Abstract
Molecular surveillance for drug-resistant malaria parasites requires reliable, timely, and scalable methods. These data may be efficiently produced by genotyping parasite populations using second-generation sequencing (SGS). We designed and validated a SGS protocol to quantify mutant allele frequencies in the Plasmodium falciparum genes dhfr and dhps in mixed isolates. We applied this new protocol to field isolates from children and compared it to standard genotyping using Sanger sequencing. The SGS protocol accurately quantified dhfr and dhps allele frequencies in a mixture of parasite strains. Using SGS of DNA that was extracted and then pooled from individual isolates, we estimated mutant allele frequencies that were closely correlated to those estimated by Sanger sequencing (correlations, >0.98). The SGS protocol obviated most molecular steps in conventional methods and is cost saving for parasite populations >50. This SGS genotyping method efficiently and reproducibly estimates parasite allele frequencies within populations of P. falciparum for molecular epidemiologic studies.
Keywords: Plasmodium falciparum, drug resistance, molecular surveillance, tropical diseases
Drug-resistant malaria parasites are widespread, undermine malaria control efforts, and continue to emerge in response to the deployment of newer drugs. Through sophisticated in vitro and animal experimentation, the molecular correlates of antimalarial resistance in Plasmodium falciparum have been characterized for multiple drugs and have been used to describe correlations between treatment response and individual genotypes [1] as well as between treatment response and prevailing genotypes [2]. The latter efforts have been aided by large-scale meta-analyses of genotype prevalence surveys intended to track resistance alleles [3–5].
The prevalence of molecular markers of drug resistance in populations of parasites can be estimated by several approaches. The most common approach is to genotype parasites from individuals using one of several polymerase chain reaction (PCR)–based protocols, classify isolates as harboring pure or mixed alleles at the loci of interest, and compute the prevalence of alleles and haplotypes. Several considerations undermine this approach's ability to reliably quantify allele frequencies: (1) field isolates are genetically complex and frequently possess parasite strains with alternate alleles; (2) mixed genotypes are usually classified as mutant, thereby excluding significant proportions of wild-type parasites; and (3) genotyping protocols cannot reliably detect minority variant genotypes, thus underestimating low-frequency subpopulations of alleles in mixed infections. Moreover, genotyping individual isolates is resource intensive, and costs are directly proportional to the number of specimens genotyped; these considerations limit large field studies. One method of statistical inference that uses maximum likelihood estimation (MLE) to compute allele frequencies can address the multiplicity of infection (MOI) [6], but this method requires additional genotype data and thus more resources.
Rapid, efficient, and quantitative genotyping approaches are needed to track the spread of molecular markers of antimalarial resistance through parasite populations. To characterize mutations conferring resistance to the antimalarial sulfadoxine-pyrimethamine (SP), in this report we describe the development and application of an approach that uses a second-generation sequencing (SGS) platform to quantify P. falciparum allele frequencies in field isolates from Tanzanian children with uncomplicated malaria using 2 distinct specimen pooling approaches. This novel approach is amenable to the efficient surveillance of drug-resistance markers and can also be applied to describe the populations of other parasite genes implicated in immunogenicity, virulence, and transmission.
METHODS
Ethics Statement
The original trial was reviewed by ethics boards of the National Institute for Medical Research, Dar es Salaam, Tanzania and the Regional Ethics Committee, Stockholm, Sweden. Molecular analyses were approved by the review board of the University of North Carolina, Chapel Hill.
Specimen Collection
Parasites were obtained from 50 children with uncomplicated P. falciparum malaria from Fukayosi, Bagamoyo District, Tanzania, enrolled in a trial of artemether-lumefantrine in 2006 [7]. Pretreatment finger-prick blood samples were collected and stored as dried blood spots (DBSs) in individual plastic bags. From these, genomic DNA (gDNA) was extracted by 2 approaches using a Chelex 100 method: (1) individually, wherein 3 0.5-cm diameter disks were punched from each DBS into separate wells of a 96-well plate and gDNA was extracted from the DBS in each well and (2) pooled, wherein a 0.5-cm disk was punched from each DBS into a single tube and gDNA was extracted from the combined DBSs.
Estimating Genotype Frequencies Using Individual Sanger Sequencing
Individual dhfr and dhps genotypes were obtained via amplification and direct sequencing from the individual gDNA specimens as described elsewhere [8], with the exception that reaction volumes were halved and 5 µL of template was input into the 12.5 µL primary reaction. Briefly, gDNA was subjected to nested amplifications for both dhfr and dhps and these amplicons were bidirectionally sequenced using ABI Big Dye Terminator chemistry (Supplementary Table 1). Reads were scored by manual inspection of chromatograms using Sequencher software (version 4.8; Gene Codes), and loci at which a secondary peak height was ≥10% of the major peak height were scored as “mixed” infections. All specimens had been previously genotyped to determine the MOI at merozoite surface protein 1 (msp1) and 2 (msp2) [7].
Estimating Genotype Frequencies Using Statistical Inference
We used MalHaploFreq to convert the prevalences of alleles into frequencies after incorporation of the MOI [6], an approach that can allow for more precise frequency estimates with limited bias [9]. We defined MOI for each individual sample as the largest number of either msp1 or msp2 alleles obtained.
