SUMMARY
The food a honey bee female larva receives determines whether she develops into a large long-lived fertile queen or a short-lived sterile worker. Through well-established nutrient-sensing and growth-promoting functions in metazoans, the insulin/insulin-like growth factor 1 signaling (IIS) pathway has become a focal topic in investigations on how differences in food environment can be translated into internal signals responsible for queen–worker determination. However, low expression levels of two insulin receptors (AmInRs) in honey bee larvae and the failure of one AmInR to influence caste differentiation are in potential conflict with such a classical growth-promoting role of IIS in queen–worker development. In view of such an apparent contradiction, and the fact that binding partners and affinities of these two AmInRs have not been worked out, we performed a functional study on insulin-like peptide genes (AmILP1 and AmILP2) in honey bee larvae by using a double-stranded RNA (dsRNA)-mediated gene knockdown approach. We found that juvenile hormone (JH) levels were diminished by AmILP1 dsRNA treatment, while the AmILP2 knockdown caused a reduction in ovary size. Blood sugar titers were not significantly affected by the treatments. From these results we conclude that AmILP2 transcript levels may influence specific organ development, such as the ovary and body mass, while more general traits of caste differentiation, such as mandibles, may require additional regulators. In addition, JH production may be regulated by AmILP1 expressed locally in the brain, similar to the function of certain ILPs in Drosophila.
KEY WORDS: queen–worker differentiation, juvenile hormone (JH), RNA interference, carbohydrate metabolism, caste development, insulin/insulin-like growth factor 1 signaling (IIS), morphological trait, ovariole number, body mass
INTRODUCTION
Eusocial insects, including honey bees, exhibit an environmentally induced caste polyphenism that promotes colony efficiency through a morphology-based division of labor: while queens are functional egg-laying machines, workers forgo reproduction and, instead, care for the brood, defend the colony and forage for food (Hölldobler and Wilson, 2008). The ecological and evolutionary success of social insects (Hölldobler and Wilson, 2008) is largely built upon such division of labor, but the mechanisms that generate alternative phenotypes (castes) in Hymenoptera and termites are not fully understood (Hartfelder and Emlen, 2012).
The most considerable progress in understanding caste development has been made in the honey bee, Apis mellifera L., where the primary trigger is differential feeding of the larvae. Queen larvae receive copious amounts of royal jelly, a glandular secretion produced by young worker bees, throughout all five larval instars (Haydak, 1970; Winston, 1987). In contrast, worker larvae are fed less frequently and receive a diet less rich in sugar (4% compared with 12% in royal jelly) during the third and fourth larval instars (Asencot and Lensky, 1985). These diets induce a series of endogenous responses that result in differential phenotypes. The most studied endocrine regulator of caste differentiation is juvenile hormone (JH) (Rachinsky et al., 1990; Rembold, 1987), which shows higher titers during the fourth to fifth instar in queen-destined larvae (Rachinsky and Hartfelder, 1990; Rachinsky et al., 1990). Functionally, JH application induces queen-like traits in larvae with a restricted diet (Goewie, 1977; Rembold et al., 1974). Although the mode of action of JH in driving queen development is still rather unclear, its effect on ovary size (i.e. ovariole number), which is one of the key morphological traits differing between queens and workers, has been revealed. JH affects ovary differentiation from the third larval instar until the onset of metamorphosis: high JH titers in queen larvae prevent autophagic programmed cell death in the ovary (Schmidt Capella and Hartfelder, 2002), thus sustaining tissue survival and differentiation into the large queen ovaries, whereas low JH titers in worker larvae cannot inhibit programmed cell death, which removes 95–99% of the ovariole primordia and leads to the small worker-type ovaries.
Ovary size defines the reproductive status of queens and workers (Winston, 1987), and regulates worker social behaviors (Wang et al., 2009; Wang et al., 2010). The adult honey bee queen has up to 150 ovarioles in each of her ovaries and is only responsible for laying eggs. In contrast, workers, which are functionally sterile, typically only have on average 2–12 ovarioles per ovary (van der Blom et al., 1994; Michener, 2000; Winston, 1987). However, the ovariole numbers may vary, and in vitro rearing experiments showed that there is a morphospace gradient in which ovary phenotypes of the queen and worker are the extremes (Leimar et al., 2012; Linksvayer et al., 2011). In addition, ovary size is also correlated with foraging behavior in workers: workers with more ovarioles perform less retinue behavior (D. Galbraith, Y.W., G.V.A., R. E. Page and C. G. Grozinger, unpublished data), initiate foraging tasks earlier in life (Wang et al., 2009; Wang et al., 2010), prefer to collect pollen over nectar, and are more sensitive to sucrose than workers with fewer ovarioles (Amdam et al., 2006; Wang et al., 2009).
The release of the honey bee genome sequence (Honeybee Genome Sequencing Consortium, 2006) greatly facilitated investigations on how the nutrient stimuli are translated into endogenous molecular signals in honey bee caste differentiation. The focus has been on two conserved eukaryotic nutrient-sensing pathways: the insulin/insulin-like growth factor 1 signaling (IIS) pathway (Mutti et al., 2011a; Wolschin et al., 2011) and the closely related and interacting target-of-rapamycin (TOR) pathway (Patel et al., 2007). Larvae subjected to RNA interference (RNAi)-mediated gene knockdown of the insulin receptor substrate (IRS) and TOR genes consistently developed into workers even when receiving a queen diet (Kamakura, 2011; Mutti et al., 2011a; Patel et al., 2007). As RNAi primarily targets the fat body (Jarosch and Moritz, 2011), a tissue functionally homologous to white adipose tissue and the liver in mammals (Chapman, 1998), these studies also provided evidence that IRS and TOR genes expressed in the fat body may remotely control JH production by the corpora allata (CA) in the retrocerebral complex (Mutti et al., 2011a). In addition, gene expression studies have revealed that the genes encoding two insulin-like peptides (AmILP1 and AmILP2) and two insulin receptors (AmInR1 and AmInR2) are differentially expressed between queen and worker larvae (de Azevedo and Hartfelder, 2008; Wheeler et al., 2006). Together with the results of a recent study showing that epidermal growth factor receptor (EGFR) gene knockdown induces the worker phenotype (Kamakura, 2011), the current evidence indicates that caste development in honey bees involves a complex interaction network composed of the IIS/TOR/EGFR pathways, JH and ecdysteroids, which are classic developmental and reproductive hormones in Drosophila (Mirth and Riddiford, 2007) and other insect species (Chapman, 1998).
Nonetheless, upon a closer look, the regulatory network of honey bee caste development is not straightforward, especially for the IIS pathway. For instance, the expression levels of AmInR1 and AmInR2 in fourth instar queen larvae decline to very low levels, just as the larvae show the highest growth rates (de Azevedo and Hartfelder, 2008). Furthermore, silencing one of the AmInR genes did not affect caste fate in honey bees (Kamakura, 2011), suggesting the effect of IRS on queen–worker phenotype differentiation may be mediated by EGFR, and not through IIS (Mutti et al., 2011a).
Clearly, based on sequence similarity, AmInR1 and AmInR2 are putative genes for receptors of insulin-like peptides, the upstream signaling factors in IIS. However, their binding partners and respective binding affinities have not been investigated. The honey bee AmILP1 and AmILP2 genes also have high sequence similarity to Drosophila ILPs (DILPs) whose roles in the IIS have been intensively investigated. Previous studies on AmILP1 and AmILP2 in honey bee brain and fat body suggested that the proteins encoded by AmILP1 and AmILP2 genes mediate nutritional signals (Ament et al., 2008; Ament et al., 2010) and regulate energy metabolism (Wang et al., 2012), which are conserved functions of ILPs across species including Drosophila. Studies on AmILP1 levels in the brain of honey bee workers (Ament et al., 2010; Corona et al., 2007) have shown that these are negatively correlated with individual nutritional status and positively related to JH titer (Ament et al., 2008). Additionally, low levels of AmILP1 transcripts in the fat body of adult bees are linked to high blood sugar levels (Wang et al., 2012). In contrast, the regulation and function of AmILP2 is less well understood as AmILP2 expression does not consistently respond to factors as AmILP1 does in adult honey bees (Amdam, 2011; Wheeler et al., 2006).
In Drosophila, silencing DILPs strongly affected larval development and carbohydrate metabolism (Brogiolo et al., 2001; Rulifson et al., 2002). Although gene expression profiles of AmILPs in honey bee larvae differ among the castes (de Azevedo and Hartfelder, 2008; Wheeler et al., 2006), actual functional data of AmILP1 and AmILP2 in larval development are still missing. Thus, to gain insight into AmILP1 and AmILP2 gene function in queen–worker differentiation we used an RNAi-mediated gene knockdown approach in larvae reared in an in vitro system (Patel et al., 2007). The treated and control larvae were assayed for transcript levels of AmILP1 and AmILP2, hemolymph sugar and JH levels, and larval body mass at the fifth larval instar, when developmental hormone titers (JH and ecdysteroids) are very different and when the caste-specific differentiation of the ovaries is in the most pronounced stage. In addition, we screened the expression of caste phenotype characters of the adults that emerged from such treatments. The results are indicative of differential roles for the AmILPs in the queen–worker differentiation process.
