Abstract
It is known that the largest developmental changes in the hippocampus take place during the prenatal period and during the first two years of postnatal life. Few studies have been conducted to address the normal developmental trajectory of the hippocampus during childhood. In this study shape analysis was applied to study the normal developing hippocampus in a group of 103 typically-developing 6 to 10 year old preadolescent children. The individual brain was normalized to a template, and then the hippocampus was manually segmented and further divided into the head, body, and tail sub-regions. Three different methods were applied for hippocampal shape analysis: radial distance mapping, surface-based template registration using the robust point matching (RPM) algorithm, and volume-based template registration using the Demons algorithm. All three methods show that the older children have bilateral expanded head segments compared to the younger children. The results analyzed based on radial distance to the centerline were consistent with those analyzed using template-based registration methods. In analyses stratified by sex, it was found that the age-associated anatomical changes were similar in boys and girls, but the age-association was strongest in girls. Total hippocampal volume and sub-regional volumes analyzed using manual segmentation did not show a significant age-association. Our results suggest that shape analysis is sensitive to detect sub-regional differences that are not revealed in volumetric analysis. The three methods presented in this study may be applied in future studies to investigate the normal developmental trajectory of the hippocampus in children. They may be further applied to detect early deviations from the normal developmental trajectory in young children for evaluating susceptibility for psychopathological disorders involving hippocampus.
Keywords: Hippocampal shape analysis, Non-rigid registration, Radial Distance Mapping, Robust Point Matching algorithm, Demons algorithm
1. Introduction
The hippocampus is a brain structure prominently involved in learning and memory. Many studies have evaluated hippocampal changes during the course of normal and pathological aging, fewer studies however, have addressed its normal developmental trajectory. The human hippocampus is identifiable between 6 – 7 gestational weeks (Humphrey, 1964) and by birth the basic neuroanatomical architecture of these regions is present (Arnold and Trojanowski, 1996a,b). Rapid hippocampal development continues over the first two postnatal years (Utsunomiya et al., 1999; Knickmeyer et al., 2008), which is reflected in gross anatomical changes. The changes after two years age have not been systematically investigated. Several studies reported developmental changes in the hippocampus from childhood to young adulthood. In general, age-associated increases in hippocampal size were observed through young adulthood (Ostby et al., 2009) with some evidence for sex-specific developmental patterns (Giedd et al., 1996; Suzuki et al., 2005).
The hippocampus is the most irregularly-shaped sub-cortical structure in the brain, and sub-regional changes are more likely to be detected using analyses that test for shape rather than volume differences. The only study that investigated longitudinal changes in hippocampal shape in 4 to 25 year old subjects concluded that the structural development of the human hippocampus is remarkably heterogeneous (Gogtay et al., 2006). In standardized space, overall hippocampal volume did not change between 4 and 25 years age but there were significant changes in hippocampal shape.
In the present study we applied three shape analysis methods to investigate hippocampal shape in a group of 6 to 10 years old children. The methods that have been applied for shape analysis are based on analyzing the boundary of the segmented hippocampus (Qiu et al., 2008&2009; Thompson et al., 2004). Two common approaches using structure-modeling and template-based registration have been reported. The structure-modeling approach analyzes the medial representation of each subject’s individual hippocampus and compares the between-group differences (Styner and Gerig, 2001). The radial distance mapping method developed by Thompson et al. (2004) is the most widely applied structure-modeling method. It maps the distance of each surface boundary point to the centerline of the hippocampus as an index of "thickness". The template-based registration approach compares the extent of transformation for each individual hippocampus to match a standard template using registration algorithms. In the present study, two different registration algorithms, the robust point matching (RPM, Chui and Rangarajan, 2003) and the Demons (Thiron, 1998), were used.
The purpose of this study was to determine age-related patterns of hippocampal shape in a cohort of typically developing children using three different methods (radial distance mapping, RPM registration and Demons registration). Results achieved by different analytical methods were compared and in addition, the volumes of the manually outlined hippocampus (total as well as head, body and tail segments) were computed and compared to the results obtained using algorithm-based shape analysis methods. Besides the main analysis on the complete group of children, separate analyses stratified by sex were performed to investigate whether age-associated anatomical changes differed in boys and girls.
2. Materials and methods
2.1. Subjects and MRI Protocol
This study was approved by the Institutional Review Board of the University of California, Irvine. The parents/guardians gave written informed consent and the children gave informed assent. The T1-weighted structural MRI scans were acquired from 103 children between the ages of 6–10 years, mean age: 7.3 ± 0.9 years (or, 74 to 119 months old, mean 87 ± 11 months old). The subjects consisted of 50 male and 53 female right-handed children. The age (in months) was used as a continuous variable in regression analysis.
All children were healthy and considered as typically-developing. All of them had a stable neonatal course and did not have any known congenital, chromosomal, or genetic anomalies (e.g., trisomy 21) or neonatal illness (e.g., respiratory distress, mechanical ventilation over 48 hours or sepsis). They had no ultrasound scanning evidence of intraventricular hemorrhage (IVH), periventricular leukomalacia, and/or low-pressure ventriculomegaly in the newborn period. At the time of enrollment, there were no emotional or physical conditions reported in a structured interview using the MacArthur Health and Behavior Questionnaire (Armstrong & Goldstein, 2003). In addition, the MR images were reviewed by a neuroradiologist and reported as normal without any structural abnormality.
Each child underwent an MRI scan conducted on a 3.0 Tesla Philips Achieva system. To minimize head motion, padding was placed around the head. Ear protection was given to all children. To further increase compliance and reduce motion, children were fitted with headphones and allowed to watch a movie of their choice while in the scanner. Following the scanner calibration and pilot scans, a high resolution T1 anatomical scan was acquired in the sagittal planewith 1mm3 isotropic voxel dimensions. A 3D Inversion-Recovery Spoiled Gradient Recalled Acquisition (IR-SPGR) sequence with the following parameters was applied: Repetition rate (TR)= 11ms, Echo Time (TE)= 3.3ms, Inversion Time (TI)= 1100ms, Turbo Field Echo factor (TFE)= 192, Number of slices: 150, no SENSE acceleration, Flip angle=18◦. Acquisition time for this protocol was seven minutes. Variations of these parameters were tested on volunteers to obtain an optimal set that provided the best gray-white matter contrast, sharpness and high resolution while ensuring that there were no discernible artifacts. The purpose was to keep the total acquisition time at a tolerable length for children.
