Abstract
Mean telomere length (TL) in blood cells is heritable and has been reported to be associated with risks of several diseases, including cancer. We conducted a meta-analysis of three GWAS for TL (total n=2240) and selected 1629 variants for replication via the “iCOGS” custom genotyping array. All ∼200 000 iCOGS variants were analysed with TL, and those displaying associations in healthy controls (n = 15 065) were further tested in breast cancer cases (n = 11 024). We found a novel TL association (Ptrend < 4 × 10−10) at 3p14.4 close to PXK and evidence (Ptrend < 7 × 10−7) for TL loci at 6p22.1 (ZNF311) and 20q11.2 (BCL2L1). We additionally confirmed (Ptrend < 5 × 10−14) the previously reported loci at 3q26.2 (TERC), 5p15.3 (TERT) and 10q24.3 (OBFC1) and found supportive evidence (Ptrend < 5 × 10−4) for the published loci at 2p16.2 (ACYP2), 4q32.2 (NAF1) and 20q13.3 (RTEL1). SNPs tagging these loci explain TL differences of up to 731 bp (corresponding to 18% of total TL in healthy individuals), however, they display little direct evidence for association with breast, ovarian or prostate cancer risks.
INTRODUCTION
Human chromosomes are capped and stabilized by telomeres, which are predominantly composed of several thousand DNA hexamer repeats (1–3). Mean telomere length (TL) is strongly heritable (4–6), but telomeres also shorten with each cell division, and consequently with age (7–11). Thus, shorter telomeres (assayed as mean length in DNA from blood cells) have been hypothesized to predispose to a number of diseases of aging, including certain cancers. Attempts to test this hypothesis in blood drawn after cancer diagnosis do indeed show cases to have shorter telomeres than their study-matched controls (12–14). However, prospectively designed studies, which use stored blood samples collected prior to cancer diagnosis, have failed to confirm this hypothesis (13,15–18). Such prospective designs are better able to address the question of whether telomeres become shorter before cancer diagnosis, and are thus predictive of cancer development. Another way of addressing the same hypothesis is to identify genetic variants that are associated with differences in TL in blood and then examine the associations of these same variants with cancer risks, both directly and in Mendelian randomization studies (19–22). If shorter telomeres are directly responsible for increased cancer risk, then genetic variants that reduce TL should also increase cancer risk and, given appropriately powered studies, this effect should be detectable.
Previously, several small GWAS with TL, as well as a meta-analysis (n = 12 000 subjects), have been published (23–26). More recently, a meta-analysis of over 37 000 individuals found seven loci affecting TL, and a fine-mapping study carried out in parallel to this work (and using the same individuals) has found variants associated with TL at the TERT locus (coding for the protein subunit of telomerase, a complex with an integral role in telomere maintenance) (27,28). Together, all these publications have identified variants at nine loci which are significantly associated with TL: TERC (3q26.2) (23,25–27) (the RNA subunit of telomerase), TERT (5p15.33) (27,28), OFBC1 (10q24.3) (24,26,27), ACYP2 (2p16.2) (27), NAF1 (4q32.2) (27), ZNF208 (19p12) (27), RTEL1 (20q13.3) (27), CTC1 (17p13.1) (26) and ZNF676 (19p12) (26).
The Illumina™ “iCOGS” custom genotyping chip (28) was designed as a collaborative project, principally involving four international consortia; the Breast Cancer Association Consortium (BCAC) (29) (http://ccge.medschl.cam.ac.uk/consortia/bcac/), the Ovarian Cancer Association Consortium (OCAC) (30) (http://ccge.medschl.cam.ac.uk/consortia/ocac/), the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) (31,32) (http://ccge.medschl.cam.ac.uk/consortia/cimba/index.html) and Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) (33,34) (http://ccge.medschl.cam.ac.uk/consortia/practical/). This chip assays ∼200 000 SNPs chosen by many researchers with different study aims. More than 200 000 participants in many cancer studies have already been typed using this chip, 26 089 of whom TL had also been determined.
