Abstract
The human apolipoprotein E (APOE) gene plays an important role in lipid metabolism. It has three common genetic variants, alleles ɛ2/ɛ3/ɛ4, which translate into three protein isoforms of apoE2, E3 and E4. These isoforms can differentially influence total serum cholesterol levels; therefore, APOE has been linked with cardiovascular disease. Additionally, its ɛ4 allele is strongly associated with the risk of Alzheimer's disease (AD), whereas the ɛ2 allele appears to have a modest protective effect for AD. Despite decades of research having illuminated multiple functional differences among the three apoE isoforms, the precise mechanisms through which different APOE alleles modify diseases risk remain incompletely understood. In this study, we examined the genomic structure of APOE in search for properties that may contribute novel biological consequences to the risk of disease. We identify one such element in the ɛ2/ɛ3/ɛ4 allele-carrying 3′-exon of APOE. We show that this exon is imbedded in a well-defined CpG island (CGI) that is highly methylated in the human postmortem brain. We demonstrate that this APOE CGI exhibits transcriptional enhancer/silencer activity. We provide evidence that this APOE CGI differentially modulates expression of genes at the APOE locus in a cell type-, DNA methylation- and ɛ2/ɛ3/ɛ4 allele-specific manner. These findings implicate a novel functional role for a 3′-exon CGI and support a modified mechanism of action for APOE in disease risk, involving not only the protein isoforms but also an epigenetically regulated transcriptional program at the APOE locus driven by the APOE CGI.
INTRODUCTION
The apolipoprotein E (APOE) gene and its protein product (apoE) play a key role in lipid metabolism, including the redistribution of lipoproteins and cholesterol. The liver produces the majority of circulating apoE that binds lipids and interacts with cell-surface membrane receptors to initiate cellular uptake of lipoprotein particles by the liver and other tissues (1). ApoE is also abundantly present in the central nervous system (2), where it promotes the transport of lipids to and from damaged neurons and thereby conducts important functions in neuronal maintenance, repair and homeostasis (3,4).
The APOE has been implicated in multiple diseases. Human apoE is a polymorphic protein, and the presence of either arginine or cysteine at amino acid positions 112 and 158 defines three common isoforms: E2, E3 and E4, which are encoded by the ɛ2, ɛ3 and ɛ4 genetic alleles, respectively (5). These isoforms are metabolically distinct and differ in both their affinity for lipoprotein particles and their binding to low-density lipoprotein (LDL) receptors (6). The isoforms influence total serum and LDL cholesterol levels (7), and thus, APOE has been linked with a higher risk of cardiovascular disease (CVD) (8–12). This ɛ2/ɛ3/ɛ4 allelic variation has also been associated with both familial and sporadic Alzheimer's disease (AD) (13). In Caucasian populations, the frequency of the ɛ4 allele is higher in AD subjects (ranging from 0.36 to 0.42) than in controls (0.14 to 0.16); this association has been consistently replicated in multiple studies including our own (13–15). Inheritance of an ɛ4 allele increases the risk of AD in a dose-dependent manner and predisposes subjects to an earlier age of onset (13,16). Conversely, the APOE ɛ2 allele appears to have a protective effect for AD.
In order to decipher APOE ɛ2/ɛ3/ɛ4 alleles' role in AD risk, mainstream research has focused on apoE isoform-specific differences in protein structure and function for the past two decades. Numerous hypotheses have been proposed for how apoE4 might increase the risk of AD, including Aβ aggregation and clearance (17,18), apoE protein domain interaction and neurotoxicity (19), neuroprotection (3,20), neuroinflammation (21), tau hyper-phosphorylation (22), apoE expression levels (23) and lipidation states (24). However, the precise mechanism(s) by which the APOE ɛ4 allele exerts its detrimental effect in AD remain elusive. For example, subjects who inherit the ɛ4 allele produce apoE4 protein early in development, yet the apoE4 isoform appears not to manifest any detrimental biological consequences until later in life. The presence of the ɛ4 allele is not necessary for AD (25) but instead modulates the risk for development of AD (26), and some individuals who inherit ɛ4 remain cognitively normal past 90 years of age (27,28). Even individuals with the rare ɛ4/ɛ4 homozygous genotype have, on average, a >50% chance of escaping AD (29), indicating that the ɛ4 allele and thus the E4 isoform alone is not sufficient to cause AD. Nevertheless, this ɛ4 allele has been consistently identified as the strongest genetic risk factor for developing late-onset AD in multiple studies and across many ethnic groups. Thus, there is a strong reason to deduce that APOE and its ɛ4 allele may play a pleiotropic role in AD, carrying additional functions distinct from its role as a coding template for the protein. Indeed multiple studies, including our own, have suggested the involvement of APOE in a complex transcriptional regulatory network that might ascribe to AD risk (30–34).
In this study, we inspected the genomic structure of APOE to search for elements that may contribute biological consequences, independent of apoE protein function. We identified a candidate element, a CpG island (CGI) located in the 3′-coding region of the APOE. We first investigated the epigenetic properties of this CGI by assessing its DNA methylation status in human postmortem brain (PMB) tissues. We then explored the function of this CGI by reporter assays to test its transcriptional regulatory activity. Finally, we used PMB and isolated brain cells from an APOE mouse model to validate this CGI's function in vivo.
RESULTS
A 3′-exon CGI is an integral epigenetic component of APOE
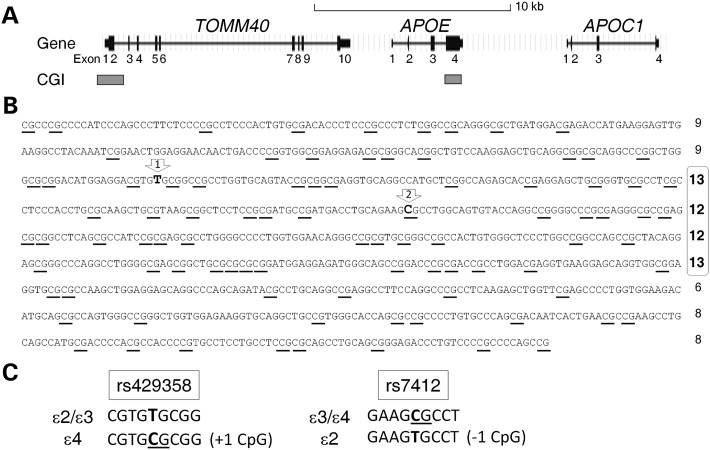
We applied in silico analysis to study the genomic features of the APOE region, using the UCSC Human Genome Browser (http://genome.ucsc.edu). Notably, whereas the APOE promoter does not contain a CGI, a single well-defined genic CGI coincides with APOE 3′-exon (Fig. 1A). This APOE CGI extends across 880 bp (hg19, chr19: 45 411 721–45 412 600; Fig. 1B), from intron 3 to the 3′-untranslated region of exon 4, which contains 90 CpG dinucleotides and carries a CG content of 72.8% with an observed-to-expected CpG ratio of 0.77. All of these features clearly surpass stringent cutoff criteria for CGI classification (35) (Table 1), indicating that this APOE region is a bona fide CGI. This CGI is also conserved in other mammals, including chimps, mice, rats, cows and dogs (UCSC Genome Browser). Detailed sequence analysis suggests the presence of a core region, occupying the center section, with a higher CpG load. The two single-nucleotide polymorphisms (SNPs) determining the ɛ2/ɛ3/ɛ4 alleles reside within this putative core region (Fig. 1B). Furthermore, when compared with ɛ2 and ɛ3, the ɛ4 allele introduces one additional CpG dinucleotide into this CGI and further saturates a small 12 bp region with four CpG sites; in contrast, the ɛ2 allele reduces one CpG and opens up a 34 bp CpG-free region (Fig. 1B and C).
Figure 1.
Gene map with CGIs at the APOE locus. (A) Genomic position of exons and CGIs. (B) Full-genomic sequence of the APOE CGI with 90 CpG dinucleotides (underlined). Numbers to the right represent CpG counts within each line of 100 nucleotides, and a potential core region (lines 3–6) is marked by numbers in bold font. (C) The two SNPs (rs429358 and rs7412) determining APOE ɛ2/ɛ3/ɛ4 alleles change the CpG content and landscape of the CGI; their positions are indicated by arrows #1 and #2 in (B).
Table 1.
