Abstract
Bacterial adhesion and biofilm formation on surfaces raises health hazard issues in the medical environment. Previous studies of bacteria adhesion have focused on observations in their natural/native environments. Recently, surface science has contributed in advancing the understanding of bacterial adhesion by providing ideal platforms that attempt to mimic the bacteria's natural environments, whilst also enabling concurrent control, selectivity and spatial control of bacterial adhesion. In this review, we will look at techniques of how nanotechnology is used to control cell adhesion on a planar scale, in addition to describing the use of nanotools for cell micropatterning. Additionally, it will provide a general background of common methods for nanoscale modification enabling biologist unfamiliar with nanotechnology to enter the field.
Keywords: bacterial adhesion, bacterial patterning, surface science, nanotechnology, self-assembled monolayers
1. Introduction
Just as biology is offering inspiration and components to nanotechnology [1,2], nanotechnology is providing new tools and technological platforms to measure, understand and control biological systems [3–9]. Nanobiotechnology [10] is the application of nanotechnology to the field of biological sciences. This area of science focuses on making molecular and nanoscale materials for biological and medical applications [8], and constructing nanometre scale tools to study natural phenomena occurring in biological systems [11]. Nanobiotechnology uses microand nanoscale science and technology in combination with the knowledge and techniques used in biological studies to manipulate molecular, genetic and cellular processes. This approach has afforded a platform for scientists to generate new devices that are fundamentally important in discovering new life science processes. One important area for which nanotechnology has contributed and will continue to help make significant advancements is in controlling bacterial adhesion and patterning. Advances in surface science have led to the creation of techniques such as self-assembled monolayers (SAMs) to fabricate nanoscale modified surfaces onto a variety of substrate materials that can control non-specific and specific interactions with bacteria [12,13]. The patterning allows novel studies to be performed such as investigating biological phenomena such as cell–cell interactions and monitoring intracellular and extracellular events [5,7,14,15].
Bacteria can exist in nature as free planktonic cells in bulk solution, but the majority prefer to live in surface-associated sessile communities known as biofilms [16]. Biofilms are generally defined as a structured community of microbial cells, enclosed in a secreted polymeric matrix on a surface [17].
2. Biological consideration for designing surfaces for controlling bacterial adhesion
Primary adhesion of bacteria onto surfaces is governed by forces such as Brownian motion, hydrodynamic forces and a variety of non-covalent interactions including van der Waals forces, electrostatic interactions and hydrophobic interactions [18]. The rate and strength of the initial adherence of microbes to surfaces depends primarily on the relationship between the (attractive or repulsive) chemical and physical properties of the aqueous phase, bacterial and substratum surfaces [19]. Researchers have devised three main theoretical models to examine bacterial adhesion to surfaces: the thermodynamic model [20,21], the DLVO model [22] (Derjaguin, Landau, Verwey and Overbeek) and the extended DVLO model [23].
The thermodynamic theory expresses adhesive forces as a measure of free energy. The thermodynamic model calculates the numerical values of free energy of the bacteria, the surrounding solution and the surface to give theoretical adhesion energy values (Gibbs adhesion energy) [24]. Adhesion is likely to occur if the free energy value is negative. The DVLO model states that initial adherence of bacteria is a balance between attractive van der Waals forces and attractive or repulsive electrostatic interactions (electrostatic tend to favour repulsion as most surfaces and bacteria are negatively charged), and their decay with separation distance [25]. However, there are some limitations to both models. The thermodynamic theory primarily takes into account hydrophobic interactions, van der Waals and somewhat excludes electrostatic interactions. Additionally, the model is based on a closed system, with no additional input or output of energy. Therefore, assumptions for bacterial free surfaces energies may be incorrect as they are living, dynamic organisms that can change energy from a system by consumption of local media, for example, and synthesis of extracellular surface features [26]. The DVLO theory does not explain a variety of different attachment behaviours, mainly overcoming an electrostatic barrier. The extended DVLO theory developed by van Oss [23] attempts to overcome these limitations.
In summary, there is not as yet a generalised initial adhesion profile valid for each and every bacterial strain and surface; however, the research to date has shown that it is a complex process involving many different interactions. In the absence of a potential docking site for bacterial adhesins, however, research has shown that generally bacteria prefer to adhere to hydrophobic surfaces over hydrophilic [27], allowing more hydrophobic interactions, and surfaces that are positively charged [28], as bacteria are typically negatively charged, therefore increasing the net electrostatic attractive force and van der Waals interactions over repulsive forces.
In order for a bacterium to exhibit irreversible attachment to surfaces following initial adhesion (unable to be removed without excessive force or rinsing), cells have evolved the ability to produce adhesins (receptors) either protruding from or attached to the cell membrane, which bind to specific molecules (ligands) on surfaces, forming a strong but non-covalent bond [18]. The process of recognition usually only involves a portion of the molecule involved and the molecular structure responsible is known as an epitope [29]. The construction of an adhesive surface with the ability to form a robust, specific, irreversible bond with bacterial adhesins is an important factor in cell–cell communication studies, and as such surfaces should be constructed so that they select for one or more of these adhesins.
Bacterial adhesins are either directly associated with the cell membrane, or they protrude outwards from the membrane in hair-like appendages [30]. Such appendages are called pili or fimbriae, and they are usually assembled from repeated proteinaceous subunits, with a terminating lectin-like subunit. Initially, pili were only identified in Gram-negative organisms such as Escherichia coli and Pseudomonas, but some species of Gram-positive bacteria have now been known to produce structurally similar appendages [30]. One of the best-studied examples of pilus assembly is the family of P-pili encoded by the ‘pap’ genes, which are expressed in most strains of uropathogenic E. coli. They are rigid helicopolymers with a terminating protein subunit called PapG, and bind repeating Gal α (1,4) Gal moieties present on glycolipids coating the surface of erythrocytes and uroepithelial cells [31], allowing the bacteria to colonise the urinary tract and cause infection [32].
Another well-studied example of protruding adhesins is type-1 fimbriae. They are expressed in most strains of enterobacteria [33], and are structurally different from P-pili. Type-1 fimbriae are flexible, rod-like fibres that bind specific mannose moieties with the subunit FimH [34]. Type-1 fimbriae are long, thin, flexible, proteinaceous appendages that protrude outside of the cell body and bind to D-mannose residues [34]. Their thickness ranges from 2 to 7 nm, and the length can be up to 2 μm [35]. First visualised by Houwink and van Iterson [36] in 1950 using electron microscopy, type-1 fimbriae are expressed in abundance (100–1000 per cell) and do not rotate independently of the cell body like flagella (Figure 1A). They are protein polymers composed mainly of identical subunits, which are held in the stable thread-like structure via hydrophobic and electrostatic interactions (Figure 1B) [37].
Figure 1.
Representations of type-1 fimbriae: (A) Electron micrograph of fimbriae on E. coli [38] and (B) the chaperone usher pathway.
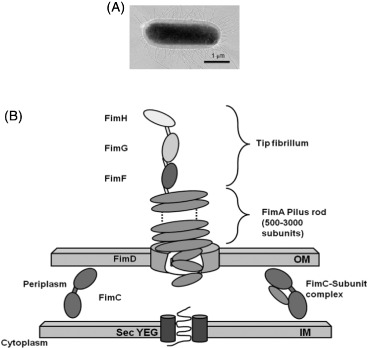
Notes: Upon translation, subunits are secreted into the periplasm via the SecYEG translocon. FimC (the ‘chaperone’) then accelerates protein folding, and delivers the subunits to the pore forming protein FimD (‘the usher’) in the outer membrane. Here, the subunits are translocated and incorporated into the growing pilus.
