Abstract
Schizophrenia genome-wide association studies (GWAS) have identified common SNPs, rare copy number variants (CNVs) and a large polygenic contribution to illness risk, but biological mechanisms remain unclear. Bioinformatic analyses of significantly associated genetic variants point to a large role for regulatory variants. To identify gene expression abnormalities in schizophrenia, we generated whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. Regression analysis identified 95 transcripts differentially expressed by affection status at a genome-wide false discovery rate (FDR) of 0.05, while simultaneously controlling for confounding effects. These transcripts represented 89 genes with functions such as neurotransmission, gene regulation, cell cycle progression, differentiation, apoptosis, microRNA (miRNA) processing and immunity. This functional diversity is consistent with schizophrenia's likely significant pathophysiological heterogeneity. The overall enrichment of immune-related genes among those differentially expressed by affection status is consistent with hypothesized immune contributions to schizophrenia risk. The observed differential expression of extended major histocompatibility complex (xMHC) region histones (HIST1H2BD, HIST1H2BC, HIST1H2BH, HIST1H2BG and HIST1H4K) converges with the genetic evidence from GWAS, which find the xMHC to be the most significant susceptibility locus. Among the differentially expressed immune-related genes, B3GNT2 is implicated in autoimmune disorders previously tied to schizophrenia risk (rheumatoid arthritis and Graves’ disease), and DICER1 is pivotal in miRNA processing potentially linking to miRNA alterations in schizophrenia (e.g. MIR137, the second strongest GWAS finding). Our analysis provides novel candidate genes for further study to assess their potential contribution to schizophrenia.
INTRODUCTION
Schizophrenia is a common and severe psychotic disorder (1,2), which presents a variety of symptoms, including hallucinations, delusions, reduced emotions, speech, interest and disorganization (3). Schizophrenia has a median lifetime prevalence of 0.40% and a morbid risk of 0.72% (4), with the typical age at onset of psychosis in adolescence or early adulthood (5). All-cause mortality is elevated ∼2.6-fold in schizophrenic individuals (4). Males show higher prevalence, severity and earlier age of onset (4–6). Schizophrenia is very heterogeneous, and many biological mechanisms have been suggested, including hypotheses where the primary event is immune, inflammatory or infectious (7,8)).
Schizophrenia is highly heritable (estimated ∼80% (9)). GWAS have shown the strongest association to be the xMHC region (6p21.32-p22.2), a genomic region of high linkage disequilibrium (LD) and numerous genes, which has not been further resolved by GWAS (10–13). Besides the xMHC region, GWAS have uncovered ∼10 other genome-wide significant individual common susceptibility loci associated with schizophrenia, and evidence for polygenes (10–17). Another strong GWAS finding is at MIR137 (microRNA 137) (13), a post-transcriptional mRNA regulator involved in neuronal development (18–20)), suggesting that mechanisms affecting differential expression in schizophrenia may potentially affect many genes (in addition to the aforementioned heritable polygenic contributions, which are especially enriched in regulatory regions, e.g. 5′UTRs (21)). CNVs have also been implicated (22). However, despite success in locus identification, our knowledge remains very tentative regarding the causative genomic sequences (individual genes or groups of genes, and their variants) and the specific biological mechanisms of schizophrenia. Since many schizophrenia GWAS hits lie outside of genes or are not in LD with obvious candidate polymorphisms likely to impact the gene structure or function such as missense SNPs (10–13), it seems likely that many variants influencing the risk are regulatory in nature, including possibly altering the level, location and timing of gene expression. Using data from HapMap LCLs, it has been shown that trait-associated SNPs are enriched for expression quantitative trait nucleotides (eQTNs), with neurological/psychiatric disorders showing similar enrichment levels to other complex traits (23). Gene expression profiling may therefore be useful—either by itself or in conjunction with linkage and association analysis—for the identification of implicated genetic variants, genes and functional gene networks, and for revealing the specific underlying disease mechanisms (24–28). Consequently, the functional dimension added by analysis of gene expression, e.g. upregulation or downregulation of genes or groups of genes in cases compared with controls, adds a layer of information that can be directly relevant to schizophrenia pathophysiology.
We sought to identify genes differentially expressed by affection status using whole-genome gene expression microarrays on LCLs from 413 schizophrenia cases and 446 controls of European ancestry (EA) (10,22,29–31). We considered three main factors in choosing to study expression in LCLs: biological validity (living cells, substantial overlap with brain expression), number/quality of available specimens, and magnitude of and control over confounding effects. LCLs present major advantages in areas of tractability, available sample size, reduced influence of state traits (including the ‘environmental’ conditions surrounding cells embedded within the whole organism), and our expectation that many (though not all) relevant transcriptional traits would not be brain specific and thus would be detectable in other tissues such as LCLs. Here, we report the findings from this LCL-based gene expression analysis of the largest schizophrenia sample analyzed for gene expression to date.
RESULTS
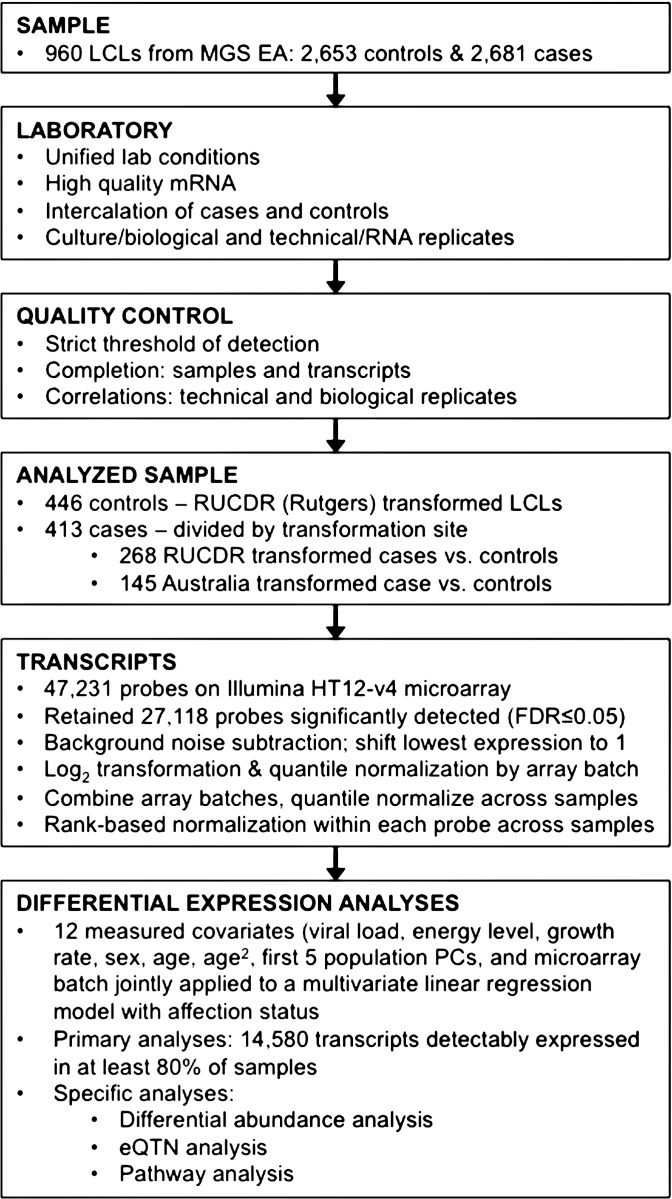
We selected 960 LCLs from the MGS case–control collection (10), matching cases and controls based on 5-year age brackets. The cases typically present chronic, severe, unremitting schizophrenia (10). Multiple epidemiological and laboratory parameters, such as viral load (Epstein-Barr virus, EBV, copy number), energy level (ATP levels adjusted for cell count) and growth rate (cell count at LCL harvest), which are known to have an effect on gene expression in LCLs (32), were similar in cases and controls, though more cases than controls were males (Table 1). A rigorous set of quality control (QC), laboratory and analytical procedures were applied, with the overall study design and data processing steps presented in Figure 1. We generated gene expression profiles (Illumina HT12v4 microarrays), and found all 960 LCL samples’ transcriptional data to be of high quality. After QC and removal of biological and technical replicates, we retained 859 unique samples (446 controls and 413 cases) and 27 118 transcripts (probes), of which 14 588 transcripts (representing 9208 genes) were then selected for further analyses since they had detectable expression (detection P ≤ 0.05) in ≥80% of the RNA samples.
Table 1.
Sample characteristics
| Samples | Controls RUCDR | Cases RUCDR | Cases Australia |
|---|---|---|---|
| Affection (N) | 446 | 268 | 145 |
| Sex (% male) | 45 | 71 | 73 |
| Age (years) | 45.7 (44.5–46.9) | 42.8 (41.5–44.0) | 41.3 (39.5–43.1) |
| EBV load (copy number) | 1.628 (1.581–1.676) | 1.461 (1.394–1.527) | 1.841 (1.749–1.934) |
| Clonality (heterozygosity) | 0.772 (0.764–0.780) | 0.785 (0.776–0.795) | 0.763 (0.749–0.776) |
| Cell count at harvest (growth rate) | 0.472 (0.463–0.482) | 0.485 (0.473–0.497) | 0.486 (0.470–0.502) |
| Energy (mean ATP/cell count) | 216 106 (210 638–221 573) | 228 691 (222 105–235 277) | 217 890 (208 870–226 911) |
Unless otherwise indicated, the values are expressed as means, with 95% confidence intervals (CIs). Cell count at harvest reflects the growth rate directly since all samples were adjusted to 250 000 cells/ml at 24 h prior to harvest. EBV load is calculated by log10(2−mfdCt), and was higher for LCLs EBV-transformed in Australia. RNA quality indices (A260/A280, A260/A230, RNA integrity number, 28s/18s rRNA ratio, cRNA yield) were all indicative of high quality, and matched well by affection and site. The cases (overall mean age 42.3) were slightly younger (Student's t-test P = 2 × 10−5) than the controls (mean age 45.7), and a larger proportion of cases were males (71%) than among controls (45%), reflecting the overall composition of the MGS sample, which in turn reflect the epidemiological trends of chronic, severe schizophrenia (4–6).
Figure 1.
Experimental and analytical procedures flowchart.
Differential expression analyses
We performed multiple linear regression analysis to identify transcripts that were differentially expressed between LCLs from schizophrenic cases and non-schizophrenic controls. To adjust for potential confounder effects, we selected 12 known or putative confounders [viral load, energy level, growth rate, sex, age, age2, the first five genotypic principal components (PCs), and microarray batch], i.e. variables likely contributing to variation in expression, for inclusion as nuisance parameters in the regression model. We then performed multiple regression analysis using these 12 measured covariates and affection status, and evaluated the relationship of affection status on each transcript's expression level.
