Abstract
The Spindle Assembly Checkpoint (SAC) is essential in mammalian mitosis to ensure the equal segregation of sister chromatids1, 2. The SAC generates a Mitotic Checkpoint Complex (MCC) to prevent the Anaphase Promoting Complex/Cyclosome (APC/C) from targeting key mitotic regulators for destruction until all the chromosomes have attached to the mitotic apparatus1, 3, 4. A single unattached kinetochore can delay anaphase for several hours5, but how it is able to block the APC/C throughout the cell is not understood. Current concepts of the SAC posit that it exhibits either an ‘all or nothing’ response6 or there is a minimum threshold sufficient to block the APC/C7. Here, we have used gene targeting to measure SAC activity and find that it does not have an ‘all or nothing’ response. Instead, the strength of the SAC depends on the amount of Mad2 recruited to kinetochores and on the amount of MCC formed. Furthermore, we show that different drugs activate the SAC to different extents, which may be relevant to their efficacy in chemotherapy.
The SAC is activated by unattached kinetochores that appear to generate an APC/C inhibitor1, 2. The exact nature of the inhibitor, or inhibitors, is not yet clear but genetic8, 9, biochemical4, 10-13 and structural data14 indicate the MCC, composed of Mad2, BubR1 and Bub3, prevents Cdc20 from activating the APC/C. This evidence is consistent with unattached kinetochores catalyzing a conformational change in Mad2 as a necessary prerequisite to binding Cdc206, 15. How a single unattached kinetochore is able to generate sufficient inhibitor to block the APC/C throughout the cell, however, is not understood. Is the inhibitory signal amplified to generate an ‘all or nothing’ response6, or is there a minimum SAC threshold sufficient to block the APC/C7? The true nature of the SAC is important both for our understanding of the mechanism behind it, and because it has been postulated that the SAC can be weakened in cancer cells16.
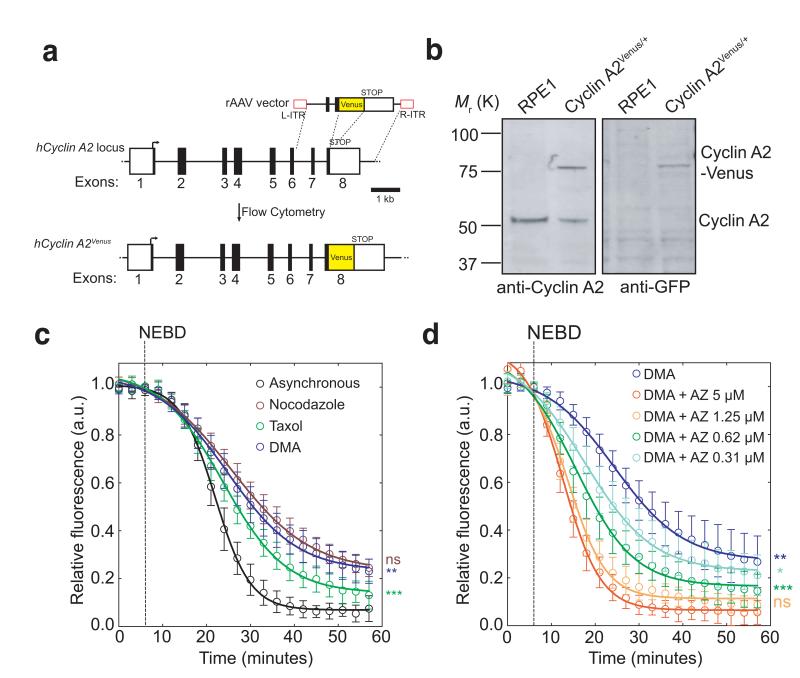
Our understanding of the SAC has been hampered by the lack of an assay to measure its activity. We recently showed that Cyclin A2 could be degraded when the SAC was active because it bound to Cdc20 in competition with the MCC17. We thought that this should provide an assay for SAC activity, i.e. the rate of MCC production, because the rate of Cyclin A2 degradation should be determined by competition for Cdc20 with the MCC. To test this we needed to measure precisely the prometaphase destruction of Cyclin A218-20. Therefore, we used recombinant adenovirus-associated virus (rAAV)-mediated gene targeting to introduce the open reading frame (ORF) of yellow fluorescent protein (Venus) into the last exon of one allele of the CCNA2 (Cyclin A2) gene in hTert-RPE1 cells (RPE1; retinal pigment epithelial) (Fig. 1a, Supplementary Fig. S1a,b). (Note that this fusion generated a functional protein18.) We chose RPE1 cells because they have a normal diploid karyotype, are not transformed and exhibit little cell death when arrested in mitosis; tagging the endogenous Cyclin A2 protein in RPE1 cells avoided the complications of mosaic protein levels in the cell population produced by ectopic expression. Immunoblotting analysis showed that cells expressed the fusion protein to the same level as the untagged protein (Fig. 1b). Time-lapse microscopy showed individual cells in the population had very similar kinetics of Cyclin A2-Venus degradation and mitotic progression (Supplementary Fig. 1c and Supplementary Movie 1).