The main length of the pilus is hollow, with an internal diameter of 2 nm and is composed of identical subunits of FimA monomers which are non-covalently associated head-to-tail and organised in a right-helical structure [34]. The helical structure is flexible and has the ability to unfold if pulled, resulting in a considerable length increase of the fimbriae. This is a very useful feature in an environment with strong hydrodynamic shear forces, allowing fimbriated bacteria to colonise many inhospitable environments that are exposed to flow, such as the urinary tract [32].
FimH is the adhesive subunit which binds mannose residues, and the minor components FimF and FimG act as adaptors for integration of the adhesin into the fimbrial structure. Recent studies have shown that FimH is able to interact with the mannosylated surface via a shear-enhanced catch bond mechanism [16,17,39,40]. This was surprising, as initially FimH was thought to act like a lectin (a protein that binds non-covalently to mono and oligosaccharides), which are thought to bind via slip bonds that are weakened under shear forces [41]. Structural simulations have shown that the FimH undergoes a conformational change when exerted to force, accompanied by an increase in binding strength. Forero et al. [42] found that by pulling fimbriae with a mannosylated tip of an atomic force microscope fimbriae could withstand intermediate force (between 25 and 60 pN) for prolonged periods of time. Tchesnokova et al. [43] found that the cysteine bond in the mannose-binding domain of FimH contributes to its adhesion strength under shear force, by creating cysteine-bond-free mutants [44]. Additionally, Aprikian et al. [44] suggested that the two FimH domains interact with each other (the main pilus and the FimH), and that the main protein has a detrimental effect on FimH binding when the two are in close contact. With shear force, the lectin domain FimH becomes separated from the main protein and allows it to switch from a low-affinity to a high-affinity state. This specific shear stress-enhanced adhesion of bacteria to mannosylated surfaces is useful for bacterial adhesion studies as it can allow the micropatterned bacterial co-cultures to be exposed to shear forces resulting from fluid flow conditions without dislodging the bacteria or causing mixing of the bacterial strains [45].
A variety of other surface-associated (non-polymeric) adhesins can also mediate the attachment of a bacterium to a host cell or surface. Bacterial surface proteins that bind to host extracellular matrix (ECM) proteins such as fibronectin, fibrinogen, vitronectin and elastin are referred to as microbial surface component recognising adhesive matrix molecules [46], and the integration of bacteria with ECM proteins is believed to contribute significantly to the virulence of a number of microorganisms, including staphylococci and streptococci [47].
3. Tailoring surface chemistry to control bacterial adhesion
3.1. Self-assembled monolayers
One of the most popular methods of bacterial cell immobilisation is through the use of functionalised SAMs [48]. SAMs possess important properties of self-organisation and adaptability to a number of technologically relevant surface substrates, providing the ideal platforms for the attachment of molecules that can increase or decrease cell adhesion.
Surfaces that are modified with SAMs are made from the spontaneous adsorption of surfactant molecules onto a surface [48]. They are used in surface chemistry to provide nanometre thick, highly ordered films that can be used as building blocks for protein [49] and carbohydrate attachment, as well as for wetting and adhesion studies [50] by tailoring the chemistry of the head group to control cell adhesion or prevent it. Each of the surfactant molecules that constitute the building blocks of the SAM can be divided into three parts, the head group (surface linking group), the backbone and the terminal (active) group (Figure 2).
Figure 2.
(Colour online) Schematic representation of a surfactant molecule.
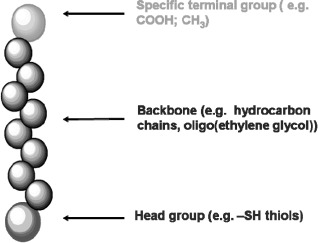
Notes: Each of the surfactant molecules can be divided into three parts, the head group (surface linking group), the backbone and the specific terminal (active) group.
A wide variety of surfactants can be used to form SAMs, including organosilanes on hydroxylated glass [51] and carboxylic acids on metal oxides [52]. However, the most popular form of SAM construction is that of thiols on gold surfaces [53]. Gold is the metal of choice for a large proportion of SAM studies as it has the lowest surface energy meaning its thermodynamically favourable for formation of gold thiol bonds [54], it is relatively inert and does not form oxides with atmospheric gases at room temperature [55]. Additionally, gold substrates are easy to prepare by physical vapour evaporation [56] of the metal onto a glass surface with a chromium or titanium adhesion layer in between (1–5 nm), allowing thin films of gold to be formed (10–200 nm). Thiols usually carry the general formula:
They consist of a thiol head group (HS–), which forms a strong, covalent bond with the gold substrate, and a specific terminating group (–X) that determines the specific physiochemical properties of the newly formed SAM, as well as providing an anchor point for further surface modification [57]. Separating the head and terminating groups is usually a hydrocarbon chain backbone ((CH2)n), which stabilises the SAM through van der Waals interactions [58]. Thiols can be deposited onto gold substrates either through vapour deposition or from a solution [59], with concentrations of 10–1000 μM. Figure 3 shows the formation of an n-alkanethiol SAM on gold.
Figure 3.
Schematic representation of SAM formation.
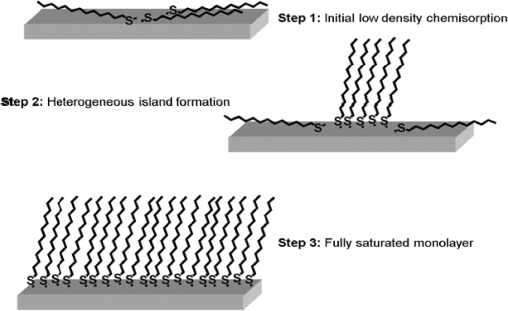
Notes: Thiols can be deposited onto gold substrates either through vapour deposition or from a solution [59], the sulphur head group of the thiol molecule forms a strong covalent bond with the gold (step 1). Low density adsorption causes the thiol to align parallel to the gold substrate, and upon a critical surface coverage, lateral pressure induces nucleation and heterogeneous island formation [53] (step 2) until full saturation is reached. The resulting SAM form packed 2D monolayers of thiol molecules, leading to effective close packed monolayers (step 3).
When a gold substrate is brought into contact with a thiol, the sulphur head group of the thiol molecule forms a strong covalent bond with the gold (chemisorption), ∼44kcal/mol [60,61] (Figure 3, step 1). The widely accepted theory of thiolate formation is that there is an oxidative adsorption of the S–H bond to the gold surface, with a reductive elimination of the hydrogen, forming an S–Au bond [27,46,62]. Scanning tunnelling microscope images have revealed that initially, low density adsorption causes the thiol to align parallel to the gold substrate, and upon a critical surface coverage lateral pressure induces nucleation and heterogeneous island formation [53] (Figure 3, step 2) until full saturation is reached. To minimise the free energy of the organic layer, the thiol molecules adopt trans conformations that allow high levels of noncovalent van der Waals bond formation between the methylene groups of the hydrocarbon backbone. The completion of this process can take several hours, depending on the nature of the backbone. The resulting SAM forms a packed 2D monolayer of thiol molecules which can have a tilt angle of (α) 30° [63]. This tilt angle occurs as it provides the parameter to maximise the van der Waals chain–chain interactions, leading to effective closed packed monolayers which is energetically favourable (Figure 3, step 3). Techniques commonly used to characterise surfaces that have been modified by SAMs, include contact angle [64], ellipsometry [65], X-ray photoelectron spectroscopy (XPS) [66], time-of-flight secondary ion mass spectrometry [64,67] surface plasmon resonance [68], fluorescence microscopy [45] and atomic force microscopy [7,15].
