Abstract
Currently there is pressing need to develop novel therapeutic agents for the treatment of infections by the human respiratory pathogens Pseudomonas aeruginosa and Streptococcus pneumoniae. The neuraminidases of these pathogens are important for host colonization in animal models of infection and are attractive targets for drug discovery. To aid in the development of inhibitors against these neuraminidases, we have determined the crystal structures of the P. aeruginosa enzyme NanPs and S. pneumoniae enzyme NanA at 1.6 and 1.7 Å resolution, respectively. In situ proteolysis with trypsin was essential for the crystallization of our recombinant NanA. The active site regions of the two enzymes are strikingly different. NanA contains a deep pocket that is similar to that in canonical neuraminidases, while the NanPs active site is much more open. The comparative studies suggest that NanPs may not be a classical neuraminidase, and may have distinct natural substrates and physiological functions. This work represents an important step in the development of drugs to prevent respiratory tract colonization by these two pathogens.
Keywords: Streptococcus pneumoniae, Pseudomonas aeruginosa, neuraminidase, crystal structure, pneumonia
INTRODUCTION
Neuraminidases are widespread among animals and microorganisms, catalyzing the release of terminal sialic acid residues from glycoconjugates [1]. Perhaps the best characterized is the influenza neuraminidase, which is required to facilitate the spread of the virus and is a proven target for anti-influenza therapeutic agents. Neuraminidases are also produced by respiratory pathogens with very different metabolic requirements, and appear to be critical for colonization and infection of the respiratory tract by these diverse pathogens. Therefore, bacterial neuraminidases may be realistic therapeutic targets for treating these pathogenic infections.
Streptococcus pneumoniae is the most common cause of bacterial pneumonia and a significant cause of otitis media in children [2]. There is a need for therapeutic strategies that are active against all serotypes, as serotypes not covered by vaccines are increasingly prevalent [3; 4]. S. pneumoniae produces at least three distinct neuraminidases [5]; NanA being the most highly expressed and active [6] that is conserved in all strains [5; 7; 8] and modifies host glycoconjugates [9; 10]. nanA mutants colonize the rodent respiratory tract less efficiently than wild type strains [6; 11; 12].
Pseudomonas aeruginosa, also a neuraminidase producer, is the major cause of respiratory tract infection in patients with cystic fibrosis [13]. The neuraminidase of P. aeruginosa (NanPs) is also important in pathogenesis and nanPs expression correlates with airway colonization [14]. In contrast to the pneumococcal enzyme, the P. aeruginosa neuraminidase does not appear to target host glycoconjugates but instead is involved in the biosynthesis of extracellular polysaccharides involved in biofilm formation [15].
We report here the crystal structures of P. aeruginosa and S. pneumoniae neuraminidases at 1.6 and 1.7 Å resolution, respectively. The structure of NanA is similar to that of canonical neuraminidases, including its interactions with inhibitors. In contrast, the structure of NanPs shows striking differences to NanA and other neuraminidases, and its active site is much more open. The structural differences between these enzymes may be related to their functional differences and their distinctive roles in pathogenesis.
Materials and Methods
Protein expression and purification
Full-length (residues 1-438) P. aeruginosa NanPs or residues 116–1035 of S. pneumoniae D39 sialidase A precursor (NanA) were sub-cloned into the pET28a vector (Novagen) and over-expressed in E. coli BL21(DE3) at 20°C as N-terminal hexa-histidine tagged proteins. The C-terminal truncated protein of NanPs represents residues 1-333 (of 438). The soluble protein was purified by nickel-agarose, anion exchange and gel filtration chromatography. Proteins were concentrated and stored at −80°C in a solution containing 20 mM Tris (pH 8.5), 200 mM NaCl and 5% (v/v) glycerol.
Protein crystallization
Crystals of NanPs free enzyme were obtained at 21 °C by the sitting-drop vapor diffusion method. The reservoir solution contained 100 mM Hepes (pH 7.0), 5% Tacsimate, 7% (w/v) PEG 5000MME, and the protein was at 10 mg/ml concentration. Selenomethionine-labeled protein was cross-microseeded with crystals from native protein to obtain adequate crystals. For data collection, the crystals were cryoprotected with the introduction of 20% (v/v) ethylene glycol, and flash-frozen in liquid nitrogen.