Validation of Genotyping Using SGS
We mixed the gDNA of parasite laboratory strains 3d7 (MRA-102G and MR4; American Type Culture Collection) and V1/S (MRA-176G and MR4), which have contrasting dhfr and dhps haplotypes: 3d7 harbors a wild-type dhfr haplotype (haplotype NCSI) and A437G substitution in dhps (haplotype SGKAA), whereas V1/S harbors 4 dhfr substitutions (haplotype IRNL) and 3 dhps substitutions (haplotype FGKAT) (mutant amino acid substitutions bolded). We amplified dilutions of each gDNA in a real-time PCR assay targeting the single-copy P. falciparum gene pfldh to generate relative quantity estimates [10]. Based on these, we mixed the gDNA of 3d7 and V1/S in a ratio of 4:1. This gDNA mixture was used as template for PCR amplification and 454 sequencing of dhfr and dhps amplicons.
For both dhfr and dhps, deep sequencing was performed in duplicate, using the same template; 5 µL of the template was input into separate primary 25-µL reactions consisting of 0.25 µL of Roche FastStart High-Fidelity Taq Polymerase, 0.5 µL of dNTPs, 2.5 µL of Reaction Buffer, 400 nmol/L each of forward and reverse primers, and 15.75 µL of water (Supplementary Table 1). Nested 25-µL reactions consisted of 2 µL of the primary round product but otherwise identical constituents. Both targets were prepared for unidirectional 454 sequencing, however a poly(A) homopolymer in dhps required that nested PCR reactions for dhps consist of 2 parallel reactions: one with the sequencing tag appended to the 5′ primer and the other with the sequencing tag appended to the 3′ primer.
Amplicons were sized using an ABI Bioanalyzer 2100 with a high-sensitivity DNA chip (Agilent Technologies) and quantified using a Quant-iT PicoGreen dsDNA assay (Life Technologies). We sequenced dhfr and dhps in separate sequencing runs; for each run, we multiplexed amplicons from 4–6 templates that had been amplified with different multiplex identifiers (MIDs), and we prepared these for sequencing following emulsion PCR protocols from Roche. Sequencing reads from the 454 GS Junior were initially filtered using the platform's shotgun analysis, then subsequently trimmed of MIDs, tags, and primers, and culled of low-quality reads based on previously determined cutoffs (eg, length and quality score) using an in-house tool [11]. High-quality reads meeting these thresholds were input into genotype frequency analyses using Lasergene Genomics Core Suite with SeqMan NGen (version 10.0; DNAStar). Unpaired readings were assembled into single contigs to respective reference sequences for dhfr (GenBank XM_001351443) or dhps (GenBank Z30654). The mutant allele frequency at single-nucleotide polymorphism (SNP) loci was defined as the number of mutant alleles divided by the coverage at that locus. We adopted the conservative approach of excluding SNPs with a frequency below 1%, owing to the reported 99.1% accuracy of 454 amplicon sequencing of P. falciparum genes [12].
Estimating Genotype Frequencies Using SGS
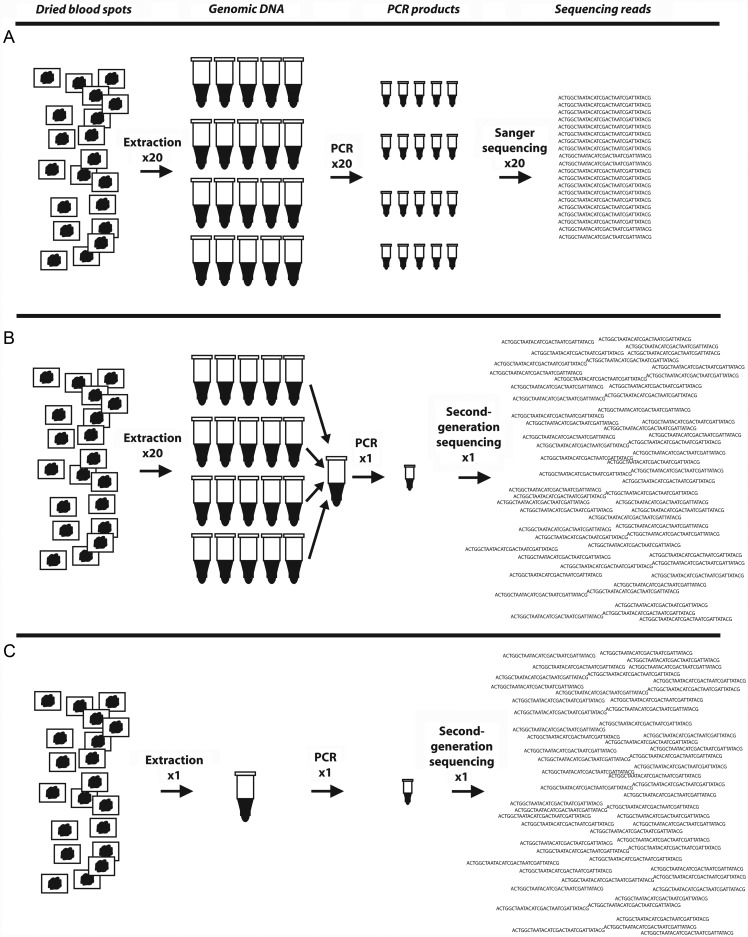
We performed 454 sequencing of dhfr and dhps codons from clinical samples using 2 different templates: (1) gDNA specimens that were extracted individually and pooled into a single aliquot (with equal volumes from each individual gDNA and (2) a gDNA specimen extracted from pooled DBSs into a single aliquot (Figure 1). Amplicons were prepared and sequenced, and reads were aligned and scored as above. In the clinical specimens, we defined novel mutations as any SNP or indel that was present in ≥1% of the total number of readings for that target [12].