Comparison of the human APOE CGI to standard and stringent CGI definitions
| Criteria | CGI definition |
APOE CGI | |
|---|---|---|---|
| Standard | Stringent | ||
| Size (bp) | ≥200 | ≥500 | 880 |
| % C or G | >50 | >55 | 72.8 |
| CpG ratio* | >0.6 | >0.65 | 0.77 |
*Ratio of observed to expected number of CpGs.
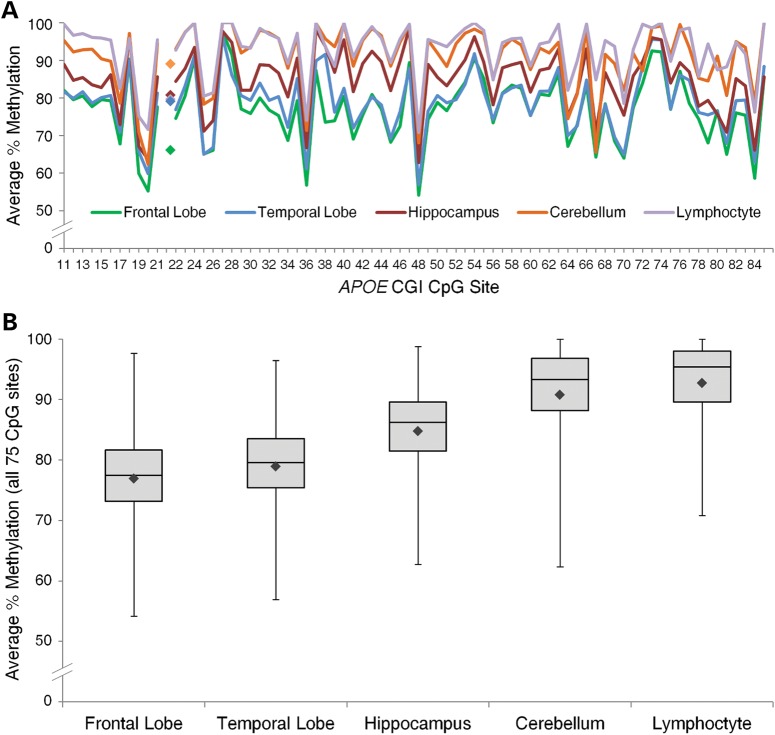
Because CGIs are highly enriched with CpG dinucleotides, and CpG sites are natural targets for DNA methylation, we assessed the methylation levels with single CpG site resolution across the APOE CGI. Using bisulfite Pyrosequencing, we quantified the percentage of methylation at 75 individual CpG sites (#11–85) in genomic DNA isolated from multiple human tissues (i.e. cerebellum, hippocampus, frontal lobe, temporal lobe and whole-blood lymphocytes) from nine AD and six age-matched control subjects (Supplementary Material, Table S1). Methylation levels were generated from 68 CpG sites with at least a 97% sample completion rate, the remaining 7 CpG sites had a completion rate slightly <80% due to lower quality of data generated by extended Pyrosequencing length from the priming site. The quality of the methylation quantification data was further assessed and validated by both experimental and technical replications (Supplementary Material, Fig. S1). All sites tested were highly methylated with mean methylation levels ranging from 54–100% at individual CpG sites (Fig. 2A). Methylation profiles were similar among all tissues tested with consistent up/down patterns between single CpGs (Fig. 2A and Supplementary Material, Fig. S2), suggesting a stringently regulated methylation and de-methylation mechanism at the level of individual CpG sites. Conversely, the overall degree of methylation varied noticeably across tissues (Fig. 2B), with all five tissue types differing significantly from each other (linear mixed effects model with Holm adjustment, P < 10−10 for all 10 pair-wise comparisons). The average methylation levels across all 75 CpG sites were lowest in frontal (76.9 ± 9.1%) and temporal lobes (78.9 ± 8.4%), with hippocampal (84.8 ± 8.4%) levels in the intermediate range and the highest levels in the cerebellum (90.8 ± 8.7%) and lymphocytes (92.8 ± 7.4%).
Figure 2.
Methylation profiles of the APOE CGI from human PMB tissues and lymphocytes. (A) The mean percentage of methylation at the 75 targeted CpG sites across tissues (n = 14 or 15) and the SNP rs429358 site for ɛ4/ɛ4 homozygous samples only (denoted by filled diamonds, n = 2 or 3). (B) For each tissue type, methylation percentages from all 15 subjects were averaged at each of the 75 CpG sites. The range and mean (denoted by filled black diamonds) methylation percentages across these 75 sites are displayed in a box-and-whisker plot. Mean methylation proportions across all 75 CpG sites were transformed, and statistically significant differences between all five tissues tested were detected (Holm-adjusted P < 10−10 for all 10 pair-wise comparisons).
Such a high methylation pattern was also observed in the ɛ4 allele (the ‘C’ variant of SNP rs429358), which is located between CpG sites 21 and 22, with average methylation levels of 68% in the frontal lobe, 82% in the temporal lobe, 83% in lymphocytes, 84% in the hippocampus, and 93% in the cerebellum (Supplementary Material, Fig. S2). Because the ɛ3 allele does not contain a CpG dinucleotide and cannot be methylated, the average percentage of methylation at this rs429358 SNP site was expectedly low in all tissues and was excluded from the CGI methylation profile analyses. Additionally, given the strong connection between APOE and AD, we extended our analysis to assess whether AD subjects have different APOE CGI methylation profiles compared with control subjects. Whereas methylation levels of several individual CpG sites were nominally statistically different between AD and controls in some tissues, significance was not maintained after correcting for multiple comparisons (except for the single case of CpG site 82 in the hippocampus, Holm-adjusted P < 0.001), and the mean methylation levels across all sites were not statistically different between the two groups. Nevertheless, such negative result might be due to the small sample size (n = 9 for AD and 6 for control) of this study. Future study with an expanded sample size should provide a more definitive conclusion.
Together, these data indicate that the APOE 3′-exon 4 is a well-defined CGI carrying a high level of DNA methylation in human brain tissues, and that the residing ɛ2/ɛ3/ɛ4 allele variants can alter the CpG load and methylation landscape of this CGI.
The APOE CGI can function as an enhancer of transcription
The presence of a CGI overlapping with the 3′-exon of a gene (3′-CGI), such as the APOE CGI, is extremely rare in the human genome (36), and the functions of these 3′-CGIs are largely unknown. Since CGIs are a critical element of the genome and the APOE CGI is highly conserved in mammals, we reasoned that this APOE CGI could be contributing novel effects to disease. Thus, we extended our study to decipher the biological function(s) of this CGI. Our previous studies have identified an extended region of the APOE locus in tight linkage disequilibrium with the ɛ2/ɛ3/ɛ4 alleles (37). This region consists of specific transcriptional regulatory haplotypes, influencing the expression levels of APOE in both cerebrospinal fluid (CSF) and PMB (33,34). Moreover, an ɛ4 allele-carrying fragment with potential enhancer activity was previously reported in a study from Chen et al. (31). These findings suggest the presence of a complex transcriptional regulatory network at the APOE locus, in which the APOE ɛ2/ɛ3/ɛ4 alleles likely play a direct role. Thus, we hypothesized that the APOE CGI is a transcriptional regulatory element and designed both ex vivo and in vivo experiments to test this putative activity.