The initial crystallization screen of NanA did not produce any hits. We therefore performed limited proteolysis to search for a stable fragment. Crystals of NanA were obtained by the sitting-drop vapor diffusion method. The reservoir solution contained 100 mM Hepes (pH 7.0) and 30% Jeffamine ED-2001 (pH 7.0), the protein was at 30 mg/ml and the drops also contained trypsin (at 5000:1 protein:trypsin ratio). For the complexes of NanA with NANA and DANA, crystals of the free enzyme were soaked overnight in a solution containing 100 mM Hepes (pH 7.0), 35% Jeffamine ED-2001 (pH 7.0), and 5 mM of each compound (NANA or DANA).
Data collection and processing
X-ray diffraction data were collected at the National Synchrotron Light Source (NSLS; Upton, NY, USA). Selenomethionyl single-wavelength anomalous diffraction (SAD) data sets at 1.9 Å and 1.7 Å resolution were collected for NanPs and NanA respectively. Native data sets at 1.6 Å resolution for NanPs and at 1.7 Å resolution for the NANA and DANA complexes of NanA were collected at the X4C beamline. The diffraction images were processed and scaled with the HKL package [16]. The NanPs crystals belong to space group P213, with cell parameters of a=b=c=125.6 Å. There is one molecule in the crystallographic asymmetric unit. The NanA crystals belong to the space group C2, with cell parameters of a=158.0 Å, b=47.7 Å, c=137.3 Å, and β=116.6 °. There are two molecules in the crystallographic asymmetric unit.
Structure determination and refinement
The positions of Se atoms were determined with the program BnP [17]. Reflection phases were calculated based on the SAD data and improved with the program SOLVE/RESOLVE [18], which also automatically located a large percentage of the residues in the molecule. The atomic model was fit into the electron density with the program O [19]. The structure refinement was carried out with the program CNS [20]. The crystallographic information is summarized in Table 1.
Table 1.
Summary of crystallographic information
| NanPs free enzyme | NanA free enzyme | NanA in complex with DANA | NanA in complex with NANA | |
|---|---|---|---|---|
| Resolution range (Å) | 30-1.6 | 30-1.7 | 30-1.7 | 30-1.7 |
| Number of observations | 377,554 | 636,912 | 279,534 | 251,113 |
| Rmerge (%)a | 8.8 (36.5) | 4.1 (9.4) | 3.0 (7.3) | 6.0 (14.2) |
| I/σI | 26.2 (4.9) | 38.8 (10.7) | 31.1 (11.5) | 25.3 (6.6) |
| Redundancy | 4.3 (3.9) | 3.4 (3.2) | 3.0 (2.5) | 2.8 (2.5) |
| Number of reflections | 85,363 | 183,283 | 93,563 | 86,772 |
| Completeness (%) | 96 (91) | 92 (90) | 93 (86) | 87 (70) |
| R factor (%) | 17.2 (21.7) | 17.2 (20.4) | 17.8 (20.5) | 17.5 (20.8) |
| Free R factor (%) | 19.1 (23.7) | 19.8 (23.4) | 20.9 (24.7) | 20.9 (24.8) |
| Residues in most favored region of the Ramachandran plot (%) | 86 | 86 | 86 | 86 |
| rmsd in bond lengths (Å) | 0.005 | 0.005 | 0.007 | 0.005 |
| rmsd in bond angles (°) | 1.4 | 1.4 | 1.3 | 1.4 |
The numbers in parenthesis are for the highest resolution shell.
Neuraminidase assay
Neuraminidase activity was performed with 1 mM of enzyme in 7.5 mM sodium chloride and 4 mM calcium chloride for 2 h as previously described [15].