Figure 1.
Schematic of different sequencing approaches used to characterize prevailing parasite genotypes. A, Specimens are collected from patients, genomic DNA (gDNA) is extracted individually, target sequences are amplified individually, and polymerase chain reaction (PCR) products are sequenced individually to obtain a sequencing read for each isolate. These reads are then used to generate a prevalence of mutant alleles or combined with an estimate of each isolate's multiplicity of infection to estimate the frequency of alleles. B, Specimens are collected from patients, gDNA is extracted individually but then combined into a gDNA pool, the target sequence is amplified from this pooled gDNA using primers adapted for second-generation sequencing (eg, 454, Illumina), and this PCR product is sequenced on a second-generation sequencing platform to yield thousands of sequencing reads. These reads are then aligned to a reference sequence and scored for mutations to estimate the frequency of mutant alleles. C, Specimens are collected from patients, but gDNA is extracted in a single pool, and the procedure follows as described for B.
Comparisons of Allele Frequencies
We computed pairwise correlation coefficients to quantify the strength of correlation between allele frequencies generated by the 4 approaches: Sanger, Sanger with MLE (Sanger-MLE), 454 sequencing of pooled gDNA (pooled gDNA), and 454 sequencing of pooled DBS (pooled DBS). Technical replicates of the 454 sequencing reactions were included as separate frequency estimations. We used a Bonferroni correction of a P value of .05 to determine significant correlations, which were computed with Stata/IC software (version 11; StataCorp).
We compared the estimated per-specimen cost of the sequencing approaches across a range of numbers of specimen. Calculations excluded labor costs, and used the following unit costs (US dollars) for steps in each protocol: gDNA extraction, $2; PCR, $2; Sanger sequencing reaction, $5; 454 GS Junior sequencing reaction, $1100.
RESULTS
Allele Estimates in Clinical Specimens Using Traditional Methods
From 50 clinical isolates, we obtained Sanger sequencing reads for 50 dhfr and 46 dhps fragments. Mutant dhfr alleles were common: 80%–88% of isolates harbored the N51I, C59R, and S108N mutations as pure or mixed genotypes, similar to findings of prior studies (Table 1) [13]. No isolates harbored the I164L mutation in dhfr. The dhps mutations A437G and K540E were present in 60.0% (95% confidence interval [CI], 44.3–74.3) and 54.3% (95% CI, 39.0–69.1) of isolates, respectively; only 1 isolate (2.2%) harbored the 581G mutation, and none harbored mutations in codons 436 or 613 (Table 1).
Table 1.
Estimates of Mutant Allele Frequencies from Clinical Specimens
| Estimate (95% CI), % |
||||
|---|---|---|---|---|
| Gene and Mutation | Sangera | Sanger–MLE | 454–Pooled gDNA | 454–Pooled DBS |
| dhfr (n = 50) | ||||
| N51I | 80 (66.2–90) | 78.92 (63.16–100) | 70.91 (70.62–71.21) | 72.70 (72.38–73.02) |
| C59R | 86 (73.3–94.2) | 80.48 (64.56–100) | 84.61 (84.38–84.85) | 45.66 (45.30–46.02) |
| S108N | 88 (75.7–95.5) | 86.16 (67.30–100) | 90.2 (89.99–90.41) | 96.62 (96.47–96.77) |
| I164L | 0 | 0 | 0 | 0 |
| dhps (n = 46) | ||||
| S436A | NA | NA | 12.78 (12.48–13.08) | 18.41 (17.54–19.28) |
| S436Y | NA | NA | 0 | 0 |
| S436F | NA | NA | 0 | 0 |
| A437G | 60 (44.3–74.3)b | 58.45 (43.89–82.89)b | 55.11 (54.67–55.56) | 54.86 (53.75–55.98) |
| K540E | 54.3 (39–69.1) | 51.14 (40.09–68.87) | 57.88 (57.47–58.29) | 60.39 (59.46–61.33) |
| A581G | 2.2 (0–11.5) | 1.09 (0–11.69) | 6.19 (5.97–6.41) | 6.35 (5.82–6.88) |
| A613S | NA | NA | 0 | 0 |
| A613T | NA | NA | 0 | 0 |
Abbreviations: CI, confidence interval; DBS, dried blood spot; gDNA, genomic DNA; MLE, maximum likelihood estimation; NA, not available (chromatograms did not adequately cover loci).
a Percentage of mixed or mutant alleles.
b n = 45.
We estimated allele frequencies using an MLE method that accounts for parasite strain multiplicity [6]. With this method, the population frequencies of mutant alleles were slightly lower than the above prevalences for all dhfr and dhps loci, probably reflecting the contribution of wild-type alleles to mixed infections (Table 1).
Validation of SGS
To test the ability of deep sequencing to accurately estimate allele frequencies, we estimated allele frequencies in a mixture of gDNA from parasite lines 3d7 and V1/S that were mixed 4:1. Technical replicates were carried through parallel amplification and sequencing steps for both targets.
