Abstract
Under field conditions, inoculated rhizobial strains are at a survival disadvantage as compared to indigenous strains. In order to out-compete native rhizobia it is not only important to develop strong nodulation efficiency but also increase their competence in the soil and rhizosphere. Competitive survival of the inoculated strain may be improved by employing strain selection and by genetic engineering of superior nitrogen fixing strains. Iron sufficiency is an important factor determining the survival and nodulation by rhizobia in soil. Siderophores, a class of ferric specific ligands that are involved in receptor specific iron transport into bacteria, constitute an important part of iron acquisition systems in rhizobia and have been shown to play a role in symbiosis as well as in saprophytic survival. Soils predominantly have iron bound to hydroxamate siderophores, a pool that is largely unavailable to catecholate-utilizing rhizobia. Outer membrane receptors for uptake of ferric hydroxamates include FhuA and FegA which are specific for ferrichrome siderophore. Increase in nodule occupancy and enhanced plant growth of the fegA and fhuA expressing engineered bioinoculants rhizobial strain have been reported. Engineering rhizobia for developing effective bioinoculants with improved ability to utilize heterologous siderophores could provide them with better iron acquisition ability and consequently, rhizospheric stability.
1. Introduction
The best known and most exploited symbiotic N2-fixing bacteria are those belonging to the family Rhizobiaceae (Rhizobia) and include the following genera: Rhizobium, Bradyrhizobium, Sinorhizobium, Azorhizobium, Mesorhizobium, and Allorhizobium [1, 2]. Bacteria of these genera have the ability to infect the roots of leguminous plants, causing the formation of a new organ called nodule and establishing a nitrogen-fixing symbiosis. Rhizobium and its allies are Gram-negative bacteria that cause the development of root and also stem nodule on plant hosts, which the bacteria inhabit as nitrogen-fixing endosymbionts. There are sequence of events that are necessary for symbiotic nitrogen fixation which includes preinfection, root colonization, root adhesion, hair branching, hair curling, infection, nodule initiation, bacterial release, bacteroid development, nodule function, nitrogen fixation, complementary functions, and nodule persistence [3]. Under natural conditions, the rhizobial strains which quickly respond to the host signals, initiating the infection and nodulation quickly and efficiently are guaranteed to have a competitive advantage over the slower ones [4]. Rhizobial bacteroids fix atmospheric nitrogen in a form that the plants can utilize and in turn the bacteroids are supplied with an environment rich in carbon as an energy source. These bacteria infect legumes and are distributed globally [5]. The rhizobial N2 fixing ability varies greatly amongst the host plant species and bacterial strains [6, 7]. Thus, rhizobial-host compatibility must be taken into account while selecting the best strain as a bioinoculant. The additional qualities required for considering a rhizobial strain for practical applications as a bioinoculant are that it must efficiently improve the chances of nodulation and have a high N2-fixing rate.
In order to efficiently improve the chances of nodulation, rhizobia must compete successfully against a variety of soil inhabitants [8]. The definition of competition can be provided in number of ways but generally is considered to be a rivalry or a conflict between microbes for nutrition, space, or any other essential limiting factors. Thus, within the rhizosphere of a susceptible plant host the rhizobia has to compete for its nutrients with the indigenous soil microflora which consists of indigenous rhizobial species. Rhizobial bioinoculants fail to nodulate their host mainly because of highly competitive indigenous rhizobia capable of nodulating the same legume species. Competitiveness between two rhizobia is thus measured by the relative ability of the individual rhizobial strain to occupy the nodule [9]. These rhizobia may nodulate the host plant but fix little or no nitrogen, thus, reducing the nitrogen-fixing process. It can be suggested that the poor nodulation could be due to the inability of an inoculant strain to replace the successfully established population of rhizobia in soil. Thus, in order to increase the probability of successful nodulation by rhizobial inoculants, it is essential to first find out the reason for the observed failure and improving the same.
From an ecological standpoint, the first step towards improving rhizobial inoculants must be to determine those aspects of the environment that can limit nodulation. There are reports from laboratory and field trials which state that there are abiotic and biotic factors like temperature, soil type, desiccation, pH, fertilizers, fungicides, resident protozoa, fungi, actinomycetes, rhizobacteria, and the nutrient supply (especially iron as it is limiting in soil) that may adversely affect nodulation of legumes by rhizobial bioinoculant strains [10].
There are two possible ways by which the effect of a desired bioinoculant could be improved (1) by modifying the environment to suit the bioinoculant strain and (2) to modify the rhizobial bioinoculant to suit the environment. The first approach is practically difficult under circumstances like modifying temperature, but the second alternative though costly and time consuming is practically possible. There may be two ways of achieving a modified rhizobial bioinoculant that suits the environment (1) by selecting a natural variant that is tolerant amongst all other rhizobial strains and (2) to genetically engineer a suitable inoculant strain. Thus, the objectives of our review was to consider the limitations that are imposed by the lack of iron as nutrition to rhizobial bioinoculant strains and to focus on ways to determine possible avenues for improving the competitive survivability of rhizobial bioinoculant strain by engineering the iron uptake machinery.
2. Competitive Survival in Soils
2.1. General Information from Various Bacteria
The rhizosphere of legume contains a large and heterogeneous community of bacteria. Interaction of these rhizosphere bacteria between each other and with other rhizobia are complex. Li and Alexander [11] observed an inhibition of the colonization and nodulation of alfalfa roots by Rhizobium meliloti in the presence of Enterobacter aerogens, Pseudomonas marginalis, Acinetobacter sp., and Klebsiella pneumoniae, whereas Bradyrhizobium sp. or Micrococcus luteus did not exert such effect on Rhizobium meliloti-alfalfa interaction. A correlation could be drawn between the growth rate of the rhizospheric bacteria and their effect on Rhizobium meliloti. The bacterial species having a faster growth rate than Rhizobium meliloti were acting as the competitors. The bacteria growing faster were preventing rhizobia from obtaining a large part of the excreted carbon needed to support cell growth. These effects may be caused due to the production of antibiotic compounds, siderophores, or plant growth hormones. The enhanced nodule occupancy by Bradyrhizobium japonicum USDA 110 in the presence of fluorescent Pseudomonas sp. was attributed to the iron availability and its influence on siderophore production by the Bradyrhizobium japonicum strains [12]. Another mechanism used by rhizobia to compete with rhizospheric bacteria, especially rhizobia noduling the same host, is by the production of bacteriocins [13].
2.1.1. Competition for Iron in Rhizosphere
In natural environments there are always competitive interactions among the microbial inhabitants with their neighbors for the available space and resources [14]. Nutritional resources are the focal point of microbial competition and the success of an organism depends upon how efficiently it can acquire limiting essential nutrients, iron being one of them.
In soils, rhizobia must not only be competent at solubilizing iron but also develop strategies to compete for this nutrient with other organisms present in the rhizosphere [15]. To meet iron requirements for growth, most microorganisms have developed specialized high affinity iron uptake system for scavenging and transporting ferric iron [16]. This includes machinery for the synthesis and secretion of low molecular weight compounds called siderophores which bind ferric iron with high affinity [17, 18] and several membrane bound and periplasmic proteins that function together in the uptake of the ferricsiderophore complex [19]. In addition to utilization of their own siderophore, S. meliloti and B. japonicum can also cross-utilize iron complex to siderophores made by other species (heterologous siderophore) [20, 21]. Most notable among the heterologous siderophores utilized by these rhizobia is ferrichrome, a prototypical hydroxamate type siderophore secreted by several fungi, including Ustilago sphaerogena [22] and Ustilago maydis [23]. Ability to utilize iron bound to ferrichrome is important from an ecological perspective since natural soils have significant concentrations of this siderophore [24, 25].
To gain an understanding of the iron distribution on a small scale on plants, Joyner and Lindow [26] developed a whole-cell biosensor to detect and measure bioavailable iron at the level of single bacterial cells. Ferric iron availability to cells of Pseudomonas syringae was assessed by quantifying the fluorescence intensity of single cells harboring a plasmid-borne transcriptional fusion of an iron-regulated promoter from a locus encoding a membrane receptor for a pyoverdine. Their results indicated that there is substantial heterogeneity of iron bioavailability to cells of P. syringae on plants, with only a small but significant subset of cells experiencing low iron availability. It is believed that chemical reactions and microbial respiration [27] can result in fluctuations in pH which in turn could affect the solubility of iron oxides, ultimately affecting the bioavailability of this element locally [28]. These results support models of competition for limiting iron resources among microbes on plants in that at least some of the cells in population encounter very low iron availability; thus, competition for this nutrient is an important aspect of rhizosphere ecology.
Competition for iron can be regarded as occurring in two stages either competition between excreted siderophores for the metal or competition between microorganisms for the Fe-siderophore complexes that are available in soil. The former is controlled by proton dissociation and formation constants of each siderophore, kinetics of exchange, and their concentration, while the latter is governed by the existence of an uptake mechanism for, and its affinity to, the Fe complex [29]. Fluorescent pseudomonads can suppress various soil borne plant diseases due to the production of siderophores and their densities in the rhizosphere [30, 31]. Pyoverdines are the major siderophores produced by fluorescent pseudomonads. They have very high affinity for Fe3+ with a stability constant of about 1032 [32] than the siderophores produced by other microorganisms that are deleterious to plant growth [33] and their membrane receptors are usually very specific [34]. These features enable the fluorescent pseudomonads to compete efficiently with the soil borne microflora. Pyoverdine-mediated iron competition has been shown to play a determinative role in the microbial antagonism performed by biocontrol strains against some pathogens.
As shown in Figure 1, siderophores may serve as Fe sources or as Fe competitors for organisms, depending on the ability of the organisms to acquire Fe from the stable Fe-siderophore complex. The siderophore mediated interactions as depicted in Figure 1 are for three organisms but may become much more complicated when more species are present.
Figure 1.
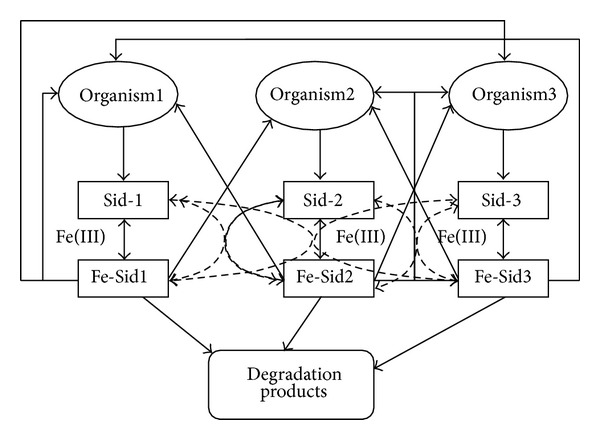
Model of siderophore mediated interactions between three organisms. The thick lines indicate transport by one organism of the Fe-Sid produced by the other organism. The dashed lines represent ligand exchange by which one Sid displaces another Sid chelating and Fe3+ iron. Degradation pathways, including biological and chemical mechanisms, are represented by the thin black arrowed lines.