2.2. Manual Segmentation of the Hippocampus
Pre-processing of the anatomic T1 images included correction for image intensity non-uniformities (Sled et al., 1998) and linear stereotaxic transformation (Collins et al., 1994) into coordinates based on the Talairach atlas (Talairach and Tournoux, 1988). Volume analyses of the hippocampus were performed using the interactive software package DISPLAY developed at the Brain Imaging Center of the Montreal Neurological Institute (MNI). All images were registered to the ICBM 152 model brain using the Affine transformation with 12 parameters (Mazziotta et al., 2001), and a standardized segmentation protocol was applied to outline the anatomical boundaries of the hippocampus (Pruessner et al., 2000). This procedure corrects for differences in head size (Collins et al., 1994). The outlined hippocampus was further separated into three segments: head, body, and tail (Pruessner et al., 2000). During late childhood (ages 6–10) the brain size reached approximately 85~95% of the adult brain. Therefore, the techniques and the template developed for the adult brain could be used for this population and the errors caused by using an adult brain template should be negligible.
All brains were segmented by the same operator (KH), who was trained to reach a certain consistency level with an experienced rater (with intraclass correlation coefficient > 0.90) before proceeding with segmentation analyses. The operator’s intra-rater reliability was 0.95 (estimate based on 30 brains that were segmented repeatedly by the operator). The segmented hippocampus on the ICBM 152 template was used in the subsequent shape analysis. Inter subject variability in brain size/geometry was quite substantial. If this was not taken into consideration by normalizing to a template, the hippocampal shape analysis results would be profoundly affected by the size and shape of the brain.
2.3. Radial Distance Mapping (RDM) Shape Analysis Method
Radial distance mapping or RDM (Thompson et al., 2004) considers the hippocampus as a 3D tube-like surface so the hippocampal shape change can be quantified by calculating the Euclidean distance between surface points and the centerline. To establish the consistent one-to-one correspondence of surface points among all subjects, a fixed grid structure is imposed on the surface of each individual subject’s hippocampus. At each surface point of this grid structure, the distance from this point to the centerline of the hippocampus is calculated, and this radial distance is used as an index of “thickness” for evaluating shape variation between two groups.
First the centerline of the hippocampus was extracted using a level-set based algorithm (Telsa and Vilanova, 2003). In order to avoid the branching problem at the beginning and ending sides of the hippocampus, the starting and ending points of the centerline was manually selected based on anatomical features. Then the Bezier curve fitting is applied to fit the control points generated by the centerline extraction algorithm. The hippocampus was then divided into 150 cross-sections along the longitudinal direction. This was done by dividing the centerline into 150 equally-spaced points, and an orthogonal plane at each point was defined. The intersection of the hippocampal surface with each orthogonal plane forms the level curve on that section. Figure 1 shows 6 cases with different hippocampal shapes, as well as the centerline and the cross-sections used for radial distance mapping. The longitudinal length, and the shapes of head, body, and tail are apparently different. On each cross-section, 100 surface points were uniformly assigned along the boundary. The starting point was determined as the outer boundary on the Y–Z projection view shown in Figure 1. For each subject, the hippocampus was represented by a matrix of 100 z-sections and 150×100 grid points on each z-section. The radial distance was obtained by calculating the distance from each surface point to the centerline and recorded into a 150×100 matrix for group comparison. A template was then generated by averaging the radial distance at each digitized grid point from all 103 subjects.
Figure 1.
Example of six hippocampi showing heterogeneous shapes. The centerline and 11 perpendicular cross-sectional planes are marked.
2.4. Surface-Based Robust Point Matching (RPM) Registration Method
Robust point matching (RPM) (Chui et al., 2001; Chui and Rangarajan, 2003) converts feature-based registration to a non-rigid point matching problem, aiming to find an optimal transformation to match the deforming set to the reference set. The sizes of the two point sets do not need to be the same, and one-to-one correspondence between the points is not mandatory. Fuzzy correspondence (Wells, 1997), which allows a point in the deforming set {xi,i=1,2,…,I} to correspond to more than one point in the reference set {yi,j=1,2,…,J}, is applied to estimate the correspondence of the two point sets during the iteration process. Given the fuzzy correspondence M with and and the transformation function f, the energy function of RPM can be calculated by:
| (1) |
where ∥Lf∥ is the smoothness measure, λ is the smoothness coefficient and ζ is the robustness coefficient. The non-rigid transformation is calculated by minimizing the energy function and applied iteratively to the deforming set. The iteration process is driven by deterministic annealing which starts at a high temperature Tinitial and stops when the process reaches a final temperature Tfinal., Specifically, the temperature T decreases through the iterations using an annealing rate (Tk=raTk–1). In our study, the non-rigid transformation is based on the Gaussian radial basis function (RBF):
| (2) |
where {cj} is a set of mapping coefficients and ϕ is a Gaussian kernel. Also, λ = λinitialT with λinitial =1, ζ =1, Tinitial = 50, Tfinal = 1.25 and ra = 0.93. The original RPM code written in MATLAB is available at: http://www.cise.ufl.edu/~anand/students/chui/rpm/TPS-RPM.zip.
The same template generated using the radial distance mapping method (150 sections, 100 points on each section) was used. It was deformed by the RPM to match the surface points of each subject as the reference set. In order to reduce the computational load, the point sets used for RPM were downsampled from the 15,000 points to 1,500. When completed, the deformations at each of the 1,500 surface points along x, y, and z directions were obtained.