To further guard against potential confounder effects, we first examined the samples (268 cases and 446 controls) transformed at Rutgers University Cell and DNA Repository (RUCDR), using highly standardized procedures. We found 95 transcripts, representing 89 genes, differentially expressed by affection status at FDR ≤0.05. See Supplementary Material, Table S1 for a full listing of regression results for these transcripts, and Supplementary Material, Figure S1 for a Manhattan plot of the full transcriptomic results. There was statistically significant enrichment for xMHC transcripts (versus transcripts located elsewhere in the genome) among the differentially expressed genes compared with all detected transcripts (Fisher's one-sided P = 6.7 × 10−3). This is intriguing since the xMHC region harbors the strongest schizophrenia locus identified by GWAS. Other significant transcripts are located in genomic regions that have not yet been tied to schizophrenia risk, though this may change, e.g. as GWAS meta-analyses (13) enlarge further. In Table 2, we list selected examples of the differentially expressed genes, with information on their potential relevance to schizophrenia based on their function and the literature. We highlight further in the discussion some of the more intriguing genes from the immune-related group (B3GNT2, DICER1) and others (MOXD1, DBNDD2, S100A10, SYT11).
Table 2.
Selected examples of transcripts differentially expressed (FDR ≤0.05) by affection status
| Gene(s) Abbreviation | Comments | References |
|---|---|---|
| B3GNT2 | A genome-wide significant association with rheumatoid arthritis has been reported, along with a less significantly association with Graves’ disease. | (49,80–84) |
| BCL2L2, BIK | These genes are involved in apoptosis; BCL2 itself (not differentially expressed) plays a crucial role in adult mouse hippocampal neurogenesis. | (131–133) |
| CYBB | A mouse knock-out prevents the typical behavioral and neurochemical abnormalities acutely induced by subanesthetic ketamine (which induces schizophrenia-like psychotic symptoms in humans). | (134,135) |
| DBNDD2 | Dysbindin domain containing 2 mediates neural differentiation and apoptosis, and dysbindin itself (not differentially expressed) has long been of interest in schizophrenia. | (106,107,136) |
| DBP | This is a transcription factor involved in the regulation of some circadian rhythm genes, and has shown potentially relevant findings of interest: (i) reduced expression in fibroblasts from bipolar disorder subjects (it is downregulated in our case LCLs), (ii) induction in rat prefrontal cortex upon methamphetamine stimulation and (iii) is among the top findings in a combined sample GWAS for bipolar disorders. | (54,137–139) |
| DICER1 | This gene is central in biogenesis of miRNAs. It is upregulated in the dorsolateral prefrontal cortex of schizophrenia cases (and in our case LCLs), and has important roles in normal central nervous system development and function, including that of dopaminergic neurons. | (86–92) |
| ELK1 | This transcription factor has been reported as showing increased protein expression in the cerebellar vermis of schizophrenic subjects (also upregulated in case LCLs). | (140) |
| FAM69A | This is a member of a FAM69A-EVI-RPL5 gene cluster implicated in the autoimmune disorder multiple sclerosis by GWASs, and had some association support in the MGS EA GWAS. | (10,141–143) |
| GBP2, GBP4 | These proinflammatory guanylate binding proteins (both upregulated in case LCLs) are induced by interferon and important for host defense against intracellular pathogens. | (144,145) |
| GLO1 | This enzyme protects against glycation by catalyzing the conversion of reactive, acyclic alpha-oxoaldehydes. It has been previously implicated in autism and schizophrenia, and shown to be protective in a mouse model of Parkinson's disease. | (146–149) |
| HERC2 | A homozygous missense mutation (Pro594Leu) in this gene is associated with a phenotypic triad of nonsyndromic intellectual disability, autism, and gait disturbance in three sibships. | (150) |
| HIST1H2BD, HIST1H2BC, HIST1H2BH, HIST1H2BG, HIST1H4K | These histones are all located in the xMHC region (which contains the most significant and best replicated GWAS association for schizophrenia). Histones have some antimicrobial activity especially lysine-rich histones, participate in the innate defense system of the human placenta particularly histone H2B (which is also found in the cytoplasm and in amniotic fluid), and there are previous reports of histone expression dysregulation in schizophrenia, Huntington's disease, and autism controls versus cases. | (10–13,37–41,46–48) |
| HNMT | This enzyme metabolizes histamine in the brain, and has been implicated in alcohol dependence (association of Thr105 functional variant with higher enzymatic activity) and Parkinson's disease (association of Ile105 functional variant with lower enzymatic activity). It is upregulated in our case LCLs. | (151–154) |
| IFITM3 | Like GBP1 (not differentially expressed), IFITM3 was upregulated in schizophrenic brains including untreated cases (IFITM3 was upregulated in case LCLs). | (155,156) |
| MOXD1 | This is a homolog of DBH (DBH is involved in the biosynthesis of norepinephrine from dopamine), and thus may be relevant for signal transduction. It is located within a previously reported schizophrenia linkage region at 6q23.2, and is the most upregulated transcript in our case LCLs. | (30,99–105) |
| NLRP1 | This is a member of the Ced-4 family of apoptosis proteins, and has been demonstrated to induce apoptosis in cells when overexpressed. Nominal associations with Alzheimer's disease and with rheumatoid arthritis have been reported. | (157,158) |
| PRKCD | This gene is involved in the terminal translocation (when dendrite maturation begins) at the final phase of neuronal migration. | (159,160) |
| PTBP3 | This regulator of cell differentiation binds RNA. | (136) |
| S100A10 | This gene is thought to be involved in the regulation of cell cycle progression and differentiation. It has been implicated in major depression (decreased expression in depressed humans and in animal models), suicide (decreased expression in peripheral blood of attempters and in prefrontal cortex of suicide completers), and bipolar disorder (increased expression in peripheral blood). It was downregulated in our case LCLs. | (108–111) |
| SGK1 | This gene participates in the regulation of neuroexcitability, inflammation, cell proliferation and apoptosis. | (161) |
| SYT11 | A genome-wide significant for association with Parkinson's Disease has been found. A functional 33 bp repeat polymorphism in its promoter region has been reported as nominally associated with schizophrenia in a small Japanese sample. Rat hippocampus studies suggest that it contributes to the regulation of neurotransmitter release in the excitatory and inhibitory presynapses, and to postsynapse-targeted membrane trafficking in dendrites. | (112–118) |
| VAMP4 and STX6 | These are components of a protein complex involved in vesicle–membrane fusion and important for cell adhesion. STX6 has been implicated in a GWAS of the tauopathy progressive supranuclear palsy. | (162–164) |
| XBP1 | This transcription factor is known to be a key regulator of MHC class II genes. It has a functional promoter variant reported as nominally associated in some studies on Asian samples, but not in other studies. | (165–170) |
Genes above are the highlighted (for potential relevance to schizophrenia) subset of the 89 genes differentially expressed (FDR ≤0.05) by affection status; the full list is given in Supplementary Material, Table S1.
Our total sample included an additional set of 145 case LCLs transformed in Australia (and shipped afterwards to RUCDR), for which we compared their expression profiles with the same 446 RUCDR controls, to check whether these results would support our prior findings in case–control analysis on RUCDR samples. Twenty-two of the 95 previously identified transcripts were also significant at FDR ≤0.05 in the analysis of the Australia-transformed cases. Of these 22 transcripts, 77% (17/22) displayed the same direction of effect (i.e. sign of regression coefficient). These 22 transcripts included four of the six previously implicated xMHC region transcripts and 18 non-xMHC transcripts, salient examples including S100A10, GBP4, MOXD1, BCL2L2 (Supplementary Material, Table S1). All remaining analyses refer to the primary comparisons of RUCDR cases versus RUCDR controls.
Enrichment analyses: categories, pathways and networks
We investigated whether the identified transcripts were enriched for various categories, such as brain-expressed or immune-related genes, both related to major pathophysiological hypotheses of schizophrenia. There was no enrichment of brain-expressed genes (77% of the 9208 analyzed genes were expressed in brain via ensembl.org, as were 76% of the 89 significant genes). However, there was an enrichment of immune-related genes (13% of the analyzed genes were immune-related, i.e. protein-coding gene function containing ‘immune’ from genecards.org, as were 22% of the 89 significant genes; Fisher's one-sided P = 0.011). Despite the fact that we included sex as a covariate in our analyses, given the differences in sex distribution between cases and controls (Table 1), we examined whether the 89 differentially expressed genes were enriched for chromosome X genes. This was not the case (4 of 89 were on chromosome X; P = 0.32). Similarly, despite including EBV copy number as a covariate, we investigated whether the identified genes are enriched for the 160 genes whose expression levels were previously reported as being significantly associated with the EBV copy number (33). Again, this was not the case (2 of 89 were EBV copy number associated genes, HIST1H2BD and FBXO32; P = 0.24). The gene ontology (GO)-term enrichment and protein–protein interaction network analyses of FDR ≤0.05 differentially expressed genes revealed some connections within and contributions mainly from the differentially expressed histones. Using the DAVID tool (34), we found no GO-terms and no KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway terms enriched at FDR ≤0.05, consistent with schizophrenia's likely heterogenous pathophysiology. However, for the protein–protein interaction network analysis using DAPPLE (Disease Association Protein-Protein Link Evaluator) (35), the gene list showed significantly higher network connectivity than expected, both direct (P = 0.001) and indirect (P = 0.001), though this was largely attributable to the network mostly containing histones.
eQTNs detection for top differentially expressed transcripts
To identify putative functional regulatory candidate variants influencing schizophrenia risk, we searched for cis-acting eQTNs of the differentially expressed transcripts by association analysis using PLINK. We used the existing GWAS SNP genotype data (Affymetrix 6.0, (10)) for the transcripts significantly differentially expressed between cases and controls at FDR ≤0.05 using a symmetrical window of size of 500 kb from the 5′ or of the 3′ end of the gene encoding for a given transcript. This resulted in 18 981 evaluated transcript–SNP pairs. Employing FDR ≤0.05 (since adjustment for genome-wide multiple testing is overly strict as we only investigated the genomic region around each gene), we found 1089 significant associations, representing 1004 different SNPs and 63 transcripts (Supplementary Material, Table S2). These SNPs are putative cis-acting regulators of their 63 transcripts with differential expression levels by schizophrenia affection status, and thus are potential functional regulatory candidate variants for influencing schizophrenia risk. We evaluated whether there was any overlap between this list of candidate SNPs (i.e. cis eQTNs of transcripts differentially expressed by schizophrenia status) and GWAS results from the same study (EA subjects from the MGS GWAS) (10), beyond the overall convergence of identifying the xMHC region. None of the candidate cis eQTNs showed significant or suggestive associations with schizophrenia in the MGS GWAS. This may be explained by the relatively small sample size (2681 EA cases and 2653 EA controls) of the MGS GWAS dataset (by standards of complex trait GWAS datasets), which did not by itself yield any genome-wide significant loci (10).
