Abstract
Oodorant receptors (ORs) are essential for insect survival in the environment and thus are ideal molecular targets for the design of insect-inspired modern green chemicals to control populations of agricultural pests and insects of medical importance. Although insect ORs are known for more than a decade, their structural biology is still in its infancy. Here, we unravel the first structural features of ORs from the malaria mosquito, the Southern house mosquito and the silkworm moth. The second extracellular loops (ECL-2s) of their predicted structures are much longer than ECL-1s and ECL-3s. The 27 amino-acid-residue-long of the ECL-2s in mosquito and the 43 amino-acid-residue-long ECL2s in moth ORs are well-conserved. About one-third of the residues are identical, including 3-4 Pro residues. Thorough examination of well-conserved residues in these structures, by point mutation and functional assay with the Xenopus oocyte recording system, strongly suggest that these “loops” include three β-turns and some degree of folding. In the Southern house mosquito three Pro residues in ECL-2 are essential for full activation of the receptor, which is finely tuned to the oviposition attractant 3-methylindole. Additionally, the “corner residues” of prolines, including Gly, Tyr, and Leu are functionally important thus suggesting that turns are stabilized not only by backbone hydrogen bonds, but also by side-chain interactions. Examination of ECL-2s from a distant taxonomical group suggests these ECL-2 loops might be functionally important in all insect ORs. Two of the four Pro residues in the predicted ECL-2 of the bombykol receptor in the silkworm moth, BmorOR1, are essential for function. Experimental evidence indicates that these loops may not be specificity determinants, but they may form a cover to the yet-to-be-identified membrane embedded binding cavities of insect ORs.
Keywords: Extracellular loop, AgamOR10, CquiOR10, BmorOR1, 3-methylindole, bombykol
1. Introduction
Insects negatively affect human society when they become agricultural pests that damage our crops and stored products or vectors of diseases that cause tragic human suffering and death [1,2,3]. With odorant receptors (ORs) housed in neurons on the antennae and other sensory organs, they perceive the world through small molecules like pheromones, attractants, and other odorants, which are essential for their success and survival [2]. Their sophisticated olfactory system may become an Achilles’ heel once we gain a better understanding of the molecular basis of odorant and OR interactions and lay the foundation for the rationale design of eco-friendly, green chemicals for controlling populations of insects of medical importance and agricultural pests while preserving beneficial insects [3].
The advent of insect genome sequences triggered an exponential growth in our knowledge of the molecular basis of insect olfaction. We now know that insect ORs are not GPCRs, as initially envisioned [4]. They are seven-transmembrane proteins with inverse topology [5], i.e., intracellular N-terminus and extracellular C-terminus. They form heteromeric ion channels [6,7,8] with a well-conserved odorant-receptor co-receptor (Orco) [9], which also has a reversed topology [10]. However, structural features of odorant-OR interactions are still terra incognita. Activity modeling and comparative analysis of Drosophila melanogaster ORs led to the hypothesis that a binding pocket is located on the extracellular halves of its transmembrane (TM) domains [11]. Additionally, it has been shown by using substitute cysteine accessibility method that a residue located at the predicted interface between the transmembrane segment-3 and extracellular loop-2 (ECL- 2) plays a role in activation of a Drosophila OR [12].
Using bioinformatics approaches and focusing on sequences and predicted toplogies of multiple ORs senstive to a common odorant, we have identified a putative moiety of the binding site in mosquito ORs, ECL-2. As described here, we probed 16 mutated and 3 wild type ORs by using the Xenopus expression system to identify functionally important residues in ECL-2 loops in ORs from the malaria mosquito, Anopheles gambiae and the Southern house mosquito, Culex quinquefasciatus, as well as the silkworm moth, Bombyx mori.
2. Materials and methods
2.1 Predictions of topology and secondary structures
Topologies of insect ORs were predicted with OCTOPUS [13] and visualized with TMRPress2D [14]. We focused on OR from three mosquito species, which have been reported to respond to indoles, particularly indole and 3-methylindole [15,16,17,18,19]: AgamOR10, CquiOR10, AaegOR10, AgamOR2, CquiOR2, and AaegOR2. For moth receptors, we compared the OR from the silkworm moth, BmorOR1, which is sensitive to the sex pheromone bombykol [20,21] with the ORs from three moth species, Heliothis virescens, Diaphania indica, and Plutella xylostella. They are HvirH13 (=HvirOR13) [22,23], DindOR1 [24], and PxylOR1 [24], respectively, which are known to be sentitive to aldyhydes with the same chain-length as bombykol but differing in the functional groups and unsaturations. Secondary structures of loops were predicted with YASPIN [25].