2.5. Volume-based Demons Registration Algorithm
Demons algorithm considers coregistration as a diffusion process of deformation grid, which can be controlled by effectors similar to Maxwell's demons. The boundaries of the static object are treated as semi-permeable membranes. Demons can control the membranes and hence adjust the diffusion (deformation) from the moving object by determining if the deformation grid is "inside" or "outside" the target object boundary (Thiron, 1998). Let S be the static object and M be the moving object, the estimated deformation is given by
| (3) |
Similar to all non-rigid registration algorithms, Demons algorithm uses an iterative process of minimizing the energy function from coarse resolution (downsampled) to fine resolution (original). The energy function of Demons algorithm is calculated by:
| (4) |
where D is the deformation field, ◦ is the composition operation and σ is the image noise ratio coefficient (the ratio between image noise and transformation uncertainty). In our study, we adopted the code developed by Kroon and Slump (2009) and it is available at: http://www.mathworks.com/matlabcentral/fileexchange/21451-multimodality-non-rigid-demon-alg orithm-image-registration. Similar to RPM registration, we used the template generated by radial distance mapping as the moving source image and each subject as the static target image. Demons algorithm refines the transformation grids locally to match the target image. After registration, the deformation of each voxel in the source image was calculated and the transformation at the 1,500 template surface points was also obtained for further evaluation.
2.6. Statistical Analyses
The radial distance (the distance of each surface point to the centerline) analyzed using the radial distance mapping method can be directly used for statistical analysis; however, this parameter could not be measured in the template-based RPM and Demons methods. Because we analyzed the surface points in all 3 methods, the concept of calculating inward / outward deformation described by Wiedenmayer et al. (2006) for hippocampal surface morphometry could be applied. The deformation at each surface point to match the template is used to calculate a “radial distance difference (RDD)” by using the centerline of the template as the reference. RDD > 0 means outward deformation (expansion) for template to match the subject, that is, the subject has an expanded hippocampus relative to the template on the cross-sectional plane. While RDD < 0 means inward deformation (shrinkage), indicating the subject has a contracted hippocampus relative to the template on the cross-sectional plane. For the radial distance mapping method, although the radial distances are readily available for comparison, the RDD value, i.e. the difference of the radial distance from the template, also can be calculated thus the same parameter obtained from three different methods can be compared.
Pearson’s linear correlation analysis was used to evaluate the association between RDD value at each surface point and child age in months as a continuous variable. The permutation test with 1 million iterations was applied to obtain the uncorrected p-value at each surface point, and then the correction for multiple comparisons was performed using the False Discovery Rate (FDR) (Genovese et al., 2002). In addition to the whole group analysis using all 103 subjects, we also performed separate analysis stratified by sex.