Fig. 1. Cyclin A2-Venus degradation as a readout of SAC activity.
(a), Venus ORF targeting into the hCCNA2 locus by rAAV-mediated homologous recombination. The rAAV vector contains the coding sequence of Venus, which was inserted between two gene-specific homology arms to replace the STOP codon at the junction between non-coding (white square) and coding (black square) regions in exon 8 and two inverted terminal repeats (L-ITR, R-ITR; red rectangles). Integrants were selected by fluorescence-activated cell sorting (FACS). (b), Western blot analysis of parental RPE1 and a Cyclin A2-Venus clone. Cell lysates were probed with anti-Cyclin A2 (left panel) and anti-GFP (right panel) antibodies. Note the two forms of Cyclin A2 in the Cyclin A2-Venus clone (the upper one uniquely recognized by the anti-GFP antibody). Molecular mass markers on the left. (c-d), Single-cell Cyclin A2-Venus destruction assays of asynchronously growing RPE1 Cyclin A2-Venus cells treated with nocodazole (n=15 cells), Taxol (n=12 cells) or DMA (n=12 cells) c) or different concentrations of AZ3146 in presence of 10 μM DMA (n=15 cells for all conditions except 5 μM AZ3146 where n= 11 cells) d). Images were acquired at 3-5 min intervals and the total cell fluorescence was quantified. Fluorescence intensities were normalized to the level at nuclear envelope breakdown (NEBD). Error bars indicate s.d. and are representative of at least two independent experiments. The time to reach half-maximum fluorescence after NEBD was used for statistical tests.
Our hypothesis predicted that the rate of Cyclin A2 degradation should be accelerated when we eliminated the production of MCC, either by depleting Mad2, or by inhibiting MPS1, an essential SAC kinase. Conversely, maintaining the SAC, e.g. by treating cells with the Eg5 spindle motor poison di-methyl anastron (DMA), should reduce the rate of Cyclin A2 degradation. Both these predictions were verified (Supplementary Fig. 1d), from which we concluded that the rate of degradation of Cyclin A2-Venus could serve as a quantitative assay for SAC activity.
Taxanes and vinca alkaloids are effective anti-cancer agents, possibly through their ability to impose activate the SAC21, 22 so understanding how such compounds affect the SAC could be therapeutically valuable. To investigate this we treated RPE1 Cyclin A2-Venus cells with Taxol, DMA or nocodazole and found that although all three drugs delayed cells in mitosis, the rate of Cyclin A2-Venus destruction was faster in Taxol than in nocodazole or DMA (Fig. 1c); therefore different microtubule (MT) poisons generated different levels of SAC activity. Furthermore, when we inhibited Mps1, the rate of Cyclin A2 degradation increased and the mitotic delay decreased in proportion to the concentration of the inhibitor (AZ3146) (Fig. 1d; Supplementary Fig.1e), showing that partially inhibiting Mps1 reduced the strength of the SAC.
Our results indicated that different spindle poisons activated the SAC to different extents, which prompted us to ask whether this determined their ability to arrest cells in mitosis. Work from a number of laboratories supports the hypothesis that the extent to which cells delay in mitosis depends on two antagonistic pathways, whereby a gradual increase in activity of a pro-apoptosis pathway is counteracted by the slow degradation of Cyclin B1 and securin, and consequent mitotic exit (called mitotic slippage)21, 23, 24. The extent of mitotic slippage varies between individual cells and between cell lines, and in response to different drugs21. We postulated that the rate of mitotic slippage could be governed by the strength of the SAC generated by different drug treatments.