MATERIALS AND METHODS
Experimental design
In this study, we used a full factorial design in which AmILP1 RNAi and AmILP2 RNAi treatments are the two independent factors. There were two levels for each of the factors: ‘0’ (no RNAi) and ‘1’ (RNAi). It is known that a factorial experimental design is ‘more efficient than one-factor-at-a-time experiments and can detect interactions’ (Montgomery, 1997). This allowed us to study the effect of each factor on the traits we are most interested in, as well as the effects of interactions between the two factors on those traits (Montgomery, 1997). Studies on ILPs in other insects have found that the functions of ILPs are usually linked (Wu and Brown, 2006). And it has been proposed that AmILP1 and AmILP2 acts as agonist and antagonist of their respective receptors, but no experimental evidence has been found so far (Nilsen et al., 2011). Therefore, determining the interaction between AmILP1 and AmILP2 should be informative for understanding how the functions of AmILP1 and AmILP2 are interconnected in honey bees.
Double-stranded RNA synthesis
DNA fragments of the AmILP1 and AmILP2 genes flanked on both sides with a T7 promoter sequence were inserted into the commercial T-easy vector (Promega, Madison, WI, USA) using the primers listed in supplementary material Table S1. Plasmids were extracted and sequenced to validate the DNA sequences of AmILP1 and AmILP2. Double-stranded RNAs (dsRNAs) of AmILP1 and AmILP2 were synthesized following a previously established protocol (Amdam et al., 2003). The gene sequence of the green fluorescent protein (GFP), which is not found in the honey bee genome, was used to produce a non-target dsRNA, serving as a control dsRNA in the RNAi assays.
In vitro rearing of honey bee larvae
Wild-type honey bees maintained at the Honeybee Research Facility at the Arizona State University Polytechnic campus (Mesa, AZ, USA) were used in these experiments. Queens from three wild-type colonies were caged for 24 h and newly hatched larvae (12–18 h old, N=1000) were grafted into Petri dishes containing a previously established nutrient-rich diet suitable for in vitro rearing of queens (Patel et al., 2007), and were kept in a cell culture incubator at 33°C and 80% humidity (Patel et al., 2007). On the second day, larvae of similar size were grafted from the Petri dishes and randomly distributed into 24-well culture plates (6 larvae per well). A full factorial design was applied on our dsRNA feeding regime, and AmILP1 dsRNA and AmILP2 dsRNA were used as two independent factors. Four experimental treatments were created: AmILP1 dsRNA, AmILP2 dsRNA, AmILP1+AmILP2 dsRNAs, and gfp dsRNA. Each well contained the larval diet supplemented with 200 μg ml−1 of each respective dsRNA. Therefore, the total dsRNA was 200 μg ml−1 for AmILP1 dsRNA, AmILP2 dsRNA and gfp dsRNA treatment groups, and 400 μg ml−1 for the AmILP1+AmILP2 dsRNA group. Using similar factorial designs in both honey bee larvae and adults, previous studies on gene knockdown have shown that the amount of dsRNA in combined treatment groups does not cause any unspecific or adverse effects (Mutti et al., 2011a; Wang et al., 2012). Every 12 h, the larvae were transferred to new diets in new plates, with changes of the position on the plate in a randomized design to minimize any location effects. After 2 days of feeding on the dsRNA-containing diet and 1.5 days of feeding on dsRNA-free diet, 20 larvae from each treatment group were collected to validate the gene knockdown and to reveal larval physiology responding to the treatments. The remaining larvae continued to be fed with dsRNA-free diet until they began defecating. Subsequently, they were transferred to filter paper-lined Petri dishes, with the filter paper being changed every day as they passed the pupal stage, and finally emerged as adult bees in the Petri dishes.
Sampling of hemolymph and larval body for gene expression, blood sugar and JH level analyses
Larvae retrieved from the experimental setup were cleaned by carefully wiping with tissue paper, and were weighed on a digital scale (VWR, Gaithersburg, MD, USA). The body of the larva was pierced with a 30 gauge BD needle, so that two samples of extruding hemolymph could be collected from each larva with glass capillaries (VWR). These hemolymph samples were used to assay sugar levels and JH titers, respectively. The hemolymph samples for carbohydrate measurement were immediately frozen on dry ice and kept at −80°C until use. The hemolymph samples for testing JH titers were collected into glass vials containing 500 μl hexane and stored at −20°C until use. The remaining carcasses of the larvae were transferred into Eppendorf tubes containing 500 μl TRIzol reagent (Invitrogen, Carlsbad, CA, USA), flash-frozen in liquid nitrogen and stored at −80°C.
RNA extraction and cDNA synthesis
After thawing and homogenization in the TRIzol reagent, RNA was extracted following the manufacturer's instructions. The quality and quantity of RNA was determined by spectrophotometry (Nanovue, GE Healthcare, Barrington, IL, USA). DNase (RNase-free, DNase kit, Applied Biosystems, Bedford. MA, USA) was added to the total RNA extract to remove trace DNA contaminants, and 1 μg of treated RNA was used for reverse transcription following an established method (Wang et al., 2009) using TaqMan reverse transcription reagents (Applied Biosystems).
Real-time quantitative PCR analyses
First-strand cDNA was used for real-time quantitative PCR (RT-qPCR) assays. Before performing the RT-qPCR, PCR amplicons from each gene were sequenced to validate the specificity of the primers (supplementary material Table S2). A dilution series of cDNA was used to establish standard curves for each gene, and amplification efficiencies were calculated based on an established method (Livak and Schmittgen, 2001; Pfaffl, 2001). After verifying that AmILP1, AmILP2 and Amrp49 primers had similar amplification efficiencies, 15 samples were randomly picked from each treatment group for expression analysis. Each biological sample was run in technical triplicate on an ABI Prism 7500 Real-Time PCR system (Applied Biosystems) for measuring AmILP1 and AmILP2 transcript levels in comparison with those of the reference gene Amrp49 by means of the ΔΔCt method (Livak and Schmittgen, 2001). Studies have shown that Amrp49, which has been renamed as rpl23 (AF441189) in the honey bee genome version Amel 4.5, is stably expressed during larval development (Lourenço et al., 2008; Reim et al., 2013) and in adults (Cameron et al., 2013). Therefore Amrp49 is commonly used as a reference gene during the larval stage (de Azevedo and Hartfelder, 2008; Martins et al., 2010) and also the adult stage of honey bees (Ben-Shahar et al., 2003; Navajas et al., 2008). RT-qPCR conditions were used as described previously for these genes (de Azevedo and Hartfelder, 2008). By monitoring negative control samples (without reverse transcriptase) and melting curve analysis, we verified that the RT-qPCR assays were not confounded by DNA contamination or primer dimers (Vandesompele et al., 2002).
Glucose and trehalose measurements
Glucose levels in the hemolymph were analyzed using a Glucose (HK) Assay Kit (Sigma, St Louis, MO, USA), following an established laboratory protocol (Hartfelder et al., 2013; Wang et al., 2012). After adding 1 ml of the glucose reagent to each hemolymph sample, these were incubated for 15 min at room temperature. A series of glucose dilutions (0, 0.5, 1, 5, 10, 30, 50 and 100 μg ml−1) was prepared to set up a standard curve. After the incubation, 100 μl of each standard and sample solution was transferred in triplicate to 96-well microplates. Absorbance at 340 nm was measured using an xMark Microplate Absorbance spectrophotometer (Bio-Rad, Hercules, CA, USA) and sample glucose concentrations were calculated by linear regression. After the glucose readings were taken, 0.5 μl of trehalase (Sigma; 0.05 U ml−1) was added to each well. The second reading for both standards and samples was taken after an overnight incubation at 37°C. Glucose produced from trehalose was calculated, by first subtracting the first glucose concentration value from the second total glucose concentration, then entering this into the equation: trehalose (μg)=glucose (μg)×342.3/(180.2×2).
JH radioimmunoassay
JH extraction from the 1 μl hemolymph aliquots in hexane was carried out following a liquid-phase separation protocol established for honey bee hemolymph (Huang et al., 1994). After addition of 1 ml NaCl (0.9%) and 1 ml hexane, the mixture was vigorously vortexed and the phases were separated by centrifugation (700 g). The hexane phase was retrieved, and the extraction was repeated twice by adding 1 ml hexane each time. The pooled hexane phases were dried by vacuum centrifugation, and the residues were redissolved in 100 μl toluene containing 0.5% (v:v) 1,2-propanediol (Sigma) and transferred to 1.5 ml glass vials. Before starting the radioimmunoassay (RIA), the solvent was removed by vacuum centrifugation.
A JH-specific antiserum (Goodman et al., 1990), previously validated for JH detection in bees (Amdam et al., 2007; Guidugli et al., 2005), was diluted 1:1250 in phosphate buffer supplemented with BSA (0.1%) and rabbit IgG (0.1%). The assays were performed with [10-3H(N)]-juvenile hormone III (specific activity 19.4 Ci mmol−1, Perkin Elmer, Boston, MA, USA) diluted in phosphate buffer to 6000–6500 c.p.m. 100 μl−1. Juvenile hormone III (Fluka, Buchs, Switzerland) was used as non-radioactive ligand. Standard curves were set up to cover a 50 pg to 10 ng range.