4. Planar control of cell adhesion using SAMs
4.1. Anti-adhesive SAMs
There has been considerable interest in the use of SAMs as model systems to prevent cell adhesion for the development of so-called ‘inert surfaces’ for biofouling applications [69,70], including developing anti-adhesive coatings for marine vessels [71], and creating biologically inert materials such as contact lenses and artificial surgical devices such as heart valves and blood vessels [72]. Certain terminating groups of SAM surfaces have been shown to resist the non-specific adsorption of proteins, and subsequently have been able to reduce cellular adhesion and biofouling. The most widely used and characterised SAMs that resist protein and cellular adsorption are those consisting of oligo(ethylene glycol) (OEG)-terminated thiols [42,73–76]. The simplified theory is that the water in the buffer solution containing the protein sticks to the –OH terminating groups of the SAM, as they are hydrogen-bond acceptor groups, forming a stable solid–liquid interface that causes steric repulsion as the protein cannot replace the bound water [77,78].
Prime and Whitesides [79] were among the first to demonstrate that OEG-SAMs reduce protein adsorption onto surfaces, by using monolayers of varying chains length (EG)n and characterising the reduction of protein adsorption by XPS and ellipsometry. They later also found that a helical form of OEG forms a more stable protein adsorption barrier than the trans form, as water binds more tightly [80]. Experimentation using OEG-SAMs has since extended to many cellular adhesion studies, showing a reduced attachment of bacterial species including staphylococcus [48,74], Helicobacter pylori [81], and Marinobacter hydrocarbonoclasticus and Cobetia marina [71].
At the other end of the spectrum, a more recent approach to anti-adhesive coatings is the development of superhydrophobic surfaces [50,51], based on the ‘Lotus effect’, whereby water drops roll-off the Lotus leaf surface under a slight force, taking with it any dissolved biofouling molecules and cells. The idea is that instead of a surface that ‘prefers’ water to the solute, prevention of biofouling could potentially occur by repelling the water altogether, removing molecules by a slight external force. However, experimentation is still in its infancy, but promises to be a potential alternative to using ethylene glycol moieties.
4.2. Pro-adhesive SAMs
In addition to anti-adhesive surfaces, SAMs can also provide platforms for the efficient immobilisation of bacterial cells on a surface. Sousa et al. [12] used SAMs with different terminating groups including carboxylic acids (COOH) and methyl (CH3) groups to determine the appropriate surface for the non-specific control of cell adhesion of staphylococcus. They found that the use of hydrophilic (COOH) groups increased the number of cells attaching to the surface when compared to the methyl groups (CH3). Additionally, they investigated the alkyl chain length and noted there was no effect, when the terminating group was a CH3; however, in the case of the terminating group being a COOH they observed that increased chain length caused significant increase in cell adhesion. The explanation for this is that to a large extent it is the wettability of the surface that controls cell adhesion.
Poly-l-lysine is also another popular method of cell immobilisation to surfaces; it is a cationic polymer and the negative surface charges of the bacterial cell wall make it an effective way of attracting cells to surfaces [82]. However, this method of bacterial adhesion is employed by using non-specific forces, rather than via specific bacterial adhesins, meaning that adhesive forces would not be robust enough to sustain immobilisation for prolonged periods of time and through shear forces. Furthermore, some researchers have found that thick layers of poly-l-lysine can actually be antimicrobial, and inhibit growth of cells [83].
By carefully designing the chemistry of terminating group of the thiol which make up the SAM, surfaces can be tailored to target specific adhesins to enable selective control of bacterial adhesion. For instance, Terrettez et al. [84] used a SAM with a terminating enzyme (colicin N) which binds with high affinity to the out membrane protein OmpF of E. coli. SAMs have also been used to exploit the FimH mannose bond that allows E. coli to adhere strongly to mannosylated surfaces. Qian et al. [85] used a mannoside derivative with an amino (NH2) group to covalently couple to COOH-terminated SAMs using carbodiimide chemistry, forming mannoside-terminated SAMs (MT-SAMs) which E. coli then adhered to via the type-1 fimbrial adhesins embedded in the cell wall. Using a similar method, Liang et al. [86] used MT-SAMs to measure the adhesion forces of uropathogenic E. coli with optical tweezers.
A variety of other approaches have been employed to selectively immobilise bacterial cells onto surfaces. One such way is by using antibodies which selectively bind antigenic epitopes that are found on the surface of bacterial cell wall/plasma membrane [87,88]. One example of immobilising bacteria in such a way was shown by Rozhok et al. [89]. They made use of three antibodies, raised from both goat and rabbit against the whole cell surface of E. coli K-12 and a specific lipopolysaccharide found on the surface of the cell wall, to control the adhesion of motile bacteria to surfaces. However, when placed in a hydrodynamic environment as demonstrated by Premkumar et al. [90] they achieved a 2% surface coverage of bacteria. This suggests that whilst adhering cells via antibody–antigen interaction in a static system is suitable, moving to a flowing system this method is not very suitable for stable adhesion. This is because the antibody–antigen binding force is relatively weak (∼50 pN), and does not have a catch bond mechanism such as the one found in bacterial fimbriae; meaning cells are not likely to withstand dislodging in a flow cell environment. Readers who are interested to know more about specific receptor–ligand interactions, can refer to reviews by Katsikogian and Missirlis [91] and Gestwicki et al. [92].
4.3. Cellular patterning
The ability to position cells on a surface with control over their spatial arrangement is being developed for fundamental biological research [93], as many studies involving interacting microorganisms, either with each other or with the environment, would benefit from simple devices able to immobilise cells in precisely defined patterns. The isolation of cells on a surface enables the study of events occurring in each individual cell, instead of relying on statistical distributions based on populations of cells. Furthermore, patterning in conjunction with adhesive surfaces prevents motile cells from migrating across the surface, and therefore makes it straightforward to observe single-cell events repeatedly.
The most common patterning procedure is microcontact printing (μCP) [94], a soft lithographic method that uses relief features (protruded features) created on stamp, to directly deposit or remove biomolecules or cells onto surfaces. Other methods of patterning cells include the use of microfluidics, inkjet printing, stencils and robotics and will be discussed in more detail in the following section.
4.4. Microcontact printing
The μCP process (Figure 4) involves the fabrication of poly(dimethoxysiloxane) (PDMS) stamps by depositing a monomeric precursor over a silicon master and subsequently curing it at 60°C (step 1). The stamp is then peeled from the master and immersed into or with a surfactant solution (steps 2 and 3). Excess surfactant is then removed from the stamp surface (step 4), leaving an ‘ink’. The stamp is then brought into conformal contact with a substrate surface (step 5), which can include a SAM surface. The ink is transferred to the substrate where it forms a patterned surface (step 6).
Figure 4.
(Colour online) The μCP process.
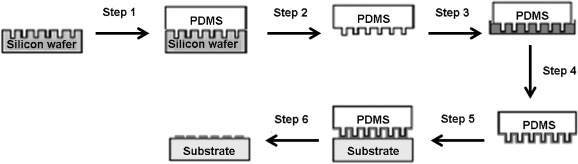
Notes: This involves the fabrication of PDMS stamps by depositing a monomeric precursor over a silicon master and cured at 60°C (step 1). The stamp is then peeled from the master (step 2) and immersed into or with a surfactant solution (step 3). Excess surfactant is then removed from the stamp surface (step 4), leaving an ‘ink’. The stamp is then brought into conformal contact with the substrate (step 5), which can include SAM surfaces. The ink is transferred to the substrate where it forms a patterned surface (step 6).
As the stamps can be constructed with almost any pattern, conformal contact can be achieved in many different geometrically controlled ways [95]. PDMS is the material most frequently used as it results in a soft polymer, and therefore conformal contact, although there is recent interest in the use of hydrogel stamps such as agarose for cellular patterning as they are generally more biocompatible. Transfer of biomolecules such as proteins via a stamp is fast; contact duration of a few seconds is needed and efficiency of transfer can exceed 99%. Patterns of biomolecules obtained in this way have high contrast and resolution because of the mechanical stability of the pattern of the stamp, and because adsorbed proteins show virtually no surface diffusion.