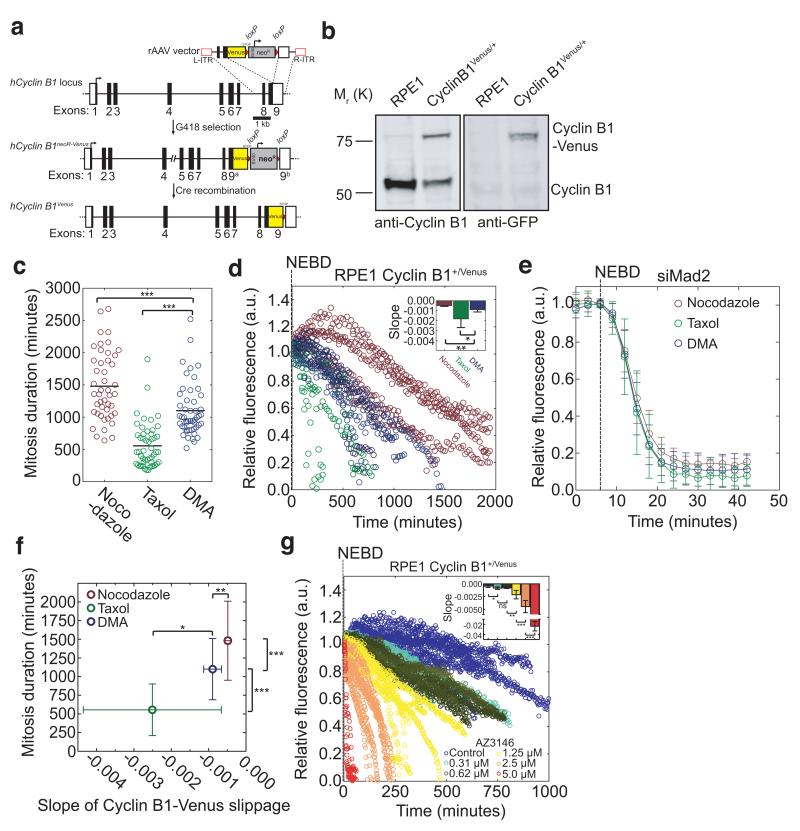
Mitotic slippage had previously been measured by the degradation of ectopic Cyclin B121, 23, but since altering the level of Cyclin B1-Cdk1 activity affected the fate of cells in mitosis21, we chose to assay the destruction of endogenous Cyclin B1 by using rAAV-mediated gene targeting to fuse exon 9 of the CCNB1 (Cyclin B1) locus to the open reading frame of Venus in RPE1 cells (Fig. 2a,b; Supplementary Fig. 2a-c), which produces a functional fusion protein25. Just as for Cyclin A2-Venus, the Cyclin B1-Venus protein accumulated and was destroyed with similar kinetics to the untagged endogenous protein (Supplementary Fig. 2d,e). Quantitative fluorescence imaging showed that Cyclin B1-Venus destruction began at metaphase (Supplementary Fig. 2f; control and movie 2) and was regulated by the SAC (Supplementary Fig. 2f; Reversine and nocodazole).
Fig. 2. Different spindle poisons activate the SAC to different extents.
(a) Venus ORF targeting into the hCCNB1 locus by rAAV-mediated homologous recombination. The rAAV vector contains both the coding sequence of Venus and a G418 resistance cassette (flanked by loxP sites, red triangles), which were inserted between two gene-specific homology arms to replace the STOP codon at the junction between non-coding (white square) and coding (black square) regions in exon 9 and two inverted terminal repeats (L-ITR, R-ITR; red rectangles). Integrants were selected by survival in G418-containing medium. (b) Western blot analysis of parental RPE1 and a Cyclin B1-Venus clone. Cell lysates were probed with anti-Cyclin B1 (left panel) and anti-GFP (right panel) antibodies. Note the two forms of Cyclin B1 in the Cyclin B1-Venus clone (the upper one uniquely recognized by the anti-GFP antibody). Molecular mass markers on the left. (c) Scatter plot displaying the duration of the mitotic arrest of RPE1 Cyclin B1-Venus cells treated with the same drugs as in Fig. 1c) (n=50 in each condition). (d) Single-cell analysis of mitotic slippage in asynchronously growing RPE1 Cyclin B1-Venus cells treated with nocodazole (n=5 cells), Taxol (n=6 cells) or DMA (n=5 cells). Images were acquired at 5 min intervals and quantified, plotted and analysed as in Fig 1c. The inset shows the slopes of the linear regressions in the main panel. Error bars indicate s.d. and are representative of at least two independent experiments. (e) Single-cell analysis of asynchronously growing RPE1 Cyclin B1-Venus cells treated with siRNA against Mad2 in presence of nocodazole (n=6 cells), Taxol (n=13 cells) or DMA (n=16 cells). Note that only cells with mitosis durations of 9-12 minutes (minimum mitosis duration in RPE1 cells) were considered. Error bars indicate s.d. and are representative of at least two independent experiments. (f) Effect of the rate of Cyclin B1-Venus slippage in d) on the duration of mitosis in different drug treatments in c). Error bars indicate s.d. and are representative of at least two independent experiments. (g) Single-cell analysis of mitotic slippage in asynchronously growing RPE1 Cyclin B1-Venus cells treated with nocodazole (n=7 cells) and increasing AZ3146 concentrations (n=5 cells for 0.31 μM; n=6 cells for 0.62 μM; n=6 cells for 1.25 μM, n=9 cells for 2.5 μM and n=7 cells for 5 μM). Images were acquired at 5 min intervals and total fluorescence was quantified. Fluorescence intensities were normalised to the level at NEBD. The insets show the slopes of the linear regressions in the respective main panel. Error bars indicate s.d. and are representative of two independent experiments.
