Abstract
Staphylococcal food poisoning is a gastrointestinal disorder caused by the consumption of food containing Staphylococcal enterotoxins. Staphylococcal enterotoxin A (SEA) is the most common enterotoxin recovered from food poisoning outbreaks in the USA. In addition to its enteric activity, SEA also acts as a potent superantigen through stimulation of T cells, although less is known about its interactions than the superantigens SEB, SEC and toxic shock syndrome toxin-1. To understand more about SEA:receptor interactions, and to develop toxin-detection systems for use in food testing, we engineered various SEA-binding receptor mutants. The extracellular domain of the receptor, a variable region of the beta chain (Vβ22) of the T-cell receptor, was engineered for stability as a soluble protein and for high affinity, using yeast-display technology. The highest affinity mutant was shown to bind SEA with a Kd value of 4 nM. This was a 25 000-fold improvement in affinity compared with the wild-type receptor, which bound to SEA with low affinity (Kd value of 100 µM), similar to other superantigen:Vβ interactions. The SEA:Vβ interface was centered around residues within the complementarity determining region 2 loop. The engineered receptor was specific for SEA, in that it did not bind to two other closely related enterotoxins SEE or SED, providing information on the SEA residues possibly involved in the interaction. The specificity and affinity of these high-affinity Vβ proteins also provide useful agents for the design of more sensitive and specific systems for SEA detection.
Keywords: directed evolution, food poisoning, Staphylococcal enterotoxin A (SEA), yeast display
Introduction
Staphylococcus aureus possesses a variety of virulence factors, including more than 20 Staphylococcal enterotoxins (SEs) and SE-like proteins (McCormick et al., 2001; Hennekinne et al., 2012). The canonical structure consists of an N-terminal, beta-barrel containing domain and a C-terminal domain that consists of a beta-grasp motif and an alpha helix which spans the center of the structure (Fields et al., 1996; Sundstrom et al., 1996a,b; Li et al., 1998, Petersson et al., 2002). In addition to their role as potent gastrointestinal toxins, S.aureus enterotoxins also mediate many hyper-inflammatory reactions associated with skin diseases, pneumonia, endocarditis and toxic shock, by acting as ‘superantigens’. The term superantigen was given to these toxins because their hyper-inflammatory properties are related to their ability to stimulate a very large fraction of the body's T cells, as compared with conventional peptide-major histocompatibility complex (MHC) antigens, resulting in massive release of inflammatory cytokines (Marrack and Kappler, 1990; Fraser and Proft, 2008). The mechanisms of this hyper-inflammatory process is now well known, involving the binding of the enterotoxins to the Vβ region of the T-cell receptor (TCR) (Fields et al., 1996; Li et al., 1998; Moza et al., 2007; Fernandez et al., 2011) and also to a class II MHC product on an antigen-presenting cell. This trimolecular interaction leads to the stimulation of a large fraction (e.g. 20%) of T cells because of the limited number of Vβ genes in human T-cell repertoire.
Characterization of various S.aureus-mediated food poisoning outbreaks throughout the world has indicated that the presence of Staphylococcal enterotoxin A (SEA) in food was most often associated with the illness (Evenson et al., 1988; Levine et al., 1996; Miwa et al., 2001; Asao et al., 2003; Ikeda et al., 2005). It has been estimated that the dose of SEA that is capable of causing the disease was a few hundred nanograms per individual (Evenson et al., 1988; Balaban and Rasooly, 2000). Although SEA was the first S.aureus enterotoxin discovered, from a strain isolated from a food poisoning outbreak (Bergdoll et al., 1959; Chu et al., 1966), the details of its interaction with its cognate Vβ receptor are largely unknown.
It is clear that other superantigens interact with their cognate Vβ receptors with low affinity (Kd in micromolar range) (Malchiodi et al., 1995; Khandekar et al., 1997; Andersen et al., 2001). In the present study, we have analyzed the binding of SEA to its cognate Vβ receptor (human Vβ22) and we have engineered soluble forms of both the wild-type receptor and a high-affinity human Vβ22. The approach followed several recent studies in which yeast surface display was coupled with mutagenesis and superantigen-binding selections to isolate mutants of various Vβ proteins (mouse Vβ8, human Vβ2), that were stabilized for expression as soluble proteins, and that had enhanced affinity for SEB, SEC or toxic shock syndrome toxin-1 (TSST-1) (Kieke et al., 2001; Buonpane et al., 2005; Buonpane et al., 2007; Wang et al., 2010; Mattis et al., 2013). These engineered Vβ proteins also enabled the structural characterization of their interactions with the cognate enterotoxin (Moza et al., 2007), as well as studies on the molecular basis of affinity improvement (Bonsor et al., 2011).
Based on earlier studies showing that human T cells expressing the Vβ22 region were selectively expanded when stimulated by SEA (Llewelyn et al., 2006), we expressed human Vβ22 in the yeast-display system. As with other Vβ domains, the wild-type Vβ22 was not expressed well on the surface of yeast. However, we selected for increased expression among random Vβ22 point mutants, and identified several framework mutations in Vβ22 that enabled surface expression, and soluble expression. Like other superantigen:Vβ interactions, the Vβ22:SEA interaction had low affinity (Kd = 100 μM). Affinity engineering by selection from a collection of site-directed Vβ22 libraries revealed the importance of the complementarity determining region 2 (CDR2) region of Vβ22 in its interaction with SEA. Mutations in CDR2 alone were capable of increasing the SEA affinity by 25 000-fold. The highest affinity mutant bound to SEA with a Kd value of 4 nM, and exhibited a high level of specificity for SEA. The results provide insight into the mode of interaction of Vβ22 with SEA, and the affinity-engineered Vβ22 represents a small specific protein domain that can be used in more sensitive detection platforms for SEA.