Based on this concept it is obvious that if plant growth promoting (PGP) organisms (free-living or symbiotic) used as bioinoculant produce siderophore of significant high affinity, it will not only succeed to establish itself but will also be able to inhibit the growth of pathogenic organisms and hence can act as a biocontrol agent [35, 36].
3. Specific Information about Rhizobia
3.1. Rhizobia Competition for Iron: Affecting Its Rhizospheric Colonization
To induce nodulation on their leguminous hosts, the members of the genus Rhizobium probably must grow in the rhizosphere as well as at the site on the root hair where invasion of the plant begins. In these environments, the rhizobia are probably growing at the expense of organic products excreted by the roots, but these bacteria must compete for the essential nutrients (like iron) with other heterotrophic inhabitants of the rhizosphere. Many rhizobial strains have the potential to increase plant growth and yields but are poor at nodulation due to weak rhizospheric colonization. Many scientific studies have been performed that show the role of competition among strains of Rhizobium sp. for nodule sites and the impact of such intraspecific competition on nodulation. These works dealt with the behavior of mixed strains of nodule bacteria towards each other and towards their legume host, where they showed that when two strains of nodule bacteria were present in the surroundings of their host's root system, active competition between them caused the strain having a higher initial growth rate to overpower the other strain outside the plant. This dominant strain was then responsible for nearly all the nodules produced in the plant. Siderophore production and cross-utilization is a major microbial mechanism of interspecies competition; many bacterial species have the ability to utilize heterologous siderophores, sequestering iron away from the siderophore producer. Difference in iron binding affinities of siderophores, produced by different species can also mediate competition [37]. Several investigators have demonstrated that individual isolates of bacteria or fungi decrease the nodulation of aseptically grown white clovers by R. trifolii [38] or the rate and extent of nodulation of aseptically grown alfalfa by R. meliloti [39]. In these studies, it was shown that the organisms causing the suppression did not produce toxins against R. trifolii in culture, indicating that the suppression of nodule formation resulted from competition with the root nodule bacteria.
There is little evidence that iron deficient soils affect the numbers of root nodule bacteria. In iron stressed soils, the proportion of siderophore-producing strains appear to increase. In alkaline soils of the midwestern United States B. japonicum serotype 135, a siderophore producer, has been shown to dominate over serotype 123, a siderophore nonproducer [40]. Similarly, 54% of S. meliloti strains isolated from alfalfa nodules from alkaline soil were siderophore producers, whereas from only 18% siderophore producing strains were obtained from an acidic soil [41]. These reports do suggest the possible advantage of siderophore production in iron deficient soil in the increased survivability and number of the producing strain in the rhizosphere.
3.1.1. Heterologous Siderophore Utilization: A Sound Strategy to Outcompete Other Rhizospheric Microflora
Complexation of Fe(III) by soil anions as well as competition for Fe(III) with other soil microorganisms are obstacles to iron acquisition commonly faced by Rhizobium sp. and all soil microflora. Rhizobia must be able to persist in the soil in the absence of their host plants and compete for iron, the availability of which is limited by its insolubility; this is one of the factors in determining how successful rhizobia are in maintaining themselves in the rhizosphere [42]. The rhizobial high affinity iron uptake systems may be of use in the competition among soil microbes for access to available iron and may enhance the survival of the free-living forms. Evidence has been presented to indicate that soil competition among rhizosphere pseudomonads may occur at the level of the uptake system of the ferric-complex specific for the individual Pseudomonas siderophore [43]. It has been shown that R. meliloti DM4 excretes and utilizes the siderophore rhizobactin, whereas several other wild-type R. meliloti strains do not [20]; thus, it is possible that a comparable competition might also occur among rhizobial strains.
Certain rhizobia have also been shown to utilize iron bound to heterologous siderophores. Bradyrhizobium japonicum USDA110 and 61A152 can utilize ferrichrome (made by numerous fungi), rhodotorulate (made by yeast), pseudobactin (made by pseudomonads), and so forth [21], and these strains are successfully used as commercial bioinoculant for soybeans. R. meliloti DM4 could utilize the hydroxamate type siderophores, ferrichrome, and ferrioxamine B (made by actinomyces) in addition to its own siderophore rhizobactin [20]. Khan et al. [37] reported that a the growth of the strain that showed higher siderophore cross-utilization got stimulated in the presence of exogenously added siderophores in comparison to a low cross-utilizing strain which did not show such effects under similar conditions. Therefore, a rhizobial strain having the ability to utilize siderophore(s) produced by another similar rhizobial strain(s) (homologous siderophore) or from a nonrhizobial strain (heterologous siderophore) should also have a competitive advantage in the rhizosphere. It could be therefore said that possession of uptake system for the siderophore produced by majority organisms in the soil and hence predominantly present in the soil can have positive implications for growth and survival of the possessing organism.
4. Improving Competitive Survival
4.1. Strain Selection and Inoculant Preparation
Legumes have the most important role in agricultural productivity. Pulse and grain legumes are grown in the agricultural land in a rotation so as to fix atmospheric nitrogen into the soil because they form nitrogen fixing symbiosomes with soil inhabiting root nodule bacteria [44]. The successful use of legumes in agriculture will be dependent upon appropriate formation of effective symbioses with root nodule bacteria. An essential component for increasing the use of legumes is the integration of plant breeding and cultivar development with appropriate research leading to the selection of elite strains of root nodule bacteria. In addition research in these areas would increase benefits to the farmers through advances in the application of appropriate inoculation technologies to deliver the new strains of bacteria to the soil with enhanced survival abilities [8, 45]. A key feature of the symbiotic relationship between root nodule bacteria and legumes is the very high degree of specificity shown for effective nodulation of a particular host legume by a strain/of root nodule bacteria. This specificity operates at both the nodulation and nitrogen fixation levels of the symbiosis and is a function of the exchange of specific chemical signals between the two partners [46, 47]. The specificity is controlled by the fact that the chemicals can only be produced if the organisms contain the information for the synthesis of the chemicals on their genes. Thus, the development and function of the symbiosis between legumes and their associated root nodule bacteria is controlled at a molecular level, and the development of effective nitrogen-fixing symbioses is conditional on both partners containing appropriate sets of genes [47–49]. Three general strategies are possible when a new legume is introduced to new lands [50]. One is that the uninoculated legume may form abundant effective nodules indicating the presence in the soil of a large population of effective root nodule bacteria. In this case a plant breeding program to select a genotype able to nodulate effectively with the indigenous root nodule bacteria would be the simplest approach for successful introduction of the legume. Secondly, there is the less frequently encountered situation where no nodules are formed on uninoculated legumes. This result indicates there is very little or in some cases no background population of root nodule bacteria in the soil that are able to nodulate the particular host legume. More often, uninoculated legumes form variably effective nodules indicating the presence of a population of ineffective or variably effective root nodule bacteria. In the latter two scenarios the clear solution to ensure success with the legume crop is to select an effective, competitive inoculant strain of root nodule bacteria adapted to the soil conditions with the following desired characteristic: like effective nitrogen-fixation with the intended host species, stress tolerance, competitive ability against the indigenous strains, genetic stability, and satisfactory growth and survival during procedures for manufacture of inoculum [51].
4.2. Genetic Manipulations
The successful performance of rhizobial inoculant strains as we know depends upon their capability to out compete the indigenous soil bacteria, survive and propagate, and enter into effective symbiosis with the host plant. The strains which fail to survive under soil conditions are most of the time ineffective in enhancing legume productivity because vast majority of nodules formed are not by the inoculated strain but by indigenous rhizobia in the soil [52, 53]. Thus, construction of genetically engineered inoculum strains of Rhizobium with an increased ability to survive under soil conditions and hence compete for nodule occupancy was considered an amicable approach to address this problem. Most of the rhizobial biofertilizer strains are poor rhizospheric colonizers due to their inability to compete with the indigenous soil microflora for nutrients; iron being one of them because of the iron rich enzymes involved in nitrogen fixation [54]. Utilization of foreign siderophores is considered to be an important mechanism to attain iron sufficiency. Pseudomonads which are known for their rhizospheric stability have diverse iron uptake systems and multiple receptor genes have been detected in their genomes [55, 56]. From the above facts it could be suggested that by increasing the number of outer membrane siderophore receptors rhizobial strains could be made more efficient with respect to iron acquisition, and hence colonizing the rhizosphere. Most of the hydroxamate siderophores present in soil are of the ferrichrome-type. Because ferrichrome is synthesized by a variety of soil fungi, it is likely iron source in the rhizosphere where hydroxamate concentrations have been estimated to be as high as 10 μM [57] and ferrichrome is found in nanomolar concentrations, as estimated by physicochemical [58] as well as bioassay methods [25]. As majority of soil bacteria are good utilizers of iron bound to hydroxamates [29], thus, the rhizobia isolates could be at a competitive disadvantage when residing free in soils. It is therefore pertinent to engineer these strains with a ferrichrome receptor to increase their iron acquisition property and hence survival.
Thus, cloning of the ferrichrome receptor genein rhizobial bioinoculant strains and understanding the effect of ferrichrome utilization on rhizobial growth and survivability under conditions, wherein ferrichrome was made available by other producer species, were achieved by heterologously expressing the E. coli fhuA gene in C. cajan rhizobia. The fhuA was engineered under the control of the lac promoter and its expression was first confirmed in E. coli by the rescue of the phenotype of ΔfhuA mutant and was subsequently introduced into the rhizobial strains and its expression monitored. The expression of E. coli fhuA in rhizobial strains imparted them the associated phenotypes, namely, the ability to utilize iron complexed with ferrichrome and sensitivity to albomycin [59].