We treated RPE1-Cyclin B1-Venus cells with various MT poisons and assayed them by time-lapse fluorescence microscopy. We reasoned that assaying Cyclin B1-Venus levels over time in cells blocked in mitosis should provide a measure of the effectiveness with which the SAC could repress APC/C activity because weaker SAC activity should result in a higher rate of Cyclin B1 destruction. In agreement with other studies21, 26, we found that Taxol-treated cells spent less time in mitosis (555.9 ±345.6 min.) compared to cells treated with DMA (1101 ±410.1 min.) or nocodazole (1481 ±529 min.) (Fig. 2c), even though DMA produces similar kinetochore attachment defects to Taxol27, 28. The reason for this discrepancy was different rates of proteolysis: Cyclin B1-Venus levels fell faster in Taxol-treated cells compared to DMA, and faster in DMA than in nocodazole (0.33 μM) (Fig. 2d). This was not caused by a direct effect of the drugs on APC/C activity because when Mad2 was depleted by siRNA, Cyclin B1-Venus was degraded with the same kinetics in the different drugs (Fig. 2e). Instead, the kinetics of Cyclin B1-Venus destruction correlated with the strength of the checkpoint we had previously measured using the disappearance of Cyclin A2-Venus. The average slope of Cyclin B1-Venus destruction in different drug treatments correlated inversely (r2=0.9482) with the time cells spent in mitosis (Fig. 2f), indicating that the length of mitotic arrest likely depended on how effectively the APC/C was inhibited by the SAC. To test this further, we used serial dilutions the MPS1 inhibitor AZ3146 in cells treated with 0.33 μM nocodazole, and found that a progressive increase in AZ3146 concentration caused an increase in the rate of Cyclin B1-Venus destruction (Fig. 2g), and, as previously observed, a reduced mitotic delay29 (Supplementary Fig. 2g).
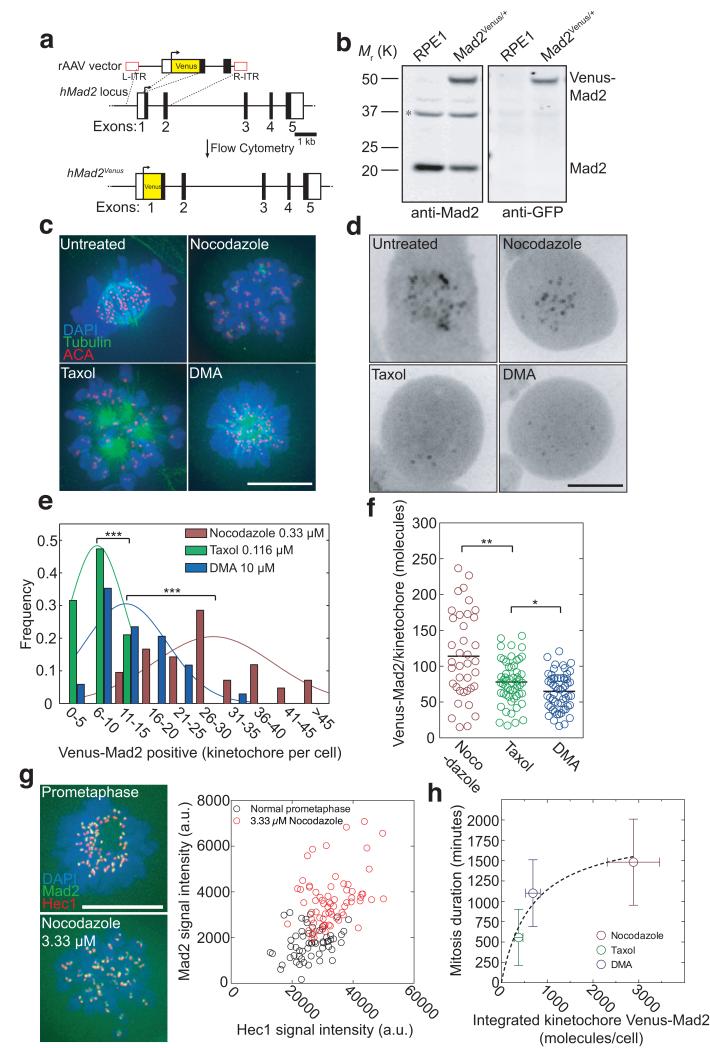
The differences in Cyclin A2 and Cyclin B1 degradation rates in response to different drugs indicated that the strength of the SAC was variable rather than ‘all or nothing’. To understand how the strength of the SAC could vary we needed a quantitative measure of the kinetochores that generated a SAC signal. To achieve this we used gene targeting to fuse the Venus ORF to the 5′ of exon 1 of one allele of the MAD2L1 gene. We tagged Mad2 at its N-terminus because a GFP-Mad2 fusion protein had previously been demonstrated to be functional in Drosophila30 Immunoblotting showed that Venus-Mad2 was expressed at a similar level to untagged Mad2 (Fig. 3a,b; Supplementary Fig. 3a,b) and high-resolution imaging confirmed that it was an accurate marker for unattached kinetochores (Supplementary Fig. 3c and d and movie 3).
Fig. 3. The amount of Mad2-positive kinetochores correlates with the length of a mitotic block.