Our aim, made possible by the Collaborative Oncology Gene-environment Study (COGS) project, was to carry out a larger GWAS than those currently published and test any confirmed TL variants for additional associations with breast, ovarian and prostate cancer risks.
RESULTS
GWAS for mean TL in blood cells
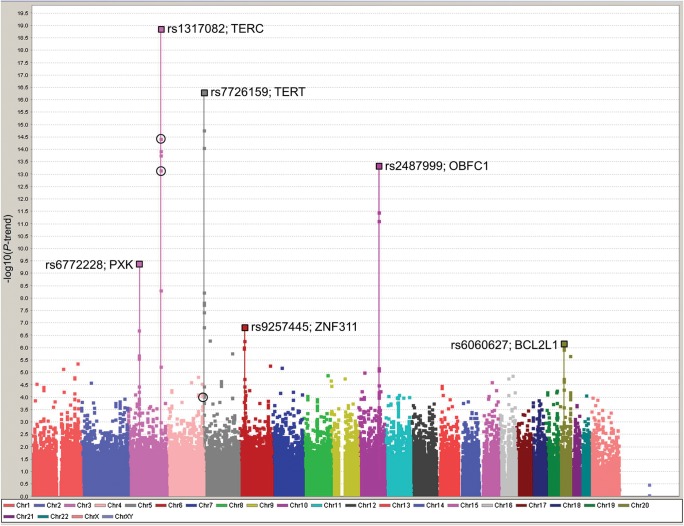
A Manhattan plot of all 187 647 SNPs on the custom “iCOGS” chip in all 26 089 participants (healthy controls and cancer cases) in whom TL was measured is shown in Figure 1. Full results of the marked ‘best SNP per peak’ analyses are given in Table 1. SNPs at four loci were associated at Ptrend < 10−7 in the analysis of healthy controls and these all increased in significance on inclusion of the cancer cases. The most significant of these mapped to the 3q26.2 locus containing the TERC gene (rs1317082, Ptrend = 1 × 10−19). This lead SNP had an effect equivalent to a TL 77 bp shorter for every minor allele carried. The second locus at 5p15.3 contains the functionally related TERT gene (rs7726159 Ptrend = 5 × 10−17) with a similarly sized per-allele effect to the TERC SNP, albeit with the minor allele associated with longer telomeres. The third most significant locus, at 10q24.3, contains the OBCF1 gene. Here, the minor allele of the lead SNP (rs2487999, Ptrend= 4 × 10−14) has a lower frequency (MAF = 0.10) but a greater effect size, a TL 100 bp longer for every minor allele carried. The fourth locus at 3p14.4, containing the PXK gene, is a novel finding (lead SNP rs6772228, Ptrend= 4 × 10−10) and has the greatest effect on TL, equivalent to 120 bp shorter TL per-allele. We also note a further two, previously unreported loci, containing SNPs displaying evidence for association with TL: 6p22.1 containing the ZNF311 gene (lead SNP rs9257445, Ptrend = 1 × 10−7) and 20q11.2 containing BCL2L1 (lead SNP rs6060627, Ptrend = 6 × 10−7), with per-minor-allele effects equivalent to a 38 bp shorter TL and a 36 bp longer TL, respectively. In addition to these six loci, SNPs at the DMRT1 locus (9p24.3) showed an association with TL at Ptrend = 1 × 10−6 in the control-only analysis, but did not replicate upon addition of the data from cancer cases (case and control analysis, Ptrend = 2 × 10−5; Supplementary Material, Table S1).
Figure 1.
Manhattan plot of all iCOGS SNPs and TL in all 26 089 participants (healthy controls and cancer cases) from the CCHS, CGPS and SEARCH studies. Solid squares represent the negative log of the per-risk-allele Ptrend against the genome position of each SNP. The lead SNPs in each of the peaks are marked with a larger square and the SNP name and gene region noted. Black open circles mark the results (in 26 089 COGS cases and controls) for the top SNPs originally selected from the GWAS meta-meta-analysis (chr3, rs10936601, Ptrend = 3.6 × 10−15; chr3, rs11709840, Ptrend = 6.5 × 10−14 and chr4, rs930306, Ptrend = 8.4 × 10−5).