Materials and methods
Cloning and expression of human Vβ22 on yeast cell surface
The gene encoding human Vβ22, synthesized and codon optimized for expression in both Escherichia coli and Saccharomyces cerevisiae (Genscript USA Inc.), was subcloned as an Aga-2 fusion (under the control of GAL1 promoter) into yeast display vector pCT302 that contains an N-terminal hemagluttinin (HA) and a C-terminal myc (c-myc) tag (Fig. 1A). AGA-2 is a yeast mating agglutinin protein and allows the expression of the protein of interest on the yeast cell surface as a fusion protein (Boder and Wittrup, 1997). Expression was induced by growth of yeast cells in medium containing galactose at 20°C for 36–48 h. Surface expression was analyzed by flow cytometry.
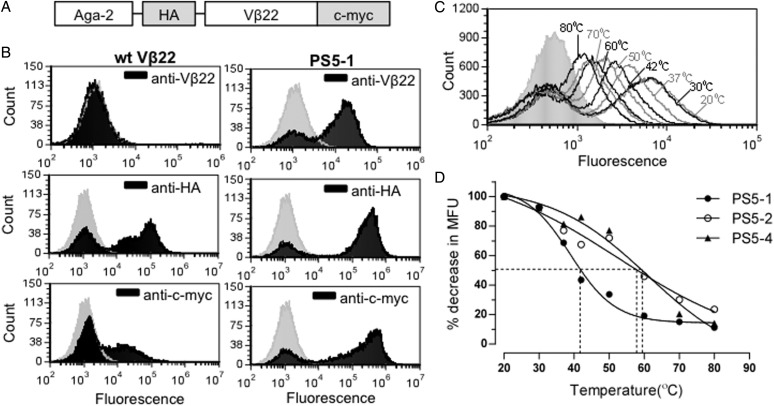
Fig. 1.
Yeast surface display of human Vβ22. (A) Schematic of yeast display construct containing human Vβ22 gene. (B) Flow cytometric analysis of induced yeast cells expressing either wt Vβ22 or PS5-1 mutant protein. Yeast cells were stained with anti-Vβ22, anti-HA or anti-c-myc (black) followed by a secondary antibody conjugated to a fluorophore. The staining profile of the cells with secondary antibody only is represented by the gray peak. Data shown are representative of three or more experiments. (C) Flow cytometric analysis of the stabilized Vβ22 mutant PS5-1 after yeast cells were incubated at various temperatures for 30 min prior to incubation with primary antibody (anti-Vβ22). (D) Thermostability analysis of stabilized Vβ22 mutants (PS5-1, PS5-2 and PS5-4) by flow cytometry, as described in (C). The point of the 50% melting temperatures of individual mutants are indicated.
Construction of random mutagenesis library and selection
Error-prone polymerase chain reaction (PCR) was used to generate PCR products with random mutations, which were then electroporated into the S.cereivisiae EBY100 strain, along with NheI and XhoI digested pCT302 vector (Richman et al., 2009). The library in yeast was generated by homologous recombination, followed by expansion in SD-CAA (Trp−) media to allow the growth of yeast cells that were transformed with pCT302 vector. After induction, the yeast library was incubated with anti-Vβ22 antibody (Clone: IMMU546, Beckman Coulter) for 1 h on ice. Cells were then washed with 0.5 ml phosphate-buffered saline (containing 0.5% bovine serum albumin), followed by incubation with goat anti-mouse IgG F(ab′)2 Alexa Fluor 488 or 647 secondary antibody (Invitrogen). Several rounds of selection were performed using a fluorescence-activated cell sorting (FACS) Aria cell sorter (BD Bioscience). Individual yeast clones obtained after plating of sorted cells on selective medium, were subjected to flow cytometry analysis with anti-Vβ22 antibody, anti-HA antibody (Clone HA.11, Covance) and anti-c-myc antibody (Invitrogen), followed by incubation with secondary antibody. Selected clones were subjected to thermostability analysis by incubating cells at higher temperatures (up to 80°C for 30 min) before incubation with anti-Vβ22 antibody.
Construction of yeast-display libraries and mutagenesis
Thermostable Vβ22 mutants obtained from the error-prone library were used as templates for site-directed mutagenesis libraries. Degenerate primers were used to introduce diversity in CDR2 region of Vβ22. PCR products were electroporated into EBY100, along with NheI and XhoI digested pCT302 vector for generation of three 4-codon libraries by homologous recombination. Site-directed mutations were generated at residues 30, 49 and 52 using Quikchange Lightning Kit (Stratagene) as described by the manufacturer.
Screening of yeast-display libraries
Yeast-display libraries were mixed in equal proportion and subjected to several rounds of flow sorting-based selection, with decreasing concentrations of biotinylated-SEA (Toxin Technology, Inc.) (1 µM to 10 nM) followed by staining with Streptavidin Alexa Fluor 488 or 647 conjugate (Invitrogen) at dilution. Individual yeast clones were isolated and analyzed by DNA sequencing and by flow cytometry for protein expression and SEA binding.
Expression of soluble Vβ proteins in E.coli
Specific Vβ22 mutants were subcloned into a pET28a expression vector (Novagen) for protein expression in E.coli BL21 (DE3) cells as inclusion bodies. Proteins were refolded (by slow dilution) from denatured inclusion bodies, followed by affinity purification with Ni agarose resin (Qiagen) and HPLC (Biocad Sprint) using a Superdex 200 (GE Healthcare) size-exclusion column as described previously (Buonpane et al., 2007).