Sequencing of dhfr returned 26 839 and 21 420 analyzable reads for the 2 replicates (Supplementary Table 2). The combined mutant allele frequencies for dhfr were 21.7% (95% CI, 21.3%–22.1%) for N51I, 21.6% (95% CI, 21.2%–22%) for C59R, 22.9% (95% CI, 22.5%–23.4%) for S108N, and 17.1% (95% CI, 16.0%–18.3%) for I164L (Table 2). Sequencing of dhps returned 1726 analyzable reads for the first replicate and 3608 for the second. The reduced yield of the first replicate was owing to the absence of reads from the 5′ end of the amplicon, reducing the yield of base calls at dhps codons 581 and 613. The combined mutant allele frequencies for dhps were 20.1% (95% CI, 21.4%–18.8%) for S436F, 100% for A437G, 0% for K540E and A581G, and 15.5% (95% CI, 13.8%–17.3%) for A613T (Table 2). Thus, all allele frequencies were close to expected frequencies, suggesting that deep sequencing the products of the PCR amplifications provides allele frequencies that accurately estimate those within the template DNA.
Table 2.
Mutant Allele Frequencies in a Mix of Genomic DNA From Parasite Clones 3d7 and V1/S
| Gene and Mutationa | Replicate 1 |
Replicate 2 |
Combined |
% Expectedb | |||
|---|---|---|---|---|---|---|---|
| Reads, No. | Mutant Allele Frequency (95% CI), % | Reads, No. | Mutant Allele Frequency (95% CI), % | Reads, No. | Mutant Allele Frequency (95% CI), % | ||
| dhfr | |||||||
| N51I | 26 838 | 21.6 (21.1–22.09) | 21 419 | 21.83 (21.27–22.38) | 48 257 | 21.70 (21.33–22.07) | 20 |
| C59R | 26 838 | 21.53 (21.03–22.02) | 21 419 | 21.72 (21.17–22.27) | 48 257 | 21.61 (21.24–21.98) | 20 |
| S108N | 19 670 | 22.10 (21.52–22.68) | 16 148 | 23.90 (23.25–24.56) | 35 818 | 22.92 (22.48–23.35) | 20 |
| I164L | 2212 | 16.5 (14.95–18.05) | 2028 | 17.9 (16.23–19.57) | 4240 | 17.17 (16.03–18.30) | 20 |
| dhps | |||||||
| S436F | 1726 | 20.57 (18.66–22.47) | 2019 | 19.71 (17.98–21.45) | 3745 | 20.11 (18.82–21.39) | 20 |
| A437G | 1726 | 100 | 2019 | 100 | 3745 | 100 | 100 |
| K540E | 1662 | 0 | 3514 | 0 | 5176 | 0 | 0 |
| A581G | 1053 | 0 | 2778 | 0 | 3831 | 0 | 0 |
| A613S | 0 | NA | 1589 | 0 | 1589 | 0 | 0 |
| A613T | 0 | NA | 1589 | 15.54 (13.76–17.33) | 1589 | 15.54 (13.76–17.33) | 20 |
Abbrevations: CI, confidence interval. NA, not available (readings were not long enough to provide coverage at this locus).
a The dhfr haplotype at codons 51, 59, 108, and 164 of 3d7 is NCSI; that of V1/S is IRNL (mutant amino acid substitutions in bold). The dhps haplotype at codons 436, 437, 540, 581, and 613 of 3d7 is SGKAA; that of V1/S is FGKAT.
b Genomic DNA from clones 3d7 and V1/S was mixed at a ratio of 4:1.
Allele Estimates in Clinical Specimens With SGS
After quality filtering, the 4 replicates of pooled gDNA from Fukayosi isolates yielded 91 157 reads of dhfr, and aggregate read coverage at loci of interest varied from 91 155 at dhfr51 to 14 636 at dhfr164 (Supplementary Table 2). The dhps sequencing of pooled Fukayosi gDNA returned 93 638 analyzable reads (47 836 5′ → 3′ reads and 45 802 3′ → 5′ reads); aggregate read coverage at the codons of interest was 47 826 (dhps437), 55 956 (dhps540), and 45 606 (dhps581).