2.2 Receptor cloning and mutagenesis
Full-length CquiOR10, CquiOrco, BmorOR1, and BmorOrco gene sequences were amplified from constructs available from previous works in our laboratory [26,27]. They were transferred into pBlueScript by standard procedures and then subcloned into pGEMHE [28], and their sequences were confirmed by DNA sequencing (Davis Sequencing Center, Davis, CA). Point mutations were made on the predicted ECL-2 region by using Phusion Site-Directed Mutagenesis Kit (Thermo Scientific, West Palm Beach, FL). With this kit, the entire plasmid pGEMHE-BmorOR1 was amplified using 5′-phosphorylated mutagenic primers (HPLC purified) that introduced the desired mutation. The following primers were used, in which the mismatched nucleotides were labeled in lower case: AgOR10-P162A-F 5′-phos-gCGCAGTACGAAATCTTC-3′, AgOR10-P162A-R 5′-phos-TGAGTCGAAATTGTTCACG -3′, AgOR10-P154A-F 5′-phos-gCGGGCGTGAACAATTTCG -3′, AgOR10- P154A-R 5′-phos-TATGAACATCCCGTACGG -3′, AgOR10-P148A-F 5′-phos-gCGTACGGTATGTTCATACC-3′, AgOR10-P148A-R 5′-phos-TAGGCCCCGCTGCCCGGTA-3′, CqOR10-P151A-F 5′-phos-gCGTACGGAATGTTCATCCCG -3′ CqOR10-P151A-R 5′-phos-GAGGCTACGTGTGCCGGTG-3′, CqOR10-P157A-F 5′-phos-CCGTACGGAATGTTCATCgCCGGGGTAAAC-3′, CqOR10-P157A-R 5′-phos-GAGGCTACGTGTGCCGGTG -3′, CqOR10-P165A-F 5′-phos-gCCCTGTACCAGGTTTTC-3′, CqOR10-P165A-R 5′-phos-CGTCTTGAAGTTGTTTACCC-3′, CqOR10-G158I- F 5′-phos-attGTAAACAACTTCAAGACGCCC-3′, CqOR10-G158I -R 5′-phos-GGGGATGAACATTCCGTAC-3′, CqOR10-F144A-F 5′-phos-gcCACCGGCACACGTAGCCT-3′, CqOR10-F144A-R 5′-phos-CAGCGGGTAGACCACGTA-3′, CqOR10-Y152A-F 5′-phos-CCGgcCGGAATGTTCATCCCCG-3′, CqOR10- Y152A -R 5′-phos-GAGGCTACGTGTGCCGGTG-3′, CqOR10-G153I- F 5′-phos-CCGTACatAATGTTCATCCCCGGGGT-3′, CqOR10-G153I-R 5′-phos-GAGGCTACGTGTGCCGGTG -3′, CqOR10-Y167A-F 5′-phos-CCCCTGgcCCAGGTTTTCTTCATCGG-3′, CqOR10-Y167A-R 5′-phos-CGTCTTGAAGTTGTTTACCC-3′, CqOR10-L150G-F 5′-phos-ggCCCGTACGGAATGTTCATCC-3′, CqOR10-L150G-R 5′-phos-GCTACGTGTGCCGGTGAA-3′, BmOR1-P165A-F 5′-phos-gCGTTGTACAACAATTACGTGTCCG-3′, BmOR1-P165A-R 5′-phos-TAGAAGATTGAACAGCCCTAGACCCATAAA-3′, BmOR1-P178A-F 5′-phos-gCTTATGGACCCAATGTAACGTTTTTCCA-3′, BmOR1-P178A-R 5′-phos-ATCCGAAAATGCCCCGGAC-3′, BmOR1-P181A-F 5′-phos-GCATTTTCGGATCCTTATGGAgCCAATGTAACGTTTTTCC-3′, BmOR1-P181A-R 5′-phos-CCCGGACACGTAATTGTTGTACAA-3′, BmOR1-P194A-F 5′-phos-GTTTATTTTGCTTTCgCCTTCGACTATTCTCACAATTTTAGG-3′, BmOR1-P194A-R 5′-phos-AGAATGGAAAAACGTTACATTGGG -3′. The amplified linear PCR products containing the desired point mutation were ligated and transformed into One Shot® TOP10 competent cells (Invitrogen). All mutations were confirmed by DNA sequencing (Davis Sequencing Center).