The RIA was conducted following a previously established procedure (Goodman et al., 1990) adapted for honey bees (Hartfelder et al., 2013). Samples were incubated overnight at 4°C, then supplemented with saturated ammonium sulfate (50% final concentration) to separate antibody-bound from free JH by centrifugation at 7500 g for 15 min. After washing the pellets with 50% ammonium sulfate and a novel precipitation/centrifugation step, the pellets were redissolved in 80 μl water before addition of 5 ml liquid scintillation cocktail (Optiphase Hisafe3, Perkin Elmer). Standard curve values were entered into a four-parameter fitting Excel spreadsheet specifically designed for enzyme-linked immunoassays (EIA) and RIA analyses (Bachem, Bubendorf, Switzerland; available from https://www.bachem.com/service-support/immunoassay-calculator/), based on the equation y={(a−d)/[1+(x/c)b]}+d, where a=maximum, b=slope, c=IC50 (the half-maximal inhibitory concentration) and d=minimum. Sample JH concentrations obtained by this polynomial regression were expressed as JH-III equivalents (pg μl−1 hemolymph).
Scoring ovariole number and additional morphological characters
Mandible and sting form, the presence of a corbicula (pollen basket) and spermatheca, and ovary size of the emerged adults were assessed under a dissecting Leica MA12 microscope (Leica, Wetzlar, Germany). Bees with more than 70 ovarioles, notched mandibles, a smooth stinger, a spermatheca and lacking a corbicula were classified as queens (Mutti et al., 2011b). Alternatively, workers were considered to have fewer than 20 ovarioles, a barbed stinger and a corbicula (Mutti et al., 2011b). Intermediates were those with 20–70 ovarioles and a mixed set of the other characters (Mutti et al., 2011b).
Statistical analysis
Gene expression data were log transformed to approximate normality (Wang et al., 2009), as verified by Bartlett and Levene's homogeneity test. A factorial ANOVA was used to test the effect of AmILP1 and AmILP2 dsRNAs on gene expression, followed by Fisher least significant difference (LSD) tests in post hoc comparisons. A Pearson correlation assay was used to reveal whether larval mass was correlated with AmILP1 and AmILP2 transcript levels and with JH titer. The factorial ANOVA was also used to test whether the treatment affected each morphological character such as ovariole number, mandible, stinger, corbicula or spermatheca. A principal component analysis (PCA) was applied on these multiple morphological characters to clarify general distribution patterns and separations of the bees from different treatment groups by reducing dimensions of variables. Then, a Kruskal–Wallis ANOVA was used to analyze the treatment effect on the values of sample bees given by the first principal component (PC1). These analyses were performed using STATISTICA 10.0 (StatSoft) software.
RESULTS
Quantitative validation of AmILP1 and AmILP2 knockdown in a full factorial experimental design
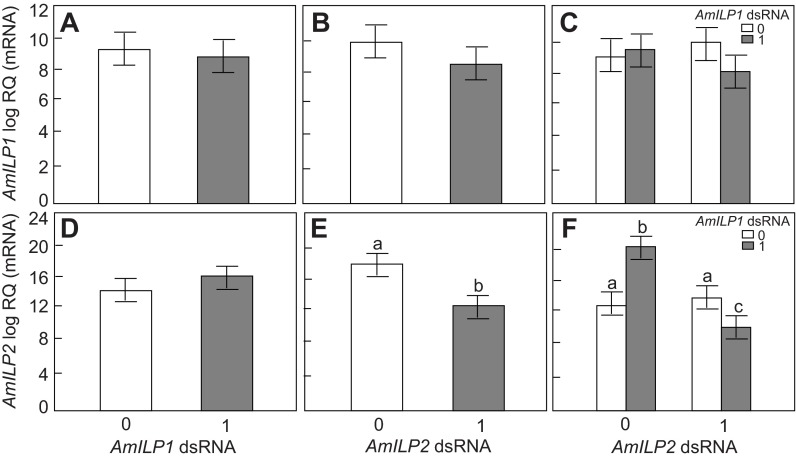
The individual whole-body RNA extracts from fifth instar larvae were assayed using an RT-qPCR protocol for AmILP1 and AmILP2 gene-knockdown verification (N=15). The overall effect of the factors (AmILP1 dsRNA treatment and AmILP2 dsRNA treatment) was determined by the main effect of a factorial ANOVA analysis. AmILP1 transcript levels were unaffected by either AmILP1 or AmILP2 dsRNA treatment (factorial ANOVA, N=15, main effect of AmILP1 dsRNA: F1,56=0.1392, P=0.6620; main effect of AmILP2 dsRNA: F1,56=1.0447, P=0.3111, Fig. 1A,B). However, there was a significant decrease in AmILP2 transcript levels in larvae with AmILP2 dsRNA treatment (factorial ANOVA, N=15, main effect of AmILP2 dsRNA: F1,56=20.9941, P<0.0001, Fig. 1E), but not in those treated with AmILP1 dsRNA (factorial ANOVA, N=15, main effect of AmILP1 dsRNA; F1,56=1.5382, P=0.2201, Fig. 1D). As the main effect of AmILP2 dsRNA on AmILP2 gene expression shown in Fig. 1E includes the effect of AmILP2 dsRNA at two levels of AmILP1 dsRNA treatment (0 and 1), this result indicates that AmILP2 dsRNA, independent or not independent of AmILP1 dsRNA, significantly downregulated the expression of its target gene at the whole-body level.
Fig. 1.
Apis mellifera insulin-like peptide AmILP1 and AmILP2 gene knockdown validation in fifth instar honey bee larvae. (A,B,D,E) The main effects of AmILP1 double-stranded (ds)RNA and AmILP2 dsRNA on AmILP1 and AmILP2 gene expression in a factorial ANOVA. (C,F) The relationships between four treatment groups, as revealed by a Fisher's least significant difference (LSD) post hoc test: the bars from left to right represent gfp (green fluorescent protein), AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. Overall, AmILP1 gene expression was not affected by AmILP1 dsRNA (A) and AmILP2 dsRNA (B). Overall, AmILP2 gene expression was not affected by AmILP1 dsRNA (D), but was significantly reduced by AmILP2 dsRNA (E). Panel F shows that compared with gfp (first bar): (i) AmILP1 dsRNA treatment increased AmILP2 expression level; (ii) AmILP2 dsRNA in the absence of AmILP1 dsRNA did not reduce the AmILP2 mRNA level; (iii) but when combined with AmILP1 dsRNA application, AmILP2 was significantly downregulated. Data are presented as means ± s.e.m. (N=15). RQ is relative quantification. Different letters (a–c) over the bars indicate significant differences between treatments. ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.
In order to determine whether AmILP1 dsRNA contributed to the main effect of AmILP2 dsRNA on AmILP2 expression, we looked at the interactions between AmILP1 dsRNA and AmILP2 dsRNA treatment. We found that there was no interaction between these two treatments on AmILP1 expression (factorial ANOVA, interaction effect: F1,56=2.1251, P=0.1505, Fig. 1C) but there was a significant interaction on AmILP2 gene expression (factorial ANOVA, F1,56=41.5698, interaction effect: P<0.0001, Fig. 1F), suggesting that the reduction of AmILP2 expression by AmILP2 dsRNA (Fig. 1E) was dependent on the level of AmILP1 dsRNA treatment. Next, we performed a Fisher's LSD post hoc test to further dissect how the four treatment groups (gfp, AmILP1 dsRNA, AmILP2 dsRNA, and AmILp1 dsRNA plus AmILP2 dsRNA) contributed to the main effects of AmILP2 dsRNA in this study. We found that, compared with gfp controls, (i) single AmILP1 dsRNA treatment actually increased AmILP2 transcript level (P<0.0001, Fig. 1F), (ii) single AmILP2 dsRNA treatment, on its own, did not significantly reduce AmILP2 mRNA levels (P=0.1985), but (iii) the combined AmILP1 dsRNA and AmILP2 dsRNA significantly decreased AmILP2 transcript abundance (P=0.0405). These results suggest that the level of AmILP1 dsRNA treatment contributed to the significant main effect of AmILP2 dsRNA in a whole larva (Fig. 1E): the application of AmILP2 dsRNA alone did not cause a reduction in the AmILP2 mRNA level, but AmILP1 dsRNA application enhanced the effect of AmILP2 dsRNA – resulting in a significant decrease in AmILP2 transcript abundance in Fig. 1E.