Several studies have shown that the utilization of heterologous siderophores by genetically introducing the receptor gene provides growth advantage to the bacteria. Brickman and Armstrong [60] found that the incorporation of alcaligin receptor gene alone could confer upon a siderophore deficient strain of P. aeruginosa the ability to utilize ferric alcaligin. Raaijmakers et al. [31] introduced the siderophore receptor for ferric pseudobactin 358 into P. fluorescens WCS374, resulting in a strain which was more competitive than the WCS374 parental strain for colonization of the radish rhizosphere. Benson et al. [61] showed that in addition to the FegA receptor required to form functional symbiosis, FegB was also necessary for B. japonicum to utilize ferrichrome. These observations reinstate the belief that among all the ligand-protein interactions of members of the bacterial iron-acquisition system, the binding of ferrisiderophores to the outer membrane receptor proteins is the most specific. For instance, in E. coli separate outer membrane receptors transport ferric iron bound to aerobactin, ferrichrome, rhodotorulic acid, and ferrioxamine, yet they all use a common set of periplasmic and inner membrane components. Genome sequences of bacteria contain numerous putative ferrisiderophore receptor genes [62, 63] but do not contain equivalent copies of the genes for periplasmic and cytoplasmic membrane-bound proteins. All the studies reported here favor the hypothesis that introduction of genetically engineered receptor genes helps in providing growth advantage to the Rhizobium species.
5. Engineering Iron Nutrition in Rhizobia
5.1. Importance of Iron in Nodulation, Survival—General Findings
Although iron is the fourth most abundant element in the Earth's crust, it is essentially unavailable in aerobic environments at biological pH, as it tends to form insoluble Fe3+ oxyhydroxide [64]. To combat iron deficiency, different organisms produce different types of siderophores. In nature the ability to utilize various sources of chelated Fe appears to be of much importance and that is the reason why microorganisms often employ more than one high affinity Fe transport system for acquiring iron. This broad transport capability may be achieved by two means: either like the E. coli model which involves many specific siderophore transporters for uptake of ferrisiderophores that it may encounter or like the Streptomyces model which involves a nonspecific receptor/transporter for its own siderophore and other siderophores that has similar coordination of iron but different structures [15].
5.1.1. Rhizobial Siderophores
Iron acquisition by Rhizobium sp. is essential for nitrogen fixation by the Rhizobium-legume root nodule symbiosis. Root nodule bacteria, form a nitrogen-fixing symbiotic interaction along with their leguminous plant hosts and have a high demand for iron [42]. Diverse types of siderophores are produced by the different rhizobial genera, for example, Rhizobium leguminosarum bv. viciae, the symbiont of peas, lentils, vetches, and some beans, synthesizes a cyclic trihydroxamate type siderophore called vicibactin, whereas Sinorhizobium meliloti 2011 under iron stress produces rhizobactin 1021 a dihydroxamate siderophore [65]. Catecholate siderophores are known to be produced by rhizobia from the cowpea group [66] and salicylic acid and dihydroxybenzoic acid are produced by Rhizobium ciceri isolated from chick pea nodules [67]; citrate as a siderophore is produced by Bradyrhizobium japonicum [68] and anthranilate is produced by Rhizobium leguminosarum.
Under natural conditions, the ability to produce siderophores has been demonstrated to confer a selective advantage to the producer organism [69]. It has also been speculated that besides capturing iron quotas necessary for growth, siderophores are also a type of iron scavengers because they can mobilize iron from weaker ferricsiderophore complexes from other species and thus are mediators of competitive interaction among organisms.
Microorganisms which themselves do not synthesize a particular type of siderophore may yet be proficient at the uptake of the iron-bound siderophore complex that they do not produce. The ability to cross-utilize heterologous siderophores may be accounted for by the presence of multiple type of siderophore receptors that are expressed for the uptake of different types of siderophores [62] or use of a low specificity (broad range) system that recognizes more than one type of siderophores [15]. Certain rhizobia have also been shown to utilize iron bound to heterologous siderophores. Rhizobium meliloti DM4 can utilize the hydroxamate type siderophores, ferrichrome (made by numerous fungi), and ferrioxamine B (made by actinomyces) in addition to its own siderophore rhizobactin [20]. Bradyrhizobium japonicum is able to utilize Fe3+ bound to two fungal siderophores rhodotorulate and ferrichrome [21].
In natural environments such as the rhizosphere, the capacity to utilize heterologous siderophores produced by other members of the rhizosphere microflora is a positive fitness factor [31, 70]. Heterologous siderophores or its producer organism may bring about a variety of responses on other target bacterial species that are present within the same niche. Growth of some species is inhibited and this has been attributed to be one of the mechanisms by which biocontrol agent's act in inhibiting the growth of pathogens in the rhizosphere [33, 71]. In certain cases, addition of heterologous siderophores results in a growth stimulatory effect [37, 72]. Presence of exogenous heterologous siderophores can induce the expression of Iron Regulated Outer Membrane Proteins (IROMPs) in target bacterial species [73] and may also positively stimulate the target organism to synthesize its native siderophore [74].
5.1.2. Iron Metabolism in Rhizobia
Siderophore-mediated high-affinity iron transport system comprises of an elaborate machinery involving genes for biosynthesis of siderophores, low-molecular weight iron chelating compounds, and specific receptors for the uptake of the siderophore-iron complex [19]. Nitrogen-fixing rhizobia, living as endosymbionts in root nodules of legume host plants, have a high demand for iron because a number of “symbiotic” proteins contain iron or heme [75]. Despite the importance of iron in these organisms, iron metabolism is relatively poorly studied in most rhizobial species. The lack of competitiveness for nodulation can be attributed to low intracellular concentration of iron in rhizobia [76]. The S. meliloti mutants, unable to acquire iron under Fe-limited conditions showed deficiency in competitive nodule occupancy. In soils, iron may be largely bound to siderophores secreted by various soil inhabitants. It has been shown that the siderophore ferrichrome, made by several fungi, is present in large amounts in soil solutions [24]. Organisms that do not synthesize ferrichrome may still be able to utilize iron bound to it, if they possess receptor for this siderophore. Thus, siderophores not only scavenge iron but also mediate antagonistic effect on other strains by depriving them of iron.
Most of the rhizobial strains fail to utilize ferrichrome and other related hydroxamate type siderophores and are at competitive disadvantage in iron sufficiency [59, 77]. Heterologous receptors like (fhuA) from Escherichia coli or (fegA) from a B. japonicum strain that are proficient at utilizing ferrichrome have been expressed in rhizobial strains nodulating Cajanus cajan [59, 78, 79] and ground nut [80] plants. Engineering rhizobial strains by incorporating genes for multiple iron-siderophore receptors potentially increased the suite of ferricsiderophores that they could utilize. Plant inoculation studies performed at our lab showed increased nodulation efficiency and nodule number per plant by the engineered strains over the parental strains (nonengineered) under natural soil conditions. The transformants clearly benefited the plant also in terms of shoot fresh weight and increased chlorophyll content. Rhizospheric colonization and survival was also increased with transformants as compared to their parent under both autoclaved and unautoclaved conditions. Thus, it was demonstrated that increased survival of the strains does improve their nodule occupancy on C. cajan which in turn had positive effects on plant growth [78]. Similarly fegA-bearing strain when inoculated in unautoclaved soils with indigenous rhizobia (that could nodulate) showed better competitive nodulation ability [79]. Whether the competitive advantage of ferrichrome receptor expressing strains is restricted at the level of saprophytic survival alone or is also important during nodulation is under investigation.
5.2. Iron Acquisition Systems: Rhizobial Machinery
5.2.1. Ferrisiderophore Uptake Machinery
Siderophores have been designed by nature in such a way that they are of sufficient size to engage the six octahedrally directed valence bonds of Fe(III); thus, they exceed the free diffusion limit of the small water-filled pores in the outer membrane of enteric bacteria. This in turn was sufficed by nature by the evolution of specific receptors for the recognition and transport of the iron-laden form of the siderophore. A siderophore system is thus comprised of two principal parts: the ligand and an arrangement for its uptake and utilization. Both parts of system are mostly subject to a common regulatory device triggered by the intracellular concentration of iron. The ferricsiderophore (Sid-Fe3+) from the extracellular medium is recognized by the N-terminal portion of a ferrisiderophore receptor, which serves two functions. First, the ferrisiderophore receptor transports Sid-Fe3+ into the periplasm, which is further transported into the cytoplasm by an ABC transporter. Both transport and induction functions require energy transduction from the TonB–ExbB–ExbD complex located in the inner membrane. Sid-Fe3+-bound ferrisiderophore receptor is believed to interact with TonB via its TonB-box motif [81].
5.2.2. Siderophore Uptake Machinery and Its Regulation in Rhizobia
Although siderophore uptake machinery of rhizobia is poorly characterized, it can be said that this group of microorganisms have similar system of siderophore mediated uptake machinery as other Gram-negative species. Relatively few reports describe the presence of Iron Regulated Outer Membrane Proteins (IROMPs) in rhizobia that are produced and bind to their specific siderophore iron complex [82]. Stevens et al. [83] identified some of the fhu genes of R. leguminosarum which are homologues of the hydroxamate siderophores uptake machinery in E. coli. One of them is FhuA that specifies the OM receptor for uptake of vicibactin and works in association with FhuCDB (inner membrane proteins). FhuC is the ABC-transporter ATPase, FhuB is the permease and FhuD is the periplasmic siderophore binding protein which brings the ferrivicibactin complex to the inner membrane machinery for its transport from the periplasm to the cytoplasm [84]. Pseudogene versions of fhuA have also been detected in several other strains of R. leguminosarum [85]. Another receptor is rhizobial ferrichrome OM receptor FegA of B. japonicum 61A152 is a hydroxamate-type siderophore receptor [86]. The fegA gene is organized in an operon with fegB which probably encodes an inner membrane protein. Mutant analysis revealed that both genes are required for utilization of the siderophore ferrichrome [61]. FegA has been shown to bind ferrichrome specifically and its homologs have been detected in other strains as well. LeVier et al. [86] compared the amino acid similarities and identities among rhizobial receptor FegA with other nonrhizobial related siderophore receptor proteins. They have shown that FegA is 53.7% similar to the E. coli Fe(III)-ferrichrome receptor FhuA and 48.3% similar to coprogen and rhodotorulic acid receptor FhuE. Like any other ferrisiderophore uptake machinery the rhizobial high affinity Fe-uptake system also requires other components other than the receptor for the transport of the ferrisiderophore-complex. RhtA is an OM receptor responsible for rhizobactin uptake in Sinorhizobium meliloti, where a specialized single permease RhtX is responsible for its transport from periplasm to cytoplasm [87]. Wexler et al. [88] showed that tonB-like gene (tonB Rl) is adjacent to an operon that specified an ABC transporter involved in heme uptake in R. leguminosarum. The two of the tonB Rl mutants, J350 and J344, had large halos on CAS plates due to higher amounts of the siderophore vicibactin that got accumulated in the extracellular medium. This was probably because the tonB Rl mutants failed to import vicibactin and thus indicated that TonB Rl was required for the Fhu transport system to operate, as found in other bacterial siderophore uptake systems. Whereas mutant analysis and complementation tests showed that the TonB system was specific for heme uptake and was dispensable for siderophore uptake proposing the existence of a second TonB homologue functioning in the uptake of Fe-chelates in Bradyrhizobium japonicum [89]. Regulation of ferrisiderophore assimilation system in rhizobia is mediated by Fur regulation which is highly conserved among most bacterial species; it is also present in Bradyrhizobium japonicum where the Fur protein has been shown to regulate the hemA gene [90] and Rhizobium leguminosarum [91]. The B. japonicum Fur proteins also have a novel function in the iron-dependent gene expression [92]. Another member of the Fur family is Irr (iron response regulator), which occurs in rhizobia. Irr was first identified in B. japonicum, as a transcriptional repressor of hemB, which specifies δ-aminolaevulinic acid dehydratase, in the heme biosynthetic pathway [93]. Irr is restricted to a few α-proteobacteria including rhizobia, Agrobacterium, the animal pathogen Brucella and Rhodopseudomonas palustris a photosynthetic bacterium very closely related to B. japonicum. It is moderately repressed in Fe-replete conditions, and this was shown to be dependent on a protein FurBj that was homologous to Fur even though the irr promoter region had no sequence similarity to the canonical fur boxes [94]. These observations suggest that rhizobial Fur differs from that of other bacteria including E. coli. Recently Yang et al. [95] had shown the control of the expression of iron transport genes and many other iron-regulated genes which are not directly involved in heme synthesis in Bradyrhizobium japonicum through Irr. The Irr was shown to have both a positive and negative effect on the gene expression. Their findings indicated that B. japonicum sensed iron via the status of heme biosynthesis in an Irr-dependent manner and thus regulated iron homeostasis and metabolism.