2.3 In vitro transcription oocyte and microinjection
In vitro transcription of cRNAs was performed by using a mMESSAGE mMACHINE T7 Kit (Ambion) according to the manufacturer’s protocol. Plasmids were linearized with Nhe I, and capped cRNA was transcribed using T7 RNA polymerase. The cRNAs were purified and re-suspended in nuclease-free water at a concentration of 0.2 μg/μl and stored at −80°C in aliquots. RNA concentrations were determined by UV spectrophotometry. cRNA were microinjected (2 ng of a receptor cRNA and 2 ng of an Orco cRNA) into Xenopus laevis oocytes on stage V or VI (EcoCyte Bioscience, Austin TX). The oocytes were then incubated at 18°C for 3–7 days in modified Barth’s solution [in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaCl2, 10 HEPES, pH 7.4] supplemented with 10 μg/ml of gentamycin, 10 μg/ml of streptomycin and 1.8 mM sodium pyruvate.
2.4 Two-electrode voltage-clamp recording
Two-electrode voltage-clamp technique (TEVC) was employed to observe odorant-induced currents at holding potential of −80mV. Signals were amplified with an OC-725C amplifier (Warner Instruments, Hamden, CT), low-pass filtered at 50 Hz and digitized at 1 kHz. Data acquisition and analysis were carried out with Digidata 1440A and software pCLAMP 10 (Molecular Devices, LLC, Sunnyvale, CA). The data were analyzed with Graphpad 6.
2.5 Odorants
3-Methylindole and bombykol were purchased from Sigma-Aldrich (St. Louis, MO) and Plant Research International (=Pherobank, Wageningen, The Netherlands), respectively. All compounds were prepared to 1M in DMSO as stock solutions stored at −20°C. 3-Methylindole was diluted in Barth’s solution, and bombykol was diluted in Barth’s solution containing 0.1% DMSO.
3. Results and discussion
3.1 ECL-2 loops in mosquito ORs are conspicuous
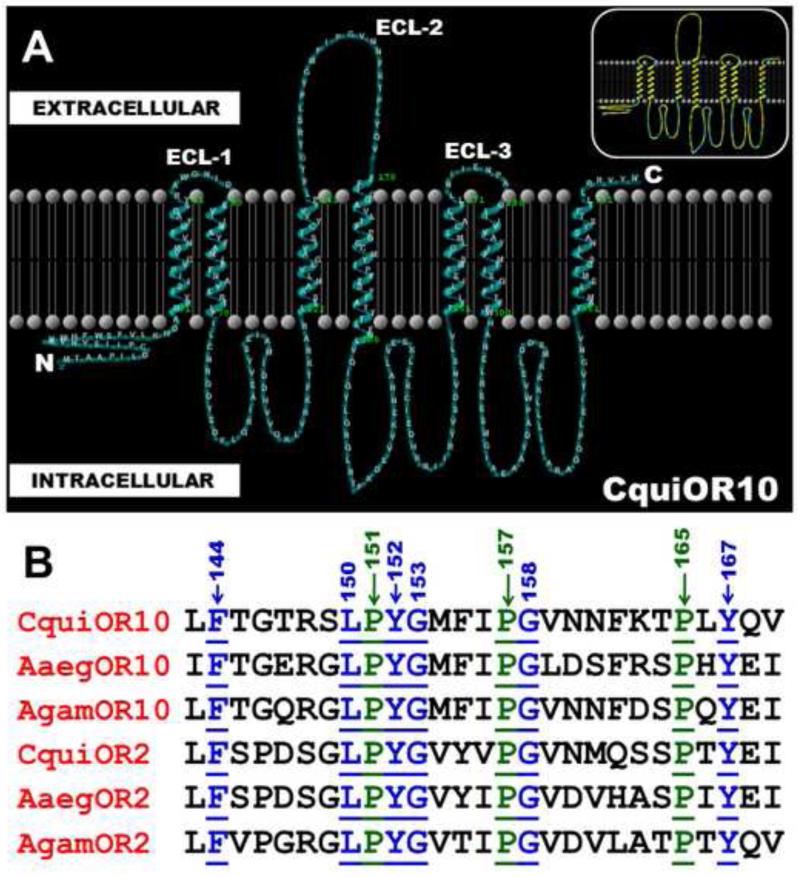
To compare their predicted topologies, we selected the largest number of insect ORs sensitive to the same ligand. Specifically, there are six ORs from mosquito known to respond to 3-methylindole (=skatole) - a common mosquito attractant [29,30]. They are AgamOR10, AgamOR2 [16,19] from An. gambiae, AaegOR10 and AaegOR2 [15] from the yellow fever mosquito Aedes aegypti, and CquiOR10 and CquiOR2 [17,18] from Cx. quinquefasciatus. Their predicted structures were obtained with OCTOPUS software [13]. As expected [31], the N and C termini were predicted to be intracellular and extracellular, respectively (Fig. 1A). We focused on the binding side of the membrane, the extracellular compartment, particularly on loops, which play important roles in the function of many proteins in building ligands’ binding sites [32]. The predicted extracellular loops in these mosquito ORs are short, with 3-8 amino acid residues (e.g.: CquiOR10, ECL-1: RAWGNID; ECL-3: LNIIENPA), except for the second extracellular loop that is conspicuously long. The 27-amino-acid-residue-long ECL-2 is highly conserved among the six ORs from three mosquito species (Fig. 1B). As opposed to the ECL-1 and ECL-3 loops, which showed high amino acid diversity, one-third of the amino acid residues in ECL-2 are well-conserved among the 6 ORs. They are devoid of potential phosphorylation sites and other posttranslational modifications. Of particular notice are three well-conserved Pro residues (Pro-151, Pro-157, and Pro-165 in CquiOR10), two Gly residues (Gly-153 and Gly-158), and three aromatic residues (Phe-144, Tyr-152, and Tyr-167). These observations prompted us to probe these well-conserved residues by using a simple and functional assay, the Xenopus oocyte recording system.