Glucose and trehalose titers in the hemolymph
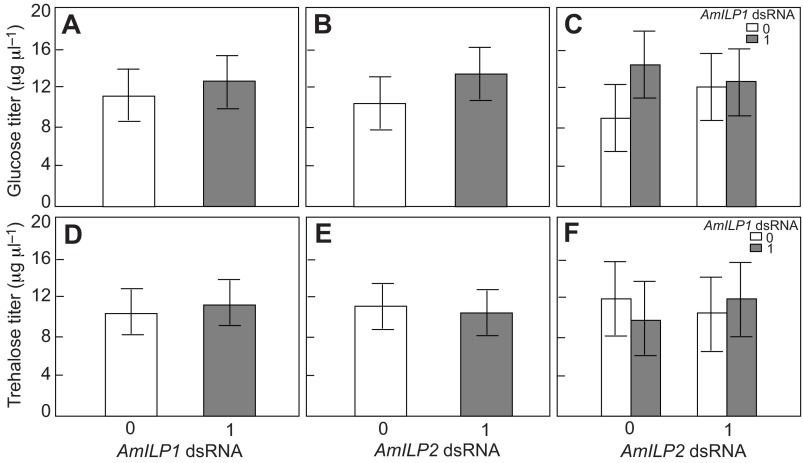
Studies in Drosophila indicated that ILPs in the brain were involved in regulating carbohydrate metabolism and blood sugar titers (Broughton et al., 2005; Saltiel and Kahn, 2001), and we previously found that AmILP1 gene expression in adult honey bees was linked with blood sugar levels (Wang et al., 2012). Here, we measured carbohydrate reserves (glucose and trehalose) in the hemolymph in order to test whether the AmILPs may directly regulate blood sugar titers during honey bee larval development. We found that neither glucose nor trehalose concentration was influenced by either AmILP1 dsRNA or AmILP2 dsRNA treatment (factorial ANOVA, main effect of AmILP1 dsRNA: N=20, F1,76,glucose=0.1310, P=0.7184 and F1,76,trehalose=0.11530, P=0.6968; main effect of AmILP2 dsRNA: F1,76,glucose=0.8100, P=0.1825 and F1,76,trehalose=0.7659, P=0.3843, Fig. 2A,B,D,E). There was also no interaction between AmILP1 dsRNA and AmILP2 dsRNA treatment on the content of either sugar (factorial ANOVA, F1,76,glucose=0.9799, P=0.3254; F1,76,trehalose=0.1190, P=0.7311, Fig. 2C,F). Considering that only AmILP2 knockdown was validated statistically at the whole-body RNA level in larvae, we infer that AmILP2 does not directly regulate hemolymph carbohydrate reserves in honey bee larvae.
Fig. 2.
Glucose and trehalose levels in the hemolymph of larvae treated with AmILP1 and AmILP2 dsRNA. (A,B,D,E) The main effects of AmILP1 dsRNA and AmILP2 dsRNA on glucose and trehalose levels in a factorial ANOVA. (C,F) The relationships between four treatment groups by LSD post hoc test: the bars from left to right represent gfp, AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. There was no main effect of either AmILP1 dsRNA or AmILP2 dsRNA on glucose (A,B) and trehalose (D,E) titers. There was no difference in the four treatment groups with respect to glucose and trehalose titers (C,F). Data are shown as means ± s.e.m. (N=20). ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.
Hemolymph JH titers in AmILP1 and AmILP2 dsRNA-treated larvae
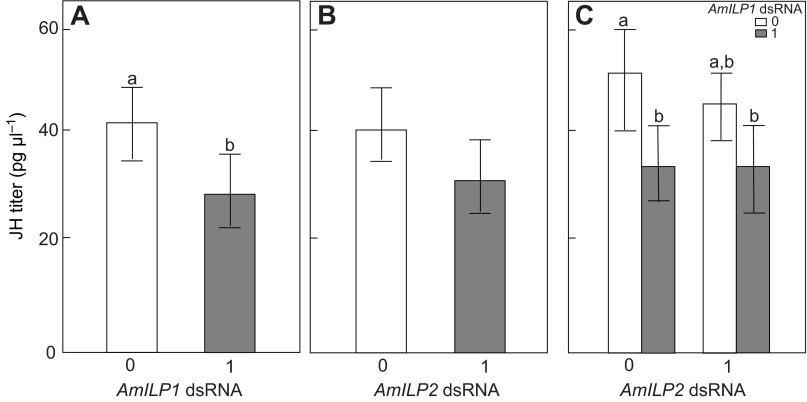
JH is a central regulator controlling queen caste development, and its levels can be regulated by EGF signaling (Kamakura, 2011) and affected by both IRS and TOR knockdown (Jin and Esteva, 2008; Mutti et al., 2011a). Therefore, examining whether JH is affected by AmILP knockdown is key to understanding the relationship between JH and IIS in honey bee larvae. By measuring the JH titers in larval hemolymph by means of a specific RIA, we found that the JH titers were significantly decreased by AmILP1 dsRNA (factorial ANOVA, N=16–19, F1,67=5.0970, P=0.0272, Fig. 3A), but not by AmILP2 RNAi (factorial ANOVA, N=16–19, F1,67=1.7474, P=0.1907, Fig. 3B). There was no significant interaction effect between AmILP1 and AmILP2 dsRNA treatments (factorial ANOVA, F1,76=1.4309, P=0.2358, Fig. 3C), indicating that the effect of AmILP1 dsRNA on JH was independent of AmILP2 dsRNA. Post hoc analysis further showed that the larvae treated with AmILP1 dsRNA (Fisher LSD: P=0.0114) and the larvae treated with combined AmILP1 and AmILP2 dsRNA (Fisher LSD: P=0.0135) had lower JH levels compared with gfp controls (Fig. 3C). As we could not verify a knockdown in terms of AmILP1 transcript level in the same individual larva, these results raised an interesting question about whether the change in JH titers in the bees treated with AmILP1 dsRNA was specific. It is, however, plausible that the whole-body RNA levels measured may have masked changes in AmILP1 transcript levels that only occurred in a small subset of cells, such as in the neuroendocrine axis.
Fig. 3.
Hemolymph juvenile hormone (JH) titer in larvae treated with AmILP1 and AmILP2 dsRNA. (A,B,D,E) The main effects of AmILP1 dsRNA and AmILP2 dsRNA on JH titer in a factorial ANOVA. (C,F) The relationships between four treatment groups as revelaed by a LSD post hoc test: the bars from left to right represent gfp, AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. There was a significant main effect of AmILP1 dsRNA on JH titer (A), but no main effect of AmILP2 dsRNA on the JH titer (B). Panel C shows that, compared with gfp, both single AmILP1 dsRNA treatment and AmILP1 dsRNA plus AmILp2 dsRNA treatment reduced JH titer, but single AmILP2 dsRNA treatment did not do so, indicating that only AmILP1 dsRNA contributed to the reduction of JH titer in A. Data are presented as means ± s.e.m. (N=16–19). Different letters indicate significant differences among treatments. ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.
Body mass
There is a correlation between body mass and ovariole number in honey bees (Linksvayer et al., 2011; Snodgrass, 1956), but a recent in vitro rearing study has shown that body size can be independent of ovariole number and other queen phenotype characters (Linksvayer et al., 2011), indicating that caste morphological traits may be regulated by separate pathways (Kamakura, 2011). In our study, the body mass of fifth instar larvae was not affected by either AmILP1 dsRNA or AmILP2 dsRNA (factorial ANOVA, N=20, main effect of AmILP1 dsRNA: F1,76=1.3834, P=0.2432; main effect of AmILP2 dsRNA, F1,76=1.6561, P=0.2020, Fig. 4A,B), but there was a significant interaction effect between AmILP1 and AmILP2 dsRNAs (factorial ANOVA, F1,76=5.5990, P=0.0205, Fig. 4C). These results suggest that the effect of AmILP1 dsRNA on the body mass of fifth instar larvae depends on the level of AmILP2 dsRNA: AmILP1 dsRNA increased the body mass in the absence of AmILP2 dsRNA, but combined AmILP1 dsRNA and AmILP2 dsRNA treatment tended to reduce body mass. Post hoc analysis further showed that larvae treated with AmILP1 dsRNA were heavier than gfp control larvae (post hoc LSD: P=0.0010) and larvae treated with both AmILP1 and AmILP2 dsRNAs (post hoc LSD: P=0.0465).
Fig. 4.
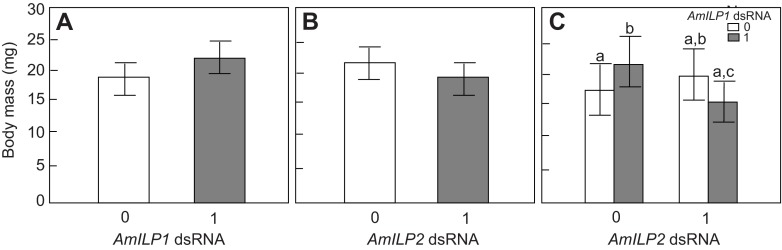
Effects of AmILP1 and AmILP2 dsRNA treatment on body mass of fifth instar larvae. (A,B,D,E) The main effects of AmILP1 dsRNA and AmILP2 dsRNA on body mass in a factorial ANOVA. (C,F) The relationships between four treatment groups as revealed by a LSD post hoc test: the bars from left to right represent gfp, AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. There was no main effect of either AmILP1 dsRNA or AmILP2 dsRNA on body mass (A,B). Panel C shows that AmILP1 dsRNA treatment increased body mass in the absence of AmILP2 dsRNA, but the combination of AmILP1 dsRNA and AmILP2 dsRNA tended to reduce body mass. Together with the result of a significant interaction between AmILP1 dsRNA and AmILP2 dsRNA treatments on body mass, these results suggest that the effect of AmILP1 dsRNA on body mass of the fifth instar larvae depends on the level of AmILP2 dsRNA. Data are represented as means ± s.e.m. (N=20). Different letters indicate significant differences among treatments. ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.