Regulation of iron responsive genes in R. leguminosarum and S. meliloti is not mediated by Fur but rather by the dissimilar RirA (rhizobial iron regulator) protein a member of the Rrf2 family is a newly found gene whose product has no sequence similarity to Fur but which has close homologues in other rhizobia, like Agrobacterium and Brucella [96]. All the above reports state that the regulation of Fe-responsive genes in rhizobia is not mediated by Fur alone. There is also positive regulation reported in rhizobia. In the heme uptake system of B. japonicum encoded by the gene cluster hmuVUT-hmuR-exbBD-tonB, transcription of the divergently oriented hmuT and hmuR genes was not only found to be induced by iron limitation but to also depend on a 21-bp promoter-upstream iron control element (ICE) which by deletion analysis was shown to be needed for positive control [97].
5.3. Importance of Iron Acquisition Systems in Nodulation: Siderophore/Transporter Mutants
5.3.1. Importance of Iron Acquisition in Rhizobial-Legume Symbiosis
There is a high demand for iron in the symbiotic interaction between legumes and rhizobia, although the mechanism by which it is provided in plant root nodules is not well understood. Recently Stacey et al. [98] studied the functional genome analysis of legume nodulation. A nodulated legume has an increased need for iron compared to a nonnodulated plant [99] since this metal is a constituent of key proteins such as nitrogenase and leghaemoglobin. Nitrogenase is made up of two proteins; both are rich in iron and essential for activity. Non-heme iron electron transfer proteins such as ferredoxin and flavodoxin are essential in nitrogen fixation [100]. Iron deficiency in nodulated legumes is very common on alkaline soils, and it affects common agricultural crops as chick pea [101], French bean [102], and peanut [103]. O'Hara et al. [104] reported the importance of iron in the legume-rhizobia symbiosis, where iron deficiency limits nodule development but not initiation in peanuts inoculated with Bradyrhizobium species. Nodulation is also drastically curtailed in lupines under iron deficiency [105]. Specific siderophore producing microorganisms stimulated the nodulation, nitrogen fixation and plant growth of leguminous plants [106, 107]. One of the possible modes of growth promotion of nodulated legumes under field conditions is production of siderophores which control the proliferation of soil-borne pathogens or facilitate the uptake of iron from environment [108, 109]. However, taking into consideration that iron stressed plants show fewer bacteroids present in the nodules, decreased amount of leghemoglobin and lower specific nitrogenase activity, and that possession of the ability to produce siderophore significantly increases the efficiency of the differentiated bacterium to fix nitrogen and induce an increase in plant growth. It can be suggested that the difference in nodule development under iron deficient conditions may be due to varying abilities of different strains of root nodule bacteria to acquire iron for nodule initiation and development.
Rhizobium leguminosarum 116, an ineffective mutant strain was shown to form white ineffective nodules on peas and had an apparent defect in iron acquisition [110]. Also the nitrogenase activity of plants inoculated with wild-type R. meliloti 1021 that produced rhizobactin 1021 and rhizobactin 1021 mutants over a 70-day period showed a significant increase in nitrogenase activity and total plant dry weight in the wild type strain as compared to the rhizobactin 1021 mutant [111]; even the efficiency in nitrogen fixation increased when the plants were inoculated with the wild type strain compared to plants inoculated with rhizobactin 1021 mutants [112]. These studies suggest that rhizobactin 1021 contributes to the efficiency of nitrogen fixation under certain conditions of plant growth. Other studies have shown conflicting results: in some cases, rhizobial mutants defective in siderophore synthesis fix N2 normally [113] but in others Sid− mutants fail to fix N2 symbiotically [114]. In a later study it was shown that, in iron-rich medium, the regulation of rhizobactin synthesis is a factor in efficient nitrogen fixation [115]. With the aim of elucidating the implication of iron and iron transport mechanisms in the three-partner interaction rhizobia- legume-soil, Fabiano et al. [116] evaluated the ability of rhizobia to express high-affinity iron transport systems and showed that iron acquisition systems are widespread in Uruguayan isolates. Studies with the native strain S. meliloti 242 demonstrated that the siderophore-mediated iron uptake systems are not essential for an efficient biological nitrogen fixation but they are involved in early steps of nodulation and in rhizobial competitiveness [117]. Besides mutants defective in siderophore biosynthesis, those defective in other aspects of iron uptake and regulation also have been shown to give similar confusing findings. A fur-null mutant of B. japonicum, under derepressed state for iron uptake, is shown to form effective symbiosis [90], whereas a manganese-resistant fur mutant strain was unable to form functional, nitrogen-fixing nodules on soybean, mung bean, or cowpea, suggesting possible roles for a Fur-regulated protein or proteins in the symbiosis at least under some circumstances [118]. Yeoman et al. [85] isolated an fhuA mutant Rhizobium leguminosarum. The mutant was shown to be defective in iron uptake and accumulated the siderophore vicibactin but did not detectably affect symbiotic N2 fixation on peas. Also fhuA::gus fusion was expressed by bacteria in the meristematic zone of pea nodules but not in mature bacteroids suggesting that vicibactin is not used for iron uptake in R. leguminosarum bacteroids. Experiments show that both fegA and fegB are required for the utilization of ferrichrome, since the fegB mutant were found to form a normal symbiosis and fegAB mutant had a dramatic phenotype in planta leading to the speculation that it might be due to the inability of the mutant to transport ferrichrome. This symbiotic defect suggested that the fegAB operon was serving a different function in planta, possibly thought to be involved in the signaling between the rhizobia and the host plant [61]. It is generally believed that siderophore biosynthesis or transport genes are not expressed when nitrogenase genes are actively expressed. How the supply of iron is provided to form nitrogenase, at a stage in nodulation when it is highly active, remains to be resolved. It may be possible that iron nutrition of the bacteroids in the nodules is through ferrous form of iron which becomes available to the bacteroids through the reductases present on the peribacteroid membrane [119]. Recently while studying the expression of 200 genes of S. meliloti upon induction with plant symbiotic elicitor luteolin during symbiosis it was found that three genes related to iron metabolism were induced by luteolin and also in nodules. One of them coded for a probable siderophore, sitA for an iron transporter and another was homologous with a gene of the tonB-hmu cluster of R. leguminosarum [120]. SitABC-transporter thus seems to be an important system for iron acquisition in planta.
5.4. Engineering Strategy
Various different strategies have been applied to improve the competitiveness of a bioinoculant in the plant environment. These are either by promoting rhizobial multiplication in the plant environment, by inhibiting the growth of competing microorganisms, or by interfering with some of the signals perceived by the competing microbes provided these signals control (at least in part) the expression of functions central to microbial fitness [121]. Because this is a triple interface (bacteria, plant, and soil) interaction, it is possible to modify one, two, or three of these factors to improve microbial colonization. An improvement of plant-microbe symbioses should involve the coordinated modifications in the partner's genotypes resulting in highly complementary combinations [122]. Genetic manipulation of the bacteria should take into account genes which can be used to increase competitivity. While much of the effort has been directed to understand genes whose deficiency leads to loss of competitivity, only few studies deal with genes whose overexpression can improve competitivity. Successful strategies in this regard are as follows: (1) construction of a chimeric Nif HDK operon under the strong NifHc promoter and expression in PHB negative mutants of R. etli [123], (2) construction of an acid-tolerant R. leguminosarum bv. trifolii strain [124], (3) expression of ACC deaminase gene in S. meliloti [125], (4) overexpression of putA [126], (5) overexpression of trehalose 6-phosphate synthase [127], (6) overexpression of rosR and pssR [128], (7) heterologous expression of ferrichrome siderophore receptor gene fegA and fhuA [78–80], and (8) overproduction of the adhesion rap1 [129]. Of these, the strategy of Peralta et al. [123] explained that R. etli strains with the chimeric nitrogenase construct assayed in greenhouse experiments had increased nitrogenase activity (58% on average), plant weight (32% on average), N content in plants (15% at 32 days after inoculation), and most importantly higher seed yield (36% on average), higher N content (25%), and higher N yield (72% on average) in seeds. Additionally, expression of the chimeric nifHDK operon in a PHB-negative R. etli strain produced an additive effect in enhancing symbiosis. Probably, this is the first report of increased seed yield and nutritional content in the common bean, obtained by using only the genetic material already present in Rhizobium. Mongiardini et al. [129] on the other hand, investigated the influence of adhesins on competitiveness of R. leguminosarum bv. trifolii using clover as test plants. In this report, the R. leguminosarum bv. trifolii adhesion protein RapA1 was overproduced from a pHC60-derived plasmid and expressed in R200 strain. When an overproducing strain and a control-carrying empty vector were coinoculated on clover plants, a positive effect of RapA1 on competition for nodule occupation was observed suggesting that optimization of RapA1 expression may be considered while improving the rhizobial competitiveness. Ability to cross-utilize heterologous siderophores is another trait that can be incorporated into bioinoculants to further improve their competitiveness. The release of genetically improved strains is often restricted due to lack of regulation and proper guidelines of its release besides its potential ecological effects, as perceived by the public. However, despite these perceptions, some recombinant rhizobial strains have been commercialized, such as S. meliloti strain RMBPC-2, which was approved by the US Environmental Protection Agency in 1997. This genetically engineered bacterium contained additional copies of nifA and dctABD to increase nitrogen fixation and when inoculated, enhanced the yield of alfalfa [130] under N-limiting conditions. Later on, inoculation of alfalfa seeds with any of the three recombinant strains of S. meliloti significantly increased overall plant biomass compared with inoculation with the wild-type strains over a 3-year period during which high proportion of nodules were occupied by the inoculum strains. However, the recombinant strains were found to be ineffective in soils where the indigenous rhizobial populations were more competitive [131].