RESULTS AND DISCUSSION
Structure of the NanA neuraminidase from S. pneumoniae
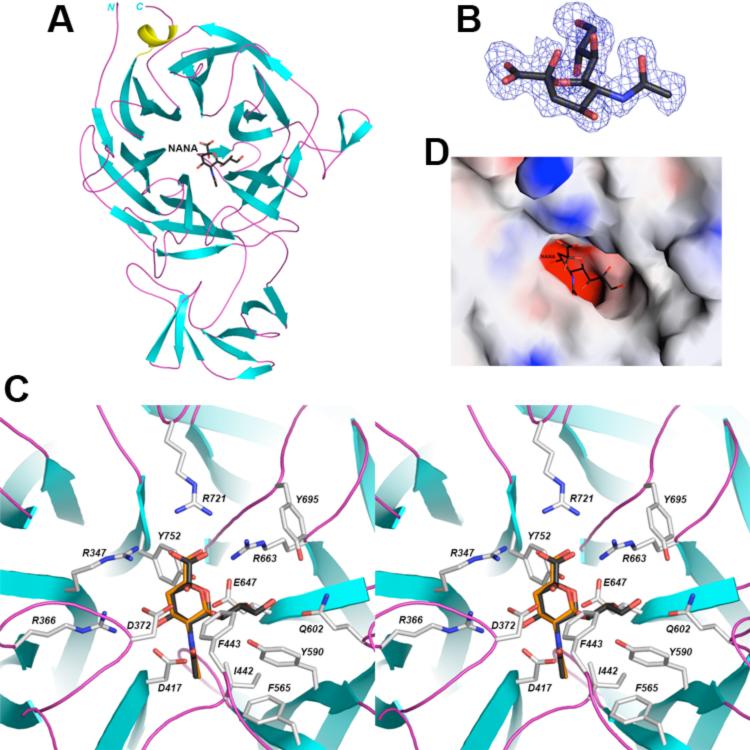
The crystal structure of the free enzyme of S. pneumoniae NanA has been determined at 1.7 Å resolution (Table 1). The bacterial expression construct contained residues 116-1035 and in situ proteolysis with trypsin was essential for the crystallization. The current atomic model contains residues 320-793 and 317-793 for the two NanA molecules in the crystallographic asymmetric unit, respectively, suggesting that roughly 200 residues from both termini of the recombinant protein were removed by trypsin during crystallization. The two NanA molecules have essentially the same conformation, with an rms distance of 0.25 Å for their equivalent Cα atoms. Purified NanA is monomeric in solution, based on gel-filtration chromatography.
The structure of NanA contains a typical six-bladed β-propeller domain, with an insertion (residues 437-535) between the second and the third β-strands of the second blade (Fig. 1A). This insertion forms a distinct domain located next to the catalytic β-propeller domain. NanA shares high structural similarity with other bacterial neuraminidases, including the Salmonella typhimurium LT2 neuraminidase [21], Vibrio cholerae neuraminidase [22], Clostridium perfringens sialidase NanI [23], and Micromonospora viridifaciens sialidase [24]. In addition, S. pneumoniae NanA in complex with the transition-state analog 2,3-dehydro-2-deoxy-N-acetylneuraminic acid (DANA) at 2.5 Å resolution [25] and NanB [26] were recently reported. The published NanA structure [25] is based on an expression construct that contains residues 319-822, although only residues 322-791 are observed in the structure and the crystals are in a different space group. Nonetheless, the overall structure and the interactions with DANA are similar to observations based on our structures. The overall rms distance in equivalent Cα positions is 0.4 Å.
Figure 1. Structure of S. pneumoniae NanA.
A. The β-strands are shown in cyan, α-helices in yellow, and connecting loops in magenta. The inhibitor NANA is shown as a stick model, in black for carbon atoms. B. The final 2Fo–Fc electron density at 1.7 Å resolution for the inhibitor NANA, contoured at 1σ, in a boat conformation. C. Stereo drawing showing detailed interactions between NANA (black) or DANA (orange) with the active site of NanA. D. Molecular surface of NanA in the active region, colored by electrostatic potential with NANA. The figures were created with PyMOL [32] and Grasp [33].
To reveal the molecular mechanism for substrate/inhibitor recognition, we determined the structures of NanA in complex with the sialic acid compound N-acetylneuraminic acid (NANA) and the transition-state analog DANA at 1.7 Å resolution (Table 1). There is essentially no conformational change in the enzyme overall or in its active site upon inhibitor binding. The rms distance in equivalent Cα atoms among the structures of the free enzyme, the NANA complex, and the DANA complex is about 0.15 Å.