Table 1.
Association between iCOGS SNPs, TL and cancer risk
| Region | Chr | SNP | Chr Position (Build 36) | MAF | TL association | TL association |
|||
|---|---|---|---|---|---|---|---|---|---|
| CCHS and SEARCH controls (n = 15 065) | CCHS, CPGS and SEARCH cases and controls (n = 26 089) | ||||||||
| Ptrend | Per-allele ΔTL (95% CI) | Ptrend | r2 (%) | F-statistic | |||||
| TERC | 3 | rs1317082 | 170 980 279 | 0.25 | 1.04E−13 | −77 (−57 to −98) | 1.33E−19 | 0.2 | 30 |
| TERT | 5 | rs7726159 | 1 335 319 | 0.34 | 6.10E−12 | 73 (55 to 92) | 4.67E−17 | 0.3 | 33 |
| OBFC1 | 10 | rs2487999 | 105 649 816 | 0.10 | 7.30E−09 | 100 (70 to 129) | 4.22E−14 | 0.2 | 23 |
| PXK | 3 | rs6772228 | 58 351 059 | 0.05 | 7.77E−08 | −120 (−83 to −158) | 3.91E−10 | 0.2 | 23 |
| ZNF311 | 6 | rs9257445 | 29 057 185 | 0.25 | 3.43E−05 | −38 (−17 to −58) | 1.38E−07 | 0.06 | 7 |
| BCL2L1 | 20 | rs6060627 | 29 725 820 | 0.30 | 8.33E−04 | 36 (17 to 55) | 6.45E−07 | 0.06 | 7 |
Single SNP estimates for the lead variants from each of the six strongest association peaks in the meta-analysis of all case and control participants (n = 26 089) for association with TL are given in Table 1 (see also Fig. 1, and for study-by-study estimates for the three control and two case populations, Supplementary Material, Table S2). The difference in TL per-minor-allele is expressed as the change in TL (ΔTL), with mean TL (95% CI), using the common homozygote as reference (see Supplementary Material) for the case and control analysis. Statistical significance (Ptrend) is also given for the healthy control only (n = 15 065) analysis (see also Supplementary Material, Table S1). r2 and F-statistics are obtained from the least-squares regression of the unranked genotype on TL residuals (after regression with age, gender, case–control status and study). All cancer risk associations are presented as ORs with 95% CI and per-allele Ptrend (Table 2). Ovarian cancer risks are broken down into serous invasive and serous LMP subgroups. MAF: minor allele frequency.
Table 2.
Association between iCOGS SNPs, TL and cancer risk
| Region | Chr | SNP | Chr Position (Build 36) | MAF | Breast cancer association |
Ovarian cancer association |
Ovarian cancer association |
Prostate cancer association |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall risk (BCAC) 46 451 cases, 42 599 controls | Serous invasive (OCAC) 8371 cases, 23 444 controls | Serous LMP (OCAC) 979 cases, 17 869 controls | Overall risk (PRACTICAL) 22 297 cases, 22 323 controls | |||||||||
| Per-allele OR (95% CI) | Ptrend | Per-allele OR (95% CI) | Ptrend | Per-allele OR (95% CI) | Ptrend | Per-allele OR (95% CI) | Ptrend | |||||
| TERC | 3 | rs1317082 | 170 980 279 | 0.25 | 0.99 (0.97–1.01) | 3.31E−01 | 1.03 (0.99–1.08) | 1.31E−01 | 0.89 (0.78–1.00) | 4.61E−02 | 0.96 (0.93–0.99) | 6.16E−03 |
| TERT | 5 | rs7726159 | 1 335 319 | 0.34 | 1.05 (1.03–1.07) | 1.52E−05 | 1.12 (1.09–1.16) | 3.36E−09 | 1.45 (1.36–1.55) | 2.00E−14 | 0.88 (0.85–0.91) | 1.84E−18 |
| OBFC1 | 10 | rs2487999 | 105 649 816 | 0.10 | 1.03 (1.00–1.06) | 6.44E−02 | 1.04 (0.98–1.10) | 2.25E−01 | 1.29 (1.14–1.43) | 5.87E−04 | 1.06 (1.02–1.11) | 1.09E−02 |