5.4.1. Bioinformatics Details about Presence/Absence of Genes in Different Rhizobia
Various techniques like mutations, deletion mapping, cloning vectors, and so forth. have facilitated the identification of genes associated with nitrogen fixation. Legume, nonlegume, and free living nitrogen fixers have a set of genes which are responsible for effective nodulation and nitrogen fixation. These are the nod, nol, noe, nif, fix, and some hydrogenase genes. The work on the genetics of nitrogen fixation was first started in Klebsiella oxytoca M5a1 and first ever detailed organization of nif genes were reported in this organism [132]. A number of studies have established that core nif genes like nifH, nifD, nifK, nifY, nifB, nifQ, nifE, nifN, nifX, nifU, nifS, nifV, nifW, and nifZ are essential for nitrogen fixation. On the basis of mutational studies the natures of different nif gene products were determined. Studies confirmed that nif HDK encodes nitrogenase while nif LA had regulatory functions. In cyanobacteria especially Anabaena sp. 7120 it was established that the organization of nif HDK differed from that of Klebsiella pneumoniae. Rao [133] reported that it was Johnston and his coworkers who discovered the presence of nodulation genes in a plasmid of Rhizobium leguminosarum and mutation of those genes rendered them useless. Like the rhizobia, Azosprillium includes a megaplasmid and sequences similar to nod genes [134] Later on studies ascertained that nod, nol, and noe genes produce nodulation signals [47, 135]. The interplay of different nod genes, triggering of the creation of root nodule, signaling cascades, and development of nodule meristem were reported by a number of researchers [136, 137]. Studies concerning nodule physiology and nodule functioning specified that the bacteria in bacteroids forms are surrounded in plant membranes resulting in the formation of symbiosomes [138]. Frankia houses a number of nif genes but researchers failed to spot nod genes in Frankia [139]. However, groundwork on Frankia genomes exposed some putative nod-like genes which did not emerge in organized clusters and failed to detect the nodA gene [140]. Rao [133] reported that fix genes were identified on the chromosomes as well as plasmids of Rhizobium and were transferable. These genes are irregular in free-living bacteria and assist in electron transfer to the nitrogenase. The amalgamation of the knowledge of plant physiology, biochemistry, genetics, and molecular biology gave idea about the understanding of the mechanism of nitrogen fixation in pregenomic era. Things changed with the accessibility of complete genome sequences of symbiotic as well as nonsymbiotic diazotrophs. Knowledge of the whole genome became the stepping stone in understanding the working principle of the bacterial cell. Mesorhizobium loti was the first sequence of a symbiotic bacterium and it was followed by Sinorhizobium meliloti [141]. The completion of the genomes of Rhizobium leguminosarum bv. viciae [142], Rhizobium etli [143], Bradyrhizobium japonicum starin USDA110 and USDA6T [144, 145] Frankia strains [146], and sequences for a number of free-living diazotrophs spanning different habitats and ecological niches [140] bolstered nitrogen fixation research. The studies on the genomes exposed new evidences pertaining to evolution and structure, interactions between plants and microbes. The detection of a number of symbiotic genes associated with nitrogen fixation has strengthened functional genomic research. Tools like DNA macro- and microarrays have been applied for studying expression at transcription level in Sinorhizobium and are being applied in other rhizobia [141]. Competitive ability of the bioinoculants strain in comparison to the indigenous microflora is governed not only by its ability to fix nitrogen (governed by nif genes) but also by the genes involved in the iron uptake system as they play a key role in proper nodule development. Pseudomonads are known for their rhizospheric stability and one of the factors contributing to this is the presence of diverse iron uptake systems. About 32 putative siderophore receptors in P. aeruginosa [55, 56], 29 in P. putida, 27 in P. fluorescens, and 23 in P. syringae [62] are reported. Analysis of protein sequences of complete genomes of Pseudomonas species using HMMER profiles revealed the presence of 45 TonB dependent siderophore receptors in the genome of P. fluorescens, 31 in P. putida, and 36 in P. aeruginosa. In contrast, a complete genome wide search in a few members of rhizobiales revealed a visible scarcity of TonB dependent siderophore receptors: 3 were present in R. etli, 3 in Mesorhizobium sp. BNC1, 2 in Mesorhizobium loti, 2 in Sinorhizobium meliloti, and 8 in Bradyrhizobium japonicum [79] Relatively high number of TonB dependent receptors present in Bradyrhizobium sp. amongst rhizobiales [147] and this could be attributed for their high rhizospheric competence and hence their reported success as commercial biofertilizers for soybean crops. In R. leguminosarum genome, the iron-siderophore uptake machinery was present on plasmid pRL, different from other rhizobia, where the iron uptake machinery is present on the genome [148] clustered together as FhuABDC (GI: pRL100325, GI: pRL100326, GI: pRL100327, and GI: pRL100328). The other FhuA homologs are not found to be associated with inner membrane machinery, similar to FhuE (rhodotorulic acid and coprogen receptor) and IutA (aerobactin receptor) of E. coli, which are also found in isolation [53]. These receptors work in association with FhuBCD (ferrichrome system) suggesting that the transport of ferrisiderophores through the inner membrane is not as specific as that through the outer membrane. It is due to this reason that a notably less number of periplasmic and cytoplasmic membrane proteins are reported to be present in Bradyrhizobium sp. and Pseudomonas sp. against a relatively large number of outer membrane receptors reported [83]. These findings led us to hypothesize that increasing the repertoire of outer membrane siderophore receptors could make rhizobial isolates more efficient with respect to iron acquisition and hence colonization of the rhizosphere.
5.4.2. Overexpression of Receptor and Findings
The successful performance of rhizobial inoculant strains depends upon their capability to out-compete the indigenous soil bacteria, survive, propagate, and enter into effective symbiosis with the host plant. Biofertilizer strains which fail to survive under soil conditions are most of the time ineffective in enhancing legume productivity because vast majority of nodules formed are not by the inoculated strain but by indigenous rhizobia in the soil [53, 104]. Rhizobium sp. ST1 was inhibited slightly in the presence of ferrichrome under lab conditions and also under natural soil conditions, but the expression of fhuA genes into Rhizobium sp. ST1 increased its rhizospheric stability, which was evident by its increase in nodule occupancy on the C. cajan plants [59, 78, 149]. The fegA gene with its native promoter when subcloned and transferred into Mesorhizobium sp. GN25 and Rhizobium sp. ST1, imparted these strains the ability to utilize ferrichrome, and increased their rhizospheric competitive abilities [79]. Similar to the above findings, where the expression of only the outer membrane receptor is shown to impart siderophore utilization, Brickman and Armstrong [60] showed that the expression of fauA gene, encoding receptor for alcaligin siderophore, imparts alcaligin utilization to a P. aeruginosa strain deficient in alcaligin production [60]. These findings provide evidence that engineering rhizobial strains with ferrichrome utilization ability provides them with a competitive edge in an environment where Fe-ferrichrome is the only available source of iron. Whether siderophore production or uptake efficiency increases the nitrogen fixation ability of nodule bacteria is not yet known, but their survival in the absence of their host definitely depends upon this characteristic.
6. Conclusion
Bacteria may become competitively successful only if armed with the appropriate tools of efficient substrate acquisition, resistance mechanisms, and competitive traits. Comparative genomics analyses have revealed the existence of competitivity genes that are conserved and many that are specific to each species. Signature-tagged mutant libraries constructed for S. meliloti and M. loti are an important and powerful resource for future functional genomics and such libraries of other strains may be useful in understanding and engineering rhizobia for better legume productivity in different agro-ecological regions of the world. Molecular biology together with screening genotypes may help to identify and concurrently develop more effective inoculants strains. Even though the use of genetically modified microbial inoculants in agriculture is controversial, future demands on agricultural produce, ever-growing populations, and decreasing cultivable lands require a regulated and lawful use of genetically engineered inoculants. A very careful assessment of genetic modification of rhizobia have to be made so that after release into soil it does not have any deleterious impact on indigenous microbial communities of agronomic importance. The competitive advantage conferred by genes involved in metabolic fitness and nutrient acquisition are perhaps relatively harmless in nature, whereas overexpression of genes coding for the synthesis of antibiotic-like molecules may affect other indigenous microbial species and may alter bacterial diversity. Recent interest in application of rhizobia to enhance growth of nonleguminous plants like rice, sugarcane, wheat, and maize either as associative symbionts or as endophytes extends the use of this group of microorganisms for plants other than legumes. Applying rhizobial inoculation technology to the nonleguminous plants, however, may cause competition problem to such plants. Therefore, the factors affecting establishment of inoculated rhizobia as endophytes need to be considered and competitive effect of native population should be addressed. Whether increasing competitiveness for nodulation also enhances endophytic competiveness could be explored.
Acknowledgments
The authors would like to thank and show their gratitude to Dr. G. Archana and Professor A. J. Desai of M. S. University of Baroda, for guidance and Department of Biotechnology (DBT), India, and University Grants Commission (UGC), India for the funding during the course of research.