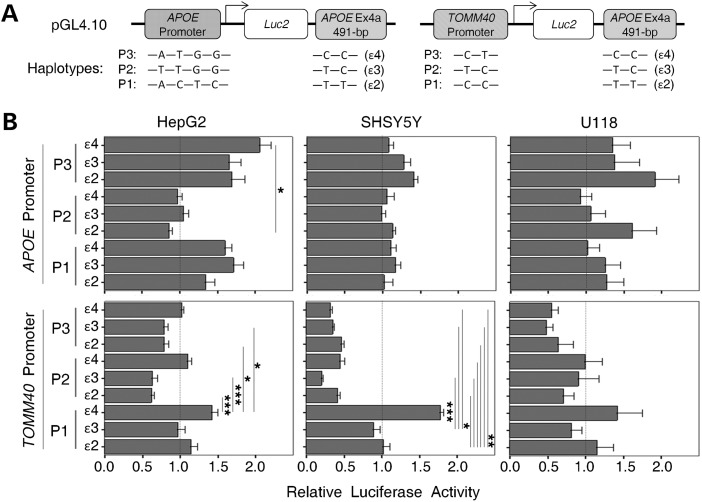
We first applied a pGL4-luciferase-based reporter system, in a cell-based transient assay, to evaluate the baseline un-methylated enhancer activity of the APOE CGI on the promoters of APOE and TOMM40. Previously, we identified three common APOE and TOMM40 promoter haplotype variants that influenced expression in human subjects (32). In this study, those promoter variants were cloned into the promoter position of reporter constructs, and a partial APOE CGI fragment (491 bp with ɛ2/ɛ3/ɛ4 allele variants) was inserted after the 3′-end of the luciferase gene (Fig. 3A). To assess effects in diverse cellular microenvironments, we evaluated luciferase expression in three human cell lines, HepG2 (hepatocytoma cells), SH-SY5Y (neuroblastoma cells) and U118 (astrocytoma cell). Hepatocyte and astrocyte cell lines were selected because both cell types are known to synthesize apoE, and we chose a neuronal cell line because of the clear association between AD and neuronal dysfunction. In these experiments, reporter constructs with the same promoter but different ɛ2/ɛ3/ɛ4 CGI variants expressed diverse luciferase activities, which were further altered by the cell type. For example, among the APOE promoter constructs, the APOE P1 and P3 promoters were enhanced by all three APOE CGI variants in HepG2 cells when compared with promoter-alone constructs (set at 1.0), and the highest reporter activity (∼2-fold increase) was observed with the P3-ɛ4 haplotype (Fig. 3B, left upper panel). In contrast, all three APOE CGI variants failed to enhance the APOE P2 promoter in HepG2. In SH-SY5Y, only modest non-significant enhancement by the three APOE CGI variants was observed regardless of APOE promoters, peaking with a ∼1.4-fold increase in luciferase activity with the APOE P3-ɛ2 haplotype (Fig. 3B, middle top panel). This P3-ɛ2 haplotype also produced the highest level of reporter activity in the U118 cells (Fig. 3B, right top panel).
Figure 3.
Enhancer/silencer effect of un-methylated APOE CGI in pGL4 reporter assays. (A) The diagram of reporter constructs with promoters of either APOE (left panel) or TOMM40 (right panel) and a 3′ APOE CGI fragment (491 bp) in the forward orientation. Haplotype compositions are also shown. (B) Constructs were transiently transfected into three cell lines (HepG2, SH-SY5Y and U118). Luciferase activity of each construct was measured and compared with its promoter-only construct counterpart (set at 1.0). Each reporter activity was generated from five experimental replicates. Expression levels between constructs were compared by one-way ANOVA [APOE promoter: HepG2, F(8, 45) = 2.99, P = 0.008; SH-SY5Y, F(8, 45) = 0.79, P = 0.61; U118, F(8, 63) = 0.39, P = 0.92; TOMM40 promoter: HepG2, F(8, 45) = 4.06, P = 0.001; SH-SY5Y, F(8, 45) = 23.81, P < 0.001; U118, F(8, 63) = 0.54, P = 0.82)]. Pair-wise comparisons were made with a post hoc Bonferroni test (*P < 0.05, **P < 0.01, ***P < 0.001).
Among the TOMM40 promoter constructs, the P1-ɛ4 haplotype in both HepG2 and SH-SY5Y cells showed the largest increase in luciferase activity (∼1.4 and ∼1.8-fold respectively), a difference that was statistically significant compared with several of the other haplotypes [Fig. 3B, left bottom and middle bottom panels, analysis of variance (ANOVA): HepG2, P = 0.001; SH-SY5Y, P < 0.001]. Conversely, all of the P2- and P3-related TOMM40 promoter constructs resulted in a >50% reduction in activity in SH-SY5Y cells. Similarly in both HepG2 and U118 cells, most haplotype constructs failed to reach the basal activity level of the promoter-alone constructs (set at 1.0). Remarkably, these results also suggest that the ɛ4 variant of the APOE CGI is able to function modestly as an enhancer on the TOMM40's P1 promoter variant in all three cell types, but in the majority of cases the TOMM40 promoter is down-regulated by the APOE CGI. Thus, the APOE CGI can influence TOMM40 promoter activity in either a positive (enhancer) or a negative (silencer) manner, based on the overall haplotype structure. This silencer effect was not observed in APOE promoter constructs, suggesting a broader impact of the APOE CGI on TOMM40 than APOE.
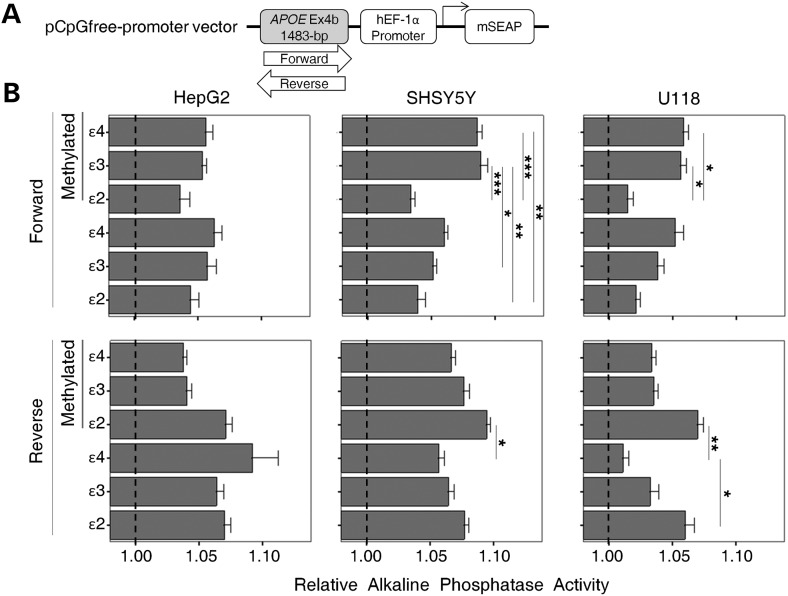
Because the APOE CGI is highly methylated in human PMB and biological effects of CGIs can be modified by DNA methylation, we further evaluated the enhancer activity of the methylated APOE CGI using a pCpG-free alkaline phosphatase (AP) reporter system (Fig. 4A). This pCpG-free promoter vector was specifically engineered by removing and replacing all the CpG dinucleotides so that DNA methylation would not influence the vector's rooted functions. The lack of CpGs within the vector restricted the in vitro CpG methylation to the inserted fragment only. For this pCpG-free vector-related reporter construct, we inserted a 1483 bp APOE CGI fragment into the 5′ site of the vector's native promoter hEF-1α (Fig. 4A). Six variants of the fragment including both forward and reverse orientations of the three ɛ2/ɛ3/ɛ4 allele variants were separately cloned. The APOE CGI of these constructs was then methylated in vitro, and both methylated and un-methylated versions of the same construct were generated. AP expression of these constructs was then evaluated in three human cell lines (i.e. HepG2, SH-SY5Y and U118).
Figure 4.
Enhancer effect of the methylated APOE CGI in pCpG-free vector reporter assays. (A) The diagram of reporter constructs with an APOE CGI fragment (1483 bp) in either the forward or reverse orientation, 5′ of the hEF-1 promoter. (B) The APOE CGI was either methylated by DNA methyltransferase treatment (methylation section) or left un-methylated and transiently transfected into three cell lines (HepG2, SH-SY5Y and U118). AP activity of each construct was measured and compared with its promoter-only construct counterpart (set at 1.0). Each reporter activity was generated from five experimental replicates. Expression levels between constructs were compared by one-way ANOVA [Forward: HepG2, F(5, 78) = 0.54, P = 0.74; SH-SY5Y, F(5, 66) = 7.39, P < 0.001; U118, F(5, 48) = 4.04, P = 0.004; Reverse: HepG2, F(5, 84) = 0.98, P = 0.43; SH-SY5Y, F(5, 66) = 2.84, P = 0.02; U118, F(5, 48) = 4.56, P = 0.002]. Pair-wise comparisons were made with a post hoc Bonferroni test (*P < 0.05, **P < 0.01, ***P < 0.001).
Increased AP activity was observed for all constructs with addition of the APOE CGI when compared with promoter-alone constructs (set at 1.0), regardless of methylation status and orientation (Fig. 4B). When stratified by orientation and methylation status, we observed an altered AP activity associated with specific allele and cell types. For example, in SH-SY5Y cells, the forward methylated ɛ3 CGI showed elevated enhancer effects, compared with its un-methylated counterpart (Fig. 4B, middle top panel, Bonferroni, P = 0.046). A similar effect was seen in the methylated ɛ4 CGI, yet this difference did not reach statistical significance (Fig. 4B, middle top panel, Bonferroni, P = 0.538). Furthermore, elevated AP activity was observed in the forward orientation for the methylated ɛ3 and ɛ4 CGIs compared with the methylated ɛ2, in both SH-SY5Y and U118 cells. (Fig. 4B, middle top and right top panels, ANOVA: SH-SY5Y, P < 0.001; U118, P = 0.004). In contrast, in the reverse orientation, the un-methylated ɛ2 CGI has significantly higher activity than the un-methylated ɛ4 in U118 cells (Fig. 4B, right bottom panel bottom section, Bonferroni, P = 0.017). These results suggest that the methylation status and orientation of the fragment also modify the functions of this CGI.