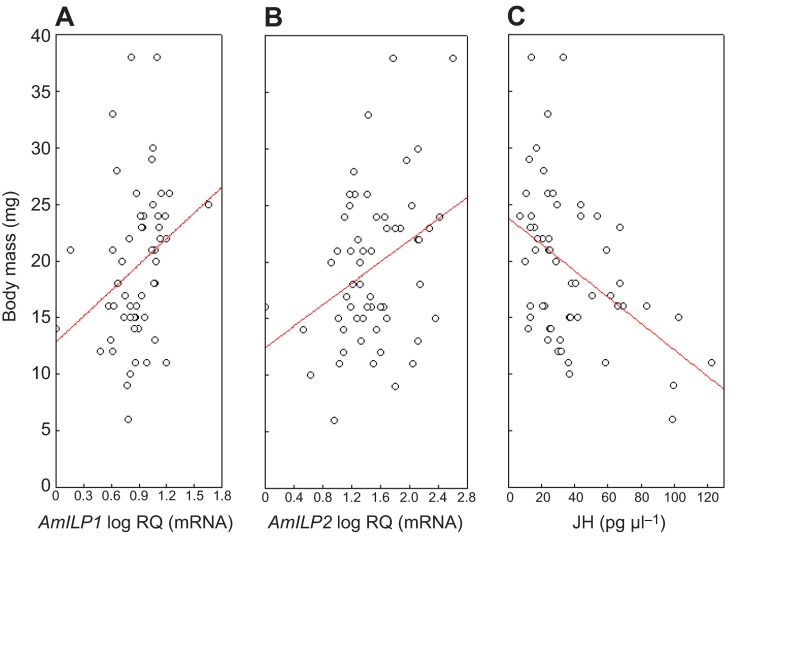
As AmILPs and JH are thought to be interconnected in the regulation of honey bee caste development, we also plotted the respective larval mass against AmILP1 and AmILP2 expression and JH titers to explore putative associations. For both AmILP1 and AmILP2, we found significant positive correlations with larval mass (Pearson correlation, N=57, AmILP1: P=0.0250; AmILP2: P=0.0092, Fig. 5A,B), suggesting that both AmILP genes are involved in regulating larval development in either a direct or an indirect way, supporting a hypothesis for general functions of AmILP1 and AmILP2 in honey bee development. In contrast, JH titers were negatively correlated with larval mass (Pearson correlation, N=57, P=0.0004, Fig. 5C). This negative correlation seems contradictory to the general role of JH in honey bee caste differentiation. However, the fifth instar is a critical stage to initiate honey bee metamorphosis, coordinated in concert by JH and ecdysteroid titers. Perhaps such dynamic changes (temporal or rapid) resulted in the negative relationship between JH and body mass. Nonetheless, further investigation is needed to test this hypothesis.
Fig. 5.
Correlation between larval body mass and AmILP expression or JH level. AmILP1 transcript abundance was positively correlated with body mass (A); AmILP2 transcript abundance was also positively correlated with body mass (B); but JH titer was negatively correlated with body mass (C). RQ is relative quantification. Circles represent individual larvae from all four RNA interference (RNAi) treatments; regression lines were obtained by Pearson correlation analysis.
Ovariole number and other morphological traits
Ovaries were dissected and ovariole number was counted after adult eclosion. We found that ovariole number was significantly reduced in bees subject to AmILP2 RNAi (factorial ANOVA, N=12–27, F1,67=5.2069, P=0.0257, Fig. 6B) but not to AmILP1 dsRNA (factorial ANOVA, N=12–27, F1,67=1.1859, P=0.2801, Fig. 6A). There was no interaction between AmILP1 and AmILP2 dsRNA treatments (factorial ANOVA, F1,67=0.0642, P=0.8008, Fig. 6C), suggesting AmILP2 dsRNA reduced ovariole number independent of AmILP1 dsRNA. Post hoc analysis showed that bees treated with AmILP2 dsRNA (Fisher LSD: P=0.0067) and the bees treated with a combination of AmILP1 and AmILP2 dsRNA (Fisher LSD: P=0.0570) had significantly fewer ovarioles than bees treated with AmILP1 dsRNA alone.
Fig. 6.
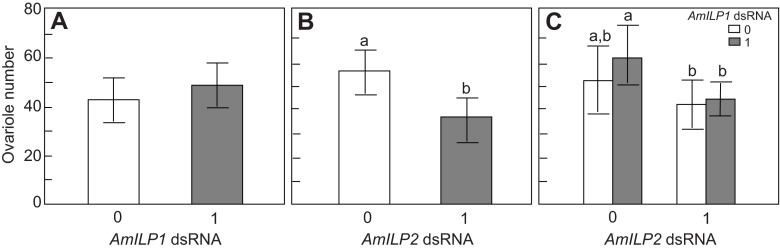
Effects of AmILP1 and AmILP2 dsRNA treatment on ovariole number of newly emerged adults. (A,B) The main effects of AmILP1 dsRNA and AmILP2 dsRNA on ovariole number in a factorial ANOVA. (C) The relationships between the four treatment groups by LSD post hoc test: the bars from left to right represent gfp, AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. Even though there was no main effect of AmILP1 dsRNA on ovariole number (A), ovariole number was significantly reduced by AmILP2 RNAi (B). Panel C shows that compared with gfp, both AmILP2 dsRNA treatment alone and AmILP1 dsRNA plus AmILP2 dsRNA treatment tended to reduce ovariole number, whereas AmILP1 dsRNA treatment tended to increase ovariole number. As we did not find a significant interaction between AmILP1 and AmILP2 dsRNA treatment on ovariole number (C), these results suggest that the main effect of AmILP2 dsRNA on ovary development in B is independent of AmILP1 dsRNA. Data are presented as means ± s.e.m. (N=12–27). Different letters indicate significant differences among treatments. ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.
Other morphological characters, such as mandible shape, stinger shape, size of spermatheca and presence/absence of a corbicula, were also monitored based on an established protocol (Mutti et al., 2011b; Patel et al., 2007). There was no main effect of either AmILP1 dsRNA or AmILP2 dsRNA, and no interaction effect on any of these morphological characters (factorial ANOVA, P>0.05; detailed results can be found in supplementary material Table S3).
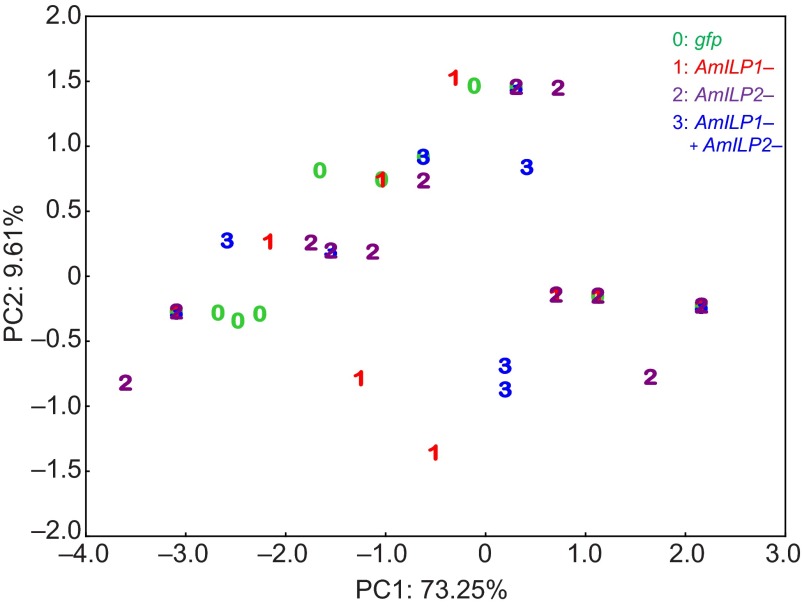
A PCA was utilized to clarify general patterns, similarities or separations of cases between the different treatment groups by reducing the dimensions of morphological variables. This revealed that 73.25% of the total variation can be explained by the first principal component (PC1), and 9.61% of the remaining total variation can be explained by the second principal component (PC2). The eigenvalue of PC1 was 3.6623, and other PCs did not exceed 1 (supplementary material Table S4), meaning that PC1 contributed more to the variance than the original variables, but other PCs could be considered as sampling noise. All the variables (morphological characters) contributed almost equally to PC1 (contributions: 17–22%, supplementary material Table S5). Analysis of PC1 versus PC2 (Fig. 7) revealed that along PC1 there was no clear separation among cases (bees) according to treatment. Next, we carried out a Kruskal–Wallis ANOVA to test whether the distribution of samples in treatment groups in PC1 differed. There was no difference in the distribution of samples among the treatment groups in PC1 (the combined variable) (Kruskal–Wallis ANOVA: χ2=4.7526, P=0.1908), suggesting that the treatments did not significantly influence queen–worker caste differentiation, which is characterized by these multiple morphological characters, though AmILP2 RNAi significantly affected ovariole number.
Fig. 7.
Score plot of PC1 and PC2 of a principal component analysis (PCA) on the multiple morphological traits. There is no clear separation of the bees among the treatment groups.
DISCUSSION
By using a gene knockdown approach, we herein performed the first functional study to investigate the role of insulin peptide-encoding genes, AmILP1 and AmILP2, in queen–worker differentiation during honey bee larval development. Our data show that AmILP2 expression was susceptible to AmILP2 RNAi when AmILP1 dsRNA was used simultaneously, which resulted in diminished transcript levels in the whole larval body. Although AmILP2 dsRNA did not cause any change in hemolymph JH levels, it had an effect on ovariole number of adult bees. In contrast, AmILP1 expression at the whole-body level was not affected by AmILP1 dsRNA treatment, but the hemolymph JH levels in these larvae were significantly reduced by the treatment. Thus, a general conclusion that can be drawn from these results is that AmILP1 and AmILP2 dsRNAs have differential efficacies to downregulate the target genes in the whole larval body.