μCP has been used in a variety of ways for bacterial patterning, usually by directly delivering adhesive or inert biomolecules (including proteins and carbohydrates) in a patterned format to surfaces for bacterial patterning. For example, Cerf et al. [96] created arrays of living bacteria by patterning inert octadecyltrichlorosilanes (OTS) using PDMS which were repulsive to bacterial adhesion, followed by backfilling with adhesive streptavidin–biotin molecules which promoted cell adhesion, thereby engineering the surface to selectively attach green fluorescence protein (GFP)-E. coli cells in patterned 10 μm circles [96] (Figure 5).
Figure 5.
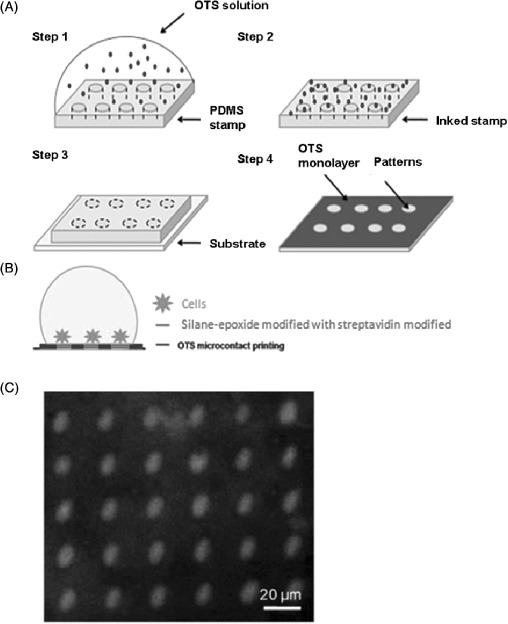
(A) Diagrammatic representation of the method used to pattern arrays of E. coli immobilised onto OTS functionalised surfaces (figures adapted from [96]). Step 1 involved the inking of the stamp with the OTS solution. The stamp was then dried with the application of a stream of nitrogen in step 2. Step 3 places the inked PDMS stamp onto the glass slide. The final step, step 4, shows that OTS molecules are transferred on the surface but not on the patterns that correspond to the carved structures of the PDMS stamp. (B) OTS μCP was followed by their modification with epoxide and streptavidin, followed by incubation with cells. (C) A fluorescence image of the selective binding of streptavidin molecules onto epoxide patterns, which allow the binding of the cells [96].
μCP has also been used to directly print thiol molecules. A SAM of an alkanethiol can be patterned onto a gold-coated surface by μCP and functionalised, followed by backfilling the un-patterned regions with an anti-adhesive OEG thiol, thus creating islands of SAMs that absorb proteins and cells, surrounded by SAMs which resist cellular adhesion. Rowan et al. [97] used a PDMS stamp to print patterns of hydrophobic SAMs and hydrophilic (mercaptoundecanoic acid) SAMs to fabricate bio-inert microstructure on gold surfaces producing ‘enclosures’ that trapped cells of E. coli. Additionally, Rozohok et al. [89] used μCP to form arrays of COOH-terminated SAMs. These were then functionalised with poly-l-lysine or with the covalent attachment of antibodies directed against E. coli.
Whilst controlling single-cell adhesion on surfaces has been discussed, understanding biofilm formation on patterned surfaces is another key aspect where nanotechnology can make a significant contribution. Towards this goal, Hou et al. [98] has used μCP to pattern surfaces with CH3-terminated SAMs which promotes the biofilm formation, surrounded by D-mannitolterminated SAMs that resist bacterial adhesion. These studies have demonstrated that biofilm formation can be confined to the CH3-terminating SAM regions for at least 26 days. These patterned surfaces can thus potentially be used to address fundamental questions such as how the physiology and gene expression of bacteria respond to restrained and well-defined microenvironments.
Direct patterning of bacteria has also been achieved with μCP. Instead of printing functional molecules to induce or inhibit cellular adhesion, stamps can be directly ‘inked’ with bacterial suspensions and printed directly onto a surface. The advantage of this method is that it is relatively rapid, and therefore limits cell exposure time to the environment; bacteria can be directly transferred to surfaces and covered in less than a minute [99]. Xu et al. [100] employed μCP to directly print bacteria using artificially hydrophilised PDMS stamps onto the surface of a nutrient-containing matrix (i.e. agarose), producing high-resolution arrays of living bacteria (Figure 6).
Figure 6.
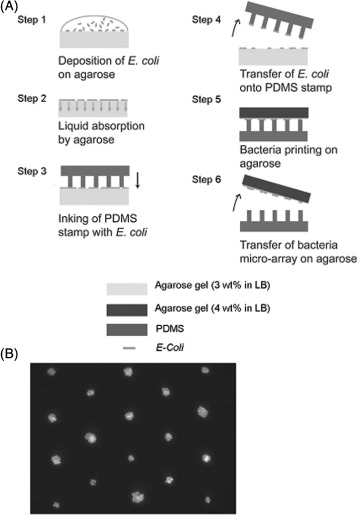
(Colour online) (A) Diagrammatic representation of the method to produce arrays of bacteria using two different types of agarose gels (figures adapted from [100]). A drop of E. coli in LB culture medium is deposited on an agarose gel (3 wt% in LB) (step 1), which absorbs the liquid (step 2). A PDMS stamp is inked by contact with bacteria covering the agarose gel (step 3). When the stamp is removed from the ‘inkpad’, a fraction of bacteria are transferred onto the stamp (step 4). The bacteria are transferred by contact with a 200 μm thick slide of agarose (4 wt% in LB) into a regular microarray (steps 5 and 6). (B) Arrays of E. coli directly patterned onto an agarose substrates [100].
Similarly, Weibel et al. [101] used micropatterned agarose stamps to print patterns of E. coli on agar plates (Figure 7). Agarose is a linear polysaccharide consisting of galactose and 3,6-anhydrogalactose subunits, and stamps can be made by casting hot solutions over PDMS masters. The agarose stamps were inked directly with suspensions of bacteria; with stamp features of 200 μm, and they found that the stamp supported many bacterial cell types when culture media was included [101].
Figure 7.
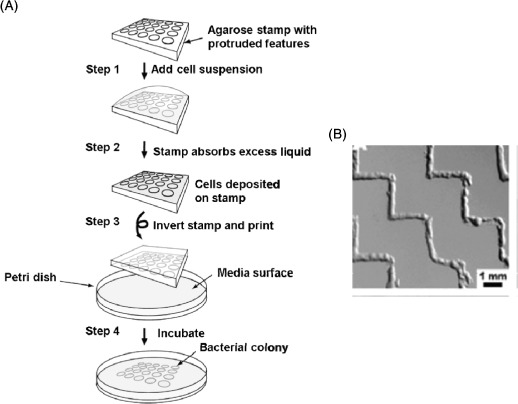
(Colour online) (A) Schematic representation of patterning bacteria using μCP (figures adapted from [101]). A cell suspension is deposited onto a 3% agarose stamp with protruded features (step 1). The excess liquid is absorbed by the stamps, leaving a layer of cells deposited on the stamp (step 2). The stamp is then brought into contact with cell culture media (containing 1.5% agar) (step 3), leading to the formation of patterns of bacterial colonies (step 4). (B) Bright field image of arrays of E. coli directly patterned onto an agarose substrate using a stamp constructed of agarose [101].
Recently, we have employed μCP in combination of surface chemistry to produce bacterial co-cultures patterns on surfaces (Figure 8) [45]. Using mannose-terminated SAMs as the adhesive surface in combination with μCP, we have produced spatially controlled and robust co-cultures that are viable and resist to detachment due to shear, which may prove useful in understanding cell–cell communication. We found that bacteria in the micropatterns fully adhere to mannose-terminated SAMs even when exposed to volumetric flow rates of 10 μL/min for 4 h. The cells remaining viable may possibly be due to the shear-enhanced catch bond mechanism of the FimH–mannose bond.
Figure 8.