Figure 1.
Predicted topology of a mosquito OR and sequences of the extracellular loops of three receptors sensitive to the same ligand. A) Topology of CquiOR10, predicted with OCTOPUS [13] and visualized with TMRPress2D [14], shows seven transmenbranes (TMs), a long intracellular N-terminus, a short extracellular C-terminus, and a long extracellular loop-2. Inset Same topology highlighting hydrophobic potential. Note the higher occurrence of polar residues (blue) in the intracellular compartment that in the biding site (extracellular compartment). B) Alignment of the second extracellular loops from six mosquito ORs. Three well-conserved Pro residues are highlighted in green, underlined and their positions are indicated with arrows. The other well conserved residues are highlighted in blue. The ECL-2 loops extend from Leu-143 to Val-169 and Leu-140 to Ile-166 in CquiOR10 and AgamOR10, respectively. Thus, well-conserved Pro residues in AgamOR10 are Pro-148, Pro-154, and Pro-162.
3.2. Proline residues are essential for AgamOR10 function
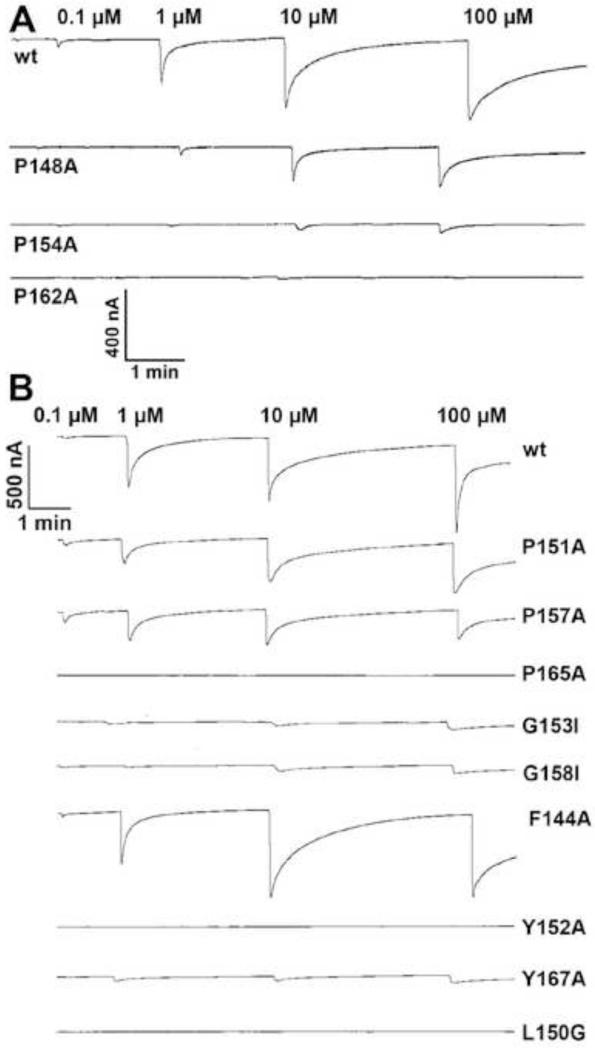
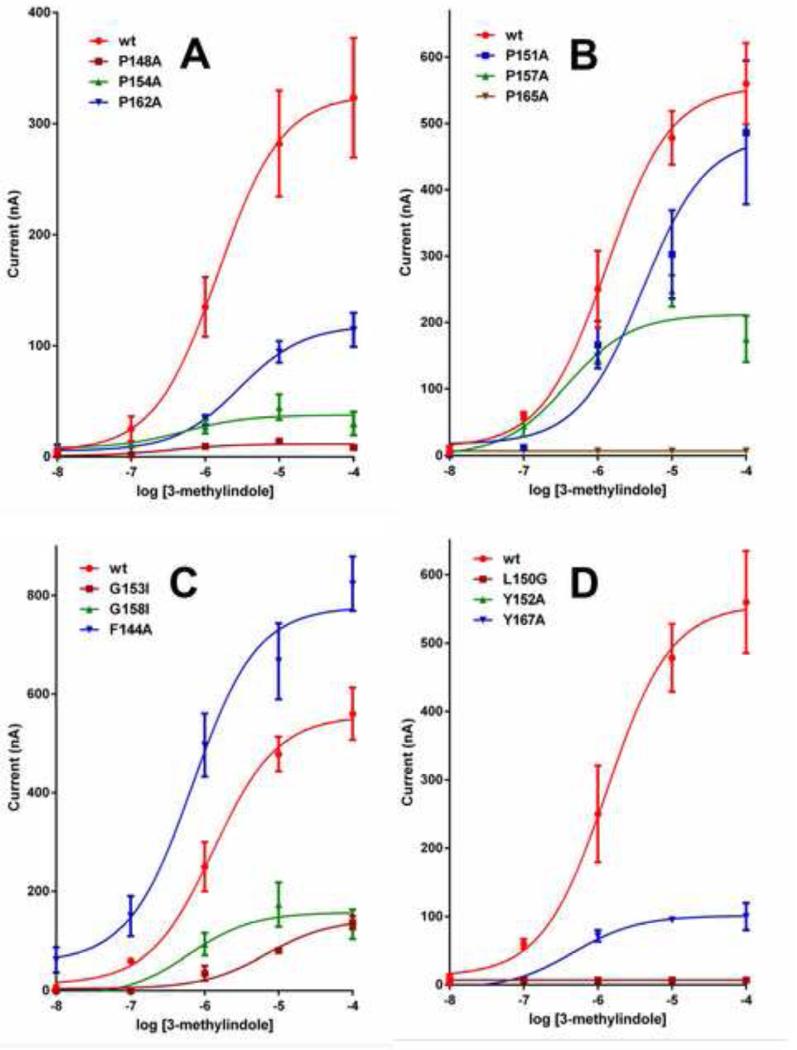
We prepared three mutants of the AgamOR10 gene and expressed each receptor with one-point mutation, along with the obligatory co-receptor AgamOrco, in the Xenopus oocyte recording system. Then, we compared the responses of mutated and wild type (wt) ORs to 3-methylindole. To minimize possible variations, these receptors were tested (n=3-5) using the same batch of eggs. Concentration response analysis showed that P162A-AgamOR10·AgamOrco-expressing oocytes were completely insensitive to 3-methylindole at all doses tested, i.e., from 0.1 to 100 μM (Fig. 2A, Fig. 3A), whereas P154A-AgamOR10·AgamOrco-expressing oocytes were slightly activated (Fig. 