A region of interest (ROI) was placed in a region that showed significant age-association on p-value maps, and the mean RDD from the ROI was calculated to evaluate the age-association using Pearson’s correlation analysis. For the volumetric data obtained using manual drawing, Pearson’s linear correlation analysis was applied to evaluate the association between hippocampal volume (head, body, tail and the whole volume) and child age. The p-values obtained from the whole group and separately in the male and female groups were reported.
3. Results
3.1. Manual Segmentation Results
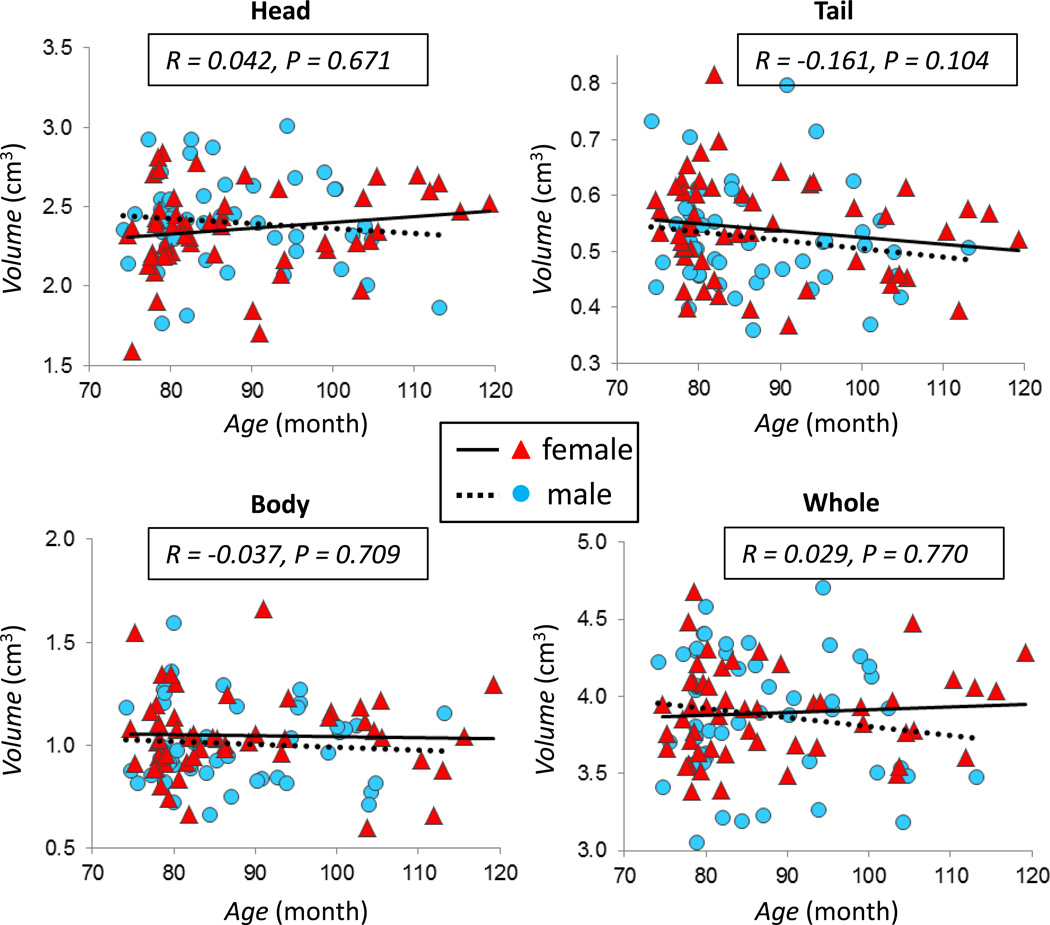
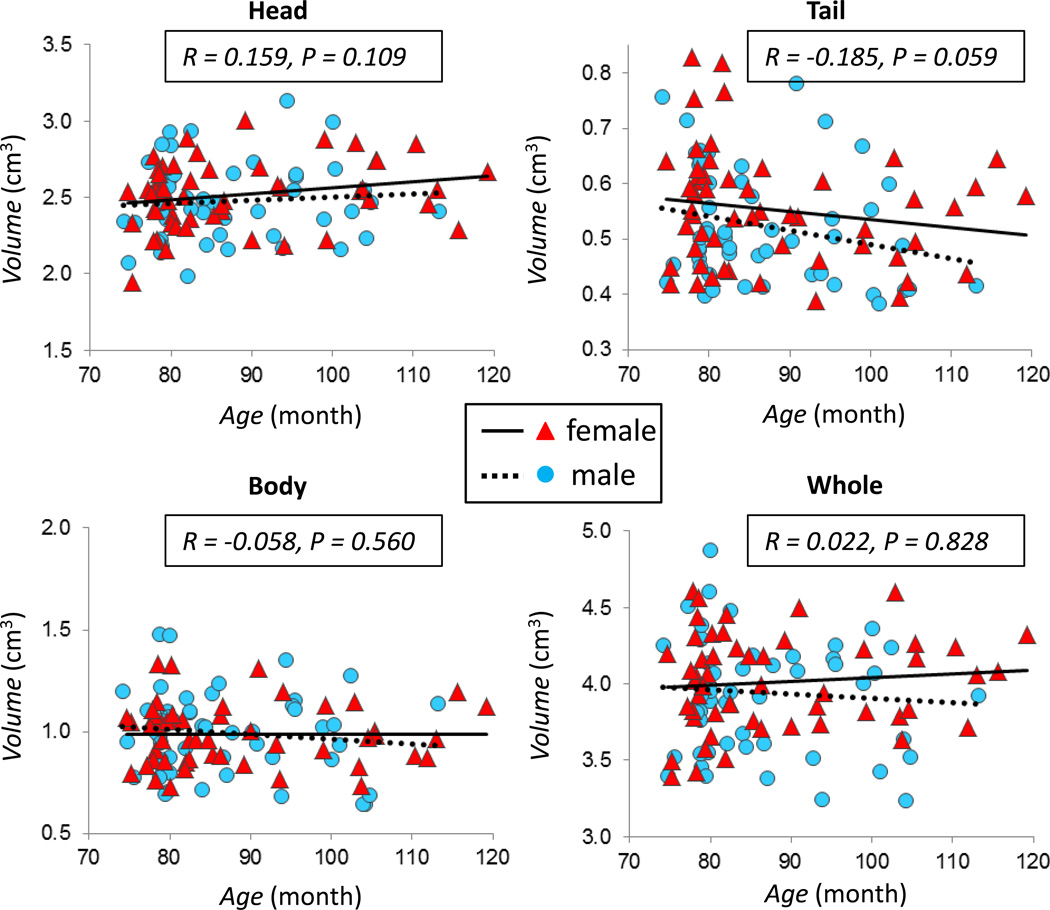
The correlation between hippocampal volume and child age is plotted in Figure 2 for the left hippocampus, and in Figure 3 for the right hippocampus. Neither the whole hippocampus nor hippocampal sub-regions were significantly associated with child age (all p’s > 0.05). The same was true when analyses were performed stratified by sex (p’s > 0.05).
Figure 2.
The correlation between child age and volume of the left hippocampus. Plots show the association between child age and the volume of the head, body, tail, and the whole left hippocampus, with the correlation coefficient r and the p value indicated in each figure for the whole group. All p values are > 0.05. Different symbols and fitting lines are used for males and females, respectively.
Figure 3.
The correlation between child age and volume of the right hippocampus. Plots show the association between child age and the volume of the head, body, tail, and the whole right hippocampus, with the correlation coefficient r and the p value for the whole group indicated in each figure. All p values are > 0.05. The volume of the tail segment is negatively correlated with age with p = 0.06 approaching the significance level. Different symbols and fitting lines are used for males and females, respectively.
3.2. Shape Analysis Based on Radial Distance Difference
Figure 4 shows the uncorrected p-value maps of the correlation between the Radial Distance Difference (RDD) value and age, Figure 4a for males, Figure 4b for females, and Figure 4c for the complete group. The display shows the top view and the bottom view by splitting the hippocampus along the longitudinal axis and opening it into two halves. The results analyzed using these different methods: the RDM method, the surface-based RPM registration, and the volume-based Demons registration, are shown in the same figure for comparison. On these three maps, significant correlations between RDD and age are mainly observed in the head segment, particularly in the bottom view. A consistent result in the head region is seen in both males and females, but the effect is strongest in females. Figure 5 shows the p value maps in the entire group after applying the FDR correction for multiple comparisons, and it can be seen that some areas in the head segment survive the FDR correction with p<0.05.
Figure 4.
The uncorrected p-value maps of the correlation between the Radial Distance Difference (RDD) value analyzed using the RDM method, the surface-based RPM registration, and the volume-based Demons registration with age in the male group(a); the female group(b); and the whole group combining males and females(c). All 3 analytic methods reveal significant correlations with child age mainly in the head segment, particularly in the bottom view. In separate sex analyses in males and females, both show significant results in the head region, but the effect is stronger in females than in males.
Figure 5.
The p-value maps after applying the FDR correction for multiple comparisons in the whole group combining males and females. It can be seen that a substantial cluster of pixels in the head segment of the bottom view survive the FDR correction with p<0.05 (showing green, yellow to red color).