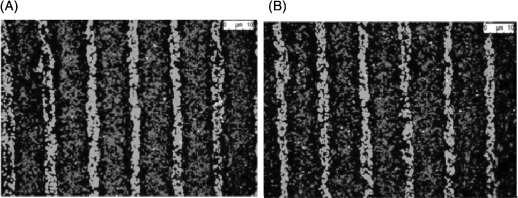
(Colour online) (A) Confocal microscope image of patterned co-cultures of E. coli. Single-strain GFP-tagged E. coli patterns on mannose-terminated SAMs were constructed, followed by an immersion with red fluorescence protein-tagged E. coli. (B) Samples placed inside the flow cell and exposed to rinsing with M9 broth at flow rates of 10 μL/min for 4 h at 37°C. The images demonstrate that the cells are tightly bound to the surface and do not detach in the conditions used [45].
5. Other patterning techniques
5.1. Microfluidics
Besides μCP, PDMS has been widely used for creating microfluidic channels and networks for which the fabricated devices have been used for cell separation and cell capture. These devices are employed in the study of cellular processes such as quorum sensing, and responses to chemical gradients. Microfluidic channels are formed by placing a layer of PDMS with channels created on the surface in contact with glass or a polymer surface that forms the roof of the channel [99]. Laminar fluid flow can then be streamed into networks of branching and recombining microchannels to produce stable gradients of nutrients and cells [102]. An interesting adaptation was employed by Balaban et al. [103], for producing motile, filamentous cells of E. coli with different shapes, by confining and growing the cells in agarose microchambers. In the presence of an antibiotic (cephalexin) that inhibits septation, the E. coli cells filamented and adopted the shape of the microchambers in which they grew [103]. An example where microfluidics has been used for cellular patterning includes work undertaken by Takayama et al. [104]. They patterned two different cell types by using multiple flow streams in capillary channels. Within these microchannels, two or more laminar flow streams can flow parallel to each other due to low convective mixing and the width of each cell pattern can be easily controlled by adjusting the flow rate [104].
5.2. Jet-based methods
Inject printers have been used to create large arrays of bacterial cells. There are two main types of inject printers, thermal and piezoelectric. In thermal printers, a resistive heating element causes air bubbles to expand and expel a liquid drop which contains a bacteria suspension [105]. Piezoelectric inkjets use a voltage-induced deformation of a rectangular piezoelectric crystal to squeeze inkjets through the nozzle and these can generally print a wider variety of solvents and are easier to clean [106]. An exciting inkjet patterning experiment employed by Merrin et al. [107] adapted a simple piezoelectric printer for patterning bacteria onto substrates, including glass slides, agar plates and nitrocellulose membranes with a printing viability of 98.5%. They were able to form patterned co-cultures as the print head contained six parallel linear banks of 32 nozzles each, with each bank connected to a different ink source (Figure 9). Connecting the inkjet to a motorised stage enabled them to vary the spacing between cultures, and allowed small drop volumes of typically less than 30 pL.
Figure 9.
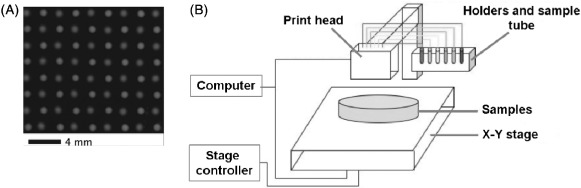
(Colour online) (A) Checkerboard pattern printed with two strains of E. coli labelled with different fluorescent proteins onto an agarose substrate using a (B) piezoelectric inkjet printer (figures adapted from [107]).
5.3. Stencils
This approach to patterning is a very simple but effective method, which uses PDMS with microengineered holes that can be deposited onto an adhesive SAM followed by immersion of cells into the holes to promote patterned deposition onto the surface. For example, Eun and Weibel [108] used freestanding, elastomeric stencils with microfabricated ‘holes’ with different shapes and dimensions, containing a positively charged amino (NH3+)-terminated SAM which promotes the adhesion of cells through electrostatic interactions, to control the spatial adhesion and growth of bacterial cells on polyelectrolyte surfaces, including Pseudomonas aeruginosa.
5.4. Robotics
Perhaps one of the most technologically advanced methods of directly patterning bacterial cells is via the use of robotic micromanipulators. Traditionally, printing techniques in laboratories are employed by hand, which can be time consuming and un-repeatable. Using micromanipulators with an X–Y–Z controlled stage controlled by computers offers a repeatable and large-scale alternative for constructing massive arrays of bacteria with micrometre resolution [109]. For example, Ingham et al., 2010 [109] used a high throughput contact printing method, employing a microscope and a stamp with massive arrays of PDMS pins with 20 μm area connected to a motorised stage. They were able to deposit viable bacteria onto porous aluminium oxide followed by effective segregation of microcolonies during out growth (Figure 10).
Figure 10.
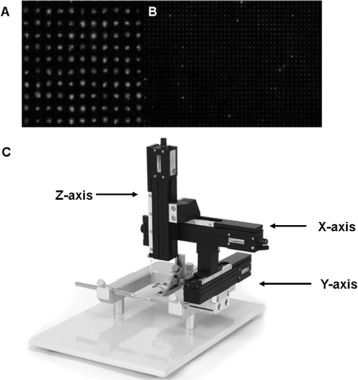
(Colour online) Arrays of Lactobacillus plantarum cells directly patterned onto a porous aluminium oxide surface using PDMS pins with (A) a cross-section of 20 × 20 μm2 and 70 μm pitch and (B) a cross-section of 10 × 10 μm2 and 35 μm pitch controlled with a (C) triple axis, motorised micromanipulator (figures adapted from [109]).
6. Concluding remarks
The images of micropatterned bacteria shown in this review, although constructed through different patterning methods, all look very similar and most have the disadvantage of not being suitable for forming co-cultured patterns. Many of the fabricated patterns have dimensions which are simply too big, creating massive arrays of bacteria and preventing analysis of individual cells. Most, however, have the shortcoming of using anti-adhesive regions to separate the cell colonies. These single-cell systems, although useful for creating large arrays of bacteria, have the disadvantage that the behaviour of isolated cells may be very different from when surrounded by other cells, and additionally makes it difficult for studying cell–cell interactions such as gene transfer. Therefore, in order for patterned arrays to more realistically mimic natural systems such as biofilms, current technologies would benefit from focus more on the formation of bacterial co-cultures, by exploiting the selective adhesive abilities of surface chemistry technologies, including SAMs. This approach involves identifying phenotypic differences between the bacterial species, thus allowing for the tailoring of the chemistry of the surface to target these phenotypic differences allowing for selective control of anti-adhesive or adhesive surfaces, which with modern patterning techniques can also be used to pattern the cells.
Acknowledgements
We thank the Leverhulme Trust grant F/00094/BD and Wellcome Trust grant WT091285MA. This research was in part supported through Birmingham Science City: Innovative Uses for Advanced Materials in the Modern World (West Midlands Centre for Advanced Materials Project 2).