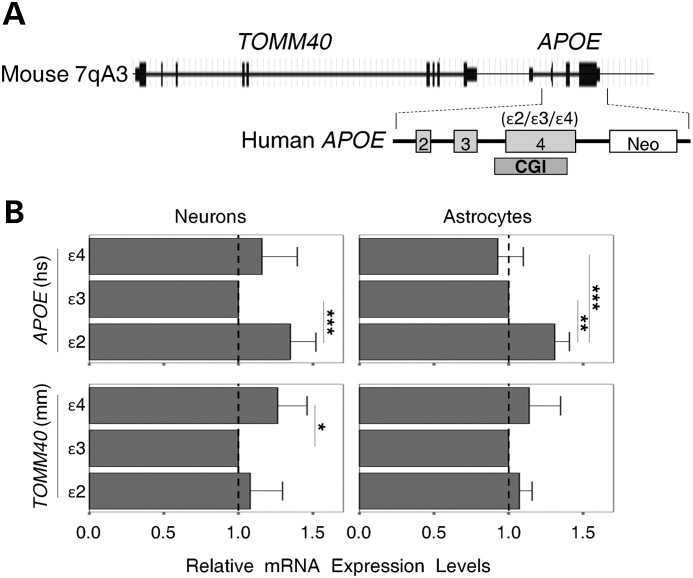
To evaluate the APOE CGI's enhancer activity in vivo, we employed a humanized APOE mouse model (38). These targeted replacement mice, ‘humanized’ at the murine APOE gene, have genomic structures similar to humans at the murine APOE locus (Fig. 5A). Primary cultures of astrocytes or neurons were prepared from the ɛ2/ɛ3/ɛ4 variants of mice, and total RNA was isolated. We then performed quantitative real-time polymerase chain reaction (qRT-PCR) to obtain messenger RNA (mRNA) expression levels of the human APOE transgene and its up-stream murine TOMM40 gene. In general, the ɛ4 and ɛ3 mice have lower APOE expression levels in both neurons and astrocytes when compared with the ɛ2 mice (Fig. 5B, top panels). In contrast, the ɛ4 mice have higher TOMM40 expression levels in neurons when compared with the ɛ3 mice [Fig. 5B, bottom panels, Tukey's honestly significant difference (HSD) test P < 0.05]. These animals were derived from the same inbred line with one solitary genetic difference (the ɛ2/ɛ3/ɛ4 alleles) located in the CGI of the human APOE transgene. Thus, these results suggest that the ɛ2/ɛ3/ɛ4 alleles alone can alter the expression of both APOE and TOMM40 genes. Such an allelic effect could be well explained by allele-dependent enhancer activity of the APOE CGI in the transgene. Notably, the APOE CGI is also highly methylated in these brain cells (data not shown), implying that, in vivo, the highly methylated APOE CGI still retains its enhancer activity.
Figure 5.
Enhancer effect of the APOE CGI in humanized APOE mice. (A) An outline of the genomic structure of the APOE locus in transgenic mice, including the human APOE gene Ex2-4. (B) Relative quantification of mRNA expression levels in neurons and astrocytes. Expression levels in each condition were normalized with an endogenous control (ACTB), and compared with ɛ3 counterparts (set at 1.0) using the ΔΔCT calculation. Each expression level was generated from at least four experimental replicates. Expression levels of APOE and TOMM40 between APOE haplotypes were compared by one-way ANOVA [APOE: neurons, F(2, 21) = 8.5, P = 0.002; astrocytes, F(2,12) = 17.07, P < 0.001; TOMM40: neurons, F(2, 21) = 5.07, P = 0.016; astrocytes, F(2, 12) = 1.46, P = 0.27]. Pair-wise comparisons were made with a post hoc Tukey honest significance test (*P < 0.05, **P < 0.01, ***P < 0.001). Error bars represent standard deviation from the mean (neurons, n = 8; astrocytes, n = 5).
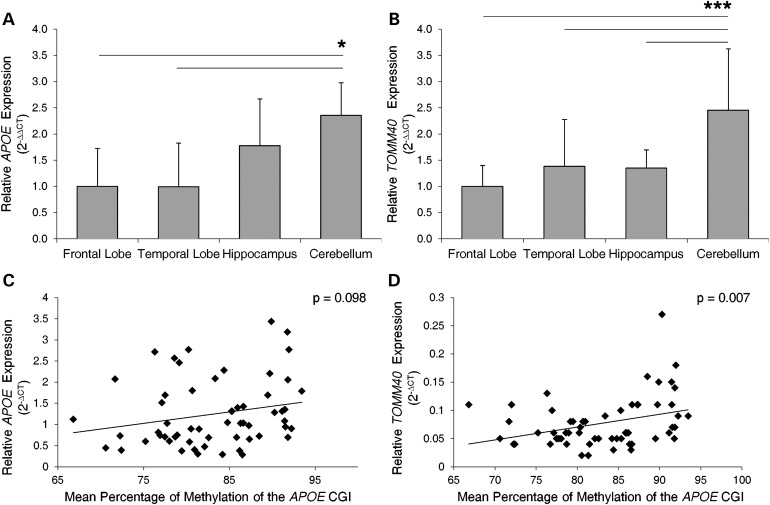
To determine whether APOE CGI methylation levels influence gene expression at the APOE locus, we assessed the mRNA expression levels of APOE and TOMM40 in PMB tissues. Overall, APOE was more highly expressed than TOMM40 (APOE average CT = 24.3 ± 0.8, TOMM40 average CT = 26.5 ± 1.3, t-test P < 10−18, with input of ∼20 ng total RNA), and its expression was significantly higher in the cerebellum compared with frontal and temporal lobes (Fig. 6A). This result is comparable with our previous study, which showed a non-significant trend of higher APOE expression in cerebellum compared with other brain tissues (34). A similar pattern was observed in TOMM40, with significantly higher expression levels in the cerebellum compared with other brain regions (Fig. 6B). Given the observed similarity between relative APOE CGI methylation levels (Fig. 2) and expression of APOE locus genes across brain regions, we ran a Pearson correlation to determine their relationship. A positive trend was observed between mean percentage of methylation of the APOE CGI and APOE expression when all samples and brain regions were analyzed together, although it did not reach the cutoff value of statistical significance (Fig. 6C, r = 0.219, n = 58, P = 0.098). A similar positive corrleation was observed for TOMM40 expression which was found to be statistically significant (Fig. 6D, r = 0.347, n = 59, P = 0.007). These results suggest that APOE and TOMM40 mRNA expression in the brain are positively correlated with methylation levels of the APOE CGI.
Figure 6.
Relative mRNA expression levels and their correlation with APOE CGI DNA methylation levels in PMB tissues. Expression levels of (A) APOE and (B) TOMM40 were normalized with an endogenous control (ACTB) and calibrated to expression levels in the frontal lobe (set to 1.0). Gene expression between tissues was evaluated by one-way ANOVA [APOE, F(3, 54) = 3.48, P = 0.022; TOMM40, F(3, 55) = 13.41, P < 0.001]. Post hoc pair-wise comparsions were made using Tukey's honest significance test (*P < 0.05, ***P ≤ 0.001). Error bars were calculated using standard propagation of error methods. The average percentage of methylation, at the APOE CGI, for each PMB sample was correlated to gene expression of (C) APOE and (D) TOMM40. A positive trend was observed between levels of methylation and APOE expression that did not reach the cutoff value for statistical significance (Pearson's correlation, r = 0.219, n = 58, P < 0.098). A positive correlation between the levels of methylation and TOMM40 expression was observed (Pearson's correlation, r = 0.347, n = 59, P < 0.007).
Together, these independent experiments and data strongly support the notion that the APOE CGI can function as a transcriptional enhancer/silencer, and can differentially modulate the promoter activity of multiple genes at the APOE locus in a cell type-, methylation level- and ɛ2/ɛ3/ɛ4 allele-specific manner.