(a) Venus ORF targeting into the hMAD2L1 locus by rAAV-mediated homologous recombination. The rAAV vector contained the coding sequence of Venus, which was inserted between two gene-specific homology arms to replace the ATG at the junction between non-coding (white square) and coding (black square) regions of exon 1 and two inverted terminal repeats (L-ITR, R-ITR; red rectangles). Integrants were selected by FACS. (b) Western blot analysis of parental RPE1 and a Venus-Mad2 clone. Cell lysates were probed with anti-Mad2 (left panel) and anti-GFP (right panel) antibodies. The asterisk indicates a cross-reacting band. Note the two forms of Mad2 in the Venus-Mad2 clone (the upper one uniquely recognized by the anti-GFP antibody). Molecular mass markers on the left. (c) Asynchronous RPE1 cells were treated for 12-16 hours with nocodazole, or Taxol or DMA, fixed with methanol and processed for immunofluorescence using anti-beta-tubulin (green) and ACA antibodies to stain the kinetochores (red). DNA was stained using DAPI (blue). Merged images are presented. Representative of 20-50 cells from at least two independent experiments. (d) Typical localization of Venus-Mad2 in living mitotic cells treated with different spindle poisons as in c). (e) Distribution of Venus-Mad2-positive kinetochores per cell following 12-16 hours of treatment with different drugs (n=42 cells for nocodazole; n=38 cells for Taxol and n=34 cells for DMA), representative of at least two independent experiments. (f) Scatter plots obtained by fluorescence correlation using data shown in Supplementary Fig. 4 of the number of Venus-Mad2 molecules per kinetochore in cells treated with different drugs (n=57 kinetochores from 8 cells for nocodazole; n=38 kinetochores from 11 cells for Taxol and n=55 kinetochores from 8 cells for DMA), representative of at least two independent experiments. The black line represents the mean. (g) Asynchronous (top) or 3.33 μM nocodazole-treated (bottom) RPE1 Venus-Mad2 cells were fixed in methanol and processed for immunofluorescence against Hec1 (red), and DNA stained with DAPI (blue). The signal intensity for Hec1, a stable kinetochore component, was used as a measure of kinetochore size. Kinetochore-associated Mad2 signals (green in image) were quantified and plotted as a function of the corresponding Hec1 fluorescence (n=56 kinetochores from 6 cells for asynchronous and n=60 kinetochores from 3 cells for 3.33 μM nocodazole). Data are representative of two independent experiments. (h) Plot of the relationship between the duration of mitosis (from Fig. 2c) and the integrated molecule number of Venus-Mad2 at kinetochores in the different poisons (from Fig. 3e,f). Error bars indicate s.d. and are representative of at least two independent experiments. Scale bars = 10 μm.
Treating RPE1 cells with Taxol or DMA or nocodazole gave the expected spindle and kinetochore phenotypes (Fig. 3c): Taxol-treated cells displayed collapsed spindles with hyperstable/elongated MTs; DMA-treated cells had monopolar spindles; nocodazole (0.33 μM)-treated cells lacked spindle MTs but contained clustered kinetochore MTs. Under these conditions, we found more kinetochores recruited Mad2 in cells treated with nocodazole (25.29 ±9.46) than with DMA (10.62 ±6.42) or Taxol (4.84 ±3.14) (Fig. 3d,e). These values were comparable to immunofluorescence measurements on the parental RPE1 cell line using an anti-Mad2 antibody (Supplementary Fig. 3d); therefore, Venus-Mad2 was an accurate marker for Mad2 recruitment to improperly attached kinetochores.
In addition to containing more Mad2-positive kinetochores, in nocodazole-treated cells the individual kinetochores recruited more Venus-Mad2 molecules (estimated as 114.0 ±60.4) than in cells treated with Taxol (77.9 ±30.1) or DMA (65 ±25.0) (Fig. 3f and Supplementary Fig 3e-j). These differences in protein numbers were not attributable to variations in the size of kinetochore since there was no correlation between the intensity of kinetochore-bound Venus-Mad2 and Hec1, a stable kinetochore protein (Fig. 3g). By contrast, the amounts of Mad2 recruited to improperly attached kinetochores inversely correlated with kinetochore microtubule abundance. The kinetochores in nocodazole-treated cells are mostly unattached (at 0.33 μM), whereas kinetochores do attach to MTs in Taxol- and DMA-treated cells but these attachments are destabilised by the error-correction machinery, thereby generating fluctuating numbers of unattached kinetochores.