Binding of soluble Vβ proteins to SEA
Binding of soluble Vβ to SEA was examined by enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR). For ELISA, soluble Vβ22 mutant proteins were coated on ELISA wells (5 μg/ml), followed by incubation with varying concentrations of biotinylated-SEA and by streptavidin-conjugated horseradish peroxidase (HRP) enzyme. Bound SEA was detected by measuring colorimetric product formed (A450 nm) upon addition of 3, 3′, 5, 5′ – Tetramethylbenzidine (TMB) substrate (KPL, Inc.). Affinity constants and kinetic parameters for Vβ:SEA binding were determined by SPR using a Biacore T100 instrument (GE Healthcare). 500RU of recombinant, unlabeled SEA was immobilized on a CM5 sensor chip. Various concentrations of soluble Vβ proteins were injected over the sensor surface and response was recorded. Global kinetic fits of the data were performed using the BiaEvaluation 4.1 software.
ELISA-based analysis for detection of unlabeled SEA, and for cross-reactivity with other superantigens
To measure binding of unlabeled SEA, an SEA detection assay was used in a ‘capture ELISA’ format wherein soluble Vβ22 mutant proteins, coated on ELISA wells (5 μg/ml), and were used to capture various concentrations of recombinant, unlabeled SEA. This was followed by incubation with anti-SEA (rabbit whole antiserum) (Sigma-Aldrich), followed by goat anti-rabbit IgG-HRP (Sigma-Aldrich), and finally TMB substrate to yield a colored product whose absorbance at 450 nm was recorded.
For cross-reactivity studies, induced yeast cells expressing Vβ protein on the surface were incubated with a mixture of 100 nM recombinant biotinylated-SEA and various concentrations of non-biotinylated SEA, SED and SEE (Toxin Technology, Inc.) for 1 h on ice. The amount of biotinylated-SEA bound to Vβ, was detected using Streptavidin-Alexa Fluor 647 conjugate.
Results
Expression of stable, human Vβ22 on the yeast cell surface
The human Vβ22 gene was cloned in frame with the yeast cell wall protein Aga-2, with flanking HA and c-myc epitope tags, in the yeast display vector pCT302 (Fig. 1A). Although yeast cells containing the wild-type Vβ22 (wt Vβ22) construct showed detectable levels of the HA and c-myc tags, an antibody to the Vβ22 region did not detect the fusion on the surface of yeast cells (Fig. 1B, left). These results suggested that the Vβ22 protein was not folded properly, as has been observed with several other Vβ domains (Kieke et al., 1999; Buonpane et al., 2005; Weber et al., 2005).
In order to determine if we could isolate a stable Vβ22 protein, we subjected the wt Vβ22 gene to random mutagenesis by generating an error-prone PCR library. The library was selected through five rounds of FACS with a saturating concentration of anti-Vβ22 antibody. A population of positive cells was selected, and various clones were analyzed by flow cytometry and sequencing. Three different mutants (PS5-1, PS5-2 and PS5-4) that were expressed on the yeast cell surface as detected with the anti-Vβ22 antibody were isolated (Fig. 1B, right panel, and data not shown). The levels of HA and c-myc were also increased in these stabilized constructs, indicating a greater number of the fusions were expressed and exported to the cell surface when the Vβ22 domain was stabilized.
To further compare the stability of the three unique Vβ22 mutants, each was subjected to an irreversible thermal denaturation assay in which yeast cells were incubated at various temperatures, followed by flow cytometry with the anti-Vβ22 antibody (Fig. 1C and D) (Orr et al., 2003). The 50% melting temperatures of the proteins ranged from 40°C (PS5-1) to 60°C (PS5-2, PS5-4). The PS5-2 and PS5-4 Vβ22 mutants exhibited the highest level of stability (Fig. 1D).
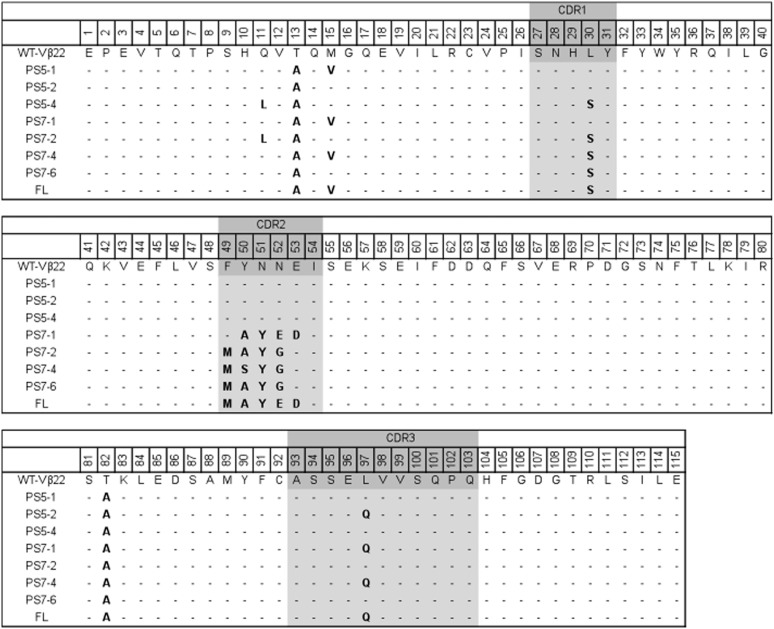
Sequence analysis of individual Vβ22 mutants (Fig. 2) indicated that all three mutants acquired two identical framework mutations, T13A and T82A, suggesting that these mutations were important in stabilization of the Vβ22 domain on the yeast surface. The increased stability of PS5-2 and PS5-4 appear to be due to one or more of the following mutations: Q11L (framework), L30S (CDR1) and/or L97Q (CDR3). Interestingly, mutations in the framework regions resulted in substitution of polar amino acids (Thr, Gln and Met) with hydrophobic amino acids (Ala, Leu and Val). In contrast, mutations in CDRs resulted in substitution of a hydrophobic amino acid (Leu) with polar amino acids (Ser and Gln). Protein modeling and structural comparison with other Vβ domains indicated that the mutations in framework regions were located on the face opposite the antigen binding face of the Vβ (CDRs) (Supplementary Fig. S1). It is interesting to note that mouse Vβ8.2 and human Vβ2.1, which have been crystallized in complex with SEB (and SEC3) and TSST-1, respectively, also contain hydrophobic residues at position 13 (Ala/Val) and position 82 (Ala).