DISCUSSION
The data presented here reveal the presence of a novel biological function of APOE, represented by an epigenetically marked CGI that manifests transcriptional enhancer/silencer activities.
We first observed that APOE has a well-defined CGI overlapping with its 3′-exon. CGIs are genomic regions comprising an unusually high frequency (i.e. observed to expected ratio) of CpG dinucleotides (39,40) and representing one of the most critical regulatory elements in the human genome with major functional roles in gene expression and regulation (41). The human genome consists of ∼50 000 CGIs, the majority (∼58%) of which overlap with repetitive sequence (42). The remaining CGIs can be divided into four subgroups according to their relative position with known genes, i.e. the 5′-(first exon/promoter overlapping), the 3′-(last exon overlapping), the intragenic and the intergenic CGIs. The majority of gene body CGIs belong to the 5′-CGI subgroup (43), which are present in virtually all housekeeping genes (44). In contrast, a 3′-CGI, such as the one in APOE, is rare in the human genome, representing <1% of total CGIs (36,43).
Here, we observed a high level of methylation across the APOE CGI. DNA methylation is an evolutionally conserved feature of the genome providing an additional layer of information content for biological processes, including embryogenesis, development, genomic imprinting, silencing of transposable elements and regulation of gene transcription (45–50). Previous studies with partial evaluations of DNA methylation at the APOE CGI have suggested a high methylation load of this CGI (51,52). In this study, we comprehensively characterized the DNA methylation profile of the APOE CGI in multiple human tissues. All of the samples analyzed were highly methylated (>75% average methylation) across the selected 75 CpG sites. The mean methylation level at each particular site ranged between 54 and 100%, with consistent up/down patterns between samples and tissues, suggesting inherent epigenetic regulation at the level of the individual CpG site. Although we found the patterns of the methylation profiles are clearly similar across PMB tissues, we detected significant brain region-specific differences in the overall degree of methylation. Methylation levels were lower in the temporal lobe, frontal lobe and hippocampus, regions affected the most in AD. In contrast, the highest methylation levels were observed in the cerebellum, where pathological changes are not as profound in AD. These results suggest that a correlation may exist between the methylation levels of the APOE CGI and the vulnerability of AD risk in the brain. Although it is also possible that variables such as medication and/or comorbidities contribute to the methylation levels in our study samples; when possible, these variables should be accounted for in future studies regarding APOE methylation in AD. One limitation of this experiment is that the methylation profiles defined here were generated from tissue with multiple mixed cell types. Since glial cells are the most prevalent cell type in the brain, it is likely that these results reflect regional differences in APOE CGI methylation within astrocytes and possibly microglia. Given that neurons are the most critical cell type in AD, and such post-mitotic cells are susceptible to biochemical manifestations of environmental influences, further studies to determine the cell type-specific methylation patterns of the APOE CGI may prove crucial in deciphering the epigenetics of AD pathogenesis.
Interestingly, the ɛ2/ɛ3/ɛ4 alleles of APOE alter the CpG load and methylation landscape of the APOE CGI. Such changes in CpG load are expected to alter the binding profiles of methyl CpG-binding domain proteins which bind specifically to the methylated DNA through their unique amino acid motif (53). Moreover, the methylation status of exons can modulate exonic protein binding, which, in turn, can affect other biological processes such as pre-mRNA processing (54). Thus, inheritance of different ɛ2/ɛ3/ɛ4 alleles in the APOE CGI might present different methylation landscapes, potentially altering protein binding, leading to diverse biological consequences, and possibly even influence the pathophysiological processes of diseases. Identifying APOE CGI-binding proteins with differential affinity for the ɛ2/ɛ3/ɛ4 variants should provide more conclusive insight.
Since a major role for CGIs involves transcriptional programming and a partial fragment of the APOE exon 4 has been shown to carry enhancer activity (31), we hypothesized that the APOE CGI is a transcriptional enhancer that can regulate multiple genes in the APOE locus. Functional studies of the un-methylated APOE CGI in a reporter assay revealed the presence of diverse enhancer activities, an effect that was modified by cell type, the ɛ2/ɛ3/ɛ4 alleles and the promoter haplotypes. This activity was more profound for the promoter of TOMM40 than for APOE, suggesting a major biological consequence of this APOE CGI in modulating expression of TOMM40.
Enhancer activity of the methylated version of the APOE CGI was also examined in a pCpG-free reporter assay system and a humanized APOE mouse model. Again, contrasting enhancer activities according to the cell type and the ɛ2/ɛ3/ɛ4 alleles were observed, suggesting that the enhancer activity is present in both the un-methylated and the methylated version of the CGI. It should be noted that most enhancers in mammals are CpG-poor and tend to have low levels of methylation (55), with methylation sometimes leading to reduced enhancer activity (56), findings that are in stark contrast to our observations. Nevertheless, our results strongly support the hypothesis that the APOE CGI is a genuine enhancer/silencer that can modulate transcriptional activity of multiple promoters (i.e. APOE, TOMM40 and hEF-1α), an effect that is present in both orientations (forward and reverse) regardless of its relative position (either upstream or downstream) of the target genes' promoter. More strikingly, the ɛ2/ɛ3/ɛ4 allele variants can modify this activity in a promoter-, haplotype- and cell type-specific manner possibly through the altered CpG content and methylation landscape.
A plausible mechanism for this enhancer activity might be DNA looping, which brings both the enhancer and promoter close together increasing the local concentration of transcriptional factors and subsequently accelerating recruitment of RNA pol II (57). Furthermore, with its potential trans-regulatory effect, the enhancer activity of the APOE CGI could extend into other diseases-risk associated genes located either distally to the APOE locus or on other chromosomes, and might explain better the APOE's strong association with diseases such as AD.
In this study, we focused on two gene targets (i.e. APOE and TOMM40) because of their known associations with AD risk. Independent studies support this line of reasoning. For example, the presence of the ɛ4 allele alone is sufficient to alter apoE expression in the human brain (58), and differential expression of the APOE transcript has been observed in different brain regions (59). Our findings support this as we observed differences in APOE expression between the cerebellum and temporal and frontal lobes in the human PMB. Additionally, multiple studies have suggested an independent role of TOMM40 in AD risk, including interaction with APP (60), influx of Aβ (61), association with AD age-at-onset (62), CSF AD biomarkers (63) and brain imaging phenotypes of AD (64). It is possible that altered TOMM40 expression levels could lead to sub-optimized protein transport of mitochondria, which could be compounded by other damaging effects associated with advancing age, leading to mitochondrial dysfunction in AD. Thus, when studies report association of the APOE ɛ2/ɛ3/ɛ4 alleles with certain disease phenotypes, one cannot automatically assume that the apoE protein isoform is the sole effector; other biological events such as the mitochondrial function, the gene regulatory network at the APOE locus and the aging-related DNA methylation drift are coming to the fore in disease association.
In conclusion, these new insights into APOE's structure and function do not diminish the importance of the apoE protein isoforms' role, but rather provide a more comprehensive picture of the complex pathophysiological involvement of APOE in diseases. Methylation of the APOE CGI and its ɛ2/ɛ3/ɛ4 variants adds another layer of embedded biological instruction, possibly involving protein binding, chromatin remodeling and specific RNA regulation in an age-dependent course. Such epigenetic modifications are potentially reversible in a cell type-specific spatial and temporal manner. Deciphering the mechanisms driving epigenetic regulation and modification of the APOE CGI may shed light on potential interventions for AD, CVD and other aging associated disorders.
MATERIALS AND METHODS
Human subjects and animals
Human genomic DNA and PMB were obtained from the University of Washington (UW) Alzheimer's Disease Research Center after approval by the human subject institutional review boards of UW and Veterans Affairs Puget Health Care System. AD subjects were volunteers who were selected by clinical diagnosis during life and neuropathologically confirmed at autopsy. All AD subjects had moderate-to-frequent neuritic plaques in the neocortex by CERAD criteria and Braak stages equal to or greater than V. Control subjects were volunteers who were ≥54 years of age, were never diagnosed with AD and lacked AD neuropathology at autopsy. The population postmortem interval (PMI), Braak stage and amyloid plaque load are all described in Supplementary Material, Table S1. Homozygous APOE ɛ2/ɛ3/ɛ4-targeted replacement mice, ‘humanized’ at the murine APOE locus (65,66), were used with the approval of the UW Institutional Animal Care and Use Committee.