References
- 1.Preece JA., Stoddart J.F. Concept transfer from biology to materials. Nanobiology. 1994;3:149–166. [Google Scholar]
- 2.Service R.F. Biology offers nanotechs a helping hand. Science. 2002;298:2322–2323. doi: 10.1126/science.298.5602.2322. [DOI] [PubMed] [Google Scholar]
- 3.Johnston S.A., Bramble J.P., Yeung C.L., Mendes P.M., Machesky L.M. Arp2/3 complex activity in filopodia of spreading cells. BMC Cell Biol. 2008;9:65–82. doi: 10.1186/1471-2121-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung C.L., Iqbal P., Allan M., Lashkor M., Preece J.A., Mendes P.M. Tuning specific biomolecular interactions using electro-switchable oligopeptide surfaces. Adv. Funct. Mater. 2010;20:2657–2663. [Google Scholar]
- 5.Costello C.M., Kreft J.U., Thomas C.M., Mendes P.M. Protein nanoarrays for high-resolution patterning of bacteria on gold surfaces. Methods Mol. Biol. 2011;790:191–200. doi: 10.1007/978-1-61779-319-6_15. [DOI] [PubMed] [Google Scholar]
- 6.Mendes P.M. Stimuli-responsive surfaces for bio-applications. Chem. Soc. Rev. 2008;37(11):2512–2529. doi: 10.1039/b714635n. [DOI] [PubMed] [Google Scholar]
- 7.Rawson F.J., Yeung C.L., Jackson S.K., Mendes P.M. Tailoring 3D single-walled carbon nanotubes anchored to indium tin oxide for natural cellular uptake and intracellular sensing. Nano Lett. 2012. doi:10.1021/nl203780d. [DOI] [PMC free article] [PubMed]
- 8.Mendes P.M., Yeung C.L., Preece J.A. Bio-nanopatterning of surfaces. Nanoscale Res. Lett. 2007;2(8):373–384. doi: 10.1007/s11671-007-9083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendes P.M., Christman K.L., Parthasarathy P., Schopf E., Ouyang J., Yang Y., Preece J.A., Maynard H.D., Chen Y., Stoddart J.F. Electrochemically controllable conjugation of proteins on surfaces. Bioconjugate Chem. 2007;18(6):1919–1923. doi: 10.1021/bc7002296. [DOI] [PubMed] [Google Scholar]
- 10.Niemeyer C.M., Mirkin C.L. Nanobiotechnology. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2004. [Google Scholar]
- 11.Mu L., Liu Y., Cai S., Kong J. A smart surface in a microfluidic chip for controlled protein separation. Chem. Eur. J. 2007;13:5113–5120. doi: 10.1002/chem.200601624. [DOI] [PubMed] [Google Scholar]
- 12.Sousa C., Teixeira P., Bordeira S., Fonseca J., Oliveira R. Staphylococcus epidermidis adhesion to modified polycarbonate surfaces: Gold and SAMs coated. J. Adhes. Sci. Technol. 2008;22(7):675–686. [Google Scholar]
- 13.Rozhok S., Holz R. Electrochemical attachment of motile bacterial cells to gold. Talanta. 2005;67(3):538–542. doi: 10.1016/j.talanta.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 14.Rawson F.J., Garrett D.J., Leech D., Downard A.J., Baronian K.H. Electron transfer from Proteus vulgaris to a covalently assembled, single walled carbon nanotube electrode functionalised with osmium bipyridine complex: Application to a whole cell biosensor. Biosens. Bioelectron. 2011;26(5):2383–2389. doi: 10.1016/j.bios.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Rawson F.J., Gross A.J., Garrett D.J., Downard A.J., Baronian K.H.R. Mediated electrochemical detection of electron transfer from the outer surface of the cell wall of Saccharomyces cerevisiae. Electrochem. Commun. 2012;15(1):85–87. [Google Scholar]
- 16.Dunne W.M. Bacterial adhesion: Seen any good biofilms lately? Clin. Microbiol. Rev. 2002;15(2):155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin-Scott H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 18.Busscher H.J., Weerkamp A.H. Specific and nonspecific interactions in bacterial adhesion to solid substrata. Fems Microbiol. Rev. 1987;46(2):165–173. [Google Scholar]
- 19.Bos R., van der Mei H.C., Busscher H.J. Physico-chemistry of initial microbial adhesive interactions - Its mechanisms and methods for study. Fems Microbiol. Rev. 1999;23(2):179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 20.Boyd E.F., Hill C.W., Rich S.M., Hartl D.L. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics. 1996;143(3):1091–1100. doi: 10.1093/genetics/143.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas C.M. Molecular-genetics of broad host range plasmid RK2. Plasmid. 1981;5(1):10–19. doi: 10.1016/0147-619x(81)90074-3. [DOI] [PubMed] [Google Scholar]
- 22.Rutter P.R., Vincent B. In: ‘Physicochemical interactions of the substratum microorganisms and the fluid phase, in Microbial Adhesion and Aggregation. Marshall K.C., editor. Berlin: Springer Verlag; 1985. pp. 21–38. [Google Scholar]
- 23.van Oss C.J. Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J. Mol. Recognit. 2003;16(4):177–190. doi: 10.1002/jmr.618. [DOI] [PubMed] [Google Scholar]
- 24.Busscher H.J., Weerkamp A.H., van der Mei H.C., van Pelt A.W., de Jong H.P., Arends J. Measurement of the surface free-energy of bacterial-cell surfaces and its relevance for adhesion. Appl. Environ. Microbiol. 1984;48(5):980–983. doi: 10.1128/aem.48.5.980-983.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An Y.H., Friedman R.J. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 1998;43(3):338–348. doi: 10.1002/(sici)1097-4636(199823)43:3<338::aid-jbm16>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 26.Morra M., Cassinelli C. Bacterial adhesion to polymer surfaces: A critical review of surface thermodynamic approaches. J. Biomater. Sci.-Polym. Ed. 1997;9(1):55–74. doi: 10.1163/156856297x00263. [DOI] [PubMed] [Google Scholar]
- 27.Cunliffe D., Smart C.A., Alexander C., Vulfson E.N. Bacterial adhesion at synthetic surfaces. Appl. Environ. Microbiol. 1999;65(11):4995–5002. doi: 10.1128/aem.65.11.4995-5002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B.K., Logan B.E. Bacterial adhesion to glass and metal-oxide surfaces. Colloids Surf. B. 2004;36(2):81–90. doi: 10.1016/j.colsurfb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Hultgren S.J., Abraham S., Caparon M., Falk P., St Geme J.W., 3rd, Normark S. Pilus and nonpilus bacterial adhesins - Assembly and function in cell recognition. Cell. 1993;73(5):887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 30.Soto G.E., Hultgren S.J. Bacterial adhesins: Common themes and variations in architecture and assembly. J. Bacteriol. 1999;181(4):1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehn M.J., Heuser J., Normark S., Hultgren S.J. P pili in uropathogenic Escherichia coli are composite fibers with distinct fibrillar adhesive tips. Nature. 1992;356(6366):252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- 32.Johnson J.R. Virulence factors in Escherichia coli urinary-tract infection. Clin. Microbiol. Rev. 1991;4(1):80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogfelt K.A. Bacterial adhesion - Genetics, biogenesis, and role in pathogenesis of fibrial adhesins of Escherichia coli. Rev. Infect. Dis. 1991;13(4):721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 34.Klemm P. Fimbrial adhesins of Escherichia coli. Rev. Infect. Dis. 1985;7(3):321–340. doi: 10.1093/clinids/7.3.321. [DOI] [PubMed] [Google Scholar]
- 35.Degraaf F.K., Mooi F.R. The fimbrial adhesins of Escherichia coli. Adv. Microb. Physiol. 1986;28:65–143. doi: 10.1016/s0065-2911(08)60237-4. [DOI] [PubMed] [Google Scholar]
- 36.Houwink A.L., van Iterson W. Electron microscopical observations on bacterial cytology. 2. A study on flagellation. Biochim. Biophys. Acta. 1950;5(1):10–44. doi: 10.1016/0006-3002(50)90144-2. [DOI] [PubMed] [Google Scholar]