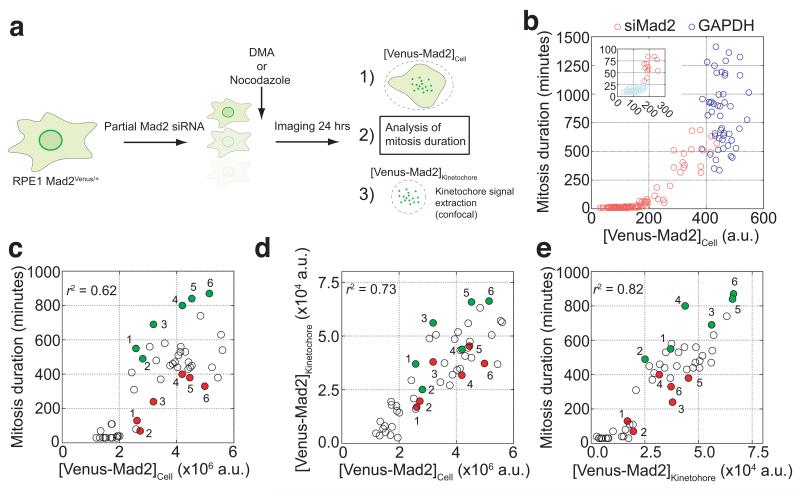
The combined numbers of Venus-Mad2 molecules and Mad2 foci per cell strongly correlated (r2=0.9123) with the extent of the mitotic delay in each drug treatment (Fig. 3h). This indicated that the recruitment of Mad2 to kinetochores was important for the strength of the SAC. To test this, we assayed the effect on the SAC of depleting Mad2 to different extents by sub-optimal siRNA treatment in RPE1 Mad2Venus/+ cells (Fig. 4a). Our endogenous targeting allowed us to determine the extent of Mad2 depletion in individual cells by measuring the total Venus-Mad2 fluorescence. Reducing total cellular Mad2 to ~30 to 40% of its normal level caused a graded reduction in the length of the checkpoint arrest (Fig, 4b), showing that under the same conditions, the amount of Mad2 did indeed influence the strength of the SAC. It was notable that once Mad2 was depleted to below 30-40% of normal the checkpoint was no longer functional and cells quickly exited mitosis (Fig. 4b,c). This high threshold amount of Mad2 required for the checkpoint surprised us because we had previously found the majority of Mad2 was free in the cell12, 31. The integrated Venus-Mad2 fluorescence at kinetochores after NEBD largely mirrored the total amount of Venus-Mad2 (Fig. 4d) and correlated with the duration of the mitotic arrest (Fig. 4e). This supported the idea that the amount of Mad2 recruited to kinetochores might determine the strength of the SAC, an idea that was strongly reinforced when we focused on cells that contained very similar amounts of total Venus-Mad2. We observed that the SAC was always stronger in those cells that recruited more Venus-Mad2 to kinetochores (Fig. 4c-e). Thus, we conclude that the amount of Mad2 recruited to kinetochores is a crucial determinant of the strength of the SAC.
Fig. 4. Reducing the level of Mad2 at kinetochores decreases the strength of the SAC.
(a) Schematic representation of experiments and analysis in b-e. RPE1 Mad2Venus/+ cells were subjected to partial siRNA against Mad2 for 24 hours before being imaged in the presence of DMA (10 μM) or nocodazole (0.33 μM). Measurements on total cellular fluorescence and thresholded kinetochore fluorescence were done 10 minutes after NEBD. (b) Graph displaying the duration of mitosis in DMA of each GAPDH (n=50 cells) or siMad2-treated cells (n=100 cells) analysed as a function of the corresponding total cellular fluorescence of Venus-Mad2 ([Venus-Mad2]Cell measured using wide-field microscopy. The inset expands the data points at the apparent threshold of Venus-Mad2, below which the checkpoint does not work (red vs green). Representative of two independent experiments. (c) Graph displaying the duration of mitosis in nocodazole of each cell analysed (n=50 cells) as a function of the corresponding total cellular fluorescence of Venus-Mad2 ([Venus-Mad2]Cell from spinning disk confocal imaging. (d) Graph displaying the relationship between total cellular fluorescence of Venus-Mad2 and integrated kinetochore fluorescence of Venus-Mad2 (n=50 cells). (e) Graph displaying the duration of mitosis of each cell analysed (n=50 cells) as a function of the corresponding integrated kinetochore fluorescence of Venus-Mad2 ([Venus-Mad2]Kinetochore. In panels c-e, cells that have similar total Mad2 levels were assigned matching numbers and the green color highlights the cell in each match that has more Venus-Mad2 at kinetochores, compared to the red cell which has less. Note that in all cases this resulted in different mitotic arrests (green, stronger checkpoint; red, weaker checkpoint. n>50 cells. Representative of two independent experiments.