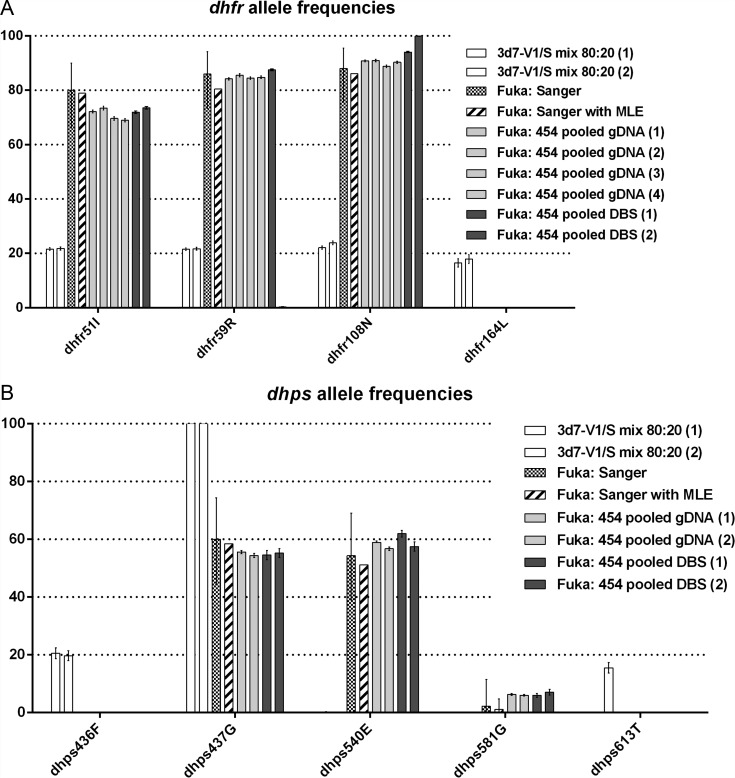
In the pooled gDNA, mutant allele frequencies in dhfr were 70.9% (95% CI, 70.6%–71.2%) for N51I, 84.6% (95% CI, 84.4%–84.9%) for C59R, 90.2% (95% CI, 90.0%–90.4%) for S108N, and 0% for I164L (Figure 2A; Table 1). In dhps, mutant allele frequencies were 55.1% (95% CI, 54.7%–55.6%) for A437G, 57.9% (95% CI, 57.5%–58.3%) for K540E, and 6.2% (95% CI, 6.0%–6.4%) for A581G (Figure 2B; Table 1).
Figure 2.
Mutant parasite allele frequencies as estimated by various methods in dhfr (A) and dhps (B). Mutant allele frequencies in dhfr and dhps as estimated by 454 second-generation sequencing (SGS) in a 80:20 mixture of parasite strains 3d7 and V1/S (white bars), by Sanger sequencing of individual parasite isolates from children in Fukayosi, Tanzania (checkerboard bars), by maximum likelihood estimation (MLE) based on Sanger sequencing of children's parasite isolates (striped bars), by SGS of pooled genomic DNA (gDNA) from children's parasite isolates (light gray bars), and by SGS of pooled dried blood spots (DBSs) from children's parasite isolates (dark gray bars). Lines indicate 95% confidence intervals based upon the proportion of alleles either within the isolates or within the reads. Numbers in parentheses indicate technical replicates of the SGS protocol.
In the amplicons generated from pooled DBS, mutant allele frequencies in dhfr were 72.7% (95% CI, 72.4%–73.0%) for N51I, 45.7% (95% CI, 45.3%–46.0%) for C59R, 96.6% (95% CI, 96.5%–96.8%) for S108N, and 0% for I164L. Mutant allele frequencies in dhps were 54.86% (95% CI, 53.8–56) for A437G, 60.39% (95% CI, 59.5–61.3) for K540E, and 6.35% (95% CI, 5.8%–6.9%) for A581G. There were no novel mutations in the sequenced fragments of dhfr or dhps from Sanger or SGS readings.
Comparative Accuracy and Cost of Genotyping Approaches
All approaches were highly correlated—with correlation coefficients >0.98—save for 1 of the replicates of the pooled DBS SGS method (Table 3). Furthermore, these pairwise positive correlations were largely significant; the nonsignificant correlations with allele frequencies generated by pooled gDNA replicates 3 and 4 are probably due to these being partial frequency estimates, because only dhfr alleles were estimated 4 independent times. Thus, pooling gDNA before SGS was highly correlated with Sanger sequencing approaches in estimating the frequency of mutant alleles in dhfr and dhps.
Table 3.
Pairwise Correlations Between Mutant Allele Frequency Estimates in Clinical Specimens Generated by Various Methodsa
| Sanger | Sanger-MLE | Pooled gDNA (1) | Pooled gDNA (2) | Pooled gDNA (3)b | Pooled gDNA (4)b | Pooled DBS (1) | Pooled DBS (2) | |
|---|---|---|---|---|---|---|---|---|
| Sanger | … | <.0001 | .0002 | .0001 | .2004 | .2686 | .0008 | 1 |
| Sanger-MLE | 0.9992 | … | .0005 | .0003 | .2935 | .3629 | .0017 | 1 |
| Pooled gDNA (1) | 0.9929 | 0.9902 | … | <.0001 | .0126 | .0219 | <.0001 | 1 |
| Pooled gDNA (2) | 0.9943 | 0.9916 | 0.9994 | … | .0130 | .0296 | <.0001 | 1 |
| Pooled gDNA (3)b | 0.9928 | 0.9895 | 0.9995 | 0.9995 | … | .0056 | .0067 | 1 |
| Pooled gDNA (4)b | 0.9904 | 0.9870 | 0.9992 | 0.9989 | 0.9998 | … | .0011 | 1 |
| Pooled DBS (1) | 0.9880 | 0.9842 | 0.9991 | 0.9983 | 0.9998 | 1 | … | 1 |
| Pooled DBS (2) | 0.6264 | 0.6503 | 0.6370 | 0.6257 | 0.5482 | 0.5510 | 0.6259 | … |
Abbreviations: DBS, dried blood spot; gDNA, genomic DNA; MLE, maximum likelihood estimation;
a Numbers in lower half are correlation coefficients between estimates of mutant allele frequencies in dhfr (codons 51, 59, 108, and 164) and dhps (codons 437, 540, and 581); numbers close to 1 indicate a positive correlation, those close to −1 indicate a negative correlation, and those close to 0 indicate no correlation between values in the 2 groups. Correlation coefficients that were significant at P < .05 after Bonferroni correction are bolded and italicized. Numbers in upper half of table are P values for the correlation coefficients. Numbers in parentheses indicate technical replicates of the second-generation sequencing reactions.
b Frequencies are available only for the 4 dhfr loci, because dhps was sequenced in only only 2 replicates.