Fig. 2.
Sequences of various Vβ22 mutants. PS5-1, PS5-2 and PS5-4 represent stabilized Vβ22 mutants isolated from error-prone library. These mutants were used as templates to generate CDR2 libraries used to isolate four different mutants, PS7-1, PS7-2, PS7-4 and PS7-6, after sorting multiple rounds with biotinylated-SEA. Two mutations (F49M and L30S) were introduced into PS7-1 to yield the mutant designated FL. Mutated residues are in bold; CDR regions are highlighted.
Generation of high-affinity, SEA-binding Vβ22 mutants by directed mutagenesis
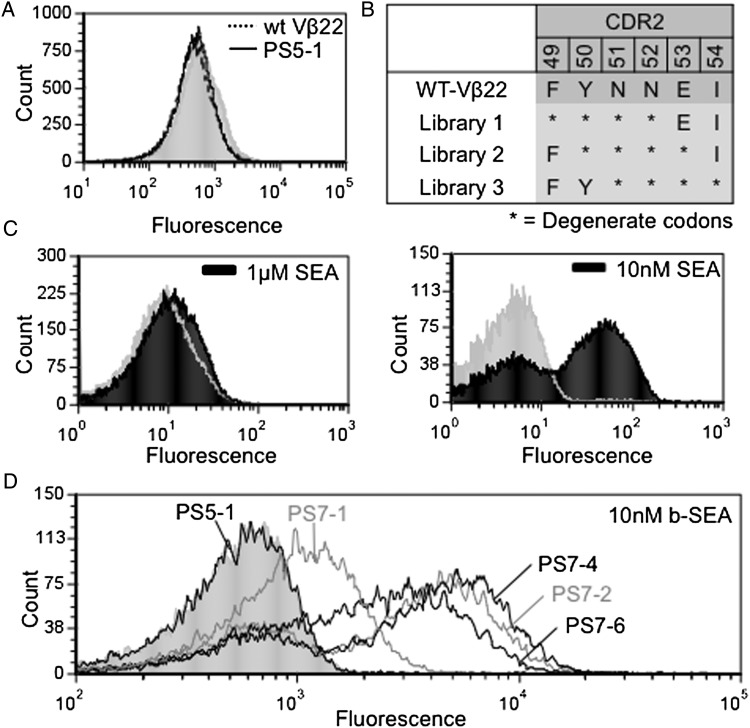
Binding analysis by flow cytometry indicated that the stabilized Vβ22 mutant, PS5-1 bound to biotinylated-SEA with low affinity (Kd > 1 μM), which is the detection limit of this technique (Fig. 3A). In order to engineer Vβ22 mutants for higher SEA-binding affinity, stabilized Vβ22 mutants PS5-1, PS5-2 and PS5-4 were used as templates for various site-directed mutant libraries. In the absence of a crystal structure of the SEA:Vβ complex, we elected to start by generating libraries in the CDR2, which is known to be central in the binding of other superantigens to their cognate Vβ receptors (Fields et al., 1996; Li et al., 1998; Fernandez et al., 2011; Nur-ur Rahman et al., 2011). Three CDR2-directed mutagenesis libraries were generated, each in a contiguous stretch of four amino acids (Fig. 3B). The three libraries, consisting of ∼2 × 107 independent clones each, were combined and subjected to consecutive rounds of sorting with decreasing concentrations of biotinylated-SEA. As expected, incubation of the pre-sorted CDR2 libraries with 1 μM biotinylated-SEA did not yield detectable staining (Fig. 3C, left). However, a distinct population of cells that bound to 10 nM SEA was obtained after the seventh sort (Fig. 3C, right).
Fig. 3.
Generation of Vβ22 mutants that bind SEA with higher affinity. (A) Flow cytometry histograms for wt Vβ22 (dotted line) and PS5-1 (solid line) with 2.5 μM biotinylated-SEA followed by Streptavidin-Alexa 488. (B) Residue positions of three site-directed mutagenesis CDR2 libraries used to select higher affinity SEA binders. (C) Flow cytometry histograms of a population of Vβ22 mutants after sorting the combined CDR2 libraries with 10 nM biotinylated-SEA (right), compared with unsorted CDR2 libraries stained with 1 μM biotinylated-SEA (left). (D) Flow cytometry histograms for individual clones (PS7-1, PS7-2, PS7-4 and PS7-6) isolated after the seventh sort of the Vβ22 CDR2 libraries, incubated with10 nM biotinylated-SEA. Fluorescence with secondary reagent only (the absence of SEA) are represented by gray (filled) peak in each histogram. Data shown are representative of three experiments.
Various clones from the sorted CDR2 libraries were analyzed by flow cytometry and sequenced. Four different mutants, designated PS7-1, PS7-2, PS7-4 and PS7-6, were verified to have improved binding of biotinylated-SEA, compared with the stabilized Vβ22 mutant, PS5-1 (Fig. 3D). The binding to biotinylated-SEA was inhibited with unlabeled SEA (see below), indicating that the binding was not due to biotin. Sequence analysis (Fig. 2) indicated that all the mutants retained the conserved T13A and T82A stabilizing mutations. The PS7-1 mutant was derived from the 50–53 CDR2 library, whereas PS7-2, PS7-4 and PS7-6 were all derived from the 49–52 CDR2 library. The latter three mutants also retained the CDR1 mutation, L30S, that was important in surface expression and stability as shown by its introduction into mutant PS7-1 and staining with either SEA (Fig. 4) or anti-Vβ22 (Supplementary Fig. S2).
Fig. 4.