2A, Fig. 3A). By contrast, P148A-AgamOR10·AgamOrco-expressing oocytes responded in a dose-dependent fashion (Fig. 2A, Fig. 3A), but with dramatically reduced sensitivity as compared to oocytes expressing the wild type OR. Interestingly, YASPIN [25] predicted β-strands within the ECL-2 segments of mosquito ORs (Fig. 1). We surmised that these Pro residues in the ECL-2 loop of AgamOR10 might be involved in β-turns, which are important for receptor function. If so, the Gly residues might confer flexibility to the backbone of the loop to accommodate the kink created by Pro [32] to change direction of the loop. Next, we tested the effect of Pro residues in the ECL-2 loop on the function of CquiOR10. As opposed to An. gambiae and Ae. aegypti, for which 3-methylindole is a ligand (odorant) of unknown ecological significance, there is solid behavioral [29,30,33] and physiological [33,34] evidence in the literature supporting that 3-methylindole, which is detected by CquiOR10 [17], plays a pivotal role in the chemical ecology of the Southern house mosquito. Thus, our in vitro experimental data with CquiOR10 can be correlated with the physiology and behavior of the animal.
Figure 2.
Mutations of conserved amino acid residues in the extracellular loop-2 and their effects on mosquito OR activity. Dose-dependent current responses elicited by 3-methylindole on oocytes expressing wt and mutated (A) AgamOR10 along with AgamOrco, or (B) CquiOR10 along with CquiOrco.
Figure 3.
Dose-dependent relationships recorded with wild type and mutated AgamOR10 and CquiOR10. Comparative dose-responses recorded from AgamOR10 (A) with mutated Pro residues. Dose-dependent relationships for CquiOR10 (B) with mutated Pro, (C) mutated Gly and Phe, and (D) mutated Leu and Tyr residues. N=3-5.