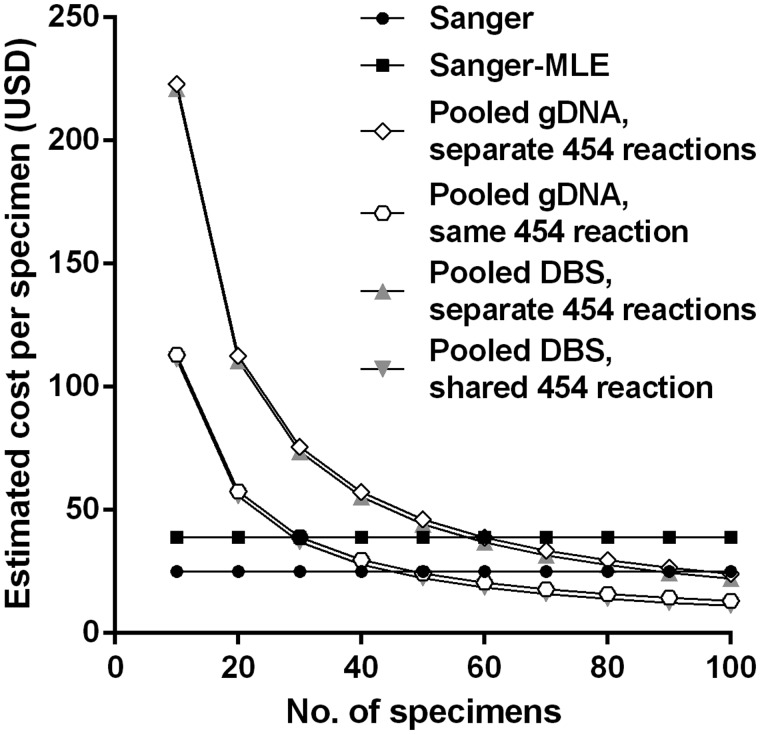
We estimated the per-specimen cost to genotype a range of numbers of isolates at both dhfr and dhps (Figure 3). Although the Sanger-sequencing methods were more cost-efficient than our methods with fewer specimens, our optimal quantitative approach (pooling gDNA specimens and sequencing amplicons of dhfr and dhps in separate 454 reactions) was less costly than Sanger sequencing at approximately 100 specimens and less costly than a Sanger-MLE approach at only 60 specimens. These costs are exclusive of labor costs; because the SGS approach obviates most PCR reactions, it substantially reduces technician time owing to fewer reactions, electrophoresis steps, and amplicon purifications. Thus, cost savings would probably be larger than estimated if labor costs were considered.
Figure 3.
Estimated cost of various methods used to genotype parasite isolates at dhfr and dhps. Estimated costs are exclusive of labor. Input costs (US dollars) per unit used in calculations were as follows: genomic DNA (gDNA) extraction, $2; polymerase chain reaction, $2; Sanger sequencing reaction, $5; 454 GS Junior second-generation sequencing reaction, $1100. Abbreviations: DBS, dried blood spot; MLE, maximum likelihood estimation.
DISCUSSION
We used a SGS platform to characterize mutant allele frequencies in a population of P. falciparum parasites from Bagamoyo district, Tanzania. In our assay validation, SGS of PCR products generated from a template of a known ratio of parasite strains returned allele frequency estimates very close to the expected frequencies. Moreover, the allele frequencies generated from SGS of field parasites were similar to those generated from conventional Sanger sequencing, at comparable cost. Because it can return reliable, replicable allele frequencies and is scalable to larger cohorts of specimens, pooled SGS of malaria parasites offers a new method by which to accurately and efficiently characterize P. falciparum genotypes for epidemiologic surveillance.
Second-generation amplicon sequencing offers the ability to characterize and quantify complex genotypes within a single specimen. Previous field studies of malaria parasites have used it to classify recurrent parasitemias after drug therapy [12], describe the diversity of vaccine-targeted parasite antigens [11, 14, 15], and quantify resistance alleles within a single isolate [16]. In these studies, when compared with conventional methods, SGS was better able to capture “minority variants” or low-frequency subpopulations. Our approach extends these applications by applying this sequencing technology to amplicons generated from pooled gDNA that represents a defined population of parasites. Therefore, the sequencing reads represent genotypes present in the original mixture and are aggregate frequency estimates across the population.