DISCUSSION
Using an LCL-based model system to profile gene expression with microarrays in a large schizophrenia case–control sample, our main results are as follows: (i) we detected 95 transcripts (89 genes) differentially expressed by schizophrenia status from analyzing RUCDR-transformed LCL samples (Supplementary Material, Table S1), including genes at the xMHC region. The xMHC findings demonstrate convergence to previous GWAS, which have repeatedly identified the xMHC region as the most significant locus (10–12), including the first round of the Psychiatric GWAS Consortium for schizophrenia (PGC-SZ1, (13)). We also detected many novel genes differentially expressed by affection status (Supplementary Material, Table S1, Table 2, and below), and found an enrichment of immune-related genes. (ii) Analysis of the Australia-transformed case LCLs (versus the RUCDR-transformed controls) showed convergence of top findings, with 22 out of 95 transcripts remaining differentially expressed, including 4 of 6 xMHC transcripts and 18 other transcripts (e.g. transcripts for S100A10, GBP4, MOXD1, BCL2L2, see Supplementary Material, Table S1). (iii) Pathway and network analyses of differentially expressed genes were predominated by histones, but otherwise without any strongly supported pathway or network, consistent with a disorder noted to be highly multifactorial, heterogenous and polygenic. (iv) 1004 cis eQTNs (FDR ≤0.05, within 500 kb, restricted to autosomes) for 63 transcripts were found for the differentially expressed genes, providing potential functional regulatory variants.
Differential expression—histone transcripts
Some of the top differentially expressed transcripts detected were located in the xMHC region, offering new, technologically independent and convergent evidence for a susceptibility locus for schizophrenia in the xMHC, which contains the most significant and best replicated GWAS association for schizophrenia (10–13). There are several possible mechanisms for histones (which constituted all the xMHC region differentially expressed genes: HIST1H2BD, HIST1H2BC, HIST1H2BH, HIST1H2BG and HIST1H4K) influencing schizophrenia risk. Covalent histone modifications are relevant to the regulation of inflammation where the chromatin architecture confers specificity to the cell type and signal in the transcriptional control of inflammation (36). Histones (and histone-derived peptides) also have antimicrobial activity, especially lysine-rich histones (reviewed in (37), including H1, H2A and H2B, the latter being the most commonly differentially expressed histone family in our study), and participate in the innate defense system of the human placenta (38), particularly histone H2B, which is also found in the cytoplasm and in amniotic fluid (39–41) and has a wide variety of roles (42). The differentially expressed xMHC region histones were all down regulated in schizophrenia cases compared with controls (the regression coefficients ranged from −0.137 to −0.155), as were all other xMHC histones (i.e. also the ones not significantly differentially expressed), suggesting a pathophysiological hypothesis of altered immunity to infections in schizophrenia, but also arguably consistent with a histone-wide artifact (see caveats), though one that would have to differentially affect cases versus controls. Some (e.g. (43,44)) have suggested that some medications used in the treatment of schizophrenia might act at an epigenetic level, such as valproic acid (a histone deacetylase inhibitor) and clozapine (an atypical antipsychotic with DNA-demethylation activity), which were found to improve behavior in a mouse model (45). Thus, perhaps some medications might partially correct an underlying histone abnormality, manifested at least in part by differential expression. It is also noteworthy that some other studies have found evidence for expression dysregulation of histones in schizophrenia (46), Huntington's disease (where the differences were found in both frontal cortex and blood prior to clinical symptoms) (47) and autism controls versus cases (48).
Differential expression—immune-related transcripts
Provocative evidence suggesting an involvement of immune mechanisms in schizophrenia, for which LCLs would be a particularly appropriate model, includes the following observations: (i) a family history of autoimmune disease is associated with increased schizophrenia risk, and autoimmune disorders modify schizophrenia risk (49,50); (ii) prospective birth cohort studies with serologically documented gestational infection and immune biomarkers show that specific infections increase the risk of schizophrenia in the offspring (7,51); (iii) co-administration of antipsychotic and anti-inflammatory drugs augments the former's antipsychotic effect (52) and (iv) schizophrenia GWAS have shown the strongest association to be the xMHC region (10–13), which is associated with immune, inflammatory and infectious disorders (53). Therefore, the involvement of primary or secondary pathogenic immune mechanisms in schizophrenia is supported by solid epidemiological data and initial genetic evidence. Interestingly, among mental disorders, there appears to be some specificity of these immune system links: (i) a genome-wide significant association at the xMHC region has not been found in GWAS of other psychiatric disorders, such as bipolar disorder (54), attempted suicide (55), attention deficit hyperactivity disorder (56), autism (57), smoking quantity (58–60) or alcohol dependence (61–63), and (ii) the familial relationship of schizophrenia to a range of autoimmune diseases extends to non-affective psychosis but not to bipolar disorder (50). A substantial proportion of expression signatures appear to be shared between different tissues (64–71), while a proportion of eQTNs (expression quantitative trait nucleotides) are tissue and cell type specific (72–75). Thus, for many (but certainly not all) genes, a peripheral tissue (e.g. LCLs) may serve as a more convenient proxy tissue (available in large numbers) to assay for disease relevant expression changes, and may be especially appropriate as a model for examining immune hypotheses of schizophrenia.
The lack of enrichment for brain-expressed genes and the enrichment of immune-related genes (P = 0.011) in the 89 genes differentially expressed by schizophrenia status suggests that our overall results may be cell model dependent (brain versus immune cells), perhaps mostly detectable in cycling cells (e.g. LCLs). Cell cycle dynamics were altered in olfactory neurosphere-derived cells in samples from schizophrenia subjects (reduced cell cycle period) (76), and such alterations in cell cycle dynamics at critical periods might affect neurogenesis and neural differentiation (77,78), Thus, as a complement to neural tissue studies (e.g. (76), or upon neural cells differentiated from induced pluripotent stem cells (79), on smaller sized samples), LCLs might provide a useful model to dissect some aspects of cell cycle differences in schizophrenia cases versus controls. Some implicated genes suggest connections to known environmental susceptibilities for schizophrenia. Besides histones, many other differentially expressed genes (e.g. ST6GAL1, SDC1, CD27, CD68, CASP1, PSTPIP2, IRAK3, RIPK3, ZBP1, GBP2, GBP4, IFITM3, BCL2L2, BIK, PRKCD, B3GNT2, DICER1, CYBB and ELK1) have established and potential connections to infection and immune response (Table 2 comments on some possible relationships to schizophrenia for selected genes), perhaps through altering susceptibility or response to various maternal infections known to increase schizophrenia risk in offspring (7), eventually leading to a psychotogenic response in the offspring.
We further highlight two especially interesting differentially expressed immune-related genes, B3GNT2 and DICER1. B3GNT2 is genome-wide significant for association with rheumatoid arthritis and less significantly associated with Graves’ disease (P = 3.5 × 10−4) in the Japanese population (80). These are both autoimmune disorders, with Graves’ disease enriched in schizophrenia cases and rheumatoid arthritis inversely associated with schizophrenia (49,81–84). Mouse knockouts of B3GNT2 display decreased adenylyl cyclase 3 activity, decreased expression of many odorant receptors, and many axon growth and guidance errors (85). DICER1 is a ribonuclease that is central in the biogenesis of microRNAs (miRNAs), short (∼22 nucleotides) noncoding RNAs; DICER1 is responsible for the processing (cleavage of the pre-miRNA hairpin structure) to form mature miRNAs. DICER1 has been reported as upregulated in the dorsolateral prefrontal cortex of schizophrenia cases (86,87), and is also upregulated in our schizophrenia cases (β = 0.126). One genome-wide scan for de novo CNVs in sporadic schizophrenia found an isolated duplication including DICER1 in one case subject (88). DICER1 plays important roles in normal central nervous system development and function including that of dopaminergic neurons (e.g. (89–92)), and miRNAs are known to be important in both neuronal function and dysfunction (e.g. see reviews (93–95)), especially MIR137 in schizophrenia (13,96). A candidate gene study of miRNA biogenesis genes found nominally significant (P = 0.006) association with a DICER1 SNP (rs3742330, located in the 3′UTR) in Chinese samples (252 schizophrenia cases and 252 controls) (97). While prefrontal cortex DICER1 expression increases over the lifespan, especially from young adulthood onwards (98), age is unlikely to explain our findings (both since we use age as a covariate and our schizophrenia cases are slightly younger than our controls, Table 1).
Differential expression—transcripts previously reported in psychosis or putatively relevant to schizophrenia pathophysiology
The differentially expressed genes also suggest involvement of various other putative mechanisms such as gene regulation, cell cycle progression, differentiation, apoptosis, miRNA processing and signal transduction. Some of these genes have been previously suggested to be involved in schizophrenia and/or in disorders presenting some symptomatic convergence with schizophrenia, or have known functions plausibly connected to its hypothesized pathophysiology. Again, Table 2 comments on some possible relationships to schizophrenia for selected genes, several of which (MOXD1, DBNDD2, S100A10 and SYT11) we further highlight here. (i) The differentially expressed MOXD1 is located within a previously reported schizophrenia linkage region at 6q23.2 (99–101), and has previously been suggested to be involved in schizophrenia. SNPs in the nearby TAAR6, while not at MOXD1 (but possibly involved in regulation of MOXD1), have been reported as associated in some (102–104) but not in other studies (e.g. (30)). MOXD1 (the most upregulated transcript in our case LCLs, β = 0.214) is a homolog of DBH, which is involved in the biosynthesis of norepinephrine from dopamine (105), and thus may be relevant for signal transduction. (ii) DBNDD2 (dysbindin domain containing 2) mediates neural differentiation and apoptosis (106), and dysbindin itself has long been of interest in schizophrenia, e.g. (107). (iii) S100A10 is thought to be involved in the regulation of cell cycle progression and differentiation, and has been implicated in major depression (decreased expression in depressed humans and in animal models) (108,109), suicide (decreased expression in peripheral blood of attempters, and in prefrontal cortex of suicide completers) (110) and bipolar disorder (increased expression in peripheral blood) (111). We find decreased expression (β = −0.166) of S100A10 in case LCLs. (iv) SYT11 is genome-wide significant for association with Parkinson's disease (112–115), and a functional 33-bp repeat polymorphism in its promoter region has been reported as nominally associated with schizophrenia in a small Japanese sample (116,117). Studies of rat hippocampus suggest that SYT11 contributes to the regulation of neurotransmitter release in the excitatory and inhibitory presynapses, and to postsynapse-targeted membrane trafficking in dendrites (118).