Tissue collection and processing
PMBs were removed, quickly dissected and flash- frozen at the time of autopsy. The PMI ranged between 2 h 15 min and 9 h 30 min and there was no significant difference between the average PMI for the AD versus control groups (Supplementary Material, Table S1). In brief, a mid-sagittal cut separated left and right hemispheres, with right-sided regions extensively sampled into tissue cassettes and immediately snap-frozen in liquid nitrogen. Frozen cerebellum, hippocampus, superior posterior temporal lobe and frontal lobe samples were stored at −80°C prior to DNA extraction and analysis. AD pathology was confirmed using histological staining of PMB. Whole-blood lymphocytes were collected at the time of clinical visit.
Cell culture
HepG2 (hepatocytoma cells), SH-SY5Y (neuroblastoma cells) and U118 (astrocytoma cell) (ATCC, Manassas, VA, USA) were cultured as previously described (32). Murine primary glial cultures were generated from 0–3-day-old mouse cortices (67). In brief, cortices were enzymatically digested and plated in 162 cm2 flasks (Corning Incorporated, Corning, NY, USA) coated with poly-l-ornithine (Sigma, St Louis, MO, USA) and maintained at 37°C with 5% CO2 in DMEM/F-12 supplemented with 10% FBS (HyClone, Logan, UT, USA), 100 U/ml penicillin and100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). Microglia were removed by gentle shaking after 2 weeks in culture; the enriched astrocyte monolayer (>96%) was then trypsinized and re-plated onto six-well poly-l-ornithine-coated culture plates. Primary neuron cultures were generated from mouse cortices of 0-day-old pups. Dissected cortices were collected in Neurobasal medium (Invitrogen), gently disrupted, digested for 20 min at 37°C using papain and DNAse I (Worthington Biochemical, Lakewood, NJ, USA), centrifuged, resuspended and passed through a 40 μm nylon strainer. Cells were resuspended in Neurobasal medium, with B27 and glutamine (Invitrogen) and plated into six-well poly-d-lysine culture plates (Sigma). Medium was replaced after 24 h followed by half-medium changes every 2 days until neurons formed extensive processes (10 days in vitro). Astrocyte and neuron cultures were gently washed with PBS and then collected using the manufacturer's isolation buffer for RNA extraction.
DNA and RNA extraction
Genomic DNA was isolated from frozen PMB tissues or whole-blood lymphocytes, using the QIAmp DNA Mini Kit and DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). Recombinant DNA plasmids were isolated and purified using the Tip-100 Ion-Exchange Column Kit (Qiagen). Total RNA was isolated from cell pellets and culture wells, using the RNeasy Extraction Kit (Qiagen). All procedures were performed according to the manufacturer's instructions.
Bisulfite Pyrosequencing
Bisulfite conversion of genomic DNA (500 ng each) was carried out using the Epitech Bisulfite Kit (Qiagen) according to the manufacturer's instruction. To control for conversion efficiency in bisulfite reactions, all DNA samples were converted in a single batch. Pyrosequencing assays were designed to evaluate the methylation status of 75 CpG sites within APOE exon 4, starting with the 11th CpG site (chr19: 45 411 858) and ending with the 85th CpG (chr19: 45 412 545). This region also covers the two SNPs (rs429358 and rs7412) that determine the ɛ2/ɛ3/ɛ4 alleles. Five sets of PCR amplicons with 7 PCR primers and 13 sequencing primers (Supplementary Material, Table S2A) were designed using the Pyromark Assay Design software, version 2.0 (Qiagen). Data quality toward the end of sequencing reads (>55 nucleotides) was low. To address this issue, we specifically designed multiple overlapping fragments with redundant Pyrosequencing reads and only reads that passed the software quality check control were included in the analyzed data set. Multiple reads at single CpG sites were accounted for in our statistical model described below. PCR was carried out with 100 ng of bisulfite-converted DNA, using the Pyromark PCR Kit (Qiagen) on GeneAmp PCR System 9700 (Applied Biosystems, CA, USA). The PCR reaction mixtures included 12.5 µl of Pyromark PCR Kit Master Mix (Qiagen), 2.5 µl of CoralLoad Concentrate (Qiagen), 1 µl (∼100 ng) of bisulfite-converted genomic DNA and 10 µm forward and reverse primers in a final reaction volume of 25 µl. PCR was carried out on GeneAmp PCR System 9700 (Applied Biosystems) with a thermal profile consisting of an initial denaturation at 94°C for 15 min, followed by 55 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s and ending at 72°C for 10 min. PCR products were purified using streptavidin-coated Sepharose beads (GE Healthcare, Piscataway, NJ, USA) to capture the biotin-labeled primer. The bound PCR product was denatured and annealed to the sequencing primer (0.3 µm) by incubation at 80°C for 2 min. Pyrosequencing was carried out using PyroMark Gold Q24 reagents (Qiagen) on a PyroMark Q24 system (Qiagen), and data were analyzed using the PyroMark Q24 software, version 2.0.6 (Qiagen).
Quantitative RT-PCR
Total RNA was reverse-transcribed into cDNA using Superscript III reverse transcriptase (Life Technologies, Carlsbad, CA, USA) with random primers. RNA expression levels were determined by qRT-PCR, using TaqMan gene expression assays, TaqMan Universal PCR master mix and a 7900 ABI real-time instrument (Applied Biosystems). TaqMan assays (Supplementary Material, Table S3) were used to quantify mRNA levels of APOE, TOMM40 and ACTB. For mouse studies, each expression level was generated from two to six technical replicates on at least 4 different days (experimental replicates) from one (astrocyte) or two (neurons) biological replicates with ACTB as an endogenous control. The comparative CT method, also referred to as the ΔΔCT method (Applied Biosystems), was used to calculate the fold changes in expression between the samples, and a relative quantification level was determined by comparing with its ɛ3 mouse counterpart (set at 1.0). For human PMB studies, 14–15 independent samples were run in triplicate per brain region with ACTB as an endogenous control. Relative expression levels were determined by normalizing all brain regions to the frontal lobe (set at 1.0) using the ΔΔCT method. In both human and mouse studies, all outlying technical replicates were omitted such that all coefficients of variation were <1%.
Reporter constructs generation and transfection
To generate reporter constructs mimicking actual human haplotypes of the APOE locus, partial APOE CGI fragments (491 bp) containing ɛ2/ɛ3/ɛ4 variants were separately cloned onto the 3′ end of the luciferase gene in pGL4.10[luc2] vector (Promega, Madison, WI, USA) using the In-Fusion Cloning Kit (Clontech, Mountain View, CA, USA). PCR fragments of both APOE and TOMM40 promoters and their variants were subsequently cloned onto the 5′ end of the luciferase gene, using the same cloning approach as previously described (32).
For the methylation-related reporter constructs, a 1483 bp APOE CGI fragment was PCR-amplified from human genomic DNA and cloned onto the 5′ end of the hEF-1α promoter on the pCpG-free promoter vector (Invivogen, San Diego, CA, USA) (Fig. 4A). Both forward and reverse orientations of the three ɛ2/ɛ3/ɛ4 haplotype variants were cloned, and all inserts were validated by full sequencing. Primers are listed in Supplementary Material, Table S2B. The pCpG-free promoter vector and its derivatives were either mock-treated or methylated in vitro using SssI CpG methylase (2 U/µg DNA, New England Biolabs, Beverly, MA, USA) with S-adenosylmethionine (160 µm) for 4 h at 37°C and purified by the GFX DNA Purification Kit (Life Technologies). A small portion was bisulfite-converted and sequenced to validate the methylation status of the inserts. PCR primers are listed in Supplementary Material, Table S2B.
To test the enhancer activity of the APOE CGI, a panel of luciferase reporter clones were generated, including 6 promoter-only constructs (APOE and TOMM40 promoters both with 3 variants: P1, P2 and P3) and 18 promoter-plus-enhancer combinations (6 promoter variants that match with the ɛ2/ɛ3/ɛ4 variants of the APOE CGI), mimicking the haplotype structure of the human APOE locus (Fig. 3A). Cells were plated in 96-well plates and transiently transfected 48 h later using the Lipofectamine 2000 reagent (Invitrogen) with the reporter plasmids (200 ng/well). Reporter activity of each construct is shown as a relative quantification calibrated against its promoter-only construct counterpart (set at 1.0) as previously described (32). For the pCpG-free reporter assay, 12 additional reporter constructs (i.e. the APOE CGI with the three ɛ2/ɛ3/ɛ4 variants in both orientations, with or without methylation) were generated. Reporter activities were determined 48 h post-transfection in the same three cell lines of HepG2, SH-SY5Y and U118, and activities were determined by mixing 50 µl of transfected cell supernatant with 200 µl of QUANTI-Blue reagent (Invitrogen, San Diego, CA, USA). After incubation at 37°C for 20 h, AP activities were measured at 620 nm with a microplate reader (SpectraMax M2E, Molecular Devices, Sunnyvale, CA, USA), according to manufacturer's specifications. Reporter activity was calibrated against original pCpG-free promoter vector (set at 1.0).