References
- 1.Vance CP. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. Legume symbiotic nitrogen fixation: agronomic aspects; pp. 509–530. [Google Scholar]
- 2.Graham PH, Vance CP. Nitrogen fixation in perspective: an overview of research and extension needs. Field Crops Research. 2000;65(2-3):93–106. [Google Scholar]
- 3.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiology and Molecular Biology Reviews. 2004;68(2):280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav AS. Genetics of competition in rhizobia for legume nodule occupancy. National Journal of Plant Improvement. 2006;8:1–6. [Google Scholar]
- 5.Dommergues YR, Subba Rao NS. Introduction of N2-fixing trees in non-N2-fixing tropical plantations. In: Subba Rao NS, Dommergues YR, editors. Microbial Interactions in Agriculture and Forestry. Vol. 2. Enfield, NH, USA: Science Publishers; 2000. pp. 131–154. [Google Scholar]
- 6.Stamford NP, Ortega AD, Temprano F, Santos DR. Effects of phosphorus fertilization and inoculation of Bradyrhizobium and mycorrhizal fungi on growth of Mimosa caesalpiniaefolia in an acid soil. Soil Biology and Biochemistry. 1997;29(5-6):959–964. [Google Scholar]
- 7.Unkovich MJ, Pate JS. An appraisal of recent field measurements of symbiotic N2 fixation by annual legumes. Field Crops Research. 2000;65(2-3):211–228. [Google Scholar]
- 8.Stephens JHG, Rask HM. Inoculant production and formulation. Field Crops Research. 2000;65(2-3):249–258. [Google Scholar]
- 9.Beattie GA, Clayton MK, Handelsman J. Quantitative comparison of the laboratory and field competitiveness of Rhizobium leguminosarum biovar phaseoli . Applied and Environmental Microbiology. 1989;55(11):2755–2761. doi: 10.1128/aem.55.11.2755-2761.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgson ALM, Stacey G. Potential for Rhizobium improvement. Critical Reviews in Biotechnology. 1986;4:1–74. [Google Scholar]
- 11.Li D-M, Alexander M. Bacterial growth rates and competition affect nodulation and root colonization by Rhizobium meliloti . Applied and Environmental Microbiology. 1986;52(4):807–811. doi: 10.1128/aem.52.4.807-811.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrmann J, Wollum AG., II Nodulation competition among Bradyrhizobium japonicum strains as influenced by rhizosphere bacteria and iron availability. Biology and Fertility of Soils. 1989;7(2):108–112. [Google Scholar]
- 13.Vlassak KM, Vanderleyden J. Factors influencing nodule occupancy by inoculant rhizobia. Critical Reviews in Plant Sciences. 1997;16(2):163–229. [Google Scholar]
- 14.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nature Reviews Microbiology. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crowley DE, Wang YC, Reid CPP, Szaniszlo PJ. Mechanisms of iron acquisition from siderophores by microorganisms and plants. Plant and Soil. 1991;130(1-2):179–198. [Google Scholar]
- 16.Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends in Biochemical Sciences. 1999;24(3):104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 17.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. Journal of Biological Chemistry. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 18.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiology and Molecular Biology Reviews. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faraldo-Gómez JD, Sansom MSP. Acquisition of siderophores in gram-negative bacteria. Nature Reviews Molecular Cell Biology. 2003;4(2):105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 20.Smith MJ, Neilands JB. Rhizobactin a siderophore from Rhizobium meliloti . Journal of Plant Nutrition. 1984;7:449–458. [Google Scholar]
- 21.Plessner O, Klapatch T, Guerinot ML. Siderophore utilization by Bradyrhizobium japonicum . Applied and Environmental Microbiology. 1993;59(5):1688–1690. doi: 10.1128/aem.59.5.1688-1690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne SM. Detection, isolation, and characterization of siderophores. Methods in Enzymology. 1994;235:329–344. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- 23.Budde AD, Leong SA. Characterization of siderophores from Ustilago maydis . Mycopathologia. 1989;108(2):125–133. doi: 10.1007/BF00436063. [DOI] [PubMed] [Google Scholar]
- 24.Powell PE, Cline GR, Reid CPP, Szaniszlo PJ. Occurrence of hydroxamate siderophore iron chelators in soils. Nature. 1980;287(5785):833–834. [Google Scholar]
- 25.Powell PE, Szaniszlo PJ, Reid CPP. Confirmation of occurrence of hydroxamate siderophores in soil by a novel Escherichia coli bioassay. Applied and Environmental Microbiology. 1983;46(5):1080–1083. doi: 10.1128/aem.46.5.1080-1083.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyner DC, Lindow SE. Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology. 2000;146(10):2435–2445. doi: 10.1099/00221287-146-10-2435. [DOI] [PubMed] [Google Scholar]
- 27.Hojberg O, Sorensen J. Microgradients of microbial oxygen consumption in a barley rhizosphere model system. Applied and Environmental Microbiology. 1993;59(2):431–437. doi: 10.1128/aem.59.2.431-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buyer JS, Sikora LJ. Rhizosphere interactions and siderophores. In: Keister DL, Cregan PB, editors. The Rhizosphere and Plant Growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 263–369. [Google Scholar]
- 29.Jurkevitch E, Hadar Y, Chen Y. Differential siderophore utilization and iron uptake by soil and rhizosphere bacteria. Applied and Environmental Microbiology. 1992;58(1):119–124. doi: 10.1128/aem.58.1.119-124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bull CT, Weller DM, Thomashow LS. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 31.Raaijmakers JM, van der Sluis I, Koster M, Bakker PAHM, Weisbeek PJ, Schippers B. Utilization of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Canadian Journal of Microbiology. 1995;41(2):126–135. [Google Scholar]
- 32.Meyer JM, Abdallah MA. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. Journal of General Microbiology. 1978;107(2):319–328. [Google Scholar]
- 33.Scher FM, Baker R. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to fusarium wilt pathogens. Phytopathology. 1982;72:1567–1573. [Google Scholar]
- 34.Hohnadel D, Meyer JM. Specificity of pyoverdine-mediated iron uptake among fluorescent Pseudomonas strains. Journal of Bacteriology. 1988;170(10):4865–4873. doi: 10.1128/jb.170.10.4865-4873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty U, Purkayastha RP. Role of rhizobitoxine in protecting soybean roots from Macrophomina phaseolina infection. Canadian Journal of Microbiology. 1984;30(3):285–289. doi: 10.1139/m84-043. [DOI] [PubMed] [Google Scholar]
- 36.Ehteshamul-Haque S, Zaki MJ, Vahidy AA, Ghaffar A. Effect of organic amendments on the efficacy of Pseudomonas aeruginosa in the control of root rot disease of sunflower. Pakistan Journal of Botany. 1998;30(1):45–50. [Google Scholar]
- 37.Khan A, Geetha R, Akolkar A, Pandya A, Archana G, Desai AJ. Differential cross-utilization of heterologous siderophores by nodule bacteria of Cajanus cajan and its possible role in growth under iron-limited conditions. Applied Soil Ecology. 2006;34(1):19–26. [Google Scholar]
- 38.Plazinski J, Rolfe BG. Influence of Azospirillum strains on the nodulation of clovers by Rhizobium strains. Applied and Environmental Microbiology. 1985;49(4):984–990. doi: 10.1128/aem.49.4.984-989.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handelsman J, Brill WJ. Erwinia herbicola isolates from alfalfa plants may play a role in nodulation of alfalfa by Rhizobium meliloti . Applied and Environmental Microbiology. 1985;49(4):818–821. doi: 10.1128/aem.49.4.818-821.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manjanatha MG, Loynachan TE, Atherly AG. Tn5 mutagenesis of Chinese Rhizobium fredii for siderophore overproduction. Soil Biology and Biochemistry. 1992;24(2):151–155. [Google Scholar]
- 41.Barran LR, Bromfield ESP. Does siderophore production influence the relative abundance of Rhizobium meliloti in two field populations? Canadian Journal of Microbiology. 1993;39(3):348–351. [Google Scholar]
- 42.Guerinot ML. Iron uptake and metabolism in the rhizobia/legume symbioses. Plant and Soil. 1991;130(1-2):199–209. [Google Scholar]
- 43.Buyer JS, Leong J. Iron transport-mediated antagonism between plant growth-promoting and plant-deleterious Pseudomonas strains. Journal of Biological Chemistry. 1986;261(2):791–794. [PubMed] [Google Scholar]
- 44.Vance CP. Enhanced agricultural sustainability through biological nitrogen fixation. In: Legocki A, Bothe H, Puhler A, editors. Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture. Berlin, Germany: Springer; 1997. pp. 179–186. [Google Scholar]
- 45.Smith RS. Legume inoculant formulation and application. Canadian Journal of Microbiology. 1992;38(6):485–492. [Google Scholar]
- 46.Denarie J, Debelle F, Rosenberg C. Signalling and host range variation in nodulation. Annual Review of Microbiology. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 47.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiology and Molecular Biology Reviews. 2000;64(1):180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips DA, Streit WR, Volpin H, Joseph CM. Plant regulation of root colonisation by Rhizobium meliloti . In: Legocki A, Bothe H, Puhler A, editors. Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture. Berlin, Germany: Springer; 1997. pp. 133–136. [Google Scholar]
- 49.Schlaman HRM, Phillips DA, Kondorosi E. Genetic organisation and transcriptional regulation of rhizobial nodulation genes. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 361–386. [Google Scholar]
- 50.Howieson JG, Loi A, O'Hara GW. The legume-rhizobia relationship in the Mediterranean Basin. In: Sulas L, editor. Legumes for Mediterranean Forage Crops, Pastures and Alternative Uses. Zaragoza, Spain: CIHEAM; 2000. pp. 305–314. (Cahiers Options Méditerranéennes, no. 45). [Google Scholar]
- 51.Howieson JG, O’Hara GW, Carr SJ. Changing roles for legumes in Mediterranean agriculture: developments from an Australian perspective. Field Crops Research. 2000;65(2-3):107–122. [Google Scholar]
- 52.Miller RH, May S. Legume inoculation: successes and failures. In: Keister DL, Cregan PB, editors. The Rhizosphere and Plant Growth. New York, NY, USA: Kluwer Academic Publishers; 1991. pp. 123–134. [Google Scholar]
- 53.Streeter JG. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Canadian Journal of Microbiology. 1994;40(7):513–522. [Google Scholar]