Efficacies of AmILP1 and AmILP2 RNAi in the fat body
RNAi efficacy is affected by many factors, such as the specificity of the dsRNA, the RNAi delivery method, the expression level of the gene, cell types in the target tissue and the nature of the regulatory machinery of RNAi. In honey bees, both AmILP-encoding genes are represented by a single copy each in the honey bee genome (de Azevedo and Hartfelder, 2008). Their transcripts are relatively short (around 400 bp), and there is no evidence for transcript variants. Additionally, the dsRNAs were designed to target 260–280 bp regions of AmILP1 and AmILP2, and no off-target matches were found by alignments against the honey bee genome. Furthermore, the final concentration of dsRNA (200 μg ml−1) in the larval diet and the in vitro rearing protocol has been validated in previous TOR and IRS knockdown studies (Mutti et al., 2011b; Patel et al., 2007). In this study, we achieved an overall 30% reduction in AmILP2 transcript levels by AmILP2 RNAi. Therefore, it is unlikely that dsRNA specificity and the protocol have issues resulting in the differential efficacies between AmILP1 and AmILP2 RNAi.
A factor that may explain why we did not achieve a significant AmILP1 gene knockdown could be the low level of AmILP1 expression in early fifth instar larvae (de Azevedo and Hartfelder, 2008). So, the difficulty encountered in achieving AmILP1 knockdown could be related to the general difficulty in downregulating a gene with low transcript abundance. Additionally, differential cell-type specificities between AmILP gene expression and dsRNA targeting may be another reason for the differential RNAi efficacies. In adult honey bees, the tissue that best responds to dsRNA treatment is the fat body (Amdam et al., 2003; Jarosch and Moritz, 2011), and it is also the predominant tissue type in larvae. The insect fat body is composed of two cell types, trophocytes and oenocytes. A recent study has revealed that the expression of AmILP1 and AmILP2 in honey bee fat body has different cell specificities: AmILP1 is highly expressed in oenocytes and AmILP2 is expressed strongly in both oenocytes and trophocytes (Nilsen et al., 2011). However, the preferential uptake characteristics of dsRNA molecules by oenocytes and trophocytes are different, with trophocytes uptaking considerably more dsRNA than oenocytes (Jarosch and Moritz, 2011). Therefore, the lack of a significant AmILP1 knockdown in our experiments may be due to the poor AmILP1 dsRNA uptake capability of oenocytes (Jarosch and Moritz, 2011), as well as the low transcript abundance of AmILP1 in the developmental stage. As gene knockdown is dose dependent, increasing the dosage or extending dsRNA feeding time may raise the success rate for knocking down the AmILP1 gene in the fat body in future studies.
In this study, the AmILP2 gene was not directly knocked down when its dsRNA was applied alone. One of the reasons could again be a low level of AmILP2 expression in fifth instar larvae (de Azevedo and Hartfelder, 2008). However, we found that AmILP2 dsRNA significantly downregulated AmILP2 gene expression when AmILP1 dsRNA was applied simultaneously, whereas AmILP1 dsRNA treatment caused an increase in AmILP2 mRNA. Though there is no simple explanation for this phenomenon, it is worthy of note that the effect of RNAi can be physiologically amplified and systemically spread in some organisms including Caenorhabditis elegans and certain insects (Tomoyasu et al., 2008; Miller et al., 2012). These processes involve RNA-directed RNA polymerase activity, which depends on high levels of expression of target RNA (Dougherty and Parks, 1995; Sijen et al., 2001). Therefore, the potency of AmILP2 dsRNA might have been enhanced once the AmILP2 transcript level was increased by AmILP1 dsRNA. As the RNAi machinery includes both transcriptional and post-transcriptional gene silencing modes (Noma et al., 2004), this can involve complex negative and positive feedback (Xie et al., 2003; Grewal and Elgin, 2007). In addition, regulatory mechanisms in RNAi vary among organisms (Tomoyasu et al., 2008), and the way in which RNAi is controlled and regulated in insects is still poorly understood. Therefore, future studies directed towards detecting and identifying regulatory mechanisms of RNAi in insects are likely to shed light on the question of how AmILP1 dsRNA could enhance the effect of AmILP2 dsRNA.
Potential relationship of brain AmILP1 to JH production and AmILP2
Interestingly, we observed a significant reduction of JH in response to AmILP1 dsRNA treatment, even though no significant downregulation was achieved for this gene at the whole-body level. It is worthy of note that the majority of ILPs are produced in the brain of most insect species (Antonova et al., 2012; Brogiolo et al., 2001; Iga and Smagghe, 2011; Riehle et al., 2006), including honey bees (Ament et al., 2008; Corona et al., 2007) and Drosophila (Brogiolo et al., 2001). And JH is synthesized in the closely associated CA of the insect retrocerebral complex (Goodman and Cusson, 2012), as also shown for honey bee larvae (Rachinsky and Hartfelder, 1990). In Drosophila, the small cluster of AmILP-producing neuroendocrine cells was shown to transmit ILPs to the JH-producing CA (Krieger et al., 2004) by axons directly projecting to the ring gland (Cao and Brown, 2001; Géminard et al., 2006). In line with these findings (Lane and Swales, 1978; Restifo et al., 1995), we hypothesize that a cluster of AmILP-producing cells in the brain of honey bee larvae may have been targeted by AmILP1 dsRNA, which consequently affected JH production in the CA. Although the adult honey bee brain has been shown to be resilient to dsRNA treatment (Farooqui et al., 2004; Jarosch and Moritz, 2011), it is possible that the larval hemolymph–brain barrier could be more leaky than that of adults (Lane and Swales, 1978; Restifo et al., 1995), especially during the onset of metamorphosis.
Furthermore, several studies have provided evidence for a positive regulation of ILP expression by JH in many insect species including Drosophila (Corona et al., 2007; Sheng et al., 2011; Tu et al., 2005). Reciprocally, it was found that JH synthesis was modulated by brain Drosophila ILPs (Tatar et al., 2003). In honey bees, treatment with methoprene (a JH analog) positively affected brain AmILP1 levels in both adult queens and workers (Corona et al., 2007). Our recent study also suggested that AmILP1 expression in the fat body is negatively linked to hemolymph JH titers in adult worker bees (Wang et al., 2012). In addition, a connection between AmILP1 expression and JH synthesis was proposed based on the temporal coincidence between the peaks of AmILP1 expression and JH titers in honey bee larvae (Wheeler et al., 2006). Finally, interference with downstream regulators of IIS and/or EGF signaling, such as the IRS (Mutti et al., 2011b) and TOR genes (Patel et al., 2007), resulted in a decrease in JH titers (Mutti et al., 2011a). Taken together, the reduction in JH levels seen as a result of AmILP1 dsRNA treatment is likely a specific effect of AmILP1 RNAi, and our study provides the first evidence that brain AmILP1 may regulate JH production in honey bee larvae.
Finally, our study indicates that the expression of brain AmILP1 and fat body AmILP2 is correlated in honey bees. In Drosophila, overexpression of insulin-like peptides (DILPs) in the fat body inhibited brain DILP secretions (Bai et al., 2012), and gene knockouts of DILPs in the brain caused synergy and compensation of expression of DILPs in the fat body (Grönke et al., 2010). In honey bees, previous studies have suggested that AmILP1 and AmILP2 act as an agonist and an antagonist, respectively, of InRs in the brain regulating JH secretion (Nilsen et al., 2011). However, how brain AmILPs connect with fat body AmILPs is poorly understood. Here, we found that AmILP1 dsRNA was able to increase AmILP2 transcript abundance in the whole body of fifth instar larvae (Fig. 1F), with the fat body making the major tissue contribution. As AmILP1 dsRNA probably affects brain AmILP1 secretion in fifth instar larvae, our findings suggest that fat body AmILP2 compensates for the downregulation of brain AmILP1, thus representing a circuitry similar to that found in Drosophila. Although AmILP1 dsRNA also induced a decrease in JH titers, we did not find any correlation between JH and fat body AmILP2 levels in these fifth instar larvae (supplementary material Fig. S1), suggesting that JH is not involved in this hypothetical compensatory response of fat body AmILP2.
Roles of JH and AmILP2 in worker caste development
Experimental and modeling evidence supports the suggestion that an elevated JH titer during the fourth and early fifth instar of honey bees inhibits the induction of autophagic cell death in the larval ovary (Schmidt Capella and Hartfelder, 2002), and rescues the queen phenotype after IRS and/or TOR gene knockdown (Mutti et al., 2011a). Therefore, a logical conclusion would be that decreasing JH levels would promote ovary degradation and induce the worker phenotype. In our study, however, the main effect of AmILP1 dsRNA was an ~40% reduction in JH titers, but there was no apparent effect on ovary degradation and caste characters in general, suggesting that other regulators in addition to a low JH titer may be required for full worker phenotype development. This is supported by the finding that the downregulation of AmILP2 transcript abundance in the fat body, in addition to an ~35% (though statistically not significant) reduction in JH titers in these larvae was associated with fewer ovarioles. Together with the fact that downregulation of IRS and TOR in the larval fat body reduces JH, this suggests that the fat body secretes regulators that modulate CA activity.