Effect of mutations at residues 30 and 49 in PS7-1: generation of FL mutant. (A) Flow cytometry histograms for binding of PS7-1 (top) and FL mutant (bottom) with various concentrations of biotinylated-SEA. Fluorescence in the absence of SEA is represented by gray (filled) peak. (B) SEA-binding titration of various mutants of PS7-1. MFUs are plotted on the y-axis and the concentrations of SEA are plotted on the x-axis. Data shown are representative of two experiments.
Importance of Vβ CDR2 mutations in high-affinity SEA binding
Conservation in the mutations (F49M, Y50S/A, N51Y, N52E/G and E53D) isolated among the four isolates suggested that they were important in high-affinity binding. In order to study the roles of several of these mutations in SEA binding, further mutagenesis studies were conducted. Since the PS7-1 mutant lacked the F49M mutation, we reasoned that its incorporation into PS7-1 might increase its affinity further. Although the maximum surface levels of PS7-1 and PS7-1-F49M were similar, and below that of L30S mutant, the F49M mutation, resulted in a modest increase in affinity, from 30 to 10 nM, for PS7-1 and PS7-1-F49M, respectively (Fig. 4B). As indicated, the introduction of the stabilizing mutation L30S into PS7-1 increased its surface expression significantly (Fig. 4B) and also had a modest effect on affinity (from 30 to 20 nM, for PS7-1 and PS7-1-L30S, respectively). Accordingly, the F49M and L30S mutations were both incorporated into PS7-1 to yield the mutant called ‘FL’. SEA-binding titrations of FL compared with PS7-1 (Fig. 4A and B) indicated that the FL mutant exhibited both higher maximum median fluorescence unit (MFU) for SEA binding (higher surface levels) and an increase in binding affinity compared with PS7-1 (Fig. 4B). Staining of the mutants with anti-Vβ22 antibody supported the finding that the F49M mutation caused a modest increase in surface expression, whereas the L30S mutation yielded a more significant increase (Supplementary Fig. S2). Thus, the FL mutant appears to exhibit both the higher surface levels and higher affinity compared with PS7-1, with an estimated apparent Kd of 15 nM.
The PS7-1 mutant contained the N52E mutation, whereas the PS7-2,-4,-6 mutants contained the N52G mutation. To examine the role of position 52 in the high-affinity state, we generated an E52G mutation in PS7-1, and the G52E mutation in the three PS7-2,-4,-6 mutants. These four mutants were examined by flow cytometry with 10 nM SEA and each of the reciprocal mutations showed a loss of SEA binding, to background levels (Supplementary Fig. S3). Thus, the glutamate at position 52 in PS7-1 and the glycine at the same position in PS7-2, PS7-4 and PS7-6 are important residues for SEA binding, but within the context of the surrounding mutations in these clones.
Finally, we suggest that the presence of the identical Y50A mutation in PS7-1 and PS7-2 may indicate a requirement for space at this position in order to accommodate the conserved tyrosine mutation at position 51. Further, the importance of an acidic residue at position 53 was exemplified by the fact that the wild-type residue is a glutamate (and present in PS7-2,-4,-6), whereas in PS7-1 it was mutated to aspartate. Accordingly, the overall charge of the CDR2 loop in PS7-1 increased to -2, compared with wt Vβ22 and the other Vβ22 mutants. This could contribute to improved electrostatic interactions with SEA, and/or the greater protein surface charge may enhance stability.
Expression of soluble human Vβ22 protein and SEA-binding analysis
Previous studies have indicated that mutants expressed at higher levels on the surface of yeast are also expressed at higher levels in secretion or refolding systems (Shusta et al., 1999; Weber et al., 2005; Jones et al., 2006). In order to study the SEA-binding properties of Vβ22, several of the Vβ mutants (PS5-1, PS7-1 and FL) were expressed in inclusion bodies in E.coli, solubilized, refolded and subjected to gel filtration. The proteins expressed and refolded from E.coli migrated on an sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gel at the expected molecular weight of about 13 kDa (Fig. 5A and data not shown).
Fig. 5.
Binding analysis of soluble Vβ22 proteins. Stabilized Vβ22 (PS5-1) and high-affinity mutant (FL) were refolded from E.coli inclusion bodies and purified by Ni-NTA affinity chromatography and size-exclusion chromatography. (A) SDS–PAGE analysis of purified Vβ22 proteins. (B) ELISA-based titration for binding of immobilized Vβ22 mutant proteins to recombinant, biotinylated SEA. Purified Vβ22 mutant proteins were coated on ELISA plates at 5 μg/ml, followed by incubation with biotinylated-SEA. Bound SEA was detected using streptavidin-conjugated HRP. Data shown are representative of two or more experiments. (C) SPR-based binding analysis for the interaction between immobilized recombinant, unlabeled SEA and PS5-1 soluble protein. Non-linear regression analysis of maximal response versus concentration is depicted. (D) SPR sensorgram for the interaction between affinity matured Vβ22 mutant FL and 500 RU immobilized recombinant SEA (unlabeled). Binding affinity constants are indicated. (E) Capture ELISA to detect unlabeled SEA. Soluble Vβ22 mutant proteins were coated on the ELISA plates at 5 μg/ml. This was used to capture various concentrations of recombinant, unlabeled SEA. Bound SEA was detected with anti-SEA antibody (whole antiserum, rabbit), followed by goat anti-rabbit IgG-HRP.
Binding of the soluble Vβ proteins to SEA was studied by ELISA and SPR. The format of the ELISA titrations do not allow an accurate Kd determination due to washes in between detection, but a minimum Kd value for proteins that have adequate dissociation kinetics can be estimated. From these results, the soluble, monomeric PS5-1 protein (equivalent to the wild-type SEA-binding domain, with the stabilizing mutations for expression) did not bind detectably (Fig. 5B). In contrast, the soluble, monomeric high-affinity mutants PS7-1 and FL bound to SEA with minimum estimated Kd values of 55 ± 20 and 30 ± 5 nM, respectively (Fig. 5B). The PS5-1 and FL mutants were also analyzed by SPR. Based on SPR equilibrium measurements, the Kd value of the PS5-1 protein was ∼100 µM (Fig. 5C), similar to the low affinities of other superantigens for their Vβ regions. The SPR-based measurement of the Kd value for the FL mutant was 4 nM, a 25 000-fold improvement in affinity (Fig. 5D).