| PXK | 3 | rs6772228 | 58 351 059 | 0.05 | 1.03 (0.98–1.07) | 2.59E−01 | 1.02 (0.93–1.10) | 6.94E−01 | 1.04 (0.82–1.25) | 7.37E−01 | 1.08 (1.02–1.14) | 1.61E−02 |
| ZNF311 | 6 | rs9257445 | 29 057 185 | 0.25 | 1.03 (1.01–1.05) | 4.86E−03 | 0.97 (0.93–1.02) | 2.36E−01 | 1.05 (0.94–1.16) | 3.61E−01 | 1.04 (1.01–1.08) | 6.58E−03 |
| BCL2L1 | 20 | rs6060627 | 29 725 820 | 0.30 | 0.98 (0.96–1.00) | 5.29E−02 | 1.03 (0.99–1.07) | 1.96E−01 | 0.93 (0.83–1.04) | 1.80E−01 | 0.97 (0.94–1.00) | 2.62E−02 |
Table 2 reveals that the minor allele of TERT rs7726159 is associated with significant protection from prostate cancer [OR = 0.88 (95% CI 0.85–0.91), Ptrend = 2 × 10−18] but significantly increased risks of breast and ovarian cancers, particularly of LMP ovarian cancer [OR = 1.45 (95% CI 1.36–1.55), Ptrend = 2 × 10−14]. We have double checked that this is not an allele-calling artefact and we also note that the minor allele of a nearby SNP, rs401681, has similarly been reported to be associated with increased risks of cancers of the lung, bladder, testes, cervix and basal cell carcinoma, but with decreased risk of melanoma (37–39). These inverted associations may be due to tissue-specific interactions that need further examination.
Codd et al. (27) reported significant TL associations with four further SNPs which were not present on the iCOGS chip. However, good surrogates for three of these were on the chip and the results for these are shown in Table 3. The best surrogate SNP for the reported 2p16.2 lead variant (rs11125529) was rs10165485 (pairwise r2 = 0.98) (35), and we find the minor allele of rs10165485 to be associated with longer TL (Ptrend = 2.4 × 10−5) in contrast to the published finding of a per-allele decrease in TL for rs11125529. The best surrogates for the published lead SNPs at 4q32.2 (rs7675998) and 20q13.3 (rs755017) are iCOGS SNPs rs2320615 and rs2738783 [r2 = 0.89 and 1.0, respectively (35)] which both have iCOGS Ptrend < 5.0 × 10−4. There were no good surrogates for the 19p12 locus lead SNPs on iCOGS.
Table 3.
The associations with TL and cancer risk of three iCOGS SNPs that are surrogates for additional peaks of TL association found by Codd et al. (26)
| Region | Chr | Codd et al. SNP | Chr Position (Build 36) | iCOGS SNP | Chr Position (Build 36) | Pairwise r2 between SNPs | TL association | Breast cancer association | Ovarian cancer association |
Prostate cancer association | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CCHS, CPGS and SEARCH | Overall risk (BCAC) | Serous invasive (OCAC) | Serous LMP (OCAC) | Overall risk (PRACTICAL) | |||||||
| Ptrend | Ptrend | Ptrend | Ptrend | Ptrend | |||||||
| ACYP2 | 2 | rs11125529 | 54 329 370 | rs10165485 | 54 335 140 | 0.98 | 2.4E−05 | 3.9E−01 | 8.2E−01 | 9.0E−02 | 6.7E−01 |
| NAF1 | 4 | rs7675998 | 164 227 270 | rs2320615 | 164 289 399 | 0.89 | 3.3E−04 | 7.6E−01 | 4.7E−01 | 4.4E−01 | 4.3E−01 |
| RTEL1 | 20 | rs755017 | 61 892 066 | rs2738783 | 61 779 056 | 1.00 | 4.5E−04 | 1.6E−01 | 2.1E−01 | 2.7E−02 | 3.2E−04 |
Ptrends for per-allele differences in TL and cancer risk are calculated as for Table 1.