Moreover, our results indicate that AmILP1 and AmILP2 have different roles during honey bee larval development, which is also consistent with their expression profiles (de Azevedo and Hartfelder, 2008). In Drosophila, different ILPs show tissue-specific functions. Whereas brain ILPs tend to regulate energy metabolism and control hemolymph sugar titers (Broughton et al., 2005; Rulifson et al., 2002), fat body ILPs are the functional equivalent of insulin-like growth factor 1 (IGF1), modulating cell proliferation and organ growth (Okamoto et al., 2009). It has already been suggested that AmILP2 may act as an IGF in the larval fat body of honey bees (Wheeler et al., 2006), but functional evidence for this hypothesis was lacking. Our data now indicate that AmILP2 knockdown in the fat body does not modulate hemolymph sugar levels during larval development, but instead AmILP2 is more related to ovary development and body mass in larvae.
Our study suggests that fat body AmILP2 more likely contributes to regulating ovariole development rather than all worker traits, as AmILP2 knockdown did not significantly affect the expression of other worker traits. Additionally, both JH and ILPs are involved in anti-apoptosis in many other insect species (Schmidt Capella and Hartfelder, 2002; Johnson et al., 2006). Therefore, AmILP2 knockdown in the fat body may mediate ovary degradation at the end of larval development. As AmILP2 knockdown did not change JH titers and the AmILP2 mRNA level was not significantly correlated with JH titers, AmILP2 may be indirectly connected with JH through other regulators such as AmILP1. Clearly, further studies are needed to test the hypothesis.
Moreover, ovary size and body size in adult bees generally are correlated (Linksvayer et al., 2011), suggesting their regulatory pathways may have common elements. Studies in Drosophila showed that fat body DILPs are involved in regulating body size (Okamoto et al., 2009) and fat cell mass (DiAngelo and Birnbaum, 2009). Here, we found that AmILP1 dsRNA treatment significantly increased AmILP2 transcript abundance at the whole-body level (mainly fat bodies), and also increased the body mass of fifth instar larvae, leading us to infer that fat body AmILP2 in honey bee larva may also play a role in determining body mass.
CONCLUSIONS
To summarize, by means of an RNAi approach we demonstrated that AmILP2 expressed in the fat body is directly involved in the expression of a queen-type ovary during honey bee caste development, whereas AmILP1 may have an indirect effect via modulation of JH production in the CA. Thus, we propose that the regulation of worker caste development is not simply a reversed pathway of queen caste development; instead, a network of regulators must cooperate with JH to drive worker development. Furthermore, rather than being an insulin-like peptide, the function of AmILP2 appears to be similar to IGF, regulating cell and organ growth. In agreement with other studies (Kamakura, 2011; Mutti et al., 2011a), our study supports the suggestion that the IIS pathway has a modulatory and probably only minor role in caste development of honey bees. Nonetheless, as binding affinities of AmILPs to AmInRs have not yet been investigated in honey bees, the exact role of AmILPs and AmInRs, especially their interactions with other local signaling and endocrine pathways, are still puzzling in our understanding of honey bee development and physiology.
Supplementary Material
ACKNOWLEDGEMENTS
We thank O. Kaftanoglu for his assistance with experiments, and A. Dolezal, K. Dolezal, J. Gibson, T. L. Zhang and three anonymous reviewers for helpful comments.
LIST OF ABBREVIATIONS
- AmILP
Apis mellifera insulin-like peptide
- AmInR
Apis mellifera insulin receptor
- CA
corpora allata
- DILP
Drosophila insulin-like peptide
- dsRNA
double-stranded RNA
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- GFP
green fluorescent protein
- IGF
insulin-like growth factor
- IIS
insulin/insulin-like growth factor 1 signaling
- ILP
insulin-like peptide
- IRS
insulin receptor substrate
- JH
juvenile hormone
- RNAi
RNA interference
- RT-qPCR
real-time quantitative PCR
- TOR
target of rapamycin
FOOTNOTES
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/216/23/4347/DC1
COMPETING INTERESTS
No competing interests declared.
FUNDING
This research was supported by the Research Council of Norway [grant nos 180504 and 185306 to G.V.A.] and the National Institute on Aging [grant no. NIA P01 AG22500 to Robert E. Page, Jr and G.V.A.). S.V.A. received a PhD fellowship from Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP 2007/04859-5]. Deposited in PMC for release after 12 months.
REFERENCES
- Amdam G. V. (2011). Social context, stress, and plasticity of aging. Aging Cell 10, 18-27 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Simões Z. L., Guidugli K. R., Norberg K., Omholt S. W. (2003). Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol. 3, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Csondes A., Fondrk M. K., Page R. E., Jr (2006). Complex social behaviour derived from maternal reproductive traits. Nature 439, 76-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Nilsen K. A., Norberg K., Fondrk M. K., Hartfelder K. (2007). Variation in endocrine signaling underlies variation in social life history. Am. Nat. 170, 37-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Corona M., Pollock H. S., Robinson G. E. (2008). Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl. Acad. Sci. USA 105, 4226-4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Wang Y., Robinson G. E. (2010). Nutritional regulation of division of labor in honey bees: toward a systems biology perspective. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 566-576 [DOI] [PubMed] [Google Scholar]
- Antonova Y., Arik A. A., Moore W., Riehle M., Brown M. R. (2012). Insulin-like Peptides: Structure, Signaling and Function. London: Academic Press; [Google Scholar]
- Asencot M., Lensky Y. (1985). The phagostimulatory effect of sugars on the induction of queenliness in female honeybee (Apis mellifera L.) larvae. Comp. Biochem. Physiol. 81A, 203-208 [Google Scholar]
- Bai H., Kang P., Tatar M. (2012). Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11, 978-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar Y., Leung H. T., Pak W. L., Sokolowski M. B., Robinson G. E. (2003). cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. J. Exp. Biol. 206, 2507-2515 [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213-221 [DOI] [PubMed] [Google Scholar]
- Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D. J., Leevers S. J., et al. (2005). Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102, 3105-3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R., Duncan E., Dearden P. K. (2013). Stable reference genes for the measurement of transcript abundance during larval caste development in the honeybee. Apidologie 44, 357-366 [Google Scholar]
- Cao C., Brown M. R. (2001). Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res. 304, 317-321 [DOI] [PubMed] [Google Scholar]
- Chapman R. F. (1998). The Insects: Structure and Function. Cambridge: Cambridge University Press; [Google Scholar]
- Corona M., Velarde R. A., Remolina S., Moran-Lauter A., Wang Y., Hughes K. A., Robinson G. E. (2007). Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 104, 7128-7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo S. V., Hartfelder K. (2008). The insulin signaling pathway in honey bee (Apis mellifera) caste development – differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 54, 1064-1071 [DOI] [PubMed] [Google Scholar]
- DiAngelo J. R., Birnbaum M. J. (2009). Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol. Cell. Biol. 29, 6341-6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty W. G., Parks T. D. (1995). Transgenes and gene suppression: telling us something new? Curr. Opin. Cell Biol. 7, 399-405 [DOI] [PubMed] [Google Scholar]
- Farooqui T., Vaessin H., Smith B. H. (2004). Octopamine receptors in the honeybee (Apis mellifera) brain and their disruption by RNA-mediated interference. J. Insect Physiol. 50, 701-713 [DOI] [PubMed] [Google Scholar]
- Géminard C., Arquier N., Layalle S., Bourouis M., Slaidina M., Delanoue R., Bjordal M., Ohanna M., Ma M., Colombani J., et al. (2006). Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes 55, S5-S8 [Google Scholar]
- Goewie E. A. (1977). Induction of caste differentiation in honey bee (Apis mellifera L.) after topical application of JH-III. Insectes Soc. 24, 265 [Google Scholar]
- Goodman W. G., Cusson M. (2012). The juvenile hormones. In Insect Endocrinology (ed. Gilbert L. I.), pp. 310-365 London: Academic Press; [Google Scholar]
- Goodman W. G., Coy D. C., Baker F. C., Xu L., Toong Y. C. (1990). Development and application of a radioimmunoassay for the juvenile hormones. Insect Biochem. 20, 357-364 [Google Scholar]
- Grewal S. I., Elgin S. C. (2007). Transcription and RNA interference in the formation of heterochromatin. Nature 447, 399-406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D. F., Broughton S., Andrews T. D., Partridge L. (2010). Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6, e1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli K. R., Nascimento A. M., Amdam G. V., Barchuk A. R., Omholt S. W., Simões Z. L. P., Hartfelder K. (2005). Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579, 4961-4965 [DOI] [PubMed] [Google Scholar]
- Hartfelder K., Emlen D. E. (2012). Endocrine control of insect polyphenism. In Insect Endocrinology (ed. Gilbert L. I.), pp. 464-522 London: Academic Press; [Google Scholar]
- Hartfelder K., Bitondi M. M. G., Brent C. S., Guidugli-Lazzarini K. R., Simões Z. L., Stabentheiner A., Tanaka E. D., Wang Y. (2013). Standard methods for physiology and biochemistry research in Apis mellifera. J. Apicult. Res. 52, doi: 10.3896/IBRA.1.52.1.06 [Google Scholar]