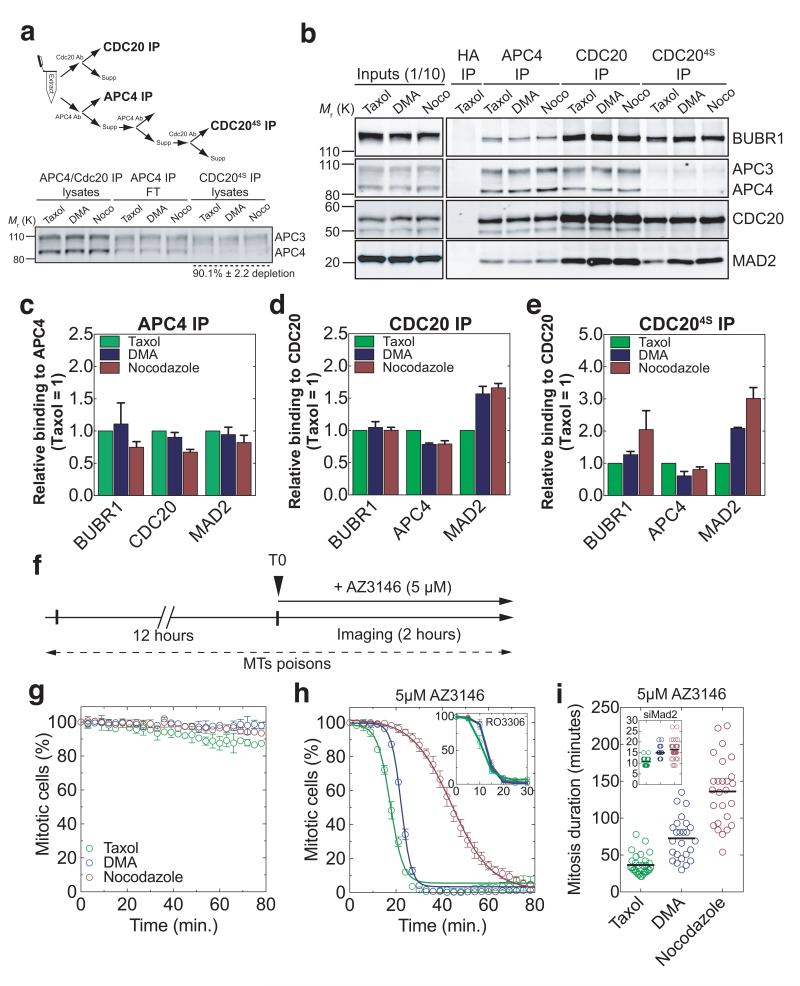
The molecular endpoint of the SAC is the incorporation of Cdc20 into the MCC4, 10, 11, 32, 33, therefore we examined the abundance of MCC components (Mad2, BubR1, and Cdc20) bound to the APC/C and Cdc20 in extracts of mitotic cells treated with different drugs (Fig. 5a). This showed that Cdc20 bound to more Mad2 in cells treated with nocodazole compared to DMA, and more in DMA compared to Taxol (Fig. 5b,d), whereas the amount of BubR1 bound to Cdc20 remained unchanged (Fig. 5b,d). We interpreted this as a change in the ratio of MCC complexes with and without Mad2, because we, and others, had previously found that Mad2 was substoichiometric to BubR1 and Cdc20 in the MCC10, 12, 13. To test this, we compared MCC that were free or bound to the APC/C by immunoprecipitation (Fig. 5a) and size exclusion chromatography (Supplementary Fig. 4a-c). This indicated that, at the molecular level, the strength of the SAC could be determined by the amount of MCC generated per unit time, and thus its concentration in the cell, and that binding to the APC/C was limiting. Whereas the levels of APC/C-bound MCC proteins remained approximately the same in all drug treatments, the levels bound to non-APC/C associated Cdc20 increased as the strength of the SAC increased. The strongest effect was seen in nocodazole-treated cells, where BubR1 accumulated ~2-fold and Mad2 ~3-fold (Fig. 5b,e & Supplementary Fig. 4b,c).
Fig. 5. Cdc20 incorporation into the MCC correlates with the strength of the SAC.
(a) Isolation of different complexes and Western blot analysis of the different lysates (n=5 extracts for nocodazole; n=4 extracts for Taxol and n=3 extracts for DMA). Error indicate s.e.m. and are representative of at least three independent experiments. (b) Anti-APC4 or anti-CDC20 immunoprecipitates (IP) from extracts of RPE1 cells treated with nocodazole (n=5 extracts), Taxol (n=4 extracts) or DMA (n=3 extracts) and isolated by mitotic shake-off 12-16 hours later. (c-e), The amount of MCC proteins that co-purified with c) APC4 or d-e) CDC20 were analysed by quantitative immunoblotting, normalised to the level in Taxol-treated cells, and displayed as bar graphs. Error bars indicate s.e.m. and are representative of at least three independent experiments. Note that the CDC20 immunoprecipitates in panel b) (last three lanes to the left) and e) were obtained from APC/C-free extracts (in panel a). (f) Schematic of experiments in panels g) and h). (g) Time-course of the percentage of mitotic cells after 12 hours of drug treatment (automatic detection, n>100 cells per condition). Error bars indicate s.d. and are representative of two independent experiments. (h) Same as g) after the addition of AZ3146 (5 μM) (automatic detection, n>100 cells per condition). Inset is the same as g) after addition of RO3306 (10 μM) (automatic detection, n>100 cells per condition). Error bars indicate s.d. and are representative of two independent experiments. (i), Scatter plot displaying the duration of mitosis of RPE1 Cyclin B1-Venus cells treated with AZ3146 (5μM) in the presence of nocodazole (n=26 cells), Taxol (n=30 cells) or DMA (n=25 cells). Inset represents similar conditions except that cells were treated with siRNA against Mad2 (n=28 cells for nocodazole; n=34 cells for Taxol and n=39 cells for DMA). Representative of at least two independent experiments.