How sensitive is this approach for the detection of low-level alleles? A concern with any sequencing approach is the possible loss of the ability to detect ”minority variant” subpopulations, which could undermine surveillance for drug-resistance alleles. Statistical genetic models predict that pooled sequencing with adequate coverage (ie, large number of reads per individual) will detect minor alleles more often than individual sequencing [17]. Indeed, our sequencing of clinical isolates consistently returned >10 000 reads covering each locus, and the corresponding clinical sensitivity of our pooled SGS approach seemed high: our application to clinical specimens from Tanzania allowed detection of the A581G mutation in dhps that was present in only 1 of 46 (2.2%) isolates by Sanger sequencing; with SGS, this mutation was present in 4.5%–6.3% in the reads from 4 replicates prepared by 2 approaches. We suspect this discrepancy reflects the ability of SGS to better capture these genotypes when present as minority variants in complex mixtures. These observations suggest that our sequencing approach can be applied to detect emerging genotypes within diverse parasite populations owing to its high clinical sensitivity.
Overall, our estimates of mutant allele frequencies are similar if slightly lower than those reported from contemporary studies in Northeastern Tanzania [13, 18, 19]. A notable exception to this is the low frequency of dhps A581G mutations in our study, which contrasts with prevalences >50% in 2 studies from the mid-2000s in towns close to our study site to the north [18, 19]. The in vivo significance of this mutation remains undetermined: although parasites bearing the A581G mutation were associated with reduced effectiveness of sulfadoxine-pyrimethamine as antenatal preventive therapy[16] and as therapy for uncomplicated childhood malaria [19], allelic exchange experiments suggest that this substitution alone does not substantially affects the parasite's in vitro susceptibility to SP [20]. This mutation's geographic range and clinical impact in ecologic studies can be further defined using our approach to genotyping parasites.
We applied SGS to 2 separate templates that were derived from field specimens: gDNA that was individually extracted and then pooled in equal volumes (pooled gDNA) and gDNA that was extracted from pooled DBSs in a single extraction (pooled DBS; Figure 1). Between approaches, mutant allele frequencies were similar at most loci except for dhfr59; at this locus, poor dhfr readings were obtained from 1 of 2 sequencing replicates obtained from the pooled DBS template, probably because homopolymers (such as that containing this substitution) are prone to return errors on SGS platforms. Because of this, the correlation of 1 of the 2 replicates of pooled DBS readings was very poor (Table 3). Aside from this subset of sequencing readings, all methods were highly correlated in their estimation of mutant allele frequencies, although this discrepancy highlights a need for technical replicates of SGS reactions. Furthermore, because most laboratories will require individual gDNA aliquots for other applications, we recommend amplification and SGS of pooled gDNA for allele frequency estimation.
For genotyping studies designed to quantify allele frequencies in >50 specimens, our approach is cost-effective when compared to the conventional approach of individual Sanger sequencing (Figure 3). A sequencing reaction on a second-generation platform is far more expensive than one on a Sanger platform, but this cost is offset by the obviation of most PCR reactions by the pooled approach, as well as by the great reduction in the number of sequencing reactions needed for pooled genotyping. Two further points merit mention. First, hands-on technician time is also greatly reduced by pooled deep sequencing, principally owing to the obviation of most PCR assays, agarose gels, and amplicon purifications. This may provide further cost savings that were not included in our calculations. Second, pooled deep sequencing can be applied to large numbers of isolates without sacrificing cost-effectiveness, and, in fact, becomes far less costly than conventional approaches with more isolates. Thus our new approach is easily scalable without sacrificing cost-effectiveness.
Pooled deep sequencing can quantify allele frequencies quickly and efficiently in large populations of malaria parasites and thus has many applications for molecular epidemiologic investigations. As we demonstrate, surveillance for mutations conferring parasite drug resistance is a manifest application and can be easily used when molecular markers of parasite drug resistance are well described. This is the case for multiple drugs. including sulfadoxine and pyrimethamine (as measured herein), chloroquine, amodiaquine, and lumefantrine. Currently, no molecular marker for prolonged parasite clearance time in response to artemisinin has been described, although recent reports have associated this parasite phenotype with mutations on P. falciparum chromosomes 10 [21] and 13 [21, 22]; PCR-based genotyping protocols have been promulgated for 2 of the mutations [23] and could be adapted to SGS to define the limits of its circulation in parasite populations. Beyond drug-resistance markers, this approach can be adapted to characterize any SNP-based parasite genotype that confers clinically significant phenotypes, such as those associated with infection severity or transmissibility.
Although promising, our study and method are subject to several limitations. Using this SGS approach, we were unable to quantify dhps haplotypes, owing to an inability to consistently sequence through a homopolymer in the dhps gene between codons 437 and 540. Second, this approach precludes the investigation of parasite mutations and individual response to therapy. Moreover, we pooled equal volumes of gDNA from isolates, and therefore allele frequencies may be biased by differences in vivo parasite densities and extraction efficiency; however, extractions were performed uniformly to minimize variability, and the exclusion of children with reported recent sulfadoxine-pyrimethamine (SP) intake would be expected to minimize potential bias of parasite density by dhfr and dhps haplotype. Finally, this approach is cost-effective in operation compared with individual genotyping; though it requires new infrastructure including access to a SGS platform and analytic software, these resources are increasingly available at academic medical centers or through outside vendors.
