Summary
p300 and CBP are multi-domain histone acetyltransferases (HATs) that regulate gene expression and are mutated in human diseases including cancer. In the August issue of NSMB, Delvecchio and colleagues report the structure of the p300 catalytic core, revealing the presence of a previously unknown RING domain that regulates the enzyme’s activity.
Reversible acetylation of lysine residues on histones, transcription factors, and transcriptional co-activators plays a central role in activating eukaryotic transcription (Eberharter and Becker, 2002). Histone acetyltransferase (HAT) enzymes, which attach an acetyl group to lysine, are grouped into three families based on sequence homology within their catalytic domains:the Gcn5 N-acetyltransferase (GNAT) family, the Morf, Ybf2, Sas2, and Tip60 (MYST) family, and the p300/CBP family (Lee and Workman, 2007). Although all three classes of HATs use acetyl-CoA as the acetyl donor and catalyze the same reaction, they differ substantially in structure and use different reaction mechanisms (Berndsen and Denu, 2008). A variety of mechanisms have evolved to regulate acetyltransferase activity and prevent inappropriate gene activation. These mechanisms include incorporation of the HAT into larger complexes that potentiate its enzymatic activity and the presence of one or more chromatin reader domains that each recognizes a specific type of histone modification (Lee and Workman, 2007). By binding to individual histone modifications such as acetylated or methylated lysine or to combinations of modifications, chromatin reader domains restrict HAT activity to the appropriate chromosomal context (Lee and Workman, 2007). Misregulation of enzymatic activity has been associated with cancer for all three HAT families, and many aggressive tumors are characterized by differences in HAT expression levels (Cohen et al., 2011).
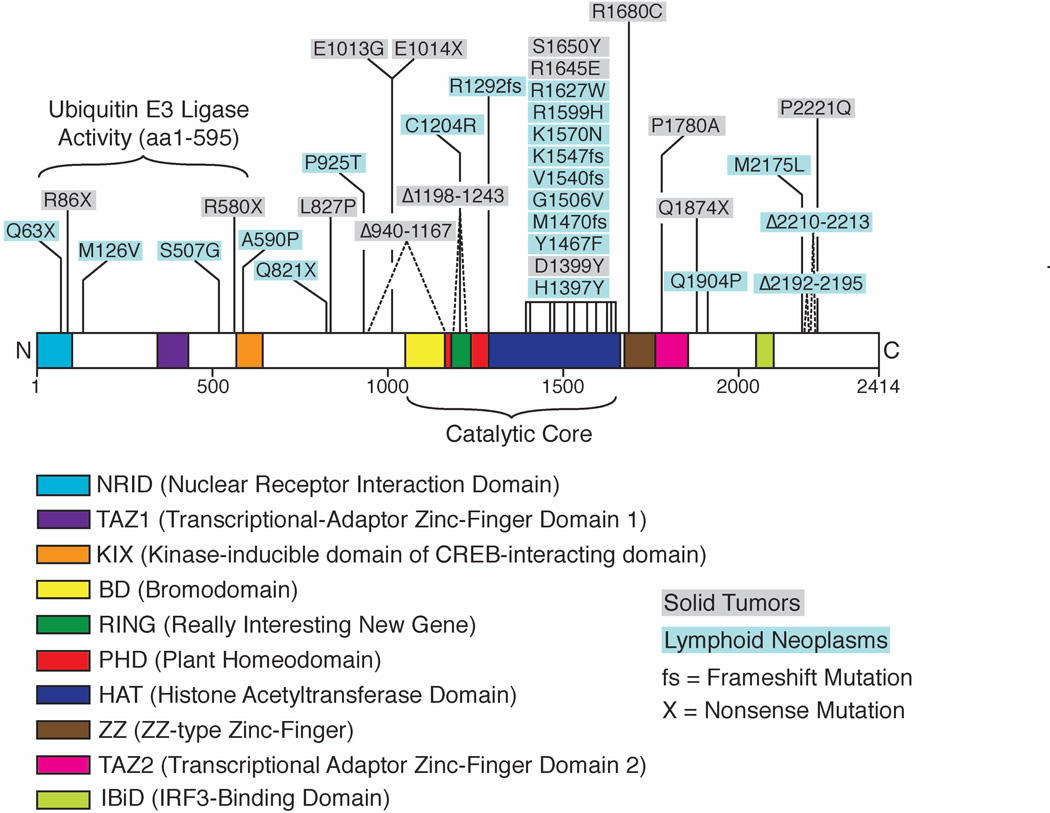
CBP and p300 are large, highly similar HATs of over 2,400 amino acid residues with overlapping cellular functions. The two proteins are ~64% identical in primary sequence, with even higher levels of conservation across their catalytic cores (Kalkhoven, 2004). p300/CBP contains at least nine annotated domains, in addition to the HAT domain, including a bromodomain, which binds acetylatedlysines (Kalkhoven, 2004), and a predicted PHD domain, which typically binds methylated lysines (Lee and Workman, 2007). In addition to associating with transcription factors such as TATA-binding protein and TFIIB, p300/CBP also interacts with the tumor suppressor proteins, p53 and BRCA1, as well as on coproteins such as fos and myb (Kalkhoven, 2004). Among HAT enzymes, p300/CBP is particularly interesting because it activates itself by auto acetylating a basic loop in the HAT domain (Thompson et al., 2004), and also acetylates at least 70 non-histone proteins, including p53 (Wang et al., 2008). Mutations in CBP give rise to the congenital development disorder, Rubinstein-Taybi Syndrome (RTS) (Kalkhoven, 2004), and multiple human cancers arise from mutations and translocations of p300/CBP (Figure 1). While an earlier structure of the isolated p300 HAT domain showed how certain oncogenic mutations disrupted catalytic activity (Liu et al., 2008), most known mutations and deletions either map outside of this domain or have no obvious impact on catalytic function (Cohen et al., 2011; Kalkhoven, 2004). Given our current understanding of p300/CBP-catalyzed acetylation, the mechanism by which these core mutations dysregulate the acetyltransferase activity of p300/CBP and contribute to carcinogenesis has remained elusive.