3.3. ROI Analysis of Radial Distance Difference with Age
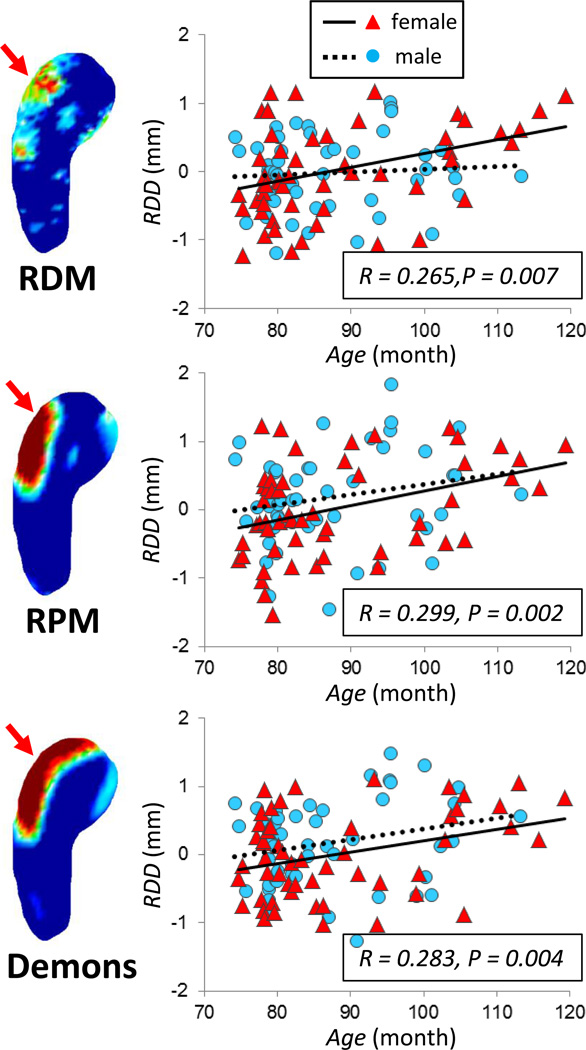
In order to further evaluate the age association in the area within the head segment that survived the FDR correction, a region of interest (ROI) was outlined based on the significant areas on the p-value maps. One ROI (approximately 7.1 mm2) was placed over the 3-dimensional surface of the head region on the left hippocampus, and another ROI (approximately 7.1 mm2) was placed on the right hippocampus. The age correlation of the mean RDD from the selected ROI in the left hippocampus is plotted in Figure 6. The Pearson correlation coefficient (r) and the p values are indicated in each figure, which are significant for all three methods in the whole group, with p= 0.001 for RDM, p= 0.004 for RPM, and p= 0.002 for Demons. The results for males and females are indicated by different symbols in the figures. In separate analysis, the results are significant in females (p= 0.001 for RDM, p= 0.012 for RPM, and p= 0.014 for Demons). In males, significant results are observed only for the Demons method (p= 0.198 for RDM, p= 0.084 for RPM, and p= 0.032 for Demons). The results analyzed from the selected ROI in the right hippocampus are shown in Figure 7. The linear age correlation p values in the whole group are p= 0.007 for RDM, p= 0.002 for RPM, and p= 0.004 for Demons. In separate analyses stratified by sex, the age-association is significant in females (p= 0.006 for RDM, p= 0.005 for RPM, and p= 0.014 for Demons), but is not significant in males (p= 0.597 for RDM, p= 0.134 for RPM, and p= 0.086 for Demons).
Figure 6.
The age correlation of the mean RDD value analyzed in a selected ROI (approximately 7.1 mm2) placed over the 3-dimensional surface of the head region on the left hippocampus. For each method using RDM, RPM and Demons registration, a mean RDD value was calculated by averaging over all pixels contained within the ROI, and used in the plot. The Pearson’s correlation coefficient r and the p values analyzed using the whole group are indicated in each figure, which are significant for all 3 methods. The older subjects are more likely to have positive RDD, indicating expanded hippocampus compared to the template in the head region of the left hippocampus. Different symbols and fitting lines are used for males and females, respectively.
Figure 7.
The age correlation of the mean RDD value analyzed in a selected ROI (approximately 7.1 mm2) placed over the 3-dimensional surface of the head region on the right hippocampus. The Pearson’s correlation coefficient r and the p values analyzed using the whole group are indicated in each figure, which are significant for all 3 methods. The older subjects are more likely to have positive RDD, indicating expanded hippocampus compared to the template in the head region of the right hippocampus. Different symbols and fitting lines are used for males and females, respectively.
RDD > 0 indicates expanded hippocampus relative to the template. It can be seen that older subjects are more likely to have a positive RDD. These figures illustrate that the results analyzed from these 2 bilateral hippocampal head ROI’s using three different methods are consistent.
4. Discussion
In this study we applied three methods to assess the shape of the hippocampus in a group of typically-developing 6 to 10 year old preadolescent children. Overall, there was high inter-individual variability in hippocampal shape, but significant age associations were noted in some sub-regions. All three methods showed a significantly expanded head segment in both the left and the right hippocampi with age. In analyses stratified by sex, it was noted that the age-associated anatomical changes were strongest in females. However, the volume of the head segment analyzed using the manual drawing was not significantly associated with age. As illustrated in Figures 4 and 5, only the shape of a sub-region in the head segment, not the entire region, was significantly associated with age. The results suggest that hippocampal shape analysis can detect anatomical differences that cannot be identified using volumetric analyses. Shape analysis of subcortical structures in the brain has been widely applied in various neurological diseases, e.g. mild cognitive impairment and Alzheimer’s disease (Thompson et al., 2004; Csernansky et al.,2005;Apostolova et al., 2006; Qiu et al., 2008&2009; Morra et al., 2009), schizophrenia (Csernansky et al., 1998; Mamah et al., 2008), Parkinson disease (Uthama et al., 2007), and depression (Zhao et al., 2008; Tamburo et al., 2009; Tae et al., 2011). Many studies have also shown that it can be used to evaluate early morphometric changes that occur before a significant volumetric atrophy is noted.
It is believed that the major growth of the hippocampus is completed before the age of two years (Utsunomiya et al., 1999; Knickmeyer et al., 2008). During the age range of 6–10 years, which is the age range of children included in this study, developmental changes related to synaptic formation and pruning of neurons in the hippocampus are expected. This is also a critical period when a new phase of neuronal rearrangements takes place in the entire brain (Andersen et al., 2003; Caviness et al., 1996). The expansion of other brain regions, e.g. the maturation of white matter tracts, may stretch or compress the hippocampus and affect its anatomical appearance (Gogtay et al., 2006; Insausti et al., 2010). Therefore, in addition to the intrinsic pruning of neurons within the hippocampus, other sources of neurological changes may also affect the shape of the hippocampus.