Potential caveats and limitations
We have chosen a cellular model of LCLs for its numerous advantages (living cells, available sample size, quality of available specimens, tractability and reduced environmental influence), coupled with substantial overlap of genes also expressed in the brain. Nevertheless, not all brain-expressed genes are expressed in LCLs, and our experiment is blind to those transcripts. We assessed the matching of our cases and controls on various parameters (Table 1), with the sex ratio being the least-matched parameter (largely reflecting the epidemiology of schizophrenia). Various known and suspected confounders may influence gene expression, and while we have carefully measured and included them (viral load, energy level, growth rate, sex, age, age2, the first five genotypic PCs and microarray batch) as nuisance covariates in the regression analysis, some confounding may still remain. However, our lack of detected enrichment for transcripts related to various confounders (e.g. sex chromosome transcripts and viral load associated transcripts) may reflect at least partial success at adjusting for such confounders. Since LCL transformation sites/protocols may differ somewhat and this was not amenable to inclusion as a covariate, we used the RUCDR-transformed case and control LCLs as the primary analysis, with the Australia-transformed case LCLs incorporated into a secondary analysis that was found to be supportive (Supplementary Material, Table S1).
While it is possible that the differentially expressed xMHC region histones are relevant to schizophrenia, they are at heightened risk (compared with other differentially expressed genes) for representing a technical artifact for various reasons. (i) Histone expression varies across the cell cycle (119), though we note that the studied LCLs were not synchronized in their cell cycle, but rather represented a mixture of cells in all the various cell cycle stages, blunting (or eliminating) such cell cycle stage-specific expression differences. (ii) The sequence similarity within the histone gene family may decrease the specificity of the sequence-based array assay. However, bioinformatic analyses (clustalw; data not shown) showed that the differentially expressed xMHC histone probes had indistinguishable alignment scores compared with randomly matched and equally sized (50 bp) non-histone probes, and were thus not obviously explained by sequence similarity. (iii) Our amplification method (i.e. reverse transcription with an oligo-dT primer) limits our detection of histone mRNAs to those that have poly(A) tails (‘variant’, or replication-independent histone mRNAs) (120); our method did not measure levels of the much more common ‘canonical’ stem-loop histone mRNAs (which are replication-dependent, i.e. highly expressed in S-phase) (121). Thus, the differential expression finding for histones relies on the small percentage with poly(A) tails (e.g. estimated to be <5% for H2A histones (120)), adding another cautionary note.
Using expression profiling as a tool for identifying genes involved in the pathogenesis of a trait of interest is in some ways a more complicated approach than linkage or association analysis, which are based on the genotypes. In contrast to genotypes, gene expression varies between tissues/cell types, is not constant during the lifetime and is influenced by myriad environmental factors and their impact on the condition of the body/tissue in which a given cell resides. In an expression profiling study, directionality of effect is conceptually uncertain, in contrast to linkage/association studies where it is generally clear that the genotype drives the phenotype. Also, confounder variables can have substantial impact, and lead to both false-negative and false-positive findings if not accounted for, which again is in contrast to genotype-based gene mapping approaches, where proper matching based on allele frequencies (i.e. ancestry) ensures proper statistical behavior, and where matching for other factors (such as sex or age) is often not necessary. To deal with these conceptual and important concerns, we have sought to match cases and controls as much as was practical, standardized our laboratory procedures, measured known and suspected confounders as part of our experiment and included them as nuisance parameters in our regression models, and focused on LCLs from a single transformation site/repository (RUCDR). Nonetheless, we cannot be 100% certain that all critical confounders have been accounted for, and each covariate that is either included in the model or not can have some influence on the set of genes whose expression is found to vary significantly between cases and controls. We have sought to gain confidence in our results by investigating the resulting gene lists in various ways, including evaluating whether there might be a logical connection (based on our present knowledge) between these genes and schizophrenia. We also compared the identified genes, and their eQTNs, with GWAS results, but we did not observe a robust overlap between both the approaches (i.e. congruence of differential expression with eQTN and MGS EA GWAS alleles). This lack of overlap should not, however, be interpreted to mean that our differential expression findings are false. There are many potential reasons for the lack of consistency in findings: (i) it is possible that the identified genes act downstream of schizophrenia (i.e. they are not involved in disease etiology, but rather in pathophysiology). We used LCLs, which are presumably fairly removed from state aspects of the individual (and their environmental exposures) from which the cell lines were derived, which casts some doubt on this possibility. (ii) It is possible that rare variation is responsible for the differing expression level, which therefore could have led to these genes not having been identified in the GWAS, while the quantified transcript levels constitute a gene-based read-out of all variants influencing a gene's expression level, regardless of the frequency. (iii) The sample sizes of our MGS expression dataset and the MGS GWAS dataset are of a limited size, and lack of power and statistical randomness can thus have a large impact. Ultimately, the future will tell whether the identified connections between schizophrenia and genes differentially expressed by schizophrenia status here are real. The present paper provides an impetus to investigate these hypotheses more deeply using different approaches.
Conclusion
Our main findings of genes differentially expressed by schizophrenia status implicate numerous genes of potential relevance to the illness (Table 2), including histones at the xMHC region showing convergence to the previous GWAS (10–13), a significant enrichment of immune-related genes supportive of immune hypotheses of schizophrenia, and many novel genes. The lack of supported gene pathways for the differentially expressed genes is consistent with heterogeneity of pathophysiology for schizophrenia. We note that many of the statistically significant differentially expressed genes (Supplementary Material, Table S1) cannot be individually connected with any of the current pathophysiological hypotheses of the disease either. While these observations might be explained by the fact that our knowledge of the pathophysiology of schizophrenia is still tentative and incomplete, we cannot rule out that our results present a higher rate of false positives than expected. Future work will be aimed at replication efforts in independent samples (including a second MGS sample to have transcription profiling via RNAseq), as well as further integration beyond transcriptomics and GWAS, such as joint analyses with future deep resequencing data. Replicated differentially expressed genes may provide insight into the pathophysiology of schizophrenia, potentially along with novel therapeutic targets.
MATERIALS AND METHODS
Subjects
The MGS sample is the most accessed National Institute of Mental Health (NIMH) GWAS database of Genotypes and Phenotypes (dbGaP, www.ncbi.nlm.nih.gov/gap) sample—467 investigator accessions as of October 2012 (294 for GAIN phs000021; 173 for nonGAIN phs000167). The collection is comprised of cases with well-characterized DSM-IV (3) schizophrenia or schizoaffective disorder, and controls screened for psychosis; for further phenotypic details, see the Supplementary Material, Text S1 and (10). We obtained the top five EA PCs (i.e. reflecting population ancestry) that were previously computed on SNP genotypes (Affymetrix 6.0) from our GWAS (10), which is found in two dbGaP entries (phs000021.v3.p2 and phs000167.v1.p1). We selected 446 case and 457 control samples from the available EA MGS participants with LCLs and GWAS data for the current study, meeting our LCL criteria (see below). We excluded from further consideration 3 samples EBV-transformed at Coriell Cell Repositories (appropriate statistical adjustment for such a small number is difficult), 26 samples with outlier sex transcript abundances (using probes for XIST, RPS4Y1, and EIF1AY) for reported sex and 15 samples without EBV copy number data in order to have full covariate data on all analyzed samples, leaving 413 cases and 446 controls for analysis (described in Table 1). We obtained institutional review board approval from NorthShore University HealthSystem and at the University of Texas Health Science Center at San Antonio (which also covers the Texas Biomedical Research Institute).
LCLs
We studied EBV-transformed (but not immortalized (122)) LCLs, which were all early stage (few cell divisions, or population doubling levels, since transformation, very far from progressing to ‘immortalization’). We measured the EBV copy number (EBV load) using a Taqman quantitative real time PCR (qPCR) assay (88,123,124) on DNA isolated from the LCLs. We excluded from consideration any LCLs known to be slowly growing (as indirectly indexed by over 100 days repository time to establish the LCL sufficiently to create adequate frozen stocks). We also excluded LCLs that displayed extreme clonality, i.e. pauciclonal or monoclonal, based on low heterozygosity (≤0.5) as determined by us using an immunoglobulin-based assay (125). We obtained the frozen LCLs from RUCDR, and after reviving the frozen LCLs per the repository guidelines we grew them in RPMI 1640 media, 15% → 25% fetal bovine serum (FBS, single lot, as was media to minimize variation), without antibiotics, at 37.0°C and 5% CO2 (both continuously monitored). We grew LCLs in batches of 40, with cases and controls intermixed, and included a culture replicate (same reference LCL, cultured de novo in each growing batch). On Day 7, we plated the LCLs at a concentration of 250 000 cells/ml, grew for 24 h, and then harvested. Harvest procedures included counting cells with an automated cell counter, using a luminescent cell viability assay for ATP levels, and placement into RNA protect preservative and −80°C for storage, all according to manufacturers' protocols, along with pelleting remaining cells for DNA extraction. Further details and values for these various measures are provided in the Supplementary Material, Text S1.
RNA
When large quantities of samples (∼500) are accumulated, we isolated total RNA with RNeasy96, ensured adequate yield and spectrophotometric ratios (A260/A280 ≥ 1.8 and A260/A230 ≥ 1.8), plated the samples (alternating case and control, inclusion of culture replicate as well as an RNA technical replicate) and shipped overnight on dry ice to the Biomedical Genomics Center, BGMC, of the University of Minnesota, which also evaluated the RNA samples with an Agilent bioanalyzer (ensuring an RNA integrity number ≥7.0 and a 28s/18s rRNA ratio ≥1.5). The BGMC performed the amplification/labeling (using the Eberwine T7 method (126), i.e. reverse transcription with an oligo-dT primer) and hybridization/scanning of these samples following manufacturers' protocols in two large continuous periods with the same personnel and single manufacturing batches of Ambion TotalPrep RNA labeling reagents and Illumina HT-12v4 BeadChips to minimize variation. The BGMC then returned the raw data (.idat) files, the loaded project files for viewing in Illumina software, and the non-normalized flat files (.txt) of expression values. The current project was completed with two equally sized groups (batches) of five 96-well plates each, with alternating case and control samples, one RNA technical replicate per plate and one culture replicate for each growing batch.