The affinity of the FL Vβ protein allows one to use it as a probe for detection of SEA in samples. Accordingly, we developed a capture ELISA assay for detection of SEA, in which immobilized high-affinity FL mutant was used to capture unlabeled SEA. Polyclonal anti-SEA was then added to detect the bound SEA. Without any optimization, this assay detected SEA at a concentration of 1–10 nM (∼100 ng/ml) (Fig. 5E).
Specificity of superantigen binding by Vβ22 mutant FL
Since the SEE and SED toxins are in the same group as SEA, and exhibit 83 and 48% sequence identity with SEA, respectively, we assessed cross-reactivity of the Vβ22 FL mutant with these related enterotoxins. For this purpose, we used a competition assay with yeast cells that expressed the FL mutant. In this assay, biotinylated-SEA was incubated with various concentrations of unlabeled recombinant SEA, SED or SEE and the mixture was added to the FL-yeast cells. Bound biotinylated-SEA was detected with streptavidin-PE by flow cytometry. SEA was able to completely inhibit the binding at higher concentrations, and detectable inhibition was observed at concentrations as low as 5 nM. In contrast, binding (competition) of SED and SEE by the FL mutant was not detected even at the highest concentration, 500 nM (Fig. 6). This result suggests that the Vβ22 FL mutant exhibits a high level of specificity for SEA.
Fig. 6.
High-affinity Vβ22 mutant FL does not cross-react with related toxins SED and SEE. Induced yeast cells expressing FL protein on the surface were incubated in a mixture of 100 nM recombinant biotinylated-SEA and various concentrations of non-biotinylated SEA, SED and SEE. Bound biotinylated-SEA was detected using streptavidin-Alexa 647 conjugate.
Discussion
Among the various exotoxins secreted by S.aureus, enterotoxin A has been most often associated with food poisoning outbreaks involving S.aureus (Evenson et al., 1988; Levine et al., 1996; Miwa et al., 2001; Asao et al., 2003; Ikeda et al., 2005). For this reason, there has been interest in developing rapid, sensitive assays for food safety testing. However, SEA is also known to act as a superantigen, inducing hyper-inflammatory reactions in conditions such as endocarditis and toxic shock (McCollister et al., 1990; Ellis et al., 2003). In the present study, our goal has been to understand more about the molecular interactions involved in the superantigen activities, but the engineered, high-affinity receptors developed in this effort also have dual use as specific, sensitive detecting agents.
Like other superantigens, the mechanism by which SEA exerts hyper-inflammatory effects involves the simultaneous binding of the enterotoxin to class II MHC molecules on antigen-presenting cells and to the Vβ domain of the TCR present on T cells. Co-crystallization of other S.aureus superantigens with Vβ receptors has revealed different topologies of binding (Fields et al., 1996; Li et al., 1998; Moza et al., 2007; Fernandez et al., 2011), but in each case the center of the interface has involved CDR2 (Nur-ur Rahman et al., 2011). Thus, our efforts with the cognate receptor for SEA, Vβ22, focused our initial libraries on this CDR2 loop. If we had not isolated higher affinity variants within these libraries, we might have concluded that the Vβ22:SEA interaction differs substantially from other superantigens (SEB, SEC, TSST-1 and SpeA), for which we have uniformly isolated higher affinity Vβ variants from CDR2 libraries (Kieke et al., 2001; Buonpane et al., 2005; Buonpane et al., 2007; Wang et al., 2010; Mattis et al., 2013). However, we were able to isolate CDR2 mutants with up to a 25 000-fold improvement in affinity, supporting the notion that this CDR2 loop of Vβ22 also resides at the interface of its interaction with SEA.
Unlike many VH domains, Vβ domains are typically unable to be expressed in soluble form, or on the surface of yeast (Kieke et al., 1999; Buonpane et al., 2005). We have suggested that one of the key reasons for this is that Vβ residues that are normally buried at the interface with the constant region domain of the β-chain, are destabilized when exposed in the isolated Vβ domain. Consistent with this suggestion, the Vβ22 domain shown here was more stable on the surface of yeast when several key mutations were made in the region opposite the CDR (Supplementary Fig. S1), where the β-chain constant region would normally be located. Somewhat surprisingly, these stabilizing mutations in the framework of Vβ22 were substitutions to hydrophobic residues. One might have predicted for a region that is normally buried (e.g. at the Vβ:Cβ interface) the residues would prefer to be more hydrophilic when exposed. Interestingly, although, some other Vβ regions do normally contain similar or identical same amino acid residues identified here in the mutated Vβ22 domains.
The yeast surface levels of the Vβ22 domain were also increased, irrespective of SEA binding, by two CDR loop mutations, L30S and L97Q. L30S was shown especially to have a significant effect on the total surface levels of Vβ22 protein. These changes may act by increasing the hydrophilicity of these exposed loops. We have shown previously that CDR1 mutations also had a significant effect on stabilization of a Vα:Vβ single-chain TCR (Weber et al., 2005), and other Vα or Vβ domains have also been stabilized through CDR mutations (Richman et al., 2009).
The low affinity of SEA for the stabilized Vβ22 domain (Kd value of 100 µM) is similar to that measured for other superantigen interactions such as SEB:Vβ8.2 (Malchiodi et al., 1995; Buonpane et al., 2007). Despite this low affinity, superantigens are capable of potent activity in part because only a small fraction of the surface TCRs on a T cell needs to be bound in order to trigger activity (Schodin et al., 1996). In addition, ternary interactions with Vβ and class II MHC can further stabilize the active complex (Andersen et al., 1999). Thus, SEA appears to follow the same principles of low-affinity interactions with the TCR as with other superantigens.