Clear electron density was observed for the NANA inhibitor, which is bound to NanA in a boat conformation (Fig. 1B). As observed in structures of NANA in complex with other neuraminidases, the compound is involved in a large network of ionic and hydrogen-bonding interactions with the active site of NanA (Fig. 1C). The carboxyl group of NANA interacts with three Arg residues (Arg347, Arg663 and Arg721), and the amide nitrogen of NANA is hydrogen-bonded to the side chain of Asp417. The two hydroxyl groups on the ring of NANA interact with the side chains of Arg366, Asp417 and Asp372, the last of which is also hydrogen-bonded to one of the hydroxyls on the glycerol group. In addition to the polar interactions, several hydrophobic side chains are located near the inhibitor. Most importantly, residues Ile442 and Phe443 in the long loop at the beginning of the insertion in blade 2 (Fig. 1A) cover up part of the inhibitor and greatly reduce the size of the active site pocket, producing a tight fit between the inhibitor and the enzyme (Fig. 1D). DANA is bound in the same position as NANA, showing the same interactions with the enzyme except for the loss of the hydrogen-bond with Asp372 due to the absence of the hydroxyl group on the ring (Fig. 1C).
Structure of the P. aeruginosa neuraminidase reveals exposed active site
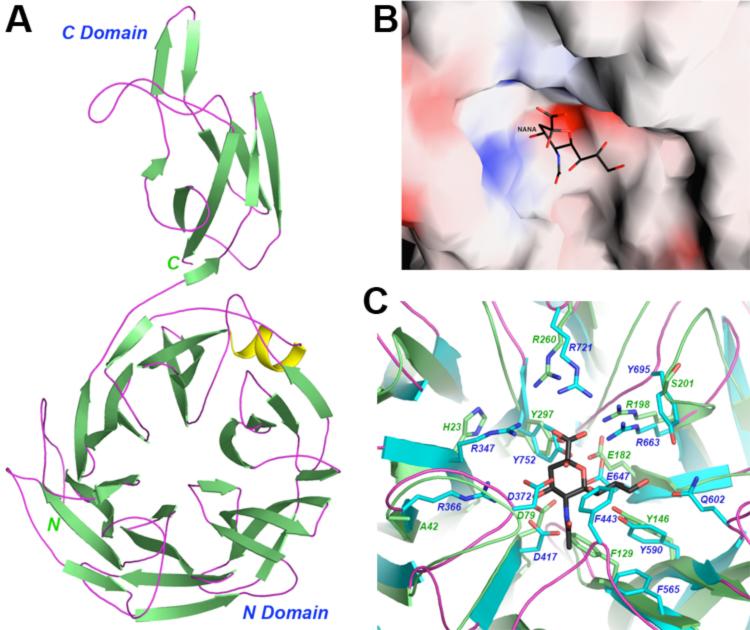
The structure of the P. aeruginosa neuraminidase (NanPs) at 1.6 Å resolution was also determined (Table 1). The enzyme, previously referred to as NanA [15], product of PA2794, has been renamed as our studies here show that it has significant differences to typical bacterial neuraminidases. The structure contains a six-bladed β-propeller in the N-terminal region (residues 1-334, Fig. 2A) and a C-terminal domain (C domain, residues 335-438) that belongs to the immunoglobulin superfamily based on comparisons with other structures in the Protein Data Bank using the SSM server [27]. A close structural homolog of the C-terminal domain is the immunoglobulin superfamily domain of the muscle protein twitchin [28]. The rms distance among equivalent Cα atoms of the two structures is 3 Å, although the amino acid sequence identity is only 6%. This domain is located about 50 Å away from the active site of the enzyme (Fig. 2A), but our mutagenesis studies showed that it is important for the catalytic activity (Fig. 3). The domain mediates the formation of a trimer in the crystal, although the protein is monomeric in solution based on gel-filtration and light scattering experiments (data not shown).
Figure 2. Structure of P. aeruginosa NanPs.