Data processing
We thus obtained expression profiles for 960 samples in two batches for 47 231 transcripts (assayed by 50 bp probes) using the Illumina HT-12v4 microarrays. We identified transcripts with significant expression at FDR ≤0.05 in both batches using a binomial test, counting the number of successful and unsuccessful detections (using Illumina's ‘detection P-value’ of ≤0.05 as the cut-off value) for each probe and computing a one-sided P-value (for a binomial test) before calculating the FDR across all probes. In our sample, this corresponded to removing any transcript significantly expressed (at P ≤ 0.05) in fewer than 62 of the 960 samples. We applied the Benjamini–Hochberg procedure (127) to generate the aforementioned FDR values, and chose FDR ≤0.05 to control the expected proportion of false positives at an ∼5% level. A total of 27 118 transcripts (∼57% of the 47 231 array probes) were significantly expressed at FDR ≤0.05. We removed all biological and technical replicate samples and only kept unrelated samples with full covariate data (413 cases and 446 controls) for analysis. Data quality, based on the average signal intensities, background noise levels, number of probes with significant signal and average correlation with other samples across expression levels, was excellent. The average correlation for the raw expression values across all significantly detected probes between 10 technical replicates, 22 culture replicates and all unrelated samples was in the expected order (0.99, 0.98 and 0.97, respectively). There was no evidence that any samples were of questionable quality. We examined [Partek and Lumi (R package) (128)] completion by RNA sample and probe to identify array outliers; this did not necessitate any sample or transcript exclusion. After removing the unexpressed transcripts, we performed a background noise subtraction. To avoid negative expression values, we shifted the expression of each batch by a constant value to shift the lowest value to 1, and then log2 transformed and quantile normalized each batch separately. We combined the batches and performed another quantile normalization. Finally, prior to further analyses, we performed a rank-based inverse normalization within each probe across samples.
Differential expression analyses
We used a multivariate linear regression analysis to relate gene expression to schizophrenia affection status, using the affection status along with various confounders as independent linear predictors of each transcript's expression level. We aimed to minimize the influence of potential confounders, which could either reduce the power to detect true differential expression by affection status and/or lead to false positives by incorporating 12 measured and suspected/known potential confounders as covariates in the analysis of differential expression: sex, age (at sample collection), age2, the top five genotypic PCs tagging ancestry (129) (previously derived from our MGS GWAS (10)), LCL energy status (ATP levels), growth rate of LCLs, LCL viral load (EBV copy number) and array batch, i.e. parameters known or strongly suspected to influence some transcripts' expression (e.g. as in (32)). To evaluate for differential expression by affection status, we performed a one-step linear regression analysis, including affection status and the 12 measured covariates to identify transcripts differently expressed by affection status (schizophrenia case versus control). We applied the Benjamini–Hochberg procedure (127) to perform multiple testing correction for all 14 588 transcripts detectable in ≥80% of the RNA samples, choosing FDR ≤0.05 to control the expected proportion of false positives at an ∼5% level for the differential expression analyses.
eQTNs
We took the 95 transcripts identified as significantly differentially expressed between cases and controls at FDR ≤0.05 in the RUCDR case–control analysis, and removed the transcripts on chromosomes X (6) and Y (0) to focus on autosomes (in mixed sex sample), two NCBI-withdrawn transcripts and nine transcripts with probes containing SNPs with MAF ≥1% in the CEU portion of 1000 genomes data. This left 78 transcripts (encoding for 75 genes). For this set of transcripts, we searched for putative cis-acting eQTNs (within 500 kb of the 5′ or of the 3′ end of, or within, the transcript) by association analysis using PLINK using the existing GWAS SNP genotype data (Affymetrix 6.0, (10)). This cis eQTNs search by association analysis used the expression data from the RUCDR-transformed sample (268 cases and 446 controls) after all processing described above, including a rank-based normalization step, and subsequent application of PLINK to perform the association analyses. We used a relatively wide 500 kb radius in both directions from a gene to detect putative cis eQTNs, given the fact that several genes are located in the extended MHC region with its extended LD structure. We focused our eQTN search on the putative cis interval around each gene, because it has been established well by many investigators, including ourselves (130), that proximal variants, presumed to act in cis, are very common (perhaps universal across genes) and often have a larger effect size than distal variants, presumed to act in trans. We applied the Benjamini–Hochberg procedure (127) to generate FDR values for eQTNs, choosing FDR ≤0.05 to control the expected proportion of false positives at an ∼5% level (since adjustment for genome-wide multiple testing, e.g. P = 5 × 10−8, is overly strict as we only investigated the genomic region around each gene).
Pathway and network analyses
We submitted our list of 89 genes differentially expressed (FDR ≤0.05) by affection to pathway and network analyses (34,35). We used DAPPLE (35) to evaluate the network connectivity of the gene list. DAPPLE identifies significant physical connectivity in direct and indirect interaction networks involving proteins encoded for input genes. It assesses the statistical significance of network connectivity and individual protein connectivity to other input proteins using a within-degree node-label permutation method. DAPPLE is based on the InWeb database that contains 428 430 reported protein interactions from multiple sources, including MINT, BIND, IntAct, KEGG annotated protein–protein interactions (PPrel), KEGG Enzymes involved in neighboring steps (ECrel) and Reactome. For input genes in the same direct network identified by DAPPLE, we then performed GO-term enrichment analysis and KEGG pathway analysis with the DAVID tool (34). We also performed GO-term and KEGG pathway enrichment analyses with DAVID for all the differentially expressed genes (i.e. not limited to those in a network as identified by DAPPLE). To explore whether the genes in schizophrenia risk CNV regions (1q21.1, 2p16.3, 15q13.3, 16p11.2, 22q11.21) might connect to the network generated by the differentially expressed transcripts in this study, we added the gene lists from these CNVs to the list of 89 genes differentially expressed in an additional DAPPLE analysis.
Data sharing
For the results of primary analyses, including cis eQTNs, we are sharing the data by depositing it into dbGaP and GEO (Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo). GWAS and phenotypic data for all subjects have already been deposited into dbGaP, and LCLs (and phenotypic data) are available through the NIMH repository (www.nimhgenetics.org) contractors (rucdr.rutgers.edu and zork.wustl.edu, respectively).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported primarily by the National Institutes of Health (NIH grant RC2MH090030 to A.R.S.); as well as for MGS by NIH grants (R01MH067257 to N.G.B., R01MH059588 to B.J.M., R01MH059571 to P.V.G., R01MH059565 to R.F., R01MH059587 to F.A., R01MH060870 to W.F.B., R01MH059566 to D.W.B., R01MH059586 to J.M.S., R01MH061675 to D.F.L., R01MH060879 to C.R.C., R01MH081800 to P.V.G., U01MH046276 to C.R.C., U01MH046289 to C. Kaufmann, U01MH046318 to M. T. Tsuang, U01MH079469 to P.V.G. and U01MH079470 to D.F.L.), the Genetic Association Information Network (GAIN, for genotyping of half of the EA sample and almost all the AA sample), and The Paul Michael Donovan Charitable Foundation. Genotyping was carried out by the Center for Genotyping and Analysis at the Broad Institute of Harvard and MIT (S.G. and D.B.M.), supported by NIH grant U54RR020278. Analyses done in San Antonio (E.I.D., H.H.H.G.) were conducted in facilities constructed with support from NIH grant RR017515, and a gift from the AT&T Foundation. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the GAIN QC team (G. R. Abecasis and J. Paschall) for making important contributions to the project; S. Purcell for assistance with PLINK; personnel at RUCDR (J. A. Tischfield, D. A. Fugman) for advice and assistance with the LCLs; C. Leites, J. Jacobi and S. Shi at NorthShore University HealthSystem for technical assistance; D. F. Levinson for helpful discussions early in the study design and for manuscript comments; T. Lehner at the NIMH for his support and scientific advice; MGS which includes P. V. Gejman, A. R. Sanders, J. Duan (NorthShore University HealthSystem, and University of Chicago, IL, USA), D. F. Levinson (Stanford University, CA, USA), J. Shi (National Cancer Institute, MD, USA), N. G. Buccola (Louisiana State University Health Sciences Center, LA, USA), B. J. Mowry (Queensland Centre for Mental Health Research, Brisbane and Queensland Brain Institute, The University of Queensland, Australia), R. Freedman, A. Olincy (University of Colorado Denver, CO, USA), F. Amin (Atlanta Veterans Affairs Medical Center and Emory University, GA, USA), D. W. Black (University of Iowa Carver College of Medicine, IA, USA), J. M. Silverman (Mount Sinai School of Medicine, NY, USA), W. F. Byerley (University of California at San Francisco, CA, USA), C. R. Cloninger (Washington University, MO, USA); and the study participants and the research staff at the study sites.