The structural basis of how high-affinity Vβ22 mutants bind to SEA remains to be determined, but it is interesting that the two adjacent aromatic residues at positions 49 and 50 in the wild-type V region, were both mutated to other residues (Met and Ala, respectively), whereas the adjacent residue at 51 was mutated to an aromatic residue (Tyr). These substitutions may allow optimal positioning of the Tyr51 side chain for stacking interactions with SEA. Additional mutations at 52 and 53 in the FL mutant yielded two adjacent negative charged residues (N52E and E52D), which could play a role in enhanced electrostatic interactions with SEA.
The putative TCR binding site on SEA has been proposed to include T21, N25, Y94, Y205, N207 and T208 (Hudson et al., 1993; Mollick et al., 1993; Swaminathan et al., 1995; Antonsson et al., 1997). These residues are closely located in the tertiary structure of SEA (Supplementary Fig. S1), and could form the binding site for Vβ. The aromatic residue (Y51) in the CDR2 region of FL could potentially be involved in π-stacking interactions with Y94 or Y205 from SEA. The only positions within or near these six SEA residues that differ between SEA and both SED and SEE are positions 206 and 207 (Fig. 7). Interestingly, both SED and SEE have negatively charged residues at position 207, which could pose an electrostatic repulsion with negative charges (residues 52 and 53) in the Vβ22 FL mutant's binding site.
Fig. 7.
Sequence alignment of SEA, SED and SEE. Putative TCR binding residues of SEA are boxed.
Finally, the Vβ22 protein FL, with a 25 000-fold increase in affinity for SEA compared with the wild-type Vβ, was capable of being expressed, purified and immobilized as a detection agent for SEA. We propose that this engineered, high-affinity Vβ22 mutant protein can be used in the development of a highly specific detection system for SEA, perhaps as a constituent in a multiplex system for simultaneous detection of multiple toxins in food (Tallent et al., 2013).
Supplementary data
Supplementary data are available at PEDS online.
Conflict of interest: D.M.K. co-founded a company called ImmuVen that has acquired rights from the University of Illinois for some of the T-cell receptors engineered in his lab.
Funding
This work was supported by National Institutes of Health (grants R43 AI102432 (to D.M.K.) and R01 AI065690 (to E.J.S.)), as well as a grant from the National Institutes of Health-supported Great Lakes Regional Center for Excellence in Biodefense and Emerging Diseases (U54 AI57153 to D.M.K).
Supplementary Material
Acknowledgements
We thank the staff of the Flow Cytometry Facility and Core DNA Sequencing Facility at University of Illinois Urbana Champaign for technical assistance and Ningyan Wang, Daiva Mattis, Jennifer Stone and Lionel Low for helpful discussions.
References
- Andersen P.S., Geisler C., Buus S., Mariuzza R.A., Karjalainen K. J. Biol. Chem. 2001;276:33452–33457. doi: 10.1074/jbc.M103750200. [DOI] [PubMed] [Google Scholar]
- Andersen P.S., Lavoie P.M., Sekaly R.P., Churchill H., Kranz D.M., Schlievert P.M., Karjalainen K., Mariuzza R.A. Immunity. 1999;10:473–483. doi: 10.1016/s1074-7613(00)80047-3. [DOI] [PubMed] [Google Scholar]
- Antonsson P., Wingren A.G., Hansson J., Kalland T., Varga M., Dohlsten M. J. Immunol. 1997;158:4245–4251. [PubMed] [Google Scholar]
- Asao T., Kumeda Y., Kawai T., Shibata T., Oda H., Haruki K., Nakazawa H., Kozaki S. Epidemiol. Infect. 2003;130:33–40. doi: 10.1017/s0950268802007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N., Rasooly A. Int. J. Food Microbiol. 2000;61:1–10. doi: 10.1016/s0168-1605(00)00377-9. [DOI] [PubMed] [Google Scholar]
- Bergdoll M.S., Sugiyama H., Dack G.M. Arch. Biochem. Biophys. 1959;85:62–69. doi: 10.1016/0003-9861(59)90447-3. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Nat. Biotech. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bonsor D.A., Postel S., Pierce B.G., Wang N., Zhu P., Buonpane R.A., Weng Z., Kranz D.M., Sundberg E.J. J. Mol. Biol. 2011;411:321–328. doi: 10.1016/j.jmb.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonpane R.A., Churchill H.R., Moza B., Sundberg E.J., Peterson M.L., Schlievert P.M., Kranz D.M. Nat. Med. 2007;13:725–729. doi: 10.1038/nm1584. [DOI] [PubMed] [Google Scholar]
- Buonpane R.A., Moza B., Sundberg E.J., Kranz D.M. J. Mol. Biol. 2005;353:308–321. doi: 10.1016/j.jmb.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Chu F.S., Thadhani K., Schantz E.J., Bergdoll M.S. Biochemistry. 1966;5:3281–3289. doi: 10.1021/bi00874a030. [DOI] [PubMed] [Google Scholar]
- Ellis M., Serreli A., Colque-Navarro P., Hedstrom U., Chacko A., Siemkowicz E., Mollby R. J. Med. Microbiol. 2003;52:109–112. doi: 10.1099/jmm.0.05003-0. [DOI] [PubMed] [Google Scholar]
- Evenson M.L., Hinds M.W., Bernstein R.S., Bergdoll M.S. Int. J. Food Microbiol. 1988;7:311–316. doi: 10.1016/0168-1605(88)90057-8. [DOI] [PubMed] [Google Scholar]
