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kim HS, Kim YB, Lee MC, Park CS, Min KW. App Immunohistochem & Mol Morph. 2004;12:205–210. doi: 10.1097/00129039-200409000-00004. [DOI] [PubMed] [Google Scholar]
- Lehmann BD, McCubrey JA, Jefferson HS, Paine MS, Chappell WH, Terrian DM. Cell Cycle. 2007;6:595–605. doi: 10.4161/cc.6.5.3901. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Gray A, Pass I, Orchiston EA, Downes CP. Biochem J. 2000;346:827–833. [PMC free article] [PubMed] [Google Scholar]
- Li DM, Sun H. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Simpson L, Takahashi M, Miliaresis C, Myers MP, Tonks N, et al. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- Lin HJ, Hsieh FC, Song H, Lin J. Br J Cancer. 2005;12:1372–1381. doi: 10.1038/sj.bjc.6602862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang X, Yu SX, et al. Oncogene. 1999;18:7034–7045. doi: 10.1038/sj.onc.1203183. [DOI] [PubMed] [Google Scholar]
- Luo J, Cantley LC. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- Lynch ED, Ostermeyer EA, Lee MK, Arena JF, Ji H, Dann J, et al. Am J Hum Genet. 1997;61:1254–1260. doi: 10.1086/301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, et al. Hu Mol Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Nyakern M, Tabellini G, Bortul R, Tazzari PL, Evangelisti C, et al. Leukemia. 2006;20:911–928. doi: 10.1038/sj.leu.2404245. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Tazzari PL, Evangelisti C, Chiarini F, Blalock WL, Billi AM, et al. Curr Med Chem. 2007;14:2009–2023. doi: 10.2174/092986707781368423. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Nimwegen MJ, Huigsloot H, Camier A, Tijdens IB, van de Water B. Mol Phamacol. 2006;70:1330–1339. doi: 10.1124/mol.106.026195. [DOI] [PubMed] [Google Scholar]
- Shelton JG, Steelman LS, Abrams SL, Bertrand FE, Franklin RA, McMahon M, et al. Cell Cycle. 2005;4:822–830. doi: 10.4161/cc.4.6.1724. [DOI] [PubMed] [Google Scholar]
- Shelton JG, Steelman LS, Abrams SL, Bertrand FE, Franklin RA, McMahon M, et al. Expert Opin Ther Targets. 2005;9:1009–1030. doi: 10.1517/14728222.9.5.1009. [DOI] [PubMed] [Google Scholar]
- Singh B, Ittmann MM, Krolewski JJ. Genes Chrom Cancer. 1998;21:166–171. [PubMed] [Google Scholar]
- Steelman LS, Bertrand FE, McCubrey JA. Expert Opin Ther Targets. 2004;8:537–550. doi: 10.1517/14728222.8.6.537. [DOI] [PubMed] [Google Scholar]
- Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, et al. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- Tokunaga E, Kimura Y, Mashino K, Oki E, Oki E, Kataoka A, Ohno S, et al. Breast Cancer. 2006;13:137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- Tsou HC, Teng DH, Ping XL, Brancolini V, Davis T, Hu R, et al. Am J Hum Genet. 1997;61:1036–1043. doi: 10.1086/301607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui S, Inoue H, Yasuda K, Suzuki K, Higashi H, Era S, et al. Oncology. 2005;68:398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Inoue H, Yasuda K, Suzuki K, Tahara K, Higashi H, et al. Cancer. 2005;104:2048–2053. doi: 10.1002/cncr.21471. [DOI] [PubMed] [Google Scholar]
- Weinstein-Oppenheimer CR, Henriquez-Roldan CF, Davis JM, Navolanic PM, Saleh OA, Steelman LS, et al. Clin Cancer Res. 2001;7:2898–2907. [PubMed] [Google Scholar]