REFERENCES
- 1.Prince M., Patel V., Saxena S., Maj M., Maselko J., Phillips M.R., Rahman A. No health without mental health. Lancet. 2007;370:859–877. doi: 10.1016/S0140-6736(07)61238-0. [DOI] [PubMed] [Google Scholar]
- 2.Wu E.Q., Birnbaum H.G., Shi L., Ball D.E., Kessler R.C., Moulis M., Aggarwal J. The economic burden of schizophrenia in the United States in 2002. J. Clin. Psychiatry. 2005;66:1122–1129. doi: 10.4088/jcp.v66n0906. [DOI] [PubMed] [Google Scholar]
- 3.APA. Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- 4.McGrath J., Saha S., Chant D., Welham J. Schizophrenia: a concise overview of incidence, prevalence and mortality. Epidemiol. Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 5.Messias E.L., Chen C.Y., Eaton W.W. Epidemiology of schizophrenia: review of findings and myths. Psychiatr. Clin. North Am. 2007;30:323–338. doi: 10.1016/j.psc.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung A., Chue P. Sex differences in schizophrenia, a review of the literature. Acta Psychiatr. Scand. Suppl. 2000;401:3–38. doi: 10.1111/j.0065-1591.2000.0ap25.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown A.S. The environment and susceptibility to schizophrenia. Prog. Neurobiol. 2011;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benros M.E., Nielsen P.R., Nordentoft M., Eaton W.W., Dalton S.O., Mortensen P.B. Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am. J. Psychiatry. 2011;168:1303–1310. doi: 10.1176/appi.ajp.2011.11030516. [DOI] [PubMed] [Google Scholar]
- 9.Cardno A.G., Gottesman I. Twin studies of schizophrenia: from bow-and-arrow concordances to Star Wars Mx and functional genomics. Am. J. Med. Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 10.Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe'er I., Dudbridge F., Holmans P.A., Whittemore A.S., Mowry B.J., et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietilainen O.P., Mors O., Mortensen P.B., et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.PGC. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wray N.R., Visscher P.M. Narrowing the boundaries of the genetic architecture of schizophrenia. Schizophr. Bull. 2010;36:14–23. doi: 10.1093/schbul/sbp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda M., Aleksic B., Kinoshita Y., Okochi T., Kawashima K., Kushima I., Ito Y., Nakamura Y., Kishi T., Okumura T., et al. Genome-wide association study of schizophrenia in a Japanese population. Biol. Psychiatry. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Lee S.H., DeCandia T.R., Ripke S., Yang J., Sullivan P.F., Goddard M.E., Keller M.C., Visscher P.M., Wray N.R. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman I.I., Shields J. A polygenic theory of schizophrenia. Proc. Natl Acad. Sci. USA. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szulwach K.E., Li X., Smrt R.D., Li Y., Luo Y., Lin L., Santistevan N.J., Li W., Zhao X., Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell. Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smrt R.D., Szulwach K.E., Pfeiffer R.L., Li X., Guo W., Pathania M., Teng Z.Q., Luo Y., Peng J., Bordey A., et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schork A.J., Thompson W.K., Pham P., Torkamani A., Roddey J.C., Sullivan P.F., Kelsoe J.R., O'Donovan M.C., Furberg H., Schork N.J., et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levinson D.F., Duan J., Oh S., Wang K., Sanders A.R., Shi J., Zhang N., Mowry B.J., Olincy A., Amin F., et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dermitzakis E.T. From gene expression to disease risk. Nat. Genet. 2008;40:492–493. doi: 10.1038/ng0508-492. [DOI] [PubMed] [Google Scholar]
- 25.Farber C.R., Lusis A.J. Integrating global gene expression analysis and genetics. Adv. Genet. 2008;60:571–601. doi: 10.1016/S0065-2660(07)00420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery S.B., Dermitzakis E.T. The resolution of the genetics of gene expression. Hum. Mol. Genet. 2009;18:R211–R215. doi: 10.1093/hmg/ddp400. [DOI] [PubMed] [Google Scholar]
- 27.Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarti A., Kapoor A. Genetics. Mendelian puzzles. Science. 2012;335:930–931. doi: 10.1126/science.1219301. [DOI] [PubMed] [Google Scholar]
- 29.Suarez B.K., Duan J., Sanders A.R., Hinrichs A.L., Jin C.H., Hou C., Buccola N.G., Hale N., Weilbaecher A.N., Nertney D.A., et al. Genomewide linkage scan of 409 European-Ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. Am. J. Hum. Genet. 2006;78:315–333. doi: 10.1086/500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders A.R., Duan J., Levinson D.F., Shi J., He D., Hou C., Burrell G.J., Rice J.P., Nertney D.A., Olincy A., et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am. J. Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 31.Sanders A.R., Levinson D.F., Duan J., Dennis J.M., Li R., Kendler K.S., Rice J.P., Shi J., Mowry B.J., Amin F., et al. The Internet-based MGS2 control sample: self-report of mental illness. Am. J. Psychiatry. 2010;167:854–865. doi: 10.1176/appi.ajp.2010.09071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choy E., Yelensky R., Bonakdar S., Plenge R.M., Saxena R., De Jager P.L., Shaw S.Y., Wolfish C.S., Slavik J.M., Cotsapas C., et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caliskan M., Cusanovich D.A., Ober C., Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum. Mol. Genet. 2011;20:1643–1652. doi: 10.1093/hmg/ddr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., Cotsapas C., Daly M.J. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medzhitov R., Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 37.Tagai C., Morita S., Shiraishi T., Miyaji K., Iwamuro S. Antimicrobial properties of arginine- and lysine-rich histones and involvement of bacterial outer membrane protease T in their differential mode of actions. Peptides. 2011;32:2003–2009. doi: 10.1016/j.peptides.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Kim H.S., Cho J.H., Park H.W., Yoon H., Kim M.S., Kim S.C. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J. Immunol. 2002;168:2356–2364. doi: 10.4049/jimmunol.168.5.2356. [DOI] [PubMed] [Google Scholar]
- 39.Witkin S.S., Linhares I.M., Bongiovanni A.M., Herway C., Skupski D. Unique alterations in infection-induced immune activation during pregnancy. BJOG. 2011;118:145–153. doi: 10.1111/j.1471-0528.2010.02773.x. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki H., Iwamuro S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug. Targets. 2008;8:195–205. doi: 10.2174/1871526510808030195. [DOI] [PubMed] [Google Scholar]
- 41.Wiesner J., Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 42.Malik H.S., Henikoff S. Phylogenomics of the nucleosome. Nat. Struct. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 43.Akbarian S. Epigenetics of schizophrenia. Curr. Top. Behav. Neurosci. 2010;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- 44.Guidotti A., Auta J., Chen Y., Davis J.M., Dong E., Gavin D.P., Grayson D.R., Matrisciano F., Pinna G., Satta R., et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2010;60:1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matrisciano F., Tueting P., Dalal I., Kadriu B., Grayson D.R., Davis J.M., Nicoletti F., Guidotti A. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–194. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Jong S., van Eijk K.R., Zeegers D.W., Strengman E., Janson E., Veldink J.H., van den Berg L.H., Cahn W., Kahn R.S., Boks M.P., et al. Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur. J. Hum. Genet. 2012;20:1004–1008. doi: 10.1038/ejhg.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y., Chopra V., Chopra R., Locascio J.J., Liao Z., Ding H., Zheng B., Matson W.R., Ferrante R.J., Rosas H.D., et al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc. Natl Acad. Sci. USA. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eaton W.W., Byrne M., Ewald H., Mors O., Chen C.Y., Agerbo E., Mortensen P.B. Association of schizophrenia and autoimmune diseases: linkage of Danish national registers. Am. J. Psychiatry. 2006;163:521–528. doi: 10.1176/appi.ajp.163.3.521. [DOI] [PubMed] [Google Scholar]
- 50.Eaton W.W., Pedersen M.G., Nielsen P.R., Mortensen P.B. Autoimmune diseases, bipolar disorder, and non-affective psychosis. Bipolar Disord. 2010;12:638–646. doi: 10.1111/j.1399-5618.2010.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown A.S., Derkits E.J. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am. J. Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laan W., Grobbee D.E., Selten J.P., Heijnen C.J., Kahn R.S., Burger H. Adjuvant aspirin therapy reduces symptoms of schizophrenia spectrum disorders: results from a randomized, double-blind, placebo-controlled trial. J. Clin. Psychiatry. 2010;71:520–527. doi: 10.4088/JCP.09m05117yel. [DOI] [PubMed] [Google Scholar]
- 53.Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Jr, Wright M.W., et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 54.Sklar P., Ripke S., Scott L.J., Andreassen O.A., Cichon S., Craddock N., Edenberg H.J., Nurnberger J.I., Jr, Rietschel M., Blackwood D., et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willour V.L., Seifuddin F., Mahon P.B., Jancic D., Pirooznia M., Steele J., Schweizer B., Goes F.S., Mondimore F.M., Mackinnon D.F., et al. A genome-wide association study of attempted suicide. Mol. Psychiatry. 2012;17:433–444. doi: 10.1038/mp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neale B.M., Medland S.E., Ripke S., Asherson P., Franke B., Lesch K.P., Faraone S.V., Nguyen T.T., Schafer H., Holmans P., et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., Sykes N., Pagnamenta A.T., et al. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bierut L.J., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O.F., Swan G.E., Rutter J., Bertelsen S., Fox L., et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tobacco_and_Genetics_Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S., et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo L., Zhang C.K., Wang F., Li C.S., Zhao H., Lu L., Zhang X.Y., Zhang H., Zhang F., Krystal J.H., et al. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS ONE. 2011;6:e26726. doi: 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bierut L.J., Agrawal A., Bucholz K.K., Doheny K.F., Laurie C., Pugh E., Fisher S., Fox L., Howells W., Bertelsen S., et al. A genome-wide association study of alcohol dependence. Proc. Natl Acad. Sci. USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edenberg H.J., Koller D.L., Xuei X., Wetherill L., McClintick J.N., Almasy L., Bierut L.J., Bucholz K.K., Goate A., Aliev F., et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 65.Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 66.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 67.Gretarsdottir S., Thorleifsson G., Reynisdottir S.T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S.M., Einarsson O.B., Gudjonsdottir H.M., et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 68.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 69.