A few studies reported the change in size of the hippocampus during childhood (Giedd et al., 1996; Suzuki et al., 2005; Ostby et al., 2009), and there was only one study reporting the change in the shape (Gogtay et al., 2006). These studies covered a much wider age-range, e.g. 4–18 years old in Giedd et al. (1996), 4–25 years old in Gogtay et al. (2006), and 8–30 years old in Ostby et al. (2009). In general, age-associated increases in hippocampal size were observed (Ostby et al., 2009) with some evidence for sex-specific patterns (Giedd et al., 1996; Suzuki et al., 2005). Since different subject cohorts spanning different large age ranges were analyzed, their results could not be directly compared to each other. Similar to these studies analyzing the size of the hippocampus, the shape analysis reported by Ostby et al., 2009 also concluded that the structural development of the human hippocampus was remarkably heterogeneous. In standardized space, the overall hippocampal volume did not change between 4 and 25 years age; but the shape analysis suggested that there were significant developmental changes in the posterior region of the hippocampus (increase over time) and the anterior region of the hippocampus (loss over time). In our study we found an age-related expansion in the head region of the hippocampus. Based on the discussion above, two possible reasons are: 1) synaptic formation and pruning of neurons within the hippocampus; and 2) mechanical force exerted by expansion of other brain structures. However, there is no sufficient data based on other imaging studies or histological evidence to suggest which one is the dominating factor. In patients with pathological conditions, detailed examination of brain specimens is needed to confirm the imaging findings- but this is almost impossible in young healthy children. Our findings may have to be validated through independent studies analyzed using different pre-adolescent cohorts.
Three methods were applied in this study, including radial distance mapping, RPM registration and Demons registration. Each of the three applied methods has specific advantages and disadvantages. The radial distance mapping method uses transparent, straightforward, analysis procedures, so the results can be easily interpreted. This method has become one of the most popular methods for hippocampal shape analyses, and has been successfully applied to study different clinical patient cohorts (Apostolova et al., 2006; Morra et al., 2009; Thompson et al., 2004; Xu et al., 2008). However, because the analysis is based on the distance to the centerline, the results depend on the ability to reliably detect the centerline in each individual subject. The automated centerline extraction algorithms are highly sensitive to boundaries, which often generate additional branches at the beginning and the ending of the structure, and require consolidation to keep only one branch (Yushkevich et al., 2006). Yushkevich et al. (2006, 2007, 2009) proposed a continuous medial representation (cm-reps) model to solve this branching problem. Other methods also have been proposed (Pizer et al., 2005; Shi et al., 2008; Hassouna and Farag, 2009), and this is still an active research area. In the present study, we manually set the beginning and ending points and used the automated algorithm to detect a unique centerline for each case. The hippocampus has a well-recognizable longitudinal axis, and the centerline extracted using different methods is expected to be pretty consistent (except at the tip of the head and tail segments) and not likely to be a main limitation affecting the obtained results.
Non-rigid registration is widely used in clinical medical imaging for different applications, such as assessing motion-related deformation for radiation therapy planning, registration of images acquired using different imaging modalities (MRI, PET, CT, ultrasound, etc.), and evaluation of structural changes related to pathological conditions. The application of RPM for hippocampal registration has been reported before by Guo et al. (2005). The volume-based fluid flow registration, similar to the Demons used here, has been applied to investigate the correlation between cortisol levels and hippocampal volumes of healthy preadolescent children by Wiedenmayer et al. (2006). Their results were similar to ours presented in this work, showing that the surface morphology analysis revealed significant correlations with cortisol in some regions, but measures of hippocampal volume were unrelated. Another method, the large deformation diffeomorphic metric mapping (LDDMM) also has been applied for hippocampal shape analysis in several studies. However, the source code is not available to the public. Yassa and Stark (2009) reported that Demons and LDDMM have similar performance for whole field registration; while for subfield analysis Demons outperforms LDDMM.
Therefore, in this study we chose to use RPM and Demons algorithms to analyze the hippocampus shape and compared the results with those analyzed by the radial distance mapping method. Since the analysis was based on the same surface grid points and the same centerline, these three methods were not totally independent. However, the underlying methodologies were different. The surface-based RPM and the volume-based Demons registration methods used different deformation models and optimization algorithms to match the template to the individual hippocampus, and it took different deformation routes to reach the same convergence results. The fact that three methods using different algorithms and different analysis approached showed similar results further enhanced our confidence on the reliability of the observed age differences. Since the registration methods did not require any manual operation or the use of the centerline, they could be directly used to evaluate the hippocampal shape.
In analyses stratified by sex, age-associated anatomical changes were strongest in females. This finding was consistent with the growing recognition of sexual dimorphism in neurological development (Bale, 2009; Lin et al, 2009, Boukouvalas et al, 2010). Compared with males, females exhibit accelerated neurological development as early as 30 weeks gestation (Buss et al, 2009; Glynn and Sandman, 2012). The differential brain development seen on MRI in preadolescent boys and girls might be related to changes in hormones (Peper et al. 2008).We have also reported sex difference in cortical thickness and sub-cortical volume of some brain structures in this same subject cohort (Muftuler et al. 2011). The next question that needs to be addressed is to find out the exact reasons leading to the detected hippocampal shape differences. In the recent two years, the study of subfields within the hippocampus has attracted more research attention - in part because of the advanced imaging technology that allows acquisition of high quality images with a sufficiently high spatial resolution and a clear division of different subfields (e.g. CA1, CA2, CA3, CA4, and dentate gyrus). We believe that with the advances in this research field, more findings will be reported that will shed new light on the specific developmental patterns that account for the changes in hippocampal shape in the pre-adolescent period.
5. Conclusions
In summary, we applied three different methods to assess hippocampal shape in a group of typically-developing children from 6 to 10 years old, and presented data demonstrating that there are developmental changes in the morphology of the hippocampus. The fact that all three methods yielded significant correlations between age and shape of the hippocampal head but not with volume of the hippocampal head, suggested that shape analyses are more sensitive in detecting subtle alterations in the anatomy of the hippocampus and may be useful in describing normal developmental trajectories of hippocampal development. Establishing the trajectories of normal brain development during childhood provides the basis for evaluating deviations that might be associated with neurological impairment and disease. Thus, these results in typically developing children may be an important reference for future studies in young children with, or at risk for, diseases of the nervous system.
The shape of normal developing hippocampus from 6–10 years old children is studied.
The radial distance mapping and two template-based registration methods were used to evaluate the age-associated shape changes.
All three methods show that the older children have bilateral expanded head segments compared to the younger children.
The manual volumetric measurement did not show significant results.