3.2. Proline, glycine, and other hydrophobic residues are essential for CquiOR10 function
When stimulated with 3-methylindole, P151A-CquiOR10·CquiOrco- and P157A-CquiOR10·CquiOrco-expressing oocytes showed lower responses than those obtained with oocytes expressing the wt receptor (Fig. 2B, Fig. 3B). More strikingly, 3-methylindole failed to active P165A-CquiOR10·CquiOrco-expressing oocytes (Fig. 2B, Fig. 3B). These results suggest that these three Pro residues in CquiOR10 might participate in the scaffold of the odorant binding site. Next, we examined the role of Gly residues, one of the typical “corner residues” [35] that forms β-turns either next to Pro [36] or in the position i+2, [32]. Here, Gly-153 and Gly-158 were mutated with a bulkier amino acid residue, Ile. Although G153I-CquiOR10·CquiOrco- and G158I-CquiOR10·CquiOrco-expressing oocytes were sensitive to 3-methylindole, the response was significantly reduced as compared to the wt receptor (Fig. 2B, Fig. 3C). This dramatic reduction in responses supports the hypothesis that these residues form a conventional β-turn with a cis-proline in the second and glycine in the fourth position. It has been argued in the light of evolution that –Pro-Xxx-Gly best fulfills the requirement for β-turns [32]. While Pro with the side chain attached to the backbone creates the kink required for the turn to change direction, Gly confers flexibility to the backbone as it is completely devoid of a side chain [32].
In addition to backbone hydrogen bonding often created between the first and the fourth residues, the entire β-turn structures are stabilized by side-chain interactions [32]. First, we examined the conserved amino acid residues close and distant from these putative β-turns. Thus, mutation of the distant and well-conserved Phe-144 showed a significant increase in response (Fig. 2B) in a dose-dependent fashion (Fig. 3C). By contrast, conserved Tyr residues close to the putative β-turns are essential for CquiOR10 function. Indeed, Y152A-CquiOR10·CquiOrco- and Y167A-CquiOR10·CquiOrco-expressing oocytes were almost completely insensitive to 3-methylindole at all doses tested (Fig. 2B, Fig. 3D). These data suggest that these aromatic residues might be important to stabilize β-turns via interaction with adjacent strands. Albeit unlikely, we cannot rule out the possibility that they are involved in the formation of the hydrophobic binding cavity necessary to accommodate the aromatic moiety of the ligand. Examination of the last conserved residue, Leu-150, showed that this hydrophobic residue too might be important for side-chain interactions. Indeed, L150G-CquiOR10·CquiOrco was completely silent (Fig. 2B, Fig. 3D), thus resembling what has been observed for Tyr-152, the other residue flanking Pro-151 in one of the putative β-turns. Taken together, these data suggest that there are multiple β-turns in ECL-2 loops and side-chain interactions. Whether these mostly hydrophobic interactions are within the ECL-2 loop or with the ECL-2 loop of the co-receptor Orco. It is worth mentioning that the predicted topology of Orco resembles that of ORs at the extracellular loop. They differ only in having a very long intracellular loop-2 (125 amino acid residues).
We postulated that ECL-2 in insect ORs may not be specificity determinant, but might form a lid covering the membrane embeded binding cavity to protect ligand from solvent. To test this hypothesis we analyzed receptors from insects in a distant taxa, which utilize a long-chain hydrophobic pheromones like bombykol (=(10E,12Z)-hexadecadien-1-ol) [21].
3.3 β-Turns in ECL-2 might be a generic feature of insect ORs
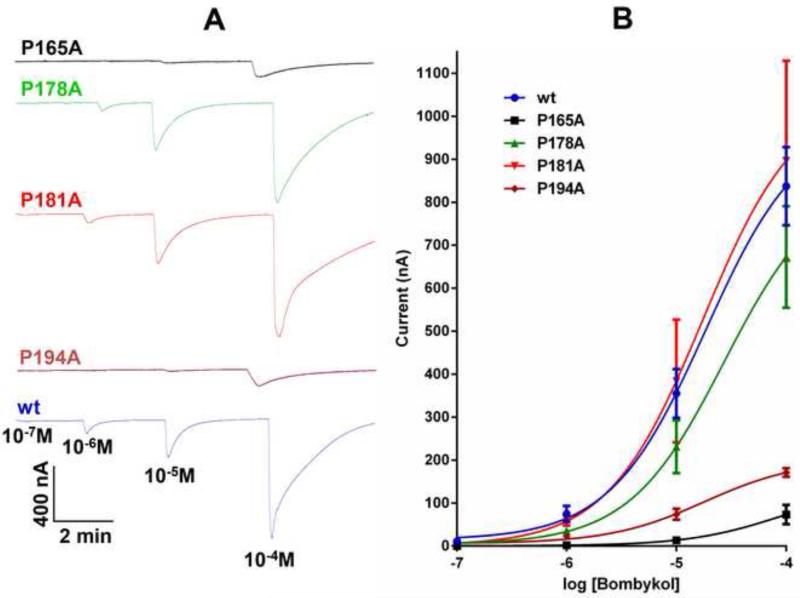