Pooled SGS of malaria parasites offers an efficient and scalable approach to quantifying drug-resistance alleles in parasite populations. With this approach, many parasites can be genotyped rapidly and quantitatively for molecular surveillance of drug-resistance alleles. Additional applications include characterizing the genotypes of parasite antigens targeted by vaccines and those associated with virulence or transmission. This approach is readily adaptable to large-scale drug-resistance surveillance, and further molecular ecologic studies will be necessary to investigate associations between prevailing genotype frequencies and in vivo drug efficacy.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Jeff Bailey (University of Massachusetts) for his assistance with read processing, and Oksana Kharabora and Marcia Peck (both from the University of North Carolina) for assistance in the laboratory. The following reagents were obtained through the MR4 as part of the BEI Resources Repository, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): P. falciparum gDNA from P. falciparum 3D7, MRA-102G, and P. falciparum V1/S, MRA-176G. We are of course indebted to the children and their guardians who participated in the clinical study in Fukayosi.
Author Contributions. S. M. T., J. J. J., and S. R. M. conceived and designed experiments. S. M. T. and C. M. P. performed the experiments. S. M. T., C. M. P., N. A., S. R. M., and J. J. J. analyzed the data. C. M. P., N. A., B. E. N., A. M., and J. J. J. contributed reagents, materials, or analysis tools. S. M. T., C. M. P., and J. J. J. wrote the paper.
Financial Support. This work was supported by the NIAID (grants K08AI100924 to S. M. T. and 5R01AI089819 to J. J. J.) and the NIH (training grant T32GM0088719 to C. M. P.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malaria J. 2009;8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ter Kuile FO, van Eijk AM, Filler SJ. Effect of sulfadoxine-pyrimethamine resistance on the efficacy of intermittent preventive therapy for malaria control during pregnancy: a systematic review. JAMA. 2007;297:2603–16. doi: 10.1001/jama.297.23.2603. [DOI] [PubMed] [Google Scholar]
- 3.Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malaria J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worldwide Antimalarial Resistance Network (WWARN) http://www.wwarn.org/ Accessed 15 April 2013. [DOI] [PMC free article] [PubMed]
- 5.Roper C. Drug resistance maps. http://www.drugresistancemaps.org/ Accessed 15 April 2013. [Google Scholar]
- 6.Hastings IM, Smith TA. MalHaploFreq: a computer programme for estimating malaria haplotype frequencies from blood samples. Malaria J. 2008;7:130. doi: 10.1186/1475-2875-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson AM, Ngasala BE, Dahlstrom S, et al. Plasmodium falciparum population dynamics during the early phase of anti-malarial drug treatment in Tanzanian children with acute uncomplicated malaria. Malaria J. 2011;10:380. doi: 10.1186/1475-2875-10-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor SM, Antonia AL, Chaluluka E, et al. Antenatal receipt of sulfadoxine-pyrimethamine does not exacerbate pregnancy-associated malaria despite the expansion of drug-resistant Plasmodium falciparum: clinical outcomes from the QuEERPAM study. Clin Infect Dis. 2012;55:42–50. doi: 10.1093/cid/cis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hastings IM, Nsanzabana C, Smith TA. A comparison of methods to detect and quantify the markers of antimalarial drug resistance. Am J Trop Med Hyg. 2010;83:489–95. doi: 10.4269/ajtmh.2010.10-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rantala AM, Taylor SM, Trottman PA, et al. Comparison of real-time PCR and microscopy for malaria parasite detection in Malawian pregnant women. Malaria J. 2010;9:269. doi: 10.1186/1475-2875-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey JA, Mvalo T, Aragam N, et al. Use of massively parallel pyrosequencing to evaluate the diversity of and selection on Plasmodium falciparum csp T-cell epitopes in Lilongwe, Malawi. J Infect Dis. 2012;206:580–7. doi: 10.1093/infdis/jis329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juliano JJ, Porter K, Mwapasa V, et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Nat Acad Sci U S A. 2010;107:20138–43. doi: 10.1073/pnas.1007068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidima W, Nkwengulila G, Premji Z, Malisa A, Mshinda H. Dhfr and dhps mutations in Plasmodium falciparum isolates in Mlandizi, Kibaha, Tanzania: association with clinical outcome. Tanzan Health Res Bull. 2006;8:50–5. [Google Scholar]
- 14.Gandhi K, Thera MA, Coulibaly D, et al. Next generation sequencing to detect variation in the Plasmodium falciparum circumsporozoite protein. Am J Trop Med Hyg. 2012;86:775–81. doi: 10.4269/ajtmh.2012.11-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takala SL, Coulibaly D, Thera MA, et al. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington WE, Mutabingwa TK, Muehlenbachs A, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Nat Acad Sci U S A. 2009;106:9027–32. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futschik A, Schlotterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–18. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alifrangis M, Lusingu JP, Mmbando B, et al. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009;80:523–7. [PubMed] [Google Scholar]
- 19.Gesase S, Gosling RD, Hashim R, et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009;4:e4569. doi: 10.1371/journal.pone.0004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triglia T, Wang P, Sims PF, Hyde JE, Cowman AF. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 1998;17:3807–15. doi: 10.1093/emboj/17.14.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takala-Harrison S, Clark TG, Jacob CG, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Nat Acad Sci U S A. 2013;110:240–5. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheeseman IH, Miller BA, Nair S, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worldwide Antimalarial Resistance Network (WWARN) PCR-RFLP for genotyping candidate P. falciparum artemisinin Resistance SNPs MAL10-688956 and MAL13-1718319 v1.0. http://www.wwarn.org/sites/default/files/WWARN%20Procedure_MOL07.pdf. Accessed 15 April 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.