To test this further, we arrested cells with nocodazole, DMA or Taxol, then inactivated the SAC with AZ3146 and monitored the kinetics of mitotic exit, predicting that cells with more MCC should take longer to exit mitosis because more time would be required for all the MCC to disassemble. In agreement with this, the rate at which cells exited mitosis was inversely proportional to the amount of the MCC in the cells: Taxol-treated cells exited faster (t1/2= 6.68 ±0.33 min.) than DMA-treated cells (t1/2= 9.00 ±0.19 min.), and DMA treated cells faster than nocodazole-treated cells (t1/2= 30.69 ±0.39 min.) (Fig. 5g,h). (Note that each half-life value was corrected by subtracting the minimum half-life for mitotic exit in the presence of a Cdk1 inhibitor RO3306 (Fig.5h, inset)). Although some of these differences might be attributed to the error-correction role of MPS129, 34, 35, 36, because cells “satisfy” the checkpoint quicker in Taxol or DMA compared to nocodazole, this would make only a minor contribution because AZ3146 reduced the mitotic delay by 95.4% for Taxol, 94.8% for DMA and 91.9% for nocodazole (Figure 2c and Figure 5i), a difference of only 3.5% between error-correction conditions and nocodazole.
Altogether, our data support a model in which the SAC does not behave as an ‘all or nothing’ toggle-switch. Instead, the SAC is like a rheostat: it can be activated to different levels, and thereby inhibit the APC/C to different extents, which dictates the length of a mitotic delay. The strength of the SAC depends upon the number of signaling centres (kinetochores), and critically upon the amount of Mad2 recruited to kinetochores, which probably determines the rate of MCC production (Supplementary Fig. 5a).
Our findings have implications for the control of mitosis. At the start of an open mitosis most kinetochores are unattached, which would produce a pulse of MCC to inhibit the APC/C. As chromosome attachments are established, the declining numbers of unattached kinetochores generate sufficient inhibitory complex to prevent premature anaphase, but since the MCC is constantly disassembled this will reduce the amount of MCC (Supplementary Fig. 5b). The progressive reduction in MCC level, allied with the role of protein phosphatases in chromosome attachment37 and SAC silencing38, 39, would contribute to the efficient coupling between correct bipolar attachment of chromosomes and rapid anaphase onset (Supplementary Fig. 5c; unperturbed). In drug-induced arrests, MCC production will be continuous, at a rate depending on the ability of the drug to prevent correct kinetochore attachments (Supplementary Fig. 5c; Taxol, DMA and nocodazole). The may explain why depleting p31Comet13, 31, 40, 41 and APC1531 , both negative regulators of the SAC, has only relatively mild effects in normally dividing cells (small pulse of MCC), while they are strongly additive in drug-induced arrests (continuous production of MCC).
Our finding that the SAC can be activated to different degrees by different drugs has implications both for anti-cancer therapies based on microtubule poisons and microtubule motor inhibitors16. Moreover, our observation that there is a critical threshold of Mad2 required for a functional SAC may be an important clue to the mechanism by which the MCC is generated and inhibits the APC/C, and for the observation that Mad2 is a haplo-insufficient tumour suppressor42.
Supplementary Material
Methods
Legends for supplementary figures and supplementary movies
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Acknowledgements
We are grateful to Prasad Jallepalli for reagents and advice, Fabien Cubizolles and Eric Nigg for generating the pAAV-Venus-Mad2 plasmid, and to Alexey Khodjakov and all our lab members laboratory for critical discussions. We would also like to thank Daniel Gerlich and Amalie Dick for sharing results prior to publication. This work was supported by a Programme grant from Cancer Research UK and a project grant from the BBSRC to JP. JP acknowledges core funding provided by the Wellcome Trust (092096) and CRUK (C6946/A14492).
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Weaver BA, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 10.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 15.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 16.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 17.Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geley S, et al. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolthuis R, et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 21.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Orth JD, et al. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–4616. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang HC, Mitchison TJ, Shi J. Stochastic competition between mechanistically independent slippage and death pathways determines cell fate during mitotic arrest. PLoS One. 2010;5:e15724. doi: 10.1371/journal.pone.0015724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Kenny AE, Brito DA, Rieder CL. Cells satisfy the mitotic checkpoint in Taxol, and do so faster in concentrations that stabilize syntelic attachments. J Cell Biol. 2009;186:675–684. doi: 10.1083/jcb.200906150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 31.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 33.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 36.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meadows JC, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagan RS, et al. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol Biol Cell. 2011;22:4236–4246. doi: 10.1091/mbc.E11-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods
Legends for supplementary figures and supplementary movies
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6