Registration methods can be used to study developmental trajectory of the shape of hippocampus.
Acknowledgements
The authors wish to thank the families who participated in this project. The assistance of Megan Blair, Natalie Hernandez and Christina Canino is gratefully acknowledged.
Funding Support: This research was supported by NIH R01 HD050662 to EPD and NIH R01HD051852 to CAS.
ABBREVIATIONS
- AD
Alzheimer’s Disease
- CT
Computed Tomography
- FDR
False Discovery Rate
- ICBM
International Consortium for Brain Mapping
- IR-SPGR
inversion-recovery spoiled gradient recalled acquisition
- LDDMM
Large Deformation Diffeomorphic Metric Mapping
- MNI
Montreal Neurological Institute
- MRI
Magnetic Resonance Imaging
- PET
Positron Emission Tomography
- RDD
Radial Distance Difference
- RDM
Radial Distance Mapping
- ROI
Region Of Interest
- RPM
Robust Point Matching
- TE
Echo Time
- TFE
Turbo Field Echo
- TI
Inversion Time
- TR
Repetition Time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neurosci. Biobehav. Rev. 2003;27:3–18. doi: 10.1016/s0149-7634(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer’s disease. Brain. 2006;129:2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Armstrong JM, Goldstein LH. Manual for the MacArthur Health and Behavior Questionnaire (HBQ 1.0) Chicago: MacArthur Foundation Research Network on Psychopathology and Development Kupfer, D. J. University of Pittsburgh; 2003. [Google Scholar]
- Arnold SE, Trojanowski JQ. Human fetal hippocampal development: I Cytoarchitecture, myeloarchitecture, and neuronal morphologic features. J Comp Neurol. 1996a;367:274–292. doi: 10.1002/(SICI)1096-9861(19960401)367:2<274::AID-CNE9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Arnold SE, Trojanowski JQ. Human fetal hippocampal development: II. The neuronal cytoskeleton. J Comp Neurol. 1996b;367:293–307. doi: 10.1002/(SICI)1096-9861(19960401)367:2<293::AID-CNE10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Bale TL. Neuroendocrine and immune influences on the CNS: it's a matter of sex. Neuron. 2009;64(1):13–16. doi: 10.1016/j.neuron.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Boukouvalas G, Gerozissis K, Markaki E, Kitraki E. High-fat feeding influences the endocrine responses of pubertal rats to an acute stress. Neuroendocrinology. 2010;92:235–245. doi: 10.1159/000321393. [DOI] [PubMed] [Google Scholar]
- Buss C, Davis EP, Class QA, Gierczak M, Pattillo C, Glynn LM, Sandman CA. Maturation of the human fetal startle response: Evidence for sex-specific maturation of the human fetus. Early Human Development. 2009;85:633–638. doi: 10.1016/j.earlhumdev.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness VS, Jr, Kennedy DN, Richelme C, Rademacher J, Filipek PA. The human brain age 7–years: a volumetric analysis based on magnetic resonance images. Cereb. Cortex. 1996;6:726–736. doi: 10.1093/cercor/6.5.726. [DOI] [PubMed] [Google Scholar]
- Chui H, Rangarajan A. A new point matching algorithm for non-rigid registration. Comput Vis Image Underst. 2003;89:114–141. [Google Scholar]
- Chui H, Win L, Schultz R, Duncan J, Rangarajan A. A unified feature registration method for brain mapping. Inf Process Med Imaging. 2001;2082:300–314. [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, Grenander U, Miller MI. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci U S A. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Vaituzis AC, Hamburger SD, Lange N, Rajapakse JC, Kaysen D, Vauss YC, Rapoport JL. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: Ages 4–18 years. J Comp Neurol. 1996;366:223–230. doi: 10.1002/(SICI)1096-9861(19960304)366:2<223::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Glynn LM, Sandman CA. Sex moderates associations between prenatal glucocorticoid exposure and human fetal neurological development. Dev Sci. 2012;15:601–610. doi: 10.1111/j.1467-7687.2012.01159.x. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Nugent TF, 3rd, Herman DH, Ordonez A, Greenstein D, Hayashi KM, Clasen L, Toga AW, Giedd JN, Rapoport JL, Thompson PM. Dynamic mapping of normal human hippocampal development. Hippocampus. 2006;6:664–672. doi: 10.1002/hipo.20193. [DOI] [PubMed] [Google Scholar]
- Guo H, Rangarajan A, Joshi S. 3D diffeomorphic shape registration using hippocampal datasets. Med Image Comput Comput Assist Interv. 2005;8:984–991. doi: 10.1007/11566489_121. [DOI] [PubMed] [Google Scholar]
- Hassouna MS, Farag AA. Variational curve skeletons using gradient vector flow. IEEE Trans Pattern Anal Mach Intell. 2009;31:2257–2274. doi: 10.1109/TPAMI.2008.271. [DOI] [PubMed] [Google Scholar]
- Humphrey T. Some observations on the development of the human hippocampal formation. Trans Am Neurol Assoc. 1964;89:207–209. [PubMed] [Google Scholar]
- Insausti R, Cebada-Sánchez S, Marcos P. Postnatal development of the human hippocampal formation. Adv Anat Embryol Cell Biol. 2010;206:1–86. [PubMed] [Google Scholar]
- Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH. A structural MRI study of human brain development from birth to 2 years. J Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon DJ, Slump CH. MRI modality transformation in demon registration. IEEE International Symposium on Biomedical Imaging (ISBI) :963–966. [Google Scholar]