Figure 1.
Domain structure of p300 overlaid with mutations and truncations found in solid tumors and lymphoid neoplasms (Iyer et al., 2004; Pasqualucci et al., 2011). The catalytic core of p300 is a hotspot for cancer mutations.
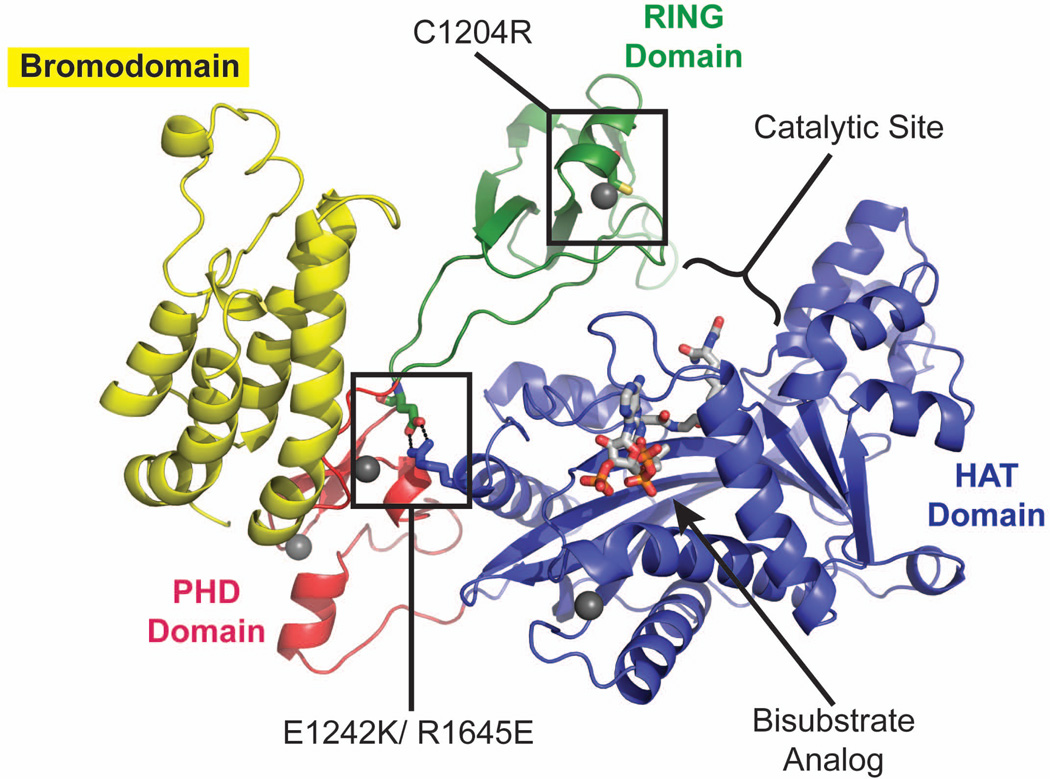
The structure reported by Delvecchio and colleagues (Delvecchio et al., 2013) of a larger p300 catalytic core fragment contains an unanticipated feature that is key to understanding p300/CBP HAT regulation. In addition to the HAT domain bound to a lysine-CoA bisubstrate analogue, the fragment also contains a bromodomain and a sequence known as the CH2 region, which contains a PHD domain. The structure quite unexpectedly revealed that the CH2 region also contains a structurally unusual RING domain, which is inserted within the PHD domain. RING domains are found in a subset of ubiquitin E3 ligases, where they mediate interactions with both E2 ubiquitin conjugating enzymes and ubiquitin (Deshaies and Joazeiro, 2009). In the new p300 structure, the RING domain contacts the active site of the HAT domain, blocking the substrate-binding cleft and thus suggesting that the RING domain might regulate HAT activity. Indeed, the authors showed that mutating key residues that perturb binding of the RING domain to the HAT domain results in a hyperactive form of the enzyme, pointing to an auto-inhibitory role for the RING domain in p300/CBP catalysis. A number of mutations and deletions that give rise to cancer and RTS also map to the RING-HAT interface, illuminating the likely mechanism by which previously uncharacterized mutations misregulate p300/CBP activity and cause disease. For example, the C1204R mutation found in some B-cell lymphomas disrupts the integrity of the RING domain by removing a zinc coordination site (Figure 2). Other curated mutations affect the RING domain as well: mutations that disturb the interaction between the RING and active site such as R1645E and E1242K are found in malignant melanoma and RTS, respectively (Figure 2), while the RING is entirely deleted in some forms of breast cancer. Accordingly, cells transfected with any of these mutant enzymes display hyperacetylation of p53 relative to a wild-type control.
Figure 2.
Structure of p300 core fragment. Individual domains colored as in figure 1. Certain p300 mutations found in cancer map to the newly discovered auto inhibitory RING domain. C1204R coordinates a zinc atom in the RING domain itself, whereas E1242 and R1645 form a salt bridge between the RING and HAT domains. (PDB ID 4BHW)