- Fernandez M.M., Cho S., De Marzi M.C., Kerzic M.C., Robinson H., Mariuzza R.A., Malchiodi E.L. J. Biol. Chem. 2011;286:1189–1195. doi: 10.1074/jbc.M110.142471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.A., Malchiodi E.L., Li H., Ysern X., Stauffacher C.V., Schlievert P.M., Karjalainen K., Mariuzza R.A. Nature. 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- Fraser J.D., Proft T. Immunol. Rev. 2008;225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- Hennekinne J.A., De Buyser M.L., Dragacci S. FEMS Microbiol. Rev. 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- Hudson K.R., Robinson H., Fraser J.D. J. Exp. Med. 1993;177:175–184. doi: 10.1084/jem.177.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Tamate N., Yamaguchi K., Makino S. Appl. Environ. Microbiol. 2005;71:2793–2795. doi: 10.1128/AEM.71.5.2793-2795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L.L., Brophy S.E., Bankovich A.J., Colf L.A., Hanick N.A., Garcia K.C., Kranz D.M. J. Biol. Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- Khandekar S.S., Bettencourt B.M., Wyss D.F., et al. J. Biol. Chem. 1997;272:32190–32197. doi: 10.1074/jbc.272.51.32190. [DOI] [PubMed] [Google Scholar]
- Kieke M.C., Shusta E.V., Boder E.T., Teyton L., Wittrup K.D., Kranz D.M. Proc. Natl Acad. Sci. USA. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieke M.C., Sundberg E., Shusta E.V., Mariuzza R.A., Wittrup K.D., Kranz D.M. J. Mol. Biol. 2001;307:1305–1315. doi: 10.1006/jmbi.2001.4560. [DOI] [PubMed] [Google Scholar]
- Levine W.C., Bennett R.W., Choi Y., Henning K.J., Rager J.R., Hendricks K.A., Hopkins D.P., Gunn R.A., Griffin P.M. J. Infect. Dis. 1996;173:1263–1267. doi: 10.1093/infdis/173.5.1263. [DOI] [PubMed] [Google Scholar]
- Li H., Llera A., Tsuchiya D., Leder L., Ysern X., Schlievert P.M., Karjalainen K., Mariuzza R.A. Immunity. 1998;9:807–816. doi: 10.1016/s1074-7613(00)80646-9. [DOI] [PubMed] [Google Scholar]
- Llewelyn M., Sriskandan S., Terrazzini N., Cohen J., Altmann D.M. Int. Immunol. 2006;18:1433–1441. doi: 10.1093/intimm/dxl076. [DOI] [PubMed] [Google Scholar]
- Malchiodi E.L., Eisenstein E., Fields B.A., Ohlendorf D.H., Schlievert P.M., Karjalainen K., Mariuzza R.A. J. Exp. Med. 1995;182:1833–1845. doi: 10.1084/jem.182.6.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P., Kappler J. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- Mattis D.M., Spaulding A.R., Chuang-Smith O.N., Sundberg E.J., Schlievert P.M., Kranz D.M. Protein Eng. Des. Sel. 2013;26:133–142. doi: 10.1093/protein/gzs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollister B.D., Kreiswirth B.N., Novick R.P., Schlievert P.M. Infect. Immun. 1990;58:2067–2070. doi: 10.1128/iai.58.7.2067-2070.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J.K., Yarwood J.M., Schlievert P.M. Annu. Rev. Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- Miwa N., Kawamura A., Masuda T., Akiyama M. Int. J. Food Microbiol. 2001;64:361–366. doi: 10.1016/s0168-1605(00)00446-3. [DOI] [PubMed] [Google Scholar]
- Mollick J.A., McMasters R.L., Grossman D., Rich R.R. J. Exp. Med. 1993;177:283–293. doi: 10.1084/jem.177.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moza B., Varma A.K., Buonpane R.A., et al. EMBO J. 2007;26:1187–1197. doi: 10.1038/sj.emboj.7601531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur-ur Rahman A.K., Bonsor D.A., Herfst C.A., et al. J. Biol. Chem. 2011;286:4871–4881. doi: 10.1074/jbc.M110.189068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B.A., Carr L.M., Wittrup K.D., Roy E.J., Kranz D.M. Biotechnol. Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- Petersson K., Thunnissen M., Forsberg G., Walse B. Structure (Camb) 2002;10:1619–1626. doi: 10.1016/s0969-2126(02)00895-x. [DOI] [PubMed] [Google Scholar]
- Richman S.A., Aggen D.H., Dossett M.L., Donermeyer D.L., Allen P.M., Greenberg P.D., Kranz D.M. Mol. Immunol. 2009;46:902–916. doi: 10.1016/j.molimm.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodin B.A., Tsomides T.J., Kranz D.M. Immunity. 1996;5:137–146. doi: 10.1016/s1074-7613(00)80490-2. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- Sundstrom M., Abrahmsen L., Antonsson P., Mehindate K., Mourad W., Dohlsten M. EMBO J. 1996a;15:6832–6840. [PMC free article] [PubMed] [Google Scholar]
- Sundstrom M., Hallen D., Svensson A., Schad E., Dohlsten M., Abrahmsen L. J. Biol. Chem. 1996b;271:32212–32216. doi: 10.1074/jbc.271.50.32212. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Furey W., Pletcher J., Sax M. Nat. Struct. Biol. 1995;2:680–686. doi: 10.1038/nsb0895-680. [DOI] [PubMed] [Google Scholar]
- Tallent S.M., Degrasse J.A., Wang N., Mattis D.M., Kranz D.M. Appl. Environ. Microbiol. 2013;79:1422–1427. doi: 10.1128/AEM.02743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Mattis D.M., Sundberg E.J., Schlievert P.M., Kranz D.M. Clin. Vaccine Immunol. 2010;17:1781–1789. doi: 10.1128/CVI.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K.S., Donermeyer D.L., Allen P.M., Kranz D.M. Proc. Natl Acad. Sci. USA. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.