A. The β-strands are shown in green, α-helices in yellow, and connecting loops in magenta. B. Molecular surface of NanPs in the active site region, colored by electrostatic potential. The view is the same as that of Figure 1D, and the position of NANA bound to NanA is shown for reference. C. Structural differences between the active site regions NanPs (in green) and NanA (in cyan). Residue numbers in green are for NanPs, and those in blue for NanA.
Figure 3. Activity of P. aeruginosa neuraminidase mutations.
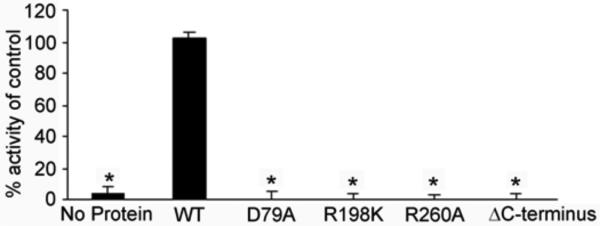
Site-directed mutations were made within the active site of P. aeruginosa neuraminidase or truncation in the C-terminus (deleting residues 334-438) and purified protein used to determine neuraminidase activity compared to wild-type enzyme (WT, control) using the fluorogenic substrate 2’-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid. *p-value <0.05.
The β-propeller domain of P. aeruginosa NanPs has the same overall structure as that of canonical neuraminidases. The rms distance among equivalent Cα atoms in P. aeruginosa NanPs and these other structures is 2.5-3 Å, but the amino acid sequence identity is 7-19%. Some of the important residues in the active site are also conserved in NanPs, consistent with our observation that it does possess weak neuraminidase activity [15]. Mutation of some of these conserved residues in the active site, including Asp79Ala (Asp417 in NanA) and Arg260Ala (Arg721 in NanA), abolished the catalytic activity (Fig. 3). Mutation of Arg292 to a lysine in the influenza neuraminidase, equivalent to Arg198 in NanPs and Arg663 in NanA, confers resistance to the inhibitor oseltamivir [29]. However, the Arg198Lys mutant of NanPs showed a loss of activity (Fig. 3).
There are dramatic differences in the shape of the active site region between NanPs and the other neuraminidases. Compared to NanA (Fig. 1D), the active site region of NanPs is much more open (Fig. 2B). An important cause of this difference is the lack of the insertion in the second blade of the propeller. The long loop at the beginning of this insertion, especially the side chains of Ile442 and Phe443 (Fig. 1C), is absent in NanPs (Fig. 2C), leading to a much more exposed active site. Additionally, variations at other amino acid residues also contribute to the different active site topography of NanPs. The side chain of Phe129 in NanPs clashes with the bound position of the acetyl group in NANA (Fig. 2C), which is the only recognizable clash between the inhibitor and the NanPs active site (Fig. 2B). The equivalent residue in NanA is Gly567.
Several other residues in the NANA binding site are not conserved in NanPs (Fig. 2C). Especially, of the four arginine residues that interact with NANA in NanA, two (Arg347 and Arg366) are replaced with other residues (His23 and Ala42) while the other two assume different conformations in NanPs (Fig. 2C). In addition, Asp372 in NanA is replaced by Gly47 in NanPs. The structural information shows that the active site of NanPs is not a good fit for the NANA inhibitor (Fig. 2B). This suggests that NanPs is probably not a conventional neuraminidase, and its natural substrate(s) remain to be identified.
The neuraminidases of S. pneumoniae and P. aeruginosa share a common role in facilitating bacterial colonization of the respiratory tract [6; 11; 12; 15]. Despite some functional similarities, our studies show that the enzymes have important structural differences. NanA is similar to canonical neuraminidases, with an active site that is a good fit for the NANA compound. In contrast, NanPs exhibits a distinctive active site surface, not accommodating NANA tightly, suggesting a distinct biochemical function. Even subtle differences in neuraminidase structure can change the specificity for substrates, as seen with the Trypanosome enzymes [30]. The neuraminidase superfamily is quite divergent with low sequence identities, even though they retain tertiary structure similarities as observed with these two neuraminidases [31]. The differences in active site and substrate binding of NanA and NanPs are also consistent with the divergence in the family. Moreover, NanPs contains a unique C-terminal domain, essential for its catalytic activity, although its exact biological function remains to be established.