The topology of BmorOR1 is predicted to have a 50-residue long intracellular N-terminus, a short extracellular C-terminus, and 7 TMs, with two short- and a 43-residue-long-extracellular loops, which includes four Pro residues (underlined), 164-LPLYNNYVSGAFSDPYGPNVTFFHSVYFAFPFDYSHNFRGYII-206, i.e., Pro-165, Pro-178, Pro-181, and Pro-194. We then compared the predicted topology of BmorOR1 with those of ORs from three moth species, which are sensitive to hydrophobic ligands with the same chain length as bombykol and differing in functional groups and unsaturations. HvirOR13 from H. virescens is sensitive to (11Z)-hexadecenal [22,23], and DindOR1 from D. indica and PxylOR1 from P. xylostella are sensitive to (11E)- and (11Z)-hexadecenal, respectively [24]. Their predicted ECL-2 loops are 43-amino-acid-residue-long, with 3-4 Pro residues, two of which are well-conserved with Pro-165 and Pro-194 in BmorOR1. Analysis of the predicted secondary structure for ECL-2 in BmorOR1 [38] suggests it forms two β-strand segments, which somewhat resembles β-hairpins – a structural feature of rhodopsin and other GPCRs [39,40,41]. In rhodopsin, the loop forms a lid over the retinal binding pocket [40], while in the mouse eugenol OR the loop is tethered by a disulfide bridge to the extracellular interface of a TM segment to become part of the receptor’s ligand binding site [2]. Since there are no cysteine residues in the predicted ECL-2 of BmorOR1, if functional, it might be folded to achieve the rigidity necessary to cover a binding cavity. Interestingly, one of the predicted β-strands in ECL-2 of BmorOR1 is flanked by Pro residues. We prepared point mutation (Pro→Ala) of the 4 Pro residues in BmorOR1, i.e., Pro-165, 178, 181, and 194, and test the hypothesis that they are functionally important (Fig. 4). Two of the four mutants, P165A- and P194A-BmorOR1, showed significant reduction in response to bombykol. While the effect on P178A-BmorOR1 was moderate, there was no significant difference in the responses recorded from P181-BmorOR1-BmorOrco-expressing oocytes compared to those from oocytes expressing the wt receptor (Fig. 4B). In short, the residue near the predicted interface between TM-3 and ECL-2 (Pro-165) as well as the residue at the other end of a predicted β-strand (Pro-194) are essential for receptor function. Interestingly, these residues are well-conserved among moth ORs, including those responding to compounds with a different functional group and differing from bombykol in the number and position of unsaturations. Thus, it is unlikely that this receptor moiety is specificity determinant. The requirements for a large hydrophobic cavity for bombykol suggest that the yet-to-be-identified binding site in BmorOR1 might be buried in the TM domain [21]. Recent evidence suggests that TM-3, the segment anchoring ECL-2 at the N-terminus contributes at least in part to the binding cavity. A single point mutation in the predicted TM-3 of another moth pheromone OR altered the recognition pattern from (11E)-tetradecenyl acetate to (12E)-tetradecenyl acetate [43]. Taken together, these findings suggest that a membrane embedded binding pocket might be covered by an ECL-2 loop not only in ORs from moths and mosquitoes, but possibly as a general feature of insect ORs.
Figure 4.
Effect of Pro residues of the extracellular loop-2 of BmorOR1 on receptor activity. (A) Current responses elicited by bombykol on BmorOR1·BmorOrco-expressing oocytes with a Pro residue mutated (top from bottom Pro165Ala (black, n=4), Pro178Ala (green, n=4), Pro184Ala (red, n=3), Pro194Ala (brown, n=3) and wt (bottom trace, blue, n=4). (B) Dose-dependent relationships.
HIGHLIGHTS.