Why was the presence of the RING domain inp300/CBP overlooked? First, the RING is embedded in a PHD domain, whose histone-interacting residues have been re-arranged as a result. Second, while canonical RING domains contain two structural zinc atoms in separate coordination sites (Deshaies and Joazeiro, 2009), the p300 RING contains a single bound zinc, with tightly packed hydrophobic interactions replacing the second metal-binding site. There is also an insertion in the p300 RING loop L2 that, in other RINGs, interacts with E2 enzymes (Deshaies and Joazeiro, 2009). Based on its established role in the ubiquitin conjugation pathway, Delvecchio and colleagues tested the p300 RING for E3 ligase activity in vitro but could detect none, at least with the panel of E2 enzymes tested. While an as-yet undiscovered role in ubiquitination cannot be ruled out, the authors’ findings indicate that the RING domain in p300 has been adapted to auto-regulate HAT activity.
The discovery of a previously unknown RING domain that gates the activity of the HAT domain is a key step forward in understanding how p300 is regulated. It remains to be seen to what degree the RING domain may co-regulate HAT activity in concert with the auto regulatory loop in the HAT domain, which was deleted in both the present and previous structural studies of p300 because it interfered with crystallization. The role of the PHD domain also remains to be determined, as it lacks features that enable other PHD domain to bind methylated lysines on histones. Perhaps this PHD domain evolved to perform a structural role, anchoring the inserted RING domain near the catalytic site and forming a compact core structure that bridges the bromodomain and catalytic domain. Although the newly identified RING domain does not appear to be an active member of the ubiquitination machinery, previous work has shown that the first ~600 residues of p300/CBP has E3ligase activity and mediate subiquitination of the tumor suppressor protein, p53(Grossman et al., 2003). Could it be that this portion of p300/CBP also contains a divergent RING that is not detectable by primary sequence analysis? More broadly, the discovery of an unanticipated domain that plays a critical role in regulating HAT activity is a reminder that other such domains likely await discovery. The inventory of regulatory mechanisms applied to HATs will surely continue to grow as structural biologists successfully tackle larger and larger protein fragments and complexes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Current opinion in structural biology. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Poręba E, Kamieniarz K, Schneider R. Histone Modifiers in Cancer Friends or Foes? Genes & cancer. 2011;2:631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. EMBO reports. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Özdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochemical pharmacology. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nature Reviews Molecular Cell Biology. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J. Regulation of the p300 HAT domain via a novel activation loop. Nature structural & molecular biology. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Current opinion in structural biology. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]