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 70.Petretto E., Mangion J., Dickens N.J., Cook S.A., Kumaran M.K., Lu H., Fischer J., Maatz H., Kren V., Pravenec M., et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Heerden J.H., Conesa A., Stein D.J., Montaner D., Russell V., Illing N. Parallel changes in gene expression in peripheral blood mononuclear cells and the brain after maternal separation in the mouse. BMC Res. Notes. 2009;2:195. doi: 10.1186/1756-0500-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilad Y., Rifkin S.A., Pritchard J.K. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gerrits A., Li Y., Tesson B.M., Bystrykh L.V., Weersing E., Ausema A., Dontje B., Wang X., Breitling R., Jansen R.C., et al. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J.H., Park I.H., Gao Y., Li J.B., Li Z., Daley G.Q., Zhang K., Church G.M. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan Y., Abrahamsen G., McGrath J.J., Mackay-Sim A. Altered cell cycle dynamics in schizophrenia. Biol. Psychiatry. 2012;71:129–135. doi: 10.1016/j.biopsych.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 77.Lange C., Huttner W.B., Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 78.Hindley C., Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem. J. 2012;444:375–382. doi: 10.1042/BJ20112040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D., et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okada Y., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Kawaguchi T., Stahl E.A., Kurreeman F.A., Nishida N., et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 2012;44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 81.Chen S.J., Chao Y.L., Chen C.Y., Chang C.M., Wu E.C., Wu C.S., Yeh H.H., Chen C.H., Tsai H.J. Prevalence of autoimmune diseases in in-patients with schizophrenia: nationwide population-based study. Br. J. Psychiatry. 2012;200:374–380. doi: 10.1192/bjp.bp.111.092098. [DOI] [PubMed] [Google Scholar]
- 82.Leucht S., Burkard T., Henderson J., Maj M., Sartorius N. Physical illness and schizophrenia: a review of the literature. Acta Psychiatr. Scand. 2007;116:317–333. doi: 10.1111/j.1600-0447.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- 83.Gorwood P., Pouchot J., Vinceneux P., Puechal X., Flipo R.M., De Bandt M., Ades J. Rheumatoid arthritis and schizophrenia: a negative association at a dimensional level. Schizophr. Res. 2004;66:21–29. doi: 10.1016/s0920-9964(03)00017-3. [DOI] [PubMed] [Google Scholar]
- 84.Gilvarry C.M., Sham P.C., Jones P.B., Cannon M., Wright P., Lewis S.W., Bebbington P., Toone B.K., Murray R.M. Family history of autoimmune diseases in psychosis. Schizophr. Res. 1996;19:33–40. doi: 10.1016/0920-9964(95)00045-3. [DOI] [PubMed] [Google Scholar]
- 85.Knott T.K., Madany P.A., Faden A.A., Xu M., Strotmann J., Henion T.R., Schwarting G.A. Olfactory discrimination largely persists in mice with defects in odorant receptor expression and axon guidance. Neural Dev. 2012;7:17. doi: 10.1186/1749-8104-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santarelli D.M., Beveridge N.J., Tooney P.A., Cairns M.J. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 87.Beveridge N.J., Gardiner E., Carroll A.P., Tooney P.A., Cairns M.J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 89.Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 90.Cuellar T.L., Davis T.H., Nelson P.T., Loeb G.B., Harfe B.D., Ullian E., McManus M.T. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl Acad. Sci. USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davis T.H., Cuellar T.L., Koch S.M., Barker A.J., Harfe B.D., McManus M.T., Ullian E.M. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang T., Liu Y., Huang M., Zhao X., Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J. Mol. Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 93.Im H.I., Kenny P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu B., Hsu P.K., Karayiorgou M., Gogos J.A. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol. Dis. 2012;46:291–301. doi: 10.1016/j.nbd.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goldie B.J., Cairns M.J. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol. Neurobiol. 2012;45:99–108. doi: 10.1007/s12035-011-8222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wright C., Turner J.A., Calhoun V.D., Perrone-Bizzozero N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Y., Wang J., Lu X., Song X., Ye Y., Zhou J., Ying B., Wang L. Evaluation of six SNPs of MicroRNA machinery genes and risk of schizophrenia. J. Mol. Neurosci. 2013;49:594–599. doi: 10.1007/s12031-012-9887-1. [DOI] [PubMed] [Google Scholar]
- 98.Beveridge N.J., Santarelli D.M., Wang X., Tooney P.A., Webster M.J., Weickert C.S., Cairns M.J. Maturation of the human dorsolateral prefrontal cortex coincides with a dynamic shift in MicroRNA expression. Schizophr. Bull. 2013 doi: 10.1093/schbul/sbs198. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cao Q., Martinez M., Zhang J., Sanders A.R., Badner J.A., Cravchik A., Markey C.J., Beshah E., Guroff J.J., Maxwell M.E., et al. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics. 1997;43:1–8. doi: 10.1006/geno.1997.4815. [DOI] [PubMed] [Google Scholar]
- 100.Martinez M., Goldin L.R., Cao Q., Zhang J., Sanders A.R., Nancarrow D.J., Taylor J.M., Levinson D.F., Kirby A., Crowe R.R., et al. Follow-up study on a susceptibility locus for schizophrenia on chromosome 6q. Am. J. Med. Genet. 1999;88:337–343. [PubMed] [Google Scholar]
- 101.Lewis C.M., Levinson D.F., Wise L.H., DeLisi L.E., Straub R.E., Hovatta I., Williams N.M., Schwab S.G., Pulver A.E., Faraone S.V., et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am. J. Hum. Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duan J., Martinez M., Sanders A.R., Hou C., Saitou N., Kitano T., Mowry B.J., Crowe R.R., Silverman J.M., Levinson D.F., et al. Polymorphisms in the trace amine receptor 4 (TRAR4) gene on chromosome 6q23.2 are associated with susceptibility to schizophrenia. Am. J. Hum. Genet. 2004;75:624–638. doi: 10.1086/424887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vladimirov V., Thiselton D.L., Kuo P.H., McClay J., Fanous A., Wormley B., Vittum J., Ribble R., Moher B., van den Oord E., et al. A region of 35 kb containing the trace amine associate receptor 6 (TAAR6) gene is associated with schizophrenia in the Irish study of high-density schizophrenia families. Mol. Psychiatry. 2007;12:842–853. doi: 10.1038/sj.mp.4001984. [DOI] [PubMed] [Google Scholar]
- 104.Pae C.U., Yu H.S., Amann D., Kim J.J., Lee C.U., Lee S.J., Jun T.Y., Lee C., Paik I.H., Patkar A.A., et al. Association of the trace amine associated receptor 6 (TAAR6) gene with schizophrenia and bipolar disorder in a Korean case–control sample. J. Psychiatr. Res. 2008;42:35–40. doi: 10.1016/j.jpsychires.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 105.Chambers K.J., Tonkin L.A., Chang E., Shelton D.N., Linskens M.H., Funk W.D. Identification and cloning of a sequence homologue of dopamine beta-hydroxylase. Gene. 1998;218:111–120. doi: 10.1016/s0378-1119(98)00344-8. [DOI] [PubMed] [Google Scholar]
- 106.Pratscher B., Friedrich C., Goger W., Allen M., Fink D., Thallinger C., Wolschek M., Frei K., Schofer C., Pehamberger H., et al. Characterization of NKIP: a novel, Na+/K+-ATPase interacting protein mediates neural differentiation and apoptosis. Exp. Cell Res. 2008;314:463–477. doi: 10.1016/j.yexcr.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 107.Straub R.E., Jiang Y., MacLean C.J., Ma Y., Webb B.T., Myakishev M.V., Harris-Kerr C., Wormley B., Sadek H., Kadambi B., et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Svenningsson P., Chergui K., Rachleff I., Flajolet M., Zhang X., El Yacoubi M., Vaugeois J.M., Nomikos G.G., Greengard P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 109.Svenningsson P., Greengard P. P11 (S100A10)–an inducible adaptor protein that modulates neuronal functions. Curr. Opin. Pharmacol. 2007;7:27–32. doi: 10.1016/j.coph.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L., Su T.P., Choi K., Maree W., Li C.T., Chung M.Y., Chen Y.S., Bai Y.M., Chou Y.H., Barker J.L., et al. P11 (S100A10) as a potential biomarker of psychiatric patients at risk of suicide. J. Psychiatr. Res. 2011;45:435–441. doi: 10.1016/j.jpsychires.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 111.Zhang L., Li C.T., Su T.P., Hu X.Z., Lanius R.A., Webster M.J., Chung M.Y., Chen Y.S., Bai Y.M., Barker J.L., et al. P11 expression and PET in bipolar disorders. J. Psychiatr. Res. 2011;45:1426–1431. doi: 10.1016/j.jpsychires.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 112.Pihlstrom L., Axelsson G., Bjornara K.A., Dizdar N., Fardell C., Forsgren L., Holmberg B., Larsen J.P., Linder J., Nissbrandt H., et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson's disease. Neurobiol. Aging. 2013;34:1708.e1707–e1713. doi: 10.1016/j.neurobiolaging.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 113.Sharma M., Ioannidis J.P., Aasly J.O., Annesi G., Brice A., Van Broeckhoven C., Bertram L., Bozi M., Crosiers D., Clarke C., et al. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79:659–667. doi: 10.1212/WNL.0b013e318264e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lill C.M., Roehr J.T., McQueen M.B., Kavvoura F.K., Bagade S., Schjeide B.M., Schjeide L.M., Meissner E., Zauft U., Allen N.C., et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: the PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nalls M.A., Plagnol V., Hernandez D.G., Sharma M., Sheerin U.M., Saad M., Simon-Sanchez J., Schulte C., Lesage S., Sveinbjornsdottir S., et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yokota H., Tsujita T., Okazaki Y., Kikuya E., Oishi M. Polymorphic 33-bp repeats with promoter-like activity in synaptotagmin 11 gene. DNA Res. 2003;10:287–289. doi: 10.1093/dnares/10.6.287. [DOI] [PubMed] [Google Scholar]
- 117.Inoue S., Imamura A., Okazaki Y., Yokota H., Arai M., Hayashi N., Furukawa A., Itokawa M., Oishi M. Synaptotagmin XI as a candidate gene for susceptibility to schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:332–340. doi: 10.1002/ajmg.b.30465. [DOI] [PubMed] [Google Scholar]
- 118.Yeo H., Kim H.W., Mo J., Lee D., Han S., Hong S., Koh M.J., Sun W., Choi S., Rhyu I.J., et al. Developmental expression and subcellular distribution of synaptotagmin 11 in rat hippocampus. Neuroscience. 2012;225:35–43. doi: 10.1016/j.neuroscience.2012.08.062. [DOI] [PubMed] [Google Scholar]
- 119.Heintz N., Sive H.L., Roeder R.G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shepard P.J., Choi E.A., Lu J., Flanagan L.A., Hertel K.J., Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marzluff W.F., Wagner E.J., Duronio R.J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sugimoto M., Furuichi Y., Ide T., Goto M. Incorrect use of ‘immortalization’ for B-lymphoblastoid cell lines transformed by Epstein-Barr virus. J. Virol. 1999;73:9690–9691. doi: 10.1128/jvi.73.11.9690-9691.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beaubier N.T., Hart A.P., Bartolo C., Willman C.L., Viswanatha D.S. Comparison of capillary electrophoresis and polyacrylamide gel electrophoresis for the evaluation of T and B cell clonality by polymerase chain reaction. Diagn. Mol. Pathol. 2000;9:121–131. doi: 10.1097/00019606-200009000-00001. [DOI] [PubMed] [Google Scholar]