Statistical analyses
We analyzed the methylation levels of 75 CpG sites from multiple tissues. The proportion of methylated cytosines was compared either across tissue types or by clinical diagnosis using linear mixed effect models (68) to account for repeated measures on a subject within tissue type across CpG sites, and also to account for possible multiple measures of methylation at a CpG site. Tissue type, CpG site and clinical diagnosis were treated as fixed effects, and subjects were treated as random effects. Proportions were transformed using the arcsine of the square root transformation to induce normality and homoscedasticity (69). Post hoc pair-wise comparisons were adjusted for multiple comparisons using the method of Holm.
In reporter assays, we were unable to test for an interaction (using a two-way ANOVA) between the promoter and the enhancer haplotypes, given that these two variables were present in a prefixed single construct. Instead, a one-way ANOVA with post hoc tests was performed using a relative to promoter value to account for different promoter effects on expression. In the assay using the pGL4.10 vector, this approach assessed a difference between promoter–enhancer haplotype constructs relative to the promoter-alone constructs while taking into account multiple comparisons. Likewise, differences in AP activity between enhancer haplotypes, orientations and methylation states were assessed by one-way ANOVA in the pCpG-free promoter vector system. All post hoc statistical tests were computed by Bonferroni correction for multiple comparisons. For qRT-PCR experiments, gene expression levels were compared by one-way ANOVA and pair-wise comparisons were made with post hoc Tukey's HSD test.
Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS), version 18 (SPSS, Inc., Chicago, IL, USA), R, version 2.15.2, and the R nlme package (70).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported in part by the US Department of Veterans Affairs Office of Research and Development Biomedical Laboratory Research Program; the National Institute of Health/NIH grants (P50 AG05136, K99 AG34214, T32 AG000258); and the Nancy and Buster Alvord Endowment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steve Millard, VA Puget Sound Health Care System, for assistance with statistical analysis of the Pyrosequencing data.
REFERENCES
- 1.Mahley R.W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. doi:10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Mahley R., Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 1999;10:207–217. doi: 10.1097/00041433-199906000-00003. doi:10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y., Weisgraber K.H., Mucke L., Mahley R.W. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J. Mol. Neurosci. 2004;23:189–204. doi: 10.1385/JMN:23:3:189. doi:10.1385/JMN:23:3:189. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Lomnitski L., Michaelson D.M., Shohami E. Motor and cognitive deficits in apolipoprotein E-deficient mice after closed head injury. Neuroscience. 1997;80:1255–1262. doi: 10.1016/s0306-4522(97)00007-9. doi:10.1016/S0306-4522(97)00007-9. [DOI] [PubMed] [Google Scholar]
- 5.Rall S.C., Jr, Weisgraber K.H., Mahley R.W. Human apolipoprotein E. The complete amino acid sequence. J. Biol. Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- 6.Hui D.Y., Innerarity T.L., Mahley R.W. Defective hepatic lipoprotein receptor binding of beta-very low density lipoproteins from type III hyperlipoproteinemic patients. Importance of apolipoprotein E. J. Biol. Chem. 1984;259:860–869. [PubMed] [Google Scholar]
- 7.Sing C.F., Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am. J. Hum. Genet. 1985;37:268–285. [PMC free article] [PubMed] [Google Scholar]
- 8.Infante-Rivard C., Levy E., Rivard G.E., Guiguet M., Feoli-Fonseca J.C. Small babies receive the cardiovascular protective apolipoprotein epsilon 2 allele less frequently than expected. J. Med. Genet. 2003;40:626–629. doi: 10.1136/jmg.40.8.626. doi:10.1136/jmg.40.8.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathiresan S., Melander O., Anevski D., Guiducci C., Burtt N.P., Roos C., Hirschhorn J.N., Berglund G., Hedblad B., Groop L., et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. doi:10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 10.Lambert J.-C., Brousseau T., Defosse V., Evans A., Arveiler D., Ruidavets J.-B., Haas B., Cambou J.-P., Luc G., Ducimetière P., et al. Independent association of an APOE gene promoter polymorphism with increased risk of myocardial infarction and decreased APOE plasma concentrations—the ECTIM Study. Hum. Mol. Genet. 2000;9:57–61. doi: 10.1093/hmg/9.1.57. doi:10.1093/hmg/9.1.57. [DOI] [PubMed] [Google Scholar]
- 11.Zannis V.I., Breslow J.L. Characterization of a unique human apolipoprotein E variant associated with type III hyperlipoproteinemia. J. Biol. Chem. 1980;255:1759–1762. [PubMed] [Google Scholar]
- 12.Stengard J.H., Pekkanen J., Sulkava R., Ehnholm C., Erkinjuntti T., Nissinen A. Apolipoprotein E polymorphism, Alzheimer's disease and vascular dementia among elderly Finnish men. Acta Neurol. Scand. 1995;92:297–298. doi: 10.1111/j.1600-0404.1995.tb00132.x. doi:10.1111/j.1600-0404.1995.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 13.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. doi:10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 14.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., Van Duijn C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. doi:10.1001/jama.1997.03550160069041. [PubMed] [Google Scholar]
- 15.Yu C.E., Payami H., Olson J.M., Boehnke M., Wijsman E.M., Orr H.T., Kukull W.A., Goddard K.A., Nemens E., White J.A. The apolipoprotein E/CI/CII gene cluster and late-onset Alzheimer disease. Am. J. Hum. Genet. 1994;54:631–642. [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer M.R., Tschanz J.T., Norton M.C., Welsh-Bohmer K.A., Steffens D.C., Wyse B.W., Breitner J.C. APOE genotype predicts when—not whether—one is predisposed to develop Alzheimer disease. Nat. Genet. 1998;19:321–322. doi: 10.1038/1206. doi:10.1038/1206. [DOI] [PubMed] [Google Scholar]
- 17.Bu G. Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. doi:10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herz J. Apolipoprotein E receptors in the nervous system. Curr. Opin. Lipidol. 2009;20:190–196. doi: 10.1097/MOL.0b013e32832d3a10. doi:10.1097/MOL.0b013e32832d3a10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J. Lipid Res. 2009;50(Suppl.):S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raber J., Wong D., Yu G.-Q., Buttini M., Mahley R.W., Pitas R.E., Mucke L. Alzheimer's disease: apolipoprotein E and cognitive performance. Nature. 2000;404:352–354. doi: 10.1038/35006165. doi:10.1038/35006165. [DOI] [PubMed] [Google Scholar]
- 21.Guo L., LaDu M.J., Van Eldik L.J. A dual role for apolipoprotein E in neuroinflammation: anti- and pro-inflammatory activity. J. Mol. Neurosci. 2004;23:205–212. doi: 10.1385/JMN:23:3:205. doi:10.1385/JMN:23:3:205. [DOI] [PubMed] [Google Scholar]
- 22.Harris F.M., Brecht W.J., Xu Q., Mahley R.W., Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J. Biol. Chem. 2004;279:44795–44801. doi: 10.1074/jbc.M408127200. doi:10.1074/jbc.M408127200. [DOI] [PubMed] [Google Scholar]
- 23.Riddell D.R., Zhou H., Atchison K., Warwick H.K., Atkinson P.J., Jefferson J., Xu L., Aschmies S., Kirksey Y., Hu Y., et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J. Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. doi:10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahrle S.E., Jiang H., Parsadanian M., Legleiter J., Han X., Fryer J.D., Kowalewski T., Holtzman D.M. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. doi:10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 25.Huang W., Qiu C., von Strauss E., Winblad B., Fratiglioni L. APOE genotype, family history of dementia, and Alzheimer disease risk: a 6-year follow-up study. Arch Neurol. 2004;61:1930–1934. doi: 10.1001/archneur.61.12.1930. doi:10.1001/archneur.61.12.1930. [DOI] [PubMed] [Google Scholar]
- 26.Corder E.H., Saunders A.M., Risch N.J., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Jr, Rimmler J.B., Locke P.A., Conneally P.M., Schmader K.E. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. doi:10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 27.Myers R.H., Schaefer E.J., Wilson P.W., D'Agostino R., Ordovas J.M., Espino A., Au R., White R.F., Knoefel J.E., Cobb J.L., et al. Apolipoprotein E epsilon4 association with dementia in a population-based study: The Framingham study. Neurology. 1996;46:673–677. doi: 10.1212/wnl.46.3.673. doi:10.1212/WNL.46.3.673. [DOI] [PubMed] [Google Scholar]