- 54.Verma DPS, Long S. The molecular biology of Rhizobium-legume symbiosis. International Review of Cytology, Supplement. 1983;14:211–245. [Google Scholar]
- 55.Dean CR, Poole K. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa . Journal of Bacteriology. 1993;175(2):317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ankenbauer RG, Quan HN. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. Journal of Bacteriology. 1994;176(2):307–319. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crowley DE, Reid CPP, Szaniszlo PJ. Microbial siderophores as iron sources for plants. In: Winklemann G, van der Helm D, Neilands JB, editors. Iron Transport in Microbes, Plants and Animals. New York, NY, USA: VCH Publishers; 1987. pp. 375–386. [Google Scholar]
- 58.Holmström SJM, Lundström US, Finlay RD, van Hees PAW. Siderophores in forest soil solution. Biogeochemistry. 2004;71(2):247–258. [Google Scholar]
- 59.Rajendran G, Mistry S, Desai AJ, Archana G. Functional expression of Escherichia coli fhuA gene in Rhizobium spp. of Cajanus cajan provides growth advantage in presence of Fe3+: ferrichrome as iron source. Archives of Microbiology. 2007;187(4):257–264. doi: 10.1007/s00203-006-0191-8. [DOI] [PubMed] [Google Scholar]
- 60.Brickman TJ, Armstrong SK. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. Journal of Bacteriology. 1999;181(19):5958–5966. doi: 10.1128/jb.181.19.5958-5966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benson HP, Boncompagni E, Guerinot ML. An iron uptake operon required for proper nodule development in the Bradyrhizobium japonicum-soybean symbiosis. Molecular Plant-Microbe Interactions. 2005;18(9):950–959. doi: 10.1094/MPMI-18-0950. [DOI] [PubMed] [Google Scholar]
- 62.Cornelis P, Matthijs S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environmental Microbiology. 2002;4(12):787–798. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 63.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Frontiers in Bioscience. 2003;8:661–686. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 64.Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiology. 1994;104(3):815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persmark M, Pittman P, Buyer JS, Schwyn B, Gill PR, Jr., Neilands JB. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. Journal of the American Chemical Society. 1993;115(10):3950–3956. [Google Scholar]
- 66.Jadhav RS, Desai AJ. Isolation and characterization of siderophore from cowpea Rhizobium (peanut isolate) Current Microbiology. 1992;24(3):137–141. [Google Scholar]
- 67.Berraho EL, Lesueur D, Diem HG, Sasson A. Iron requirement and siderophore production in Rhizobium ciceri during growth on an iron-deficient medium. World Journal of Microbiology and Biotechnology. 1997;13(5):501–510. [Google Scholar]
- 68.Guerinot ML, Meidl EJ, Plessner O. Citrate as a siderophore in Bradyrhizobium japonicum . Journal of Bacteriology. 1990;172(6):3298–3303. doi: 10.1128/jb.172.6.3298-3303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Bellis P, Ercolani GL. Growth interactions during bacterial colonization of seedling rootlets. Applied and Environmental Microbiology. 2001;67(4):1945–1948. doi: 10.1128/AEM.67.4.1945-1948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loper JE, Henkels MD. Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Applied and Environmental Microbiology. 1999;65(12):5357–5363. doi: 10.1128/aem.65.12.5357-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah S, Karkhanis V, Desai A. Isolation and characterization of siderophore, with antimicrobial activity, from Azospirillum lipoferum M. Current Microbiology. 1992;25(6):347–351. [Google Scholar]
- 72.Guan LL, Onuki H, Kamino K. Bacterial growth stimulation with exogenous siderophore and synthetic N-acyl homoserine lactone autoinducers under iron-limited and low-nutrient conditions. Applied and Environmental Microbiology. 2000;66(7):2797–2803. doi: 10.1128/aem.66.7.2797-2803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiology and Molecular Biology Reviews. 1997;61(3):319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guan LL, Kanoh K, Kamino K. Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Applied and Environmental Microbiology. 2001;67(4):1710–1717. doi: 10.1128/AEM.67.4.1710-1717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guerinot ML. Iron and the nodule. In: Barton LB, Hemming BC, editors. Iron Chelation in Plants and Soil Microorganisms. New York, NY, USA: Academic Press; 1993. pp. 197–217. [Google Scholar]
- 76.Battistoni F, Platero R, Noya F, Arias A, Fabiano E. Intracellular Fe content influences nodulation competitiveness of Sinorhizobium meliloti strains as inocula of alfalfa. Soil Biology and Biochemistry. 2002;34(5):593–597. [Google Scholar]
- 77.Carlton TM, Sullivan JT, Stuart GS, Hutt K, Lamont IL, Ronson CW. Ferrichrome utilization in a mesorhizobial population: microevolution of a three-locus system. Environmental Microbiology. 2007;9(12):2923–2932. doi: 10.1111/j.1462-2920.2007.01402.x. [DOI] [PubMed] [Google Scholar]
- 78.Geetha R, Desai AJ, Archana G. Effect of the expression of Escherichia coli fhuA gene in Rhizobium sp. IC3123 and ST1 in planta: its role in increased nodule occupancy and function in Pigeon pea . Applied Soil Ecology. 2009;43(2-3):185–190. [Google Scholar]
- 79.Joshi FR, Desai DK, Archana G, Desai AJ. Enhanced survival and nodule occupancy of Pigeon pea nodulating Rhizobium sp. ST1 expressing fegA gene of Bradyrhizobium japonicum 61A152. OnLine Journal of Biological Sciences. 2009;9(2):40–51. [Google Scholar]
- 80.Joshi F, Chaudhari A, Joglekar P, Archana G, Desai A. Effect of expression of Bradyrhizobium japonicum 61A152 fegA gene in Mesorhizobium sp., on its competitive survival and nodule occupancy on Arachis hypogea . Applied Soil Ecology. 2008;40(2):338–347. [Google Scholar]
- 81.Chakraborty R, Storey E, van der Helm D. Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli . BioMetals. 2007;20(3-4):263–274. doi: 10.1007/s10534-006-9060-9. [DOI] [PubMed] [Google Scholar]
- 82.Jadhav RS, Desai A. Role of siderophore in iron uptake in cowpea Rhizobium GN1 (peanut isolate): possible involvement of iron repressible outer membrane proteins. FEMS Microbiology Letters. 1994;115(2-3):185–190. [Google Scholar]
- 83.Stevens JB, Carter RA, Hussain H, Carson KC, Dilworth MJ, Johnston AWB. The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: fhuCDB are adjacent to a pseudogene version of fhuA . Microbiology. 1999;145(3):593–601. doi: 10.1099/13500872-145-3-593. [DOI] [PubMed] [Google Scholar]
- 84.Braun V, Braun M. Active transport of iron and siderophore antibiotics. Current Opinion in Microbiology. 2002;5(2):194–201. doi: 10.1016/s1369-5274(02)00298-9. [DOI] [PubMed] [Google Scholar]
- 85.Yeoman KH, Wisniewski-Dye F, Timony C, et al. Analysis of the Rhizobium leguminosarum siderophore-uptake gene fhuA: differential expression in free-living bacteria and nitrogen-fixing bacteroids and distribution of an fhuA pseudogene in different strains. Microbiology. 2000;146(4):829–837. doi: 10.1099/00221287-146-4-829. [DOI] [PubMed] [Google Scholar]
- 86.LeVier K, Day DA, Guerinot ML. Iron uptake by symbiosomes from soybean root nodules. Plant Physiology. 1996;111(3):893–900. doi: 10.1104/pp.111.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cuív PÓ, Clarke P, Lynch D, O’Connell M. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. Journal of Bacteriology. 2004;186(10):2996–3005. doi: 10.1128/JB.186.10.2996-3005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wexler M, Yeoman KH, Stevens JB, de Luca NG, Sawers G, Johnston AWB. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Molecular Microbiology. 2001;41(4):801–816. doi: 10.1046/j.1365-2958.2001.02556.x. [DOI] [PubMed] [Google Scholar]
- 89.Nienaber A, Hennecke H, Fischer H-M. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum . Molecular Microbiology. 2001;41(4):787–800. doi: 10.1046/j.1365-2958.2001.02555.x. [DOI] [PubMed] [Google Scholar]
- 90.Hamza I, Hassett R, O’Brian MR. Identification of a functional fur gene in Bradyrhizobium japonicum . Journal of Bacteriology. 1999;181(18):5843–5846. doi: 10.1128/jb.181.18.5843-5846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Luca NG, Wexler M, Pereira MJ, Yeoman KH, Johnston AWB. Is the fur gene of Rhizobium leguminosarum essential? FEMS Microbiology Letters. 1998;168(2):289–295. doi: 10.1016/s0378-1097(98)00454-6. [DOI] [PubMed] [Google Scholar]
- 92.Yang J, Sangwan I, O’Brian MR. The Bradyrhizobium japonicum Fur protein is an iron-responsive regulator in vivo. Molecular Genetics and Genomics. 2006;276(6):555–564. doi: 10.1007/s00438-006-0162-4. [DOI] [PubMed] [Google Scholar]
- 93.Hamza I, Chauhan S, Hassett R, O’Brian MR. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. Journal of Biological Chemistry. 1998;273(34):21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- 94.Hamza I, Qi Z, King ND, O’Brian MR. Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum . Microbiology. 2000;146(3):669–676. doi: 10.1099/00221287-146-3-669. [DOI] [PubMed] [Google Scholar]
- 95.Yang J, Sangwan I, Lindemann A, et al. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Molecular Microbiology. 2006;60(2):427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AWB. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum . Microbiology. 2002;148(12):4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- 97.Rudolph G, Semini G, Hauser F, et al. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. Journal of Bacteriology. 2006;188(2):733–744. doi: 10.1128/JB.188.2.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stacey G, Libault M, Brechenmacher L, Wan J, May GD. Genetics and functional genomics of legume nodulation. Current Opinion in Plant Biology. 2006;9(2):110–121. doi: 10.1016/j.pbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 99.Deryto M, Skorupska A. Rhizobial siderophore as an iron source for clover. Physiologia Plantarum. 1992;85:549–553. [Google Scholar]
- 100.Eady RR, Postgate JR. Nitrogenase. Nature. 1974;249(5460):805–809. doi: 10.1038/249805a0. [DOI] [PubMed] [Google Scholar]
- 101.Rai R, Singh SN, Prasad V. Effect of presmued amended pyrite on symbiotic N2-fixation, active iron content of nodules, grain yield and quality of chickpea (Cicer arietinum Linn:) genotypes in calcareous soil. Journal of Plant Nutrition. 1982;5:905–913. [Google Scholar]