ACKNOWLEDEMENTS
D.P. was the recipient of an NHRMC Biomedical Overseas Fellowship. This work was supported by the Thrasher Research Fund (D.P.), Deans Pilot Grant Columbia University (A.P.) and Cystic Fibrosis Foundation Grant (A.P.).
REFERENCES
- 1.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol. 1996;6:830–7. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organisation (WHO) Pneumococcal conjugate vaccine for childhood immunization - WHO position paper, Weekly Epidemiological Record. WHO; Geneva: 2007. pp. 93–104. [PubMed] [Google Scholar]
- 3.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. Jama. 2007;297:1784–92. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 5.Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect Immun. 2006;74:3360–5. doi: 10.1128/IAI.01442-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun. 2006;74:4014–20. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly RT, Farmer S, Greiff D. Neuraminidase activities of clinical isolates of Diplococcus pneumoniae. J Bacteriol. 1967;94:272–3. doi: 10.1128/jb.94.1.272-273.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King SJ, Whatmore AM, Dowson CG. NanA, a neuraminidase from Streptococcus pneumoniae, shows high levels of sequence diversity, at least in part through recombination with Streptococcus oralis. J Bacteriol. 2005;187:5376–86. doi: 10.1128/JB.187.15.5376-5386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol. 2004;54:159–71. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 10.King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–74. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 11.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–9. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 12.Tong HH, Blue LE, James MA, DeMaria TF. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–4. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171:1209–23. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, Quentin R. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 15.Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kaneko Y, Wolfgang MC, Hsiao YS, Tong L, Prince A. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Weeks CM, Miller R. The design and implementation of SnB v2.0. J. Appl. Cryst. 1999;32:120–124. [Google Scholar]
- 18.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth. Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Crennell SJ, Garman EF, Philippon C, Vasella A, Laver WG, Vimr ER, Taylor GL. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J. Mol. Biol. 1996;259:264–280. doi: 10.1006/jmbi.1996.0318. [DOI] [PubMed] [Google Scholar]
- 22.Crennell S, Garman E, Laver G, Vimr E, Taylor G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure. 1994;2:535–44. doi: 10.1016/s0969-2126(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 23.Newstead SL, Potter JA, Wilson JC, Xu G, Chien CH, Watts AG, Withers SG, Taylor GL. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J Biol Chem. 2008;283:9080–8. doi: 10.1074/jbc.M710247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaskell A, Crennell S, Taylor G. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 25.Xu G, Li X, Andrew PW, Taylor GL. Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA. Acta Cryst. 2008;F64:772–775. doi: 10.1107/S1744309108024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gut H, King SJ, Walsh MA. Structural and functional studies of Streptococcus pneumoniae neuraminidase B: An intramolecular trans-sialidase. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 28.Fong S, Hamill SJ, Proctoer M, Freund SMV, Benian GM, Chothia C, Bycroft M, Clarke J. Structure and stability of an immunoglobulin superfamily domain from twitchin, a muscle protein of the nematode Caenorhabditis elegans. J. Mol. Biol. 1996;264:624–639. doi: 10.1006/jmbi.1996.0665. [DOI] [PubMed] [Google Scholar]
- 29.Carr J, Ives J, Kelly L, Lambkin R, Oxford J, Mendel D, Tai L, Roberts N. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 2002;54:79–88. doi: 10.1016/s0166-3542(01)00215-7. [DOI] [PubMed] [Google Scholar]
- 30.Amaya MF, Buschiazzo A, Nguyen T, Alzari PM. The high resolution structures of free and inhibitor-bound Trypanosoma rangeli sialidase and its comparison with T. cruzi trans-sialidase. J Mol Biol. 2003;325:773–84. doi: 10.1016/s0022-2836(02)01306-2. [DOI] [PubMed] [Google Scholar]
- 31.Roggentin P, Schauer R, Hoyer LL, Vimr ER. The sialidase superfamily and its spread by horizontal gene transfer. Mol Microbiol. 1993;9:915–21. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 33.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]