- 37.Knight S., Bouckaert J. In: ‘Structure, function, and assembly of type 1 fimbriae, in Glycoscience and Microbial Adhesion. Lindhorst T.K., Oscarson S., editors. Berlin/Heidelberg: Springer; 2009. pp. 67–107. [Google Scholar]
- 38.Capitani G., Eidam O., Glockshuber R., Grütter M.G. Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect. 2006;8(8):2284–2290. doi: 10.1016/j.micinf.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Bruinsma G.M., van der Mei H.C., Busscher H.J. Bacterial adhesion to surface hydrophilic and hydrophobic contact lenses. Biomaterials. 2001;22(24):3217–3224. doi: 10.1016/s0142-9612(01)00159-4. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt D.L., Brady Jr R.F., Lam K., Schmidt D.C., Chaudhury M.K. Contact angle hysteresis, adhesion, and marine biofouling. Langmuir. 2004;20(7):2830–2836. doi: 10.1021/la035385o. [DOI] [PubMed] [Google Scholar]
- 41.Bell G.I. Models for specific and adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 42.Forero M., Yakovenko O., Sokurenko E.V., Thomas W.E., Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006;4(9):e298, 1509–1516. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tchesnokova V., Aprikian P., Yakovenko O., Larock C., Kidd B., Vogel V., Thomas W., Sokurenko E. Integrin-like allosteric properties of the catch bond-forming FimH adhesin of Escherichia coli. J. Biol. Chem. 2008;283(12):7823–7833. doi: 10.1074/jbc.M707804200. [DOI] [PubMed] [Google Scholar]
- 44.Aprikian P., Tchesnokova V., Kidd B., Yakovenko O., Yarov-Yarovoy V., Trinchina E., Vogel V., Thomas W., Sokurenko E. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 2007;282(32):23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- 45.Costello C.M., Kreft J.-U., Thomas C.M., Hammes D.M., Bao P., Evans S.D., Mendes P.M. Exploiting additive and subtractive patterning for spatially controlled and robust bacterial co-cultures. Soft Matter. 2012;8:9147–9155. [Google Scholar]
- 46.Falkow S., Isberg R.R., Portnoy D.A. The interaction of bacteria with mammalian-cells. Annu. Rev. Cell. Biol. 1992;8:333–363. doi: 10.1146/annurev.cb.08.110192.002001. [DOI] [PubMed] [Google Scholar]
- 47.Chhatwal G.S. Anchorless adhesins and invasins of Gram-positive bacteria: A new class of virulence factors. Trends Microbiol. 2002;10(5):205–208. doi: 10.1016/s0966-842x(02)02351-x. [DOI] [PubMed] [Google Scholar]
- 48.Ulman A. Formation and structure of self-assembled monolayers. Chem. Rev. 1996;96(4):1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 49.Prime K.L., Whitesides G.M. Self-assembled organic monolayers - Model systems for studying adsorption of proteins at surfaces. Science. 1991;252(5009):1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 50.Mrksich M. Using self-assembled monolayers to understand the biomaterials interface. Curr. Opin. Colloid Interface Sci. 1997;2(1):83–88. [Google Scholar]
- 51.Fang J.Y., Knobler C.M. Phase-separated two-component self-assembled organosilane monolayers and their use in selective adsorption of a protein. Langmuir. 1996;12(5):1368–1374. [Google Scholar]
- 52.Gao W., Dickinson L., Grozinger C., Morin F.G., Reven L. Self-assembled monolayers of alkylphosphonic acids on metal oxides. Langmuir. 1996;12(26):6429–6435. [Google Scholar]
- 53.Poirier G.E., Pylant E.D. The self-assembly mechanism of alkanethiols on Au(111) Science. 1996;272(5265):1145–1148. doi: 10.1126/science.272.5265.1145. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber F. Structure and growth of self-assembling monolayers. Progr. Surf. Sci. 2000;65(5–8):151–256. [Google Scholar]
- 55.Love J.C., Estroff L.A., Kriebel J.K., Nuzzo R.G., Whitesides G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005;105(4):1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 56.Choy K.L. Chemical vapour deposition of coatings. Progr. Mater. Sci. 2003;48(2):57–170. [Google Scholar]
- 57.Fendler J.H. Self-assembled nanostructured materials. Chem. Mater. 1996;8(8):1616–1624. [Google Scholar]
- 58.Delamarche E., Michel B., Biebuyck H.A., Gerber C. Golden interfaces: The surface of self-assembled monolayers. Adv. Mater. 1996;8(9):719–729. [Google Scholar]
- 59.Wink T., van Zuilen S.J., Bult A., van Bennkom W.P. Self-assembled monolayers for biosensors. Analyst. 1997;122(4):R43–R50. doi: 10.1039/a606964i. [DOI] [PubMed] [Google Scholar]
- 60.Schwartz D.K. Mechanisms and kinetics of self-assembled monolayer formation. Annu. Rev. Phys. Chem. 2001;52:107–137. doi: 10.1146/annurev.physchem.52.1.107. [DOI] [PubMed] [Google Scholar]
- 61.Dubois L.H., Nuzzo R.G. Synthesis, structure, and properties of model organic-surfaces. Annu. Rev. Phys. Chem. 1992;43:437–463. [Google Scholar]
- 62.Mol O., Oudega B. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. Fems Microbiol. Rev. 1996;19(1):25–52. doi: 10.1111/j.1574-6976.1996.tb00252.x. [DOI] [PubMed] [Google Scholar]
- 63.Nuzzo R.G., Zegarski B.R., Dubois L.H. Fundamental-studies of the chemisorption of organosulfur compounds on Au(111) - Implications for molecular self-assembly on gold surfaces. J. Am. Chem. Soc. 1987;109(3):733–740. [Google Scholar]
- 64.Mathieu H.J. Surface characterization in biomaterials applications. J. Surf. Anal. 2008;14(4):293–298. [Google Scholar]
- 65.Tompkins H.G., Irene E.A. Handbook of Ellipsometry. Berlin: Springer-Verlag GmbH & Co. Publishing; 2005. [Google Scholar]
- 66.Chastain J. Handbook of X-ray Photoelectron Spectroscopy. Minnesota, MN: Perkin-Elmer Corporation; 1992. [Google Scholar]
- 67.Banoub J.H., El Aneed A., Cohen A.M., Joly N. Structural investigation of bacterial lipopolysaccharides by mass spectrometry and tandem mass spectrometry. Mass Spectrom. Rev. 2010;29:606–650. doi: 10.1002/mas.20258. [DOI] [PubMed] [Google Scholar]
- 68.Wegner G.J., Lee H.J., Corn R.M. Characterization and optimization of peptide arrays for the study of epitopeantibody interactions using surface plasmon resonance imaging. Anal. Chem. 2002;74:5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 69.Lewis A.L. Phosphorylcholine-based polymers and their use in the prevention of biofouling. Colloids Surf. B. 2000;18(3–4):261–275. doi: 10.1016/s0927-7765(99)00152-6. [DOI] [PubMed] [Google Scholar]
- 70.Wisniewski N., Reichert M. Methods for reducing biosensor membrane biofouling. Colloids Surf. B. 2000;18(3–4):197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 71.Pranzetti A., Salaün S., Mieszkin S., Callow M.E., Callow J.A., Preece J.A., Mendes P.M. Model organic surfaces to probe marine bacterial adhesion kinetics by surface plasmon resonance. Adv. Funct. Mater. 2012;22(17):3672–3681. [Google Scholar]
- 72.Lee D.H., Kim D., Oh T., Cho K. Phase state effect on adhesion behavior of self-assembled monolayers. Langmuir. 2004;20(19):8124–8130. doi: 10.1021/la049448u. [DOI] [PubMed] [Google Scholar]
- 73.Mendes P.M., Chen Y., Palmer R.E., Nikitin K., Fitzmaurice D., Preece J.A. Nanostructures from nanoparticles. J. Phys. Condens. Matter. 2003;15(42):S3047–S3063. [Google Scholar]
- 74.Whitesides G.M., Grzybowski B. Self-assembly at all scales. Science. 2002;295(5564):2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 75.Berg J.M., Tymoczko J.L., Stryer L. Biochemistry. 5th ed. New York: W.H. Freeman and Company; 2002. [Google Scholar]