Probing Insect Odorant Receptors with their Cognate Ligands: Insights into Structural Features Pingxi Xu and Walter S. Leal
The predicted extracellular loop-2 of insect odorant receptors are well-conserved
Mutation of Pro residues impaired function of mosquito and moth ORs
Pro and corner Gly residues seem to form Pro-xxx-Gly and Pro-Gly β-turns
Tyr and Leu residues contribute to side-chain interactions
ECL-2 is not specificity determinant but may cover odorant binding cavity
Acknowledgments
We thank lab members for insighful discussions and Dr. Lawrence J. Zwiebel for sharing clones of AgamOR10 and AgamOrco. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI095514. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Abbreviations
- OR
odorant receptor
- Orco
odorant receptor co-receptor
- ECL
extracellular loop
- AgamOR
Anopheles gambiae OR
- CquiOR
Culex quinquefasciatus OR
- BmorOR
Bombyx mori OR
- wt
wild type
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Leal WS. An acute olfactory system is essential for reproduction - the raison d’etre for adult insects. Proc Am Phyl Soc. 2012;156:295–302. [Google Scholar]
- [2].Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- [3].Leal WS. Healing power of honey. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306617110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- [5].Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- [7].Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- [8].Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- [9].Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- [10].Lundin C, Kall L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, Heijne G, Nilsson I. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581:5601–5604. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo S, Kim J. Dissecting the molecular mechanism of Drosophila odorant receptors through activity modeling and comparative analysis. Proteins. 2010;78:381–399. doi: 10.1002/prot.22556. [DOI] [PubMed] [Google Scholar]
- [12].Nichols AS, Luetje CW. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J Biol Chem. 2010;285:11854–11862. doi: 10.1074/jbc.M109.058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Viklund H, Elofsson A. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 2008;24:1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- [14].Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics. 2004;20:3258–3260. doi: 10.1093/bioinformatics/bth358. [DOI] [PubMed] [Google Scholar]
- [15].Bohbot JD, Jones PL, Wang G, Pitts RJ, Pask GM, Zwiebel LJ. Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem Senses. 2011;36:149–160. doi: 10.1093/chemse/bjq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J Chem Ecol. 2010;36:797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the Southern House mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS ONE. 2010;5:e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- [21].Xu P, Hooper AM, Pickett JA, Leal WS. Specificity determinants of the silkworm moth sex pheromone. PLoS ONE. 2012;7:e44190. doi: 10.1371/journal.pone.0044190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- [23].Wang G, Vasquez GM, Schal C, Zwiebel LJ, Gould F. Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol Biol. 2011;20:125–133. doi: 10.1111/j.1365-2583.2010.01045.x. [DOI] [PubMed] [Google Scholar]
- [24].Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R, Toyohara H, Takabayashi J, Miyoshi H, Nishioka T. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur J Neurosci. 2008;28:893–902. doi: 10.1111/j.1460-9568.2008.06429.x. [DOI] [PubMed] [Google Scholar]
- [25].Lin K, Simossis VA, Taylor WR, Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics. 2005;21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- [26].Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci USA. 2006;103:16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Syed Z, Kopp A, Kimbrell DA, Leal WS. Bombykol receptors in the silkworm moth and the fruit fly. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1003881107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- [29].Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE. 2008;3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Millar JG, Chaney JD, Mulla MS. Identification of oviposition attractants for Culex quinquefasciatus from fermented Bermuda grass infusions. J Am Mosq Control Assoc. 1992;8:11–17. [PubMed] [Google Scholar]
- [31].Tsitoura P, Andronopoulou E, Tsikou D, Agalou A, Papakonstantinou MP, Kotzia GA, Labropoulou V, Swevers L, Georgoussi Z, Iatrou K. Expression and membrane topology of Anopheles gambiae odorant receptors in lepidopteran insect cells. PLoS ONE. 2010;5:e15428. doi: 10.1371/journal.pone.0015428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kessel A, Ben-Tal N. Introduction to Proteins: Structure, Function, and Motion. CRC Press; Boca Raton, FL: 2011. [Google Scholar]
- [33].Blackwell A, Mordue AJ, Hansson BS, Wadhams LJ, Pickett JA. A behavioral and electrophysiological study of oviposition cues for Culex quinquefasciatus. Physiological Entomology. 1993;18:343–348. [Google Scholar]
- [34].Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc Natl Acad Sci U S A. 2009;106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marcelino AM, Gierasch LM. Roles of beta-turns in protein folding: from peptide models to protein engineering. Biopolymers. 2008;89:380–391. doi: 10.1002/bip.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guruprasad K, Rajkumar S. Beta-and gamma-turns in proteins revisited: a new set of amino acid turn-type dependent positional preferences and potentials. J Biosci. 2000;25:143–156. [PubMed] [Google Scholar]
- [37].Hsu HJ, Chang HJ, Peng HP, Huang SS, Lin MY, Yang AS. Assessing computational amino acid beta-turn propensities with a phage-displayed combinatorial library and directed evolution. Structure. 2006;14:1499–1510. doi: 10.1016/j.str.2006.08.006. [DOI] [PubMed] [Google Scholar]
- [38].McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- [39].Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012 doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tobin AB. G-protein-coupled receptor structure: what can we learn? F1000 Biol Rep. 2009;1:11. doi: 10.3410/B1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012 doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baud O, Etter S, Spreafico M, Bordoli L, Schwede T, Vogel H, Pick H. The mouse eugenol odorant receptor: structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry. 2011;50:843–853. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]
- [43].Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Jr., Macallister IE, Kavanaugh MP, Wanner KW. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]