- Lin Y, Ter Horst GJ, Wichmann R, Bakker P, Liu A, Li X, Westenbroek C. Sex differences in the effects of acute and chronic stress and recovery after long-term stress on stress-related brain regions of rats. Cereb Cortex. 2009;19(9):1978–1989. doi: 10.1093/cercor/bhn225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamah D, Harms MP, Wang L, Barch D, Thompson P, Kim J, Miller MI, Csernansky JG. Basal ganglia shape abnormalities in the unaffected siblings of schizophrenia patients. Biol Psychiatry. 2008;64:111–120. doi: 10.1016/j.biopsych.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Feidler J, Smith K, Boomsma D, Hulshoff Pol H, Cannon T, Kawashima R, Mazoyer B. A four-dimensional probabilistic atlas of the human brain. J Am Med Inform Assoc. 2001;8:401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM Alzheimer's Disease Neuroimaging Initiative. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Map. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuler LT, Davis EP, Buss C, Head K, Hasso AN, Sandman CA. Cortical and subcortical changes in typically developing preadolescent children. Brain Res. 2011;1399:15–24. doi: 10.1016/j.brainres.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostby Y, Tamnes CK, Fjell AM, Westlye LT, Due-Tønnessen P, Walhovd KB. Heterogeneity in subcortical brain development: A structural magnetic resonance imaging study of brain maturation from 8 to 30 years. J Neurosci. 2009;29:11772–11782. doi: 10.1523/JNEUROSCI.1242-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peper JS, Brouwer RM, Schnack HG, van Baal GC, van Leeuwen M, van den Berg SM, Delemarre-Van de Waal HA, Janke AL, Collins DL, Evans AC, Boomsma DI, Kahn RS, Hulshoff Pol HE. Cerebral white matter in early puberty is associated with luteinizing hormone concentrations. Psychoneuroendocrinology. 2008;33(7):909–915. doi: 10.1016/j.psyneuen.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Pizer SM, Fletcher PT, Joshi S, Gash AG, Stough J, Thall A, Tracton G, Chaney EL. A method and software for segmentation of anatomic object ensembles by deformable m-reps. Med Phys. 2005;32:1335–1345. doi: 10.1118/1.1869872. [DOI] [PubMed] [Google Scholar]
- Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb Cortex. 2000;10:433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- Qiu A, Younes L, Miller MI, Csernansky JG. Parallel transport in diffeomorphisms distinguishes the time-dependent pattern of hippocampal surface deformation due to healthy aging and the dementia of the Alzheimer's type. Neuroimage. 2008;40:68–76. doi: 10.1016/j.neuroimage.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Fennema-Notestine C, Dale AM, Miller MI Alzheimer's Disease Neuroimaging Initiative. Regional shape abnormalities in mild cognitive impairment and Alzheimer's disease. Neuroimage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lai R, Krishna S, Sicotte N, Dinov I, Toga AW. Anisotropic Laplace-Beltrami Eigenmaps: Bridging Reeb Graphs and Skeletons. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2008;2008:1–7. doi: 10.1109/CVPRW.2008.4563018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Styner M, Gerig G. Proc. Info. Proc. Med. Imag. Lect. Notes Comput. Sci. vol. 2082. Berlin: Springer Press; 2001. Medial models incorporating object variability for 3D shape analysis; pp. 502–516. [Google Scholar]
- Suzuki M, Hagino H, Nohara S, Zhou SY, Kawasaki Y, Takahashi T, Matsui M, Seto H, Ono T, Kurachi M. Male-specific volume expansion of the human hippocampus during adolescence. Cereb Cortex. 2005;15:187–193. doi: 10.1093/cercor/bhh121. [DOI] [PubMed] [Google Scholar]
- Tae WS, Kim SS, Lee KU, Nam EC, Choi JW, Park JI. Hippocampal shape deformation in female patients with unremitting major depressive disorder. AJNR Am J Neuroradiol. 2011;32:671–676. doi: 10.3174/ajnr.A2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. Stuttgart: Thieme; 1988. [Google Scholar]
- Tamburo RJ, Siegle GJ, Stetten GD, Cois CA, Butters MA, Reynolds CF, 3rd, Aizenstein HJ. Amygdalae morphometry in late-life depression. Int J Geriatr Psychiatry. 2009;24:837–846. doi: 10.1002/gps.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telsa A, Vilanova A. A robust level-set algorithm for centerline extraction. Proc Sym Data Vis. 2003:185–194. [Google Scholar]
- Thirion JP. Image matching as a diffusion process: an analogy with Maxwell's demons. Med Image Anal. 1998;2:243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Uthama A, Abugharbieh R, Traboulsee A, McKeown MJ. Invariant SPHARM shape descriptors for complex geometry in MR region of interest analysis. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:1322–1325. doi: 10.1109/IEMBS.2007.4352541. [DOI] [PubMed] [Google Scholar]
- Utsunomiya H, Takano K, Okazaki M, Mitsudome A. Development of the temporal lobe in infants and children: analysis by MR-based volumetry. AJNR Am J Neuroradiol. 1999;20:717–723. [PMC free article] [PubMed] [Google Scholar]
- Wells WM., III Statistical approaches to features-based object recognition. Int J Comput Vis. 1997;21:63–98. [Google Scholar]
- Wiedenmayer CP, Bansal R, Anderson GM, Zhu H, Amat J, Whiteman R, Peterson BS. Cortisol levels and hippocampus volumes in healthy preadolescent children. Biol Psychiatry. 2006;60:856–861. doi: 10.1016/j.biopsych.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Valentino DJ, Scher AI, Dinov I, White LR, Thompson PM, Launer LJ, Toga AW. Age effects on hippocampal structural changes in old men: the HAAS. Neuroimage. 2008;40:1003–1015. doi: 10.1016/j.neuroimage.2007.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa M, Stark CE. A quantitative evaluation of cross-participant registration techniques for MRI studies of the medial temporal lobe. Neuroimage. 2009;44:319–327. doi: 10.1016/j.neuroimage.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Zhang H, Gee JC. Continuous medial representation for anatomical structures. IEEE Trans Med Imaging. 2006;25:1547–1564. doi: 10.1109/tmi.2006.884634. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Detre JA, Mechanic-Hamilton D, Fernández-Seara MA, Tang KZ, Hoang A, Korczykowski M, Zhang H, Gee JC. Hippocampus-specific fMRI group activation analysis using the continuous medial representation. Neuroimage. 2007;35:1516–1530. doi: 10.1016/j.neuroimage.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA. Continuous medial representation of brain structures using the biharmonic PDE. Neuroimage. 2009;45:99–110. doi: 10.1016/j.neuroimage.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Taylor WD, Styner M, Steffens DC, Krishnan KR, MacFall JR. Hippocampus shape analysis and late-life depression. PLoS One. 3:e1837. doi: 10.1371/journal.pone.0001837. [DOI] [PMC free article] [PubMed] [Google Scholar]