- 28.Khachaturian A.S., Corcoran C.D., Mayer L.S., Zandi P.P., Breitner J.C. Apolipoprotein E epsilon4 count affects age at onset of Alzheimer disease, but not lifetime susceptibility: The Cache County Study. Arch. Gen. Psychiatry. 2004;61:518–524. doi: 10.1001/archpsyc.61.5.518. doi:10.1001/archpsyc.61.5.518. [DOI] [PubMed] [Google Scholar]
- 29.Liddell M.B., Lovestone S., Owen M.J. Genetic risk of Alzheimer's disease: advising relatives. Br. J. Psychiatry. 2001;178:7–11. doi: 10.1192/bjp.178.1.7. doi:10.1192/bjp.178.1.7. [DOI] [PubMed] [Google Scholar]
- 30.Zheng P., Pennacchio L.A., Le Goff W., Rubin E.M., Smith J.D. Identification of a novel enhancer of brain expression near the apoE gene cluster by comparative genomics. Biochim. Biophys. Acta. 2004;1676:41–50. doi: 10.1016/j.bbaexp.2003.10.007. doi:10.1016/j.bbaexp.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Chen H.P., Lin A., Bloom J.S., Khan A.H., Park C.C., Smith D.J. Screening reveals conserved and nonconserved transcriptional regulatory elements including an E3/E4 allele-dependent APOE coding region enhancer. Genomics. 2008;92:292–300. doi: 10.1016/j.ygeno.2008.07.009. doi:10.1016/j.ygeno.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Bekris L.M., Lutz F., Yu C.-E. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J. Hum. Genet. 2012;57:18–25. doi: 10.1038/jhg.2011.123. doi:10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bekris L.M., Millard S.P., Galloway N.M., Vuletic S., Albers J.J., Li G., Galasko D.R., DeCarli C., Farlow M.R., Clark C.M., et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J. Alzheimers Dis. 2008;13:255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bekris L.M., Galloway N.M., Montine T.J., Schellenberg G.D., Yu C.E. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:409–417. doi: 10.1002/ajmg.b.30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai D., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. doi:10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. doi:10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C.E., Seltman H., Peskind E.R., Galloway N., Zhou P.X., Rosenthal E., Wijsman E.M., Tsuang D.W., Devlin B., Schellenberg G.D. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. doi:10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan P.M., Mace B.E., Maeda N., Schmechel D.E. Marked regional differences of brain human apolipoprotein E expression in targeted replacement mice. Neuroscience. 2004;124:725–733. doi: 10.1016/j.neuroscience.2003.10.011. doi:10.1016/j.neuroscience.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. doi:10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 40.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. doi:10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 41.Laird P.W. Cancer epigenetics. Hum. Mol. Genet. 2005;14(Spec. No. 1):R65–R76. doi: 10.1093/hmg/ddi113. doi:10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 42.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 43.Medvedeva Y.A., Fridman M.V., Oparina N.J., Malko D.B., Ermakova E.O., Kulakovskiy I.V., Heinzel A., Makeev V.J. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics. 2010;11:48. doi: 10.1186/1471-2164-11-48. doi:10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J., He F., Hu S., Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. doi:10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. doi:10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 46.Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. doi:10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 47.Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. doi:10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 48.Hisano M., Ohta H., Nishimune Y., Nozaki M. Methylation of CpG dinucleotides in the open reading frame of a testicular germ cell-specific intronless gene, Tact1/Actl7b, represses its expression in somatic cells. Nucleic Acids Res. 2003;31:4797–4804. doi: 10.1093/nar/gkg670. doi:10.1093/nar/gkg670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagase H., Ghosh S. Epigenetics: differential DNA methylation in mammalian somatic tissues. FEBS J. 2008;275:1617–1623. doi: 10.1111/j.1742-4658.2008.06330.x. doi:10.1111/j.1742-4658.2008.06330.x. [DOI] [PubMed] [Google Scholar]
- 50.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. doi:10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larsen F., Solheim J., Prydz H. A methylated CpG island 3′ in the apolipoprotein-E gene does not repress its transcription. Hum. Mol. Genet. 1993;2:775–780. doi: 10.1093/hmg/2.6.775. doi:10.1093/hmg/2.6.775. [DOI] [PubMed] [Google Scholar]
- 52.Wang S.C., Oelze B., Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. doi:10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. doi:10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Scholer A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D., et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 56.Schmidl C., Klug M., Boeld T.J., Andreesen R., Hoffmann P., Edinger M., Rehli M. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. doi:10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson K.D., Christensen H.M., Zhao B., Bresnick E.H. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. doi:10.1016/S1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 58.Bray N.J., Jehu L., Moskvina V., Buxbaum J.D., Dracheva S., Haroutunian V., Williams J., Buckland P.R., Owen M.J., O'Donovan M.C. Allelic expression of APOE in human brain: effects of epsilon status and promoter haplotypes. Hum. Mol. Genet. 2004;13:2885–2892. doi: 10.1093/hmg/ddh299. doi:10.1093/hmg/ddh299. [DOI] [PubMed] [Google Scholar]
- 59.Conejero-Goldberg C., Hyde T.M., Chen S., Dreses-Werringloer U., Herman M.M., Kleinman J.E., Davies P., Goldberg T.E. Molecular signatures in post-mortem brain tissue of younger individuals at high risk for Alzheimer's disease as based on APOE genotype. Mol. Psychiatry. 2011;16:836–847. doi: 10.1038/mp.2010.57. doi:10.1038/mp.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Devi L., Prabhu B.M., Galati D.F., Avadhani N.G., Anandatheerthavarada H.K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. doi:10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddy P.H. Amyloid beta, mitochondrial structural and functional dynamics in Alzheimer's disease. Exp. Neurol. 2009;218:286–292. doi: 10.1016/j.expneurol.2009.03.042. doi:10.1016/j.expneurol.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roses A.D., Lutz M.W., Amrine-Madsen H., Saunders A.M., Crenshaw D.G., Sundseth S.S., Huentelman M.J., Welsh-Bohmer K.A., Reiman E.M. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2010;10:375–384. doi: 10.1038/tpj.2009.69. doi:10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S., Swaminathan S., Shen L., Risacher S.L., Nho K., Foroud T., Shaw L.M., Trojanowski J.Q., Potkin S.G., Huentelman M.J., et al. Genome-wide association study of CSF biomarkers Aβ1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. doi:10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen L., Kim S., Risacher S.L., Nho K., Swaminathan S., West J.D., Foroud T., Pankratz N., Moore J.H., Sloan C.D., et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. NeuroImage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. doi:10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu P.T., Schmechel D., Rothrock-Christian T., Burkhart D.S., Qiu H.L., Popko B., Sullivan P., Maeda N., Saunders A.M., Roses A.D., et al. Human apolipoprotein E2, E3, and E4 isoform-specific transgenic mice: human-like pattern of glial and neuronal immunoreactivity in central nervous system not observed in wild-type mice. Neurobiol. Dis. 1996;3:229–245. doi: 10.1006/nbdi.1996.0023. doi:10.1006/nbdi.1996.0023. [DOI] [PubMed] [Google Scholar]
- 66.Sullivan P.M., Mezdour H., Aratani Y., Knouff C., Najib J., Reddick R.L., Quarfordt S.H., Maeda N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 1997;272:17972–17980. doi: 10.1074/jbc.272.29.17972. doi:10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 67.Cudaback E., Li X., Yang Y., Yoo T., Montine K.S., Craft S., Montine T.J., Keene C.D. Apolipoprotein C-I is an APOE genotype-dependent suppressor of glial activation. J. Neuroinflammation. 2012;9:192. doi: 10.1186/1742-2094-9-192. doi:10.1186/1742-2094-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinheiro J., Bates D. Mixed Effects Models in S and S-Plus. New York, NY: Springer; 2000. [Google Scholar]
- 69.Zar J. Biostatistical Analysis. 5th edn. Upper Saddle, NJ: Prentice Hall; 2010. [Google Scholar]
- 70.R-Core-Team. Vienna, Austria. ISBN: 3-900051-07-0.: R Foundation for Statistical Computing; 2012. R: A Language and Environment for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.