- 126.Van Gelder R.N., von Zastrow M.E., Yool A., Dement W.C., Barchas J.D., Eberwine J.H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 128.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 129.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 130.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 131.Wyllie A.H. Where, O death, is thy sting?’ A brief review of apoptosis biology. Mol. Neurobiol. 2010;42:4–9. doi: 10.1007/s12035-010-8125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chinnadurai G., Vijayalingam S., Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27(Suppl 1)):S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sasaki T., Kitagawa K., Yagita Y., Sugiura S., Omura-Matsuoka E., Tanaka S., Matsushita K., Okano H., Tsujimoto Y., Hori M. Bcl2 enhances survival of newborn neurons in the normal and ischemic hippocampus. J. Neurosci. Res. 2006;84:1187–1196. doi: 10.1002/jnr.21036. [DOI] [PubMed] [Google Scholar]
- 134.Sorce S., Schiavone S., Tucci P., Colaianna M., Jaquet V., Cuomo V., Dubois-Dauphin M., Trabace L., Krause K.H. The NADPH oxidase NOX2 controls glutamate release: a novel mechanism involved in psychosis-like ketamine responses. J. Neurosci. 2010;30:11317–11325. doi: 10.1523/JNEUROSCI.1491-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krystal J.H., Karper L.P., Seibyl J.P., Freeman G.K., Delaney R., Bremner J.D., Heninger G.R., Bowers M.B., Jr, Charney D.S. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch. Gen. Psychiatry. 1994;51:199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- 136.Sadvakassova G., Dobocan M.C., Difalco M.R., Congote L.F. Regulator of differentiation 1 (ROD1) binds to the amphipathic C-terminal peptide of thrombospondin-4 and is involved in its mitogenic activity. J. Cell Physiol. 2009;220:672–679. doi: 10.1002/jcp.21817. [DOI] [PubMed] [Google Scholar]
- 137.Franken P., Lopez-Molina L., Marcacci L., Schibler U., Tafti M. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J. Neurosci. 2000;20:617–625. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang S., Van Dongen H.P., Wang K., Berrettini W., Bucan M. Assessment of circadian function in fibroblasts of patients with bipolar disorder. Mol. Psychiatry. 2009;14:143–155. doi: 10.1038/mp.2008.10. [DOI] [PubMed] [Google Scholar]
- 139.Niculescu A.B., 3rd, Segal D.S., Kuczenski R., Barrett T., Hauger R.L., Kelsoe J.R. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol. Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- 140.Kyosseva S.V., Elbein A.D., Hutton T.L., Griffin S.T., Mrak R.E., Sturner W.Q., Karson C.N. Increased levels of transcription factors Elk-1, cyclic adenosine monophosphate response element-binding protein, and activating transcription factor 2 in the cerebellar vermis of schizophrenic patients. Arch. Gen. Psychiatry. 2000;57:685–691. doi: 10.1001/archpsyc.57.7.685. [DOI] [PubMed] [Google Scholar]
- 141.Oksenberg J.R., Baranzini S.E., Sawcer S., Hauser S.L. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat. Rev. Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 142.Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., Ivinson A.J., et al. Risk alleles for multiple sclerosis identified by a genome-wide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 143.Hoppenbrouwers I.A., Aulchenko Y.S., Ebers G.C., Ramagopalan S.V., Oostra B.A., van Duijn C.M., Hintzen R.Q. EVI5 Is a risk gene for multiple sclerosis. Genes Immun. 2008;9:334–337. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- 144.Vestal D.J., Jeyaratnam J.A. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hu Y., Wang J., Yang B., Zheng N., Qin M., Ji Y., Lin G., Tian L., Wu X., Wu L., et al. Guanylate binding protein 4 negatively regulates virus-induced type I IFN and antiviral response by targeting IFN regulatory factor 7. J. Immunol. 2011;187:6456–6462. doi: 10.4049/jimmunol.1003691. [DOI] [PubMed] [Google Scholar]
- 146.Thornalley P.J. Glyoxalase I—structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 147.Toyosima M., Maekawa M., Toyota T., Iwayama Y., Arai M., Ichikawa T., Miyashita M., Arinami T., Itokawa M., Yoshikawa T. Schizophrenia with the 22q11.2 deletion and additional genetic defects: case history. Br. J. Psychiatry. 2011;199:245–246. doi: 10.1192/bjp.bp.111.093849. [DOI] [PubMed] [Google Scholar]
- 148.Barua M., Jenkins E.C., Chen W., Kuizon S., Pullarkat R.K., Junaid M.A. Glyoxalase I polymorphism rs2736654 causing the Ala111Glu substitution modulates enzyme activity—implications for autism. Autism Res. 2011;4:262–270. doi: 10.1002/aur.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kurz A., Rabbani N., Walter M., Bonin M., Thornalley P., Auburger G., Gispert S. Alpha-synuclein deficiency leads to increased glyoxalase I expression and glycation stress. Cell Mol. Life Sci. 2011;68:721–733. doi: 10.1007/s00018-010-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Puffenberger E.G., Jinks R.N., Wang H., Xin B., Fiorentini C., Sherman E.A., Degrazio D., Shaw C., Sougnez C., Cibulskis K., et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 2012;33:1639–1646. doi: 10.1002/humu.22237. [DOI] [PubMed] [Google Scholar]
- 151.Barnes W.G., Hough L.B. Membrane-bound histamine N-methyltransferase in mouse brain: possible role in the synaptic inactivation of neuronal histamine. J. Neurochem. 2002;82:1262–1271. doi: 10.1046/j.1471-4159.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 152.Tiligada E., Kyriakidis K., Chazot P.L., Passani M.B. Histamine pharmacology and new CNS drug targets. CNS Neurosci. Ther. 2011;17:620–628. doi: 10.1111/j.1755-5949.2010.00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oroszi G., Enoch M.A., Chun J., Virkkunen M., Goldman D. Thr105ile, a functional polymorphism of histamine N-methyltransferase, is associated with alcoholism in two independent populations. Alcohol. Clin. Exp. Res. 2005;29:303–309. doi: 10.1097/01.alc.0000156128.28257.2e. [DOI] [PubMed] [Google Scholar]
- 154.Palada V., Terzic J., Mazzulli J., Bwala G., Hagenah J., Peterlin B., Hung A.Y., Klein C., Krainc D. Histamine N-methyltransferase Thr105Ile polymorphism is associated with Parkinson's disease. Neurobiol. Aging. 2012;33:836.e831–836.e833. doi: 10.1016/j.neurobiolaging.2011.06.015. –. [DOI] [PubMed] [Google Scholar]
- 155.Saetre P., Emilsson L., Axelsson E., Kreuger J., Lindholm E., Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Arion D., Unger T., Lewis D.A., Levitt P., Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol. Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pontillo A., Catamo E., Arosio B., Mari D., Crovella S. NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2012;26:277–281. doi: 10.1097/WAD.0b013e318231a8ac. [DOI] [PubMed] [Google Scholar]
- 158.Sui J., Li H., Fang Y., Liu Y., Li M., Zhong B., Yang F., Zou Q., Wu Y. NLRP1 gene polymorphism influences gene transcription and is a risk factor for rheumatoid arthritis in Han Chinese. Arthritis Rheum. 2012;64:647–654. doi: 10.1002/art.33370. [DOI] [PubMed] [Google Scholar]
- 159.Kawauchi T., Shikanai M., Kosodo Y. Extra-cell cycle regulatory functions of cyclin-dependent kinases (CDK) and CDK inhibitor proteins contribute to brain development and neurological disorders. Genes Cells. 2013;18:176–194. doi: 10.1111/gtc.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nishimura Y.V., Sekine K., Chihama K., Nakajima K., Hoshino M., Nabeshima Y., Kawauchi T. Dissecting the factors involved in the locomotion mode of neuronal migration in the developing cerebral cortex. J. Biol. Chem. 2010;285:5878–5887. doi: 10.1074/jbc.M109.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lang F., Strutz-Seebohm N., Seebohm G., Lang U.E. Significance of SGK1 in the regulation of neuronal function. J. Physiol. 2010;588:3349–3354. doi: 10.1113/jphysiol.2010.190926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scales S.J., Chen Y.A., Yoo B.Y., Patel S.M., Doung Y.C., Scheller R.H. SNAREs contribute to the specificity of membrane fusion. Neuron. 2000;26:457–464. doi: 10.1016/s0896-6273(00)81177-0. [DOI] [PubMed] [Google Scholar]
- 163.Zhang Y., Shu L., Chen X. Syntaxin 6, a regulator of the protein trafficking machinery and a target of the p53 family, is required for cell adhesion and survival. J. Biol. Chem. 2008;283:30689–30698. doi: 10.1074/jbc.M801711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoglinger G.U., Melhem N.M., Dickson D.W., Sleiman P.M., Wang L.S., Klei L., Rademakers R., de Silva R., Litvan I., Riley D.E., et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.He Y., Sun S., Sha H., Liu Z., Yang L., Xue Z., Chen H., Qi L. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15:13–25. doi: 10.3727/105221610x12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen W., Duan S., Zhou J., Sun Y., Zheng Y., Gu N., Feng G., He L. A case–control study provides evidence of association for a functional polymorphism-197C/G in XBP1 to schizophrenia and suggests a sex-dependent effect. Biochem. Biophys. Res. Commun. 2004;319:866–870. doi: 10.1016/j.bbrc.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 167.Kakiuchi C., Ishiwata M., Umekage T., Tochigi M., Kohda K., Sasaki T., Kato T. Association of the XBP1–116C/G polymorphism with schizophrenia in the Japanese population. Psychiatry Clin. Neurosci. 2004;58:438–440. doi: 10.1111/j.1440-1819.2004.01280.x. [DOI] [PubMed] [Google Scholar]
- 168.Jonsson E.G., Cichon S., Schumacher J., Abou Jamra R., Schulze T.G., Deschner M., Forslund K., Hall H., Propping P., Czerski P.M., et al. Association study of a functional promoter polymorphism in the XBP1 gene and schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:71–75. doi: 10.1002/ajmg.b.30262. [DOI] [PubMed] [Google Scholar]
- 169.Watanabe Y., Fukui N., Muratake T., Amagane H., Kaneko N., Nunokawa A., Someya T. Association study of a functional promoter polymorphism of the X-box binding protein 1 gene in Japanese patients with schizophrenia. Psychiatry Clin. Neurosci. 2006;60:633–635. doi: 10.1111/j.1440-1819.2006.01570.x. [DOI] [PubMed] [Google Scholar]
- 170.Carter C.J. eIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia? Schizophr. Bull. 2007;33:1343–1353. doi: 10.1093/schbul/sbm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leckman J.F., Sholomskas D., Thompson W.D., Belanger A., Weissman M.M. Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch. Gen. Psychiatry. 1982;39:879–883. doi: 10.1001/archpsyc.1982.04290080001001. [DOI] [PubMed] [Google Scholar]
- 172.Kessler R.C., Andrews G., Mroczek D., Ustun B., Wittchen H.U. The World Health Organization composite international diagnostic interview short-form (CIDI-SF) Int. J. Methods Psychiatr. Res. 1998;7:171–185. [Google Scholar]
- 173.Nelson C.B., Kessler R.C., Mroczek D. 2001. Scoring the World Health Organization's Composite International Diagnostic Interview Short Form (CIDI-SF; v1.0 NOV98 for all disorders except OCD which is from v1.1 MAR99) www.hcp.med.harvard.edu/ncs/ftpdir/cidisf_readme.pdf. accessed March 18, 2009)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.