- 102.Hemantaranjan A, Garg OK. Introduction of nitrogen fixing nodules through iron and zinc fertilization in the non-nodule forming French bean (Phaseolus vulgaris L) Journal of Plant Nutrition. 1986;9:281–288. [Google Scholar]
- 103.O'Hara GW, Hartzook A, Bell RW, Loneragan JF. Response to Bradyrhizobium strains of peanut cultivars grown under iron stress. Journal of Plant Nutrition. 1988;11:843–852. [Google Scholar]
- 104.O'Hara GW, Dilworth MJ, Boonkerd N, Parkpian P. Iron deficiency specifically limits nodule development in peanut inoculated with Bradyrhizobium sp. New Phytologist. 1988;108(1):51–57. doi: 10.1111/j.1469-8137.1988.tb00203.x. [DOI] [PubMed] [Google Scholar]
- 105.Tang C, Robson AD, Dilworth MJ. The role of iron in nodulation and nitrogen fixation in Lupinus angustifolius L. New Phytologist. 1990;114:173–182. [Google Scholar]
- 106.Grimes HD, Mount MS. Influence oF Pseudomonas putida on nodulation of Phaseolus vulgaris . Soil Biology and Biochemistry. 1984;16(1):27–30. [Google Scholar]
- 107.Shenker M, Hadar Y, Chen Y. Kinetics of iron complexing and metal exchange in solutions by rhizoferrin, a fungal siderophore. Soil Science Society of America Journal. 1999;63(6):1681–1687. [Google Scholar]
- 108.Schippers B, Lugtenberg B, Weisbeek PJ. Plant growth control by fluorescent pseudomonads. In: Chet I, editor. Innovative Approaches to Plant Disease Control. New York, NY, USA: John Wiley & Sons; 1987. pp. 19–39. [Google Scholar]
- 109.Omar SA, Abd-Alla MH. Biocontrol of fungal root rot diseases of crop plants by the use of rhizobia and bradyrhizobia. Folia Microbiologica. 1998;43(4):431–437. [Google Scholar]
- 110.Nadler KD, Johnston AWB, Chen J-W, John TR. A Rhizobium leguminosarum mutant defective in symbiotic iron acquisition. Journal of Bacteriology. 1990;172(2):670–677. doi: 10.1128/jb.172.2.670-677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gill PR, Jr., Barton LL, Scoble MD, Neilands JB. A high-affinity iron transport system of Rhizobium meliloti may be required for efficient nitrogen fixation in planta. Plant and Soil. 1991;130(1-2):211–217. [Google Scholar]
- 112.Barton LL, Fekete FA, Vester CR, Gill PR, Neilands JB. Physiological characteristics of Rhizobium meliloti 1021 Tn5 mutants with altered rhizobactin activities. Journal of Plant Nutrition. 1992;15:2145–2156. [Google Scholar]
- 113.Reigh G, O’Connell M. Siderophore-mediated iron transport correlates with the presence of specific iron-regulated proteins in the outer membrane of Rhizobium meliloti . Journal of Bacteriology. 1993;175(1):94–102. doi: 10.1128/jb.175.1.94-102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barsomian GD, Urzainqui A, Lohman K, Walker GC. Rhizobium meliloti mutants unable to synthesize anthranilate display a novel symbiotic phenotype. Journal of Bacteriology. 1992;174(13):4416–4426. doi: 10.1128/jb.174.13.4416-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barton LL, Johnson GV, Schitoskey K, Wertz M. Siderophore-mediated iron metabolism in growth and nitrogen fixation by alfalfa nodulated with Rhizobium meliloti . Journal of Plant Nutrition. 1996;19(8-9):1201–1210. [Google Scholar]
- 116.Fabiano E, Gualtieri G, Pritsch C, Polla G, Arias A. Extent of high-affinity iron transport systems in field isolates of rhizobia. Plant and Soil. 1994;164(2):177–185. [Google Scholar]
- 117.Fabiano E, Gill PR, Noya F, Bagnasco P, De La Fuente L, Arias A. Siderophore-mediated iron acquisition mutants in Rhizobium meliloti 242 and its effect on the nodulation kinetic on alfalfa nodules. Symbiosis. 1995;19:197–211. [Google Scholar]
- 118.Benson HP, LeVier K, Guerinot ML. A dominant-negative fur mutation in Bradyrhizobium japonicum . Journal of Bacteriology. 2004;186(5):1409–1414. doi: 10.1128/JB.186.5.1409-1414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moreau S, Day DA, Puppo A. Ferrous iron is transported across the peribacteroid membrane of soybean nodules. Planta. 1998;207(1):83–87. [Google Scholar]
- 120.Ampe F, Kiss E, Sabourdy F, Batut J. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biology. 2003;4(2, article R15) doi: 10.1186/gb-2003-4-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Savka MA, Dessaux Y, Oger P, Rossbach S. Engineering bacterial competitiveness and persistence in the phytosphere. Molecular Plant-Microbe Interactions. 2002;15(9):866–874. doi: 10.1094/MPMI.2002.15.9.866. [DOI] [PubMed] [Google Scholar]
- 122.Tikhonovich IA, Provorov NA. Cooperation of plants and microorganisms: getting closer to the genetic construction of sustainable agro-systems. Biotechnology Journal. 2007;2(7):833–848. doi: 10.1002/biot.200700014. [DOI] [PubMed] [Google Scholar]
- 123.Peralta H, Mora Y, Salazar E, Encarnación S, Palacios R, Mora J. Engineering the nifH promoter region and abolishing poly-β -hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris . Applied and Environmental Microbiology. 2004;70(6):3272–3281. doi: 10.1128/AEM.70.6.3272-3281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen H, Richardson AE, Gartner E, Djordjevic MA, Roughley RJ, Rolfe BG. Construction of an acid-tolerant Rhizobium leguminosarum biovar trifolii strain with enhanced capacity for nitrogen fixation. Applied and Environmental Microbiology. 1991;57(7):2005–2011. doi: 10.1128/aem.57.7.2005-2011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma W, Charles TC, Glick BR. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Applied and Environmental Microbiology. 2004;70(10):5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Dillewijn P, Soto MJ, Villadas PJ, Toro N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Applied and Environmental Microbiology. 2001;67(9):3860–3865. doi: 10.1128/AEM.67.9.3860-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suárez R, Wong A, Ramírez M, et al. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Molecular Plant-Microbe Interactions. 2008;21(7):958–966. doi: 10.1094/MPMI-21-7-0958. [DOI] [PubMed] [Google Scholar]
- 128.Janczarek M, Jaroszuk-Ściseł J, Skorupska A. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii . Antonie van Leeuwenhoek. 2009;96(4):471–486. doi: 10.1007/s10482-009-9362-3. [DOI] [PubMed] [Google Scholar]
- 129.Mongiardini EJ, Pérez-Giménez J, Althabegoiti MJ, et al. Overproduction of the rhizobial adhesin RapA1 increases competitiveness for nodulation. Soil Biology and Biochemistry. 2009;41(9):2017–2020. [Google Scholar]
- 130.Bosworth AH, Williams MK, Albrecht KA, et al. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti with an extra copy of dctABD and/or modified nifA expression. Applied and Environmental Microbiology. 1994;60(10):3815–3832. doi: 10.1128/aem.60.10.3815-3832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scupham AJ, Bosworth AH, Ellis WR, Wacek TJ, Albrecht KA, Triplett EW. Inoculation with Sinorhizobium meliloti RMBPC-2 increases alfalfa yield compared with inoculation with a nonengineered wild-type strain. Applied and Environmental Microbiology. 1996;62(11):4260–4262. doi: 10.1128/aem.62.11.4260-4262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arnold W, Rump A, Klipp W, Priefer UB, Pühler A. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae . Journal of Molecular Biology. 1988;203(3):715–738. doi: 10.1016/0022-2836(88)90205-7. [DOI] [PubMed] [Google Scholar]
- 133.Rao NSS. Ten decades of research on biological nitrogen fixation. Current Science. 1988;57:710–713. [Google Scholar]
- 134.Elmerich C. Molecular biology and ecology of diazotrophs associated with non-leguminous plants. Nature Biotechnology. 1984;2(11):967–978. [Google Scholar]
- 135.Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annual Review of Genetics. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 136.Long SR. Genes and signals in the rhizobium-legumes symbiosis. Plant Physiology. 2001;125(1):69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Geurts R, Bisseling T. Rhizobium nod factor perception and signalling. Plant Cell. 2002;14:S239–S249. doi: 10.1105/tpc.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.White J, Prell J, James EK, Poole P. Nutrient sharing between symbionts. Plant Physiology. 2007;144(2):604–614. doi: 10.1104/pp.107.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cérémonie H, Cournoyer B, Maillet F, Normand P, Fernandez MP. Genetic complementation of rhizobial nod mutants with Frankia DNA: artifact or reality? Molecular and General Genetics. 1998;260(1):115–119. doi: 10.1007/s004380050877. [DOI] [PubMed] [Google Scholar]
- 140.Franche C, Lindström K, Elmerich C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant and Soil. 2009;321(1-2):35–59. [Google Scholar]
- 141.Pühler A, Arlat M, Becker A, Göttfert M, Morrissey JP, O’Gara F. What can bacterial genome research teach us about bacteria-plant interactions? Current Opinion in Plant Biology. 2004;7(2):137–147. doi: 10.1016/j.pbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 142.Young JPW, Crossman LC, Johnston AWB, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biology. 2006;7(4, article R34) doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.González V, Santamaría RI, Bustos P, et al. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3834–3839. doi: 10.1073/pnas.0508502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kaneko T, Nakamura Y, Sato S, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Research. 2002;9(6):189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 145.Kaneko T, Maita H, Hirakawa H, et al. Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum strain USDA6T . Genes. 2011;2(4):763–787. doi: 10.3390/genes2040763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Normand P, Lapierre P, Tisa LS, et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Research. 2007;17(1):7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hume DJ, Shelp BJ. Superior performance of the Hup- Bradyrhizobium japonicum strains 532C in Ontario soybean field trials. Canadian Journal of Plant Science. 1990;70:661–666. [Google Scholar]
- 148.Lynch D, O’Brien J, Welch T, et al. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti . Journal of Bacteriology. 2001;183(8):2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rajendran G, Sing F, Desai AJ, Archana G. Enhanced growth and nodulation of Pigeon pea by co-inoculation of Bacillus strains with Rhizobium spp. Bioresource Technology. 2008;99(11):4544–4550. doi: 10.1016/j.biortech.2007.06.057. [DOI] [PubMed] [Google Scholar]