- 76.Zhang S.G. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003;21(10):1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 77.Whitesides G.M., Boncheva M. Beyond molecules: Self-assembly of mesoscopic and macroscopic components. Proc. Natl Acad. Sci. USA. 2002;99(8):4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stang P.J., Olenyuk B. Self-assembly, symmetry, and molecular architecture: Coordination as the motif in the rational design of supramolecular metallacyclic polygons and polyhedra. Accounts Chem. Res. 1997;30(12):502–518. [Google Scholar]
- 79.Prime K.L., Whitesides G.M. Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide) - A model system using self-assembled monolayers. J. Am. Chem. Soc. 1993;115(23):10714–10721. [Google Scholar]
- 80.Harder P., Grunze M., Dahint R., Whitesides G.M., Laibinis P.E. Molecular conformation in oligo(ethylene glycol)-terminated self-assembled monolayers on gold and silver surfaces determines their ability to resist protein adsorption. J. Phys. Chem. B. 1998;102(2):426–436. [Google Scholar]
- 81.Parreira P., Magalhães A., Gonçalves I.C., Gomes J., Vidal R., Reis C.A., Leckband D.E., Martins M.C. Effect of surface chemistry on bacterial adhesion, viability, and morphology. J. Biomed. Mater. Res. Part A. 2011;99(3):344–353. doi: 10.1002/jbm.a.33178. [DOI] [PubMed] [Google Scholar]
- 82.Meyer R.L., Zhou X., Tang L., Arpanaei A., Kingshott P., Besenbacher F. Immobilisation of living bacteria for AFM imaging under physiological conditions. Ultramicroscopy. 2010;110(11):1349–1357. doi: 10.1016/j.ultramic.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 83.Colville K., Tompkins N., Rutenberg A.D., Jericho M.H. Effects of poly(l-lysine) substrates on attached Escherichia coli bacteria. Langmuir. 2010;26(4):2639–2644. doi: 10.1021/la902826n. [DOI] [PubMed] [Google Scholar]
- 84.Terrettaz S., Ulrich W.P., Vogel H., Hong Q., Dover L.G., Lakey J.H. Stable self-assembly of a protein engineering scaffold on gold surfaces. Protein Sci. 2002;11(8):1917–1925. doi: 10.1110/ps.0206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qian X.P., Metallo S.J., Choi I.S., Wu H., Liang M.N., Whitesides G.M. Arrays of self-assembled monolayers for studying inhibition of bacterial adhesion. Anal. Chem. 2002;74(8):1805–1810. doi: 10.1021/ac011042o. [DOI] [PubMed] [Google Scholar]
- 86.Liang M.N., Smith S.P., Metallo S.J., Choi I.S., Prentiss M., Whitesides G.M. Measuring the forces involved in polyvalent adhesion of uropathogenic Escherichia coli to mannose-presenting surfaces. Proc. Natl Acad. Sci. USA. 2000;97(24):13092–13096. doi: 10.1073/pnas.230451697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pinegin B.V., Kulakov A.V., Yarilin D.A., Klimova S.V., Khaitov R.M. Competitive specificity analysis of natural antibodies against epitope of bacterial cEll wall peptidoglycoan: Glucosaminylmuramyl dipeptide carrying adjuvant activity. Russ. J. Immunol. 1998;3(2):141–146. [PubMed] [Google Scholar]
- 88.Lee S.Y., Choi J.H., Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21(1):45–52. doi: 10.1016/s0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 89.Rozhok S., Shen C.K., Littler P.L., Fan Z., Liu C., Mirkin C.A., Holz R.C. Methods for fabricating microarrays of motile bacteria. Small. 2005;1(4):445–451. doi: 10.1002/smll.200400072. [DOI] [PubMed] [Google Scholar]
- 90.Premkumar J.R., Lev O., Marks R.S., Polyak B., Rosen R., Belkin S. Antibody-based immobilization of bioluminescent bacterial sensor cells. Talanta. 2001;55(5):1029–1038. doi: 10.1016/s0039-9140(01)00533-1. [DOI] [PubMed] [Google Scholar]
- 91.Katsikogianni M., Missirlis Y.F. Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cells Mater. 2004;8:37–57. doi: 10.22203/ecm.v008a05. [DOI] [PubMed] [Google Scholar]
- 92.Gestwicki J.E., Cairo C.W., Strong L.E., Oetjen K.A., Kiessling L.L. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. JACS. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 93.Whitesides G.M., Ostuni E., Takayama S., Jiang X., Ingber D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 94.Xia Y.N., Whitesides G.M. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 95.Michel B., Bernard A., Bietsch A., Delamarche E., Geissler M., Juncker D., Kind H., Renault J.-P., Rothuizen H., Schmid H., Schmidt-Winkel P., Stutz R., Wolf H. Printing meets lithography: Soft approaches to high-resolution printing. IBM J. Res. Dev. 2001;45(5):697–719. [Google Scholar]
- 96.Cerf A., Cau J.C., Vieu C. Controlled assembly of bacteria on chemical patterns using soft lithography. Colloids Surf. B. 2008;65(2):285–291. doi: 10.1016/j.colsurfb.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 97.Rowan B., Wheeler M.A., Crooks R.M. Patterning bacteria within hyperbranched polymer film templates. Langmuir. 2002;18(25):9914–9917. [Google Scholar]
- 98.Hou S., Burton E.A., Wu R.L., Luk Y.Y., Ren D. Prolonged control of patterned biofilm formation by bio-inert surface chemistry. Chem. Commun. 2009;10:1207–1209. doi: 10.1039/b822197a. [DOI] [PubMed] [Google Scholar]
- 99.Weibel D.B., DiLuzio W.R., Whitesides G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007;5(3):209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 100.Xu L.P., Robert L., Ouyang Q., Taddei F., Chen Y., Lindner A.B., Baigl D. Microcontact printing of living bacteria arrays with cellular resolution. Nano Lett. 2007;7(7):2068–2072. doi: 10.1021/nl070983z. [DOI] [PubMed] [Google Scholar]
- 101.Weibel D.B., Lee A., Mayer M., Brady S.F., Bruzewicz D., Yang J., Diluzio W.R., Clardy J., Whitesides G.M. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Langmuir. 2005;21(14):6436–6442. doi: 10.1021/la047173c. [DOI] [PubMed] [Google Scholar]
- 102.Unger M.A., Chou H.P., Thorsen T., Scherer A., Quake S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 103.Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 104.Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D.E., Whitesides G.M. Selective chemical treatment of cellular microdomains using multiple laminar streams. Chem. Biol. 2003;10(2):123–130. doi: 10.1016/s1074-5521(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 105.Sirringhaus H., Kawase T., Friend R.H., Shimoda T., Inbasekaran M., Wu W., Woo E.P. High-resolution inkjet printing of all-polymer transistor circuits. Science. 2000;290(5499):2123–2126. doi: 10.1126/science.290.5499.2123. [DOI] [PubMed] [Google Scholar]
- 106.Tekin E., Smith P.J., Schubert U.S. Inkjet printing as a deposition and patterning tool for polymers and inorganic particles. Soft Matter. 2008;4(4):703–713. doi: 10.1039/b711984d. [DOI] [PubMed] [Google Scholar]
- 107.Merrin J., Leibler S., Chuang J.S. Printing multistrain bacterial patterns with a piezoelectric inkjet printer. Plos One. 2007;2(7):e663, 1–7. doi: 10.1371/journal.pone.0000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eun Y.-J., Weibel D.B. Fabrication of microbial biofilm arrays by geometric control of cell adhesion. Langmuir. 2009;25(8):4643–4654. doi: 10.1021/la803985a. [DOI] [PubMed] [Google Scholar]
- 109.Ingham C., Bomer J., Sprenkels A., van den Berg A., de Vos W., van Hylckama Vlieg J. High-resolution microcontact printing and transfer of massive arrays of microorganisms on planar and compartmentalized nanoporous aluminium oxide. Lab. Chip. 2010;10(11):1410–1416. doi: 10.1039/b925796a. [DOI] [PubMed] [Google Scholar]