- Haydak M. H. (1970). Honey bee nutrition. Annu. Rev. Entomol. 15, 143-156 [Google Scholar]
- Hölldobler B., Wilson E. O. (2008). The Superorganism: The Beauty, Elegance and Strangeness of Insect Societies. New York, NY; London: W. W. Norton; [Google Scholar]
- Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Y., Robinson G. E., Borst D. W. (1994). Physiological correlates of division of labor among similarly aged honey bees. J. Comp. Physiol. A 174, 731-739 [DOI] [PubMed] [Google Scholar]
- Iga M., Smagghe G. (2011). Relationship between larval–pupal metamorphosis and transcript expression of insulin-like peptide and insulin receptor in Spodoptera littoralis. Peptides 32, 531-538 [DOI] [PubMed] [Google Scholar]
- Jarosch A., Moritz R. F. A. (2011). Systemic RNA-interference in the honeybee Apis mellifera: tissue dependent uptake of fluorescent siRNA after intra-abdominal application observed by laser-scanning microscopy. J. Insect Physiol. 57, 851-857 [DOI] [PubMed] [Google Scholar]
- Jin Q., Esteva F. J. (2008). Cross-talk between the ErbB/HER family and the type I insulin-like growth factor receptor signaling pathway in breast cancer. J. Mammary Gland Biol. Neoplasia 13, 485-498 [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Bernal-Mizrachi E., Alejandro E. U., Han Z., Kalynyak T. B., Li H., Beith J. L., Gross J., Warnock G. L., Townsend R. R., et al. (2006). Insulin protects islets from apoptosis via Pdx1 and specific changes in the human islet proteome. Proc. Natl. Acad. Sci. USA 103, 19575-19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura M. (2011). Royalactin induces queen differentiation in honeybees. Nature 473, 478-483 [DOI] [PubMed] [Google Scholar]
- Krieger M. J., Jahan N., Riehle M. A., Cao C., Brown M. R. (2004). Molecular characterization of insulin-like peptide genes and their expression in the African malaria mosquito, Anopheles gambiae. Insect Mol. Biol. 13, 305-315 [DOI] [PubMed] [Google Scholar]
- Lane N. J., Swales L. S. (1978). Changes in the blood-brain barrier of the central nervous system in the blowfly during development, with special reference to the formation and disaggregation of gap and tight junctions. I. Larval development. Dev. Biol. 62, 389-414 [DOI] [PubMed] [Google Scholar]
- Leimar O., Hartfelder K., Laubichler M. D., Page R. E., Jr (2012). Development and evolution of caste dimorphism in honeybees – a modeling approach. Ecol. Evol. 2, 3098-3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer T. A., Kaftanoglu O., Akyol E., Blatch S., Amdam G. V., Page R. E., Jr (2011). Larval and nurse worker control of developmental plasticity and the evolution of honey bee queen–worker dimorphism. J. Evol. Biol. 24, 1939-1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402-408 [DOI] [PubMed] [Google Scholar]
- Lourenço A. P., Mackert A., Cristino A. D., Simões Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie (Celle) 39, 372-385 [Google Scholar]
- Martins J. R., Nunes F. M. F., Cristino A. S., Simões Z. L., Bitondi M. M. G. (2010). The four hexamerin genes in the honey bee: structure, molecular evolution and function deduced from expression patterns in queens, workers and drones. BMC Mol. Biol. 11, 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener C. D. (2000). The Bees of the World. Baltimore, MD: Johns Hopkins University Press; [Google Scholar]
- Miller S. C., Miyata K., Brown S. J., Tomoyasu Y. (2012). Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE 7, e47431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., Riddiford L. M. (2007). Size assessment and growth control: how adult size is determined in insects. Bioessays 29, 344-355 [DOI] [PubMed] [Google Scholar]
- Montgomery D. C. (1997). Design And Analysis Of Experiments. Hoboken, NJ: John Wiley; [Google Scholar]
- Mutti N. S., Dolezal A. G., Wolschin F., Mutti J. S., Gill K. S., Amdam G. V. (2011a). IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 214, 3977-3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N. S., Wang Y., Kaftanoglu O., Amdam G. V. (2011b). Honey bee PTEN—description, developmental knockdown, and tissue-specific expression of splice-variants correlated with alternative social phenotypes. PLoS ONE 6, e22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navajas M., Migeon A., Alaux C., Martin-Magniette M., Robinson G., Evans J., Cros-Arteil S., Crauser D., Le Conte Y. (2008). Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics 9, 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen K. A., Ihle K. E., Frederick K., Fondrk M. K., Smedal B., Hartfelder K., Amdam G. V. (2011). Insulin-like peptide genes in honey bee fat body respond differently to manipulation of social behavioral physiology. J. Exp. Biol. 214, 1488-1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S. I. (2004). RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36, 1174-1180 [DOI] [PubMed] [Google Scholar]
- Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H., O'Connor M. B., Mizoguchi A. (2009). A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17, 885-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Fondrk M. K., Kaftanoglu O., Emore C., Hunt G., Frederick K., Amdam G. V. (2007). The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE 2, e509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinsky A., Hartfelder K. (1990). Corpora allata activity, a prime regulating element for caste-specific juvenile hormone titre in honey bee larvae (Apis mellifera carnica). J. Insect Physiol. 36, 189-194 [Google Scholar]
- Rachinsky A., Strambi C., Strambi A., Hartfelder K. (1990). Caste and metamorphosis: hemolymph titers of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen. Comp. Endocrinol. 79, 31-38 [DOI] [PubMed] [Google Scholar]
- Reim T., Thamm M., Rolke D., Blenau W., Scheiner R. (2013). Suitability of three common reference genes for quantitative real-time PCR in honey bees. Apidologie (Celle) 44, 342-350 [Google Scholar]
- Rembold H. (1987). Caste specific modulation of juvenile hormone titers in Apis mellifera. Insect Biochem. 17, 1003-1006 [Google Scholar]
- Rembold H., Czoppelt C., Rao P. J. (1974). Effect of juvenile hormone treatment on caste differentiation in the honeybee, Apis mellifera. J. Insect Physiol. 20, 1193-1202 [DOI] [PubMed] [Google Scholar]
- Restifo L. L., Estes P. S., Russo C. D. (1995). Genetics of ecdysteroid-regulated central nervous system metamorphosis in Drosophila (Diptera: Drosophilidae). Eur. J. Entomol. 92, 169-187 [Google Scholar]
- Riehle M. A., Fan Y., Cao C., Brown M. R. (2006). Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides 27, 2547-2560 [DOI] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118-1120 [DOI] [PubMed] [Google Scholar]
- Saltiel A. R., Kahn C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799-806 [DOI] [PubMed] [Google Scholar]
- Schmidt Capella I. C., Hartfelder K. (2002). Juvenile-hormone-dependent interaction of actin and spectrin is crucial for polymorphic differentiation of the larval honey bee ovary. Cell Tissue Res. 307, 265-272 [DOI] [PubMed] [Google Scholar]
- Sheng Z. T., Xu J. J., Bai H., Zhu F., Palli S. R. (2011). Juvenile hormone regulates vitellogenin gene expression through insulin-like peptide signaling pathway in the red flour beetle, Tribolium castaneum. J. Biol. Chem. 286, 41924-41936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H., Fire A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465-476 [DOI] [PubMed] [Google Scholar]
- Snodgrass R. E. (1956). Anatomy of the Honey Bee. New York, NY: Comstock Publishing Associates; [Google Scholar]
- Tatar M., Bartke A., Antebi A. (2003). The endocrine regulation of aging by insulin-like signals. Science 299, 1346-1351 [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y., Miller S. C., Tomita S., Schoppmeier M., Grossmann D., Bucher G. (2008). Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9, R10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M. P., Yin C. M., Tatar M. (2005). Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen. Comp. Endocrinol. 142, 347-356 [DOI] [PubMed] [Google Scholar]
- van der Bloom J. D., Boot W. J., Velthuis H. H. W. (1994). Simultaneous queen raising and egg laying by workers in Africanized honeybee colonies (Apis mellifera L) in Costa Rica. Apidologie (Celle) 25, 367-374 [Google Scholar]
- Vandesompele J., De Paepe A., Speleman F. (2002). Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 303, 95-98 [DOI] [PubMed] [Google Scholar]
- Wang Y., Amdam G. V., Rueppell O., Wallrichs M. A., Fondrk M. K., Kaftanoglu O., Page R. E., Jr (2009). PDK1 and HR46 gene homologs tie social behavior to ovary signals. PLoS ONE 4, e4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kaftanoglu O., Siegel A. J., Page R. E., Jr, Amdam G. V. (2010). Surgically increased ovarian mass in the honey bee confirms link between reproductive physiology and worker behavior. J. Insect Physiol. 56, 1816-1824 [DOI] [PubMed] [Google Scholar]
- Wang Y., Brent C. S., Fennern E., Amdam G. V. (2012). Gustatory perception and fat body energy metabolism are jointly affected by vitellogenin and juvenile hormone in honey bees. PLoS Genet. 8, e1002779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. E., Buck N., Evans J. D. (2006). Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol. Biol. 15, 597-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston M. L. (1987). The Biology of the Honey Bee. Cambridge MA: Harvard University Press; [Google Scholar]
- Wolschin F., Mutti N. S., Amdam G. V. (2011). Insulin receptor substrate influences female caste development in honeybees. Biol. Lett. 7, 112-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Brown M. R. (2006). Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51, 1-24 [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau K. D., Carrington J. C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784-789 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.