Proportion of variance in mean TL explained
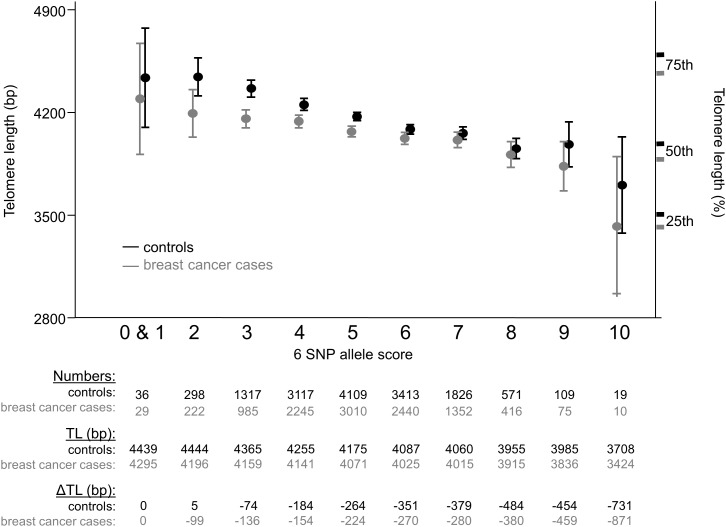
Least-squares regression of the unranked genotype on TL residuals (after regression with age, gender, case–control status and study) led to estimates that SNPs tagging loci associated with TL at Ptrend < 7 × 10−7 individually explained from 0.06 to 0.2% of the total variation in TL and had an F-statistics from 7–33 (Table 1). This indicates that these SNPs are likely to be valid instruments for Mendelian randomization studies as an F-statistics ≥10 is considered to be a valid instrument (36). The distribution of the allele scores—the sum of the number of ‘shorter telomere’ alleles, be they the minor or major allele—with these six SNPs approaches a normal distribution and mean TL decreases considerably with increasing numbers of shorter TL-associated alleles, both for cases and controls (Fig. 2).
Figure 2.
The cumulative effect of the top six SNPs on TL. Mean TL and associated 95% CI in base pairs (bp) is illustrated corresponding to the number of ‘shorter telomere’ alleles carried of the six SNPs. The results for the 10 784 breast cancer cases and 14 815 controls are shown in grey and black, respectively. An additional y-axis on the right-hand side of the figure marks the TL at the 25th percentile (3438 for cases, 3525 for controls), 50th (median) (3903 for cases, 4007 for controls) and 75% percentile of length (4492 for cases, 4616 for controls), with cases and controls in grey and black, respectively. Tabulated below the graph is the distribution of the six SNP ‘shorter telomere’ allele scores among 14 815 controls by fraction of total number of individuals (numbers), the mean TL by six SNP allele score (TL) and the difference in TL compared with the lowest allele score group (0 and 1) (ΔTL). There were no individuals present with an allele score of 11 or 12. Data for breast cancer cases are again shown in grey and control data in black.
Associations with cancer risks
We examined the top SNP from each of our confirmed TL loci for additional associations with risks of all hormone-related cancers among the consortia participating in COGS (Table 2). All analyses were confined to participants of European ancestry, thus we examined breast cancer risk in 41 BCAC studies (n = 89 050 cases and controls), ovarian cancer risk in up to 34 OCAC studies (n = 39 774), of whom 8372 cases had serous epithelial ovarian cancer and 979 had serous low malignant potential (LMP) ovarian neoplasias and prostate cancer risk in 32 studies participating in the PRACTICAL Consortium (n = 44 620) (29,30,32).
None of the six SNPs tested displayed strong evidence for association with cancer, with the exception of TERT variant, rs7726159, which was significantly associated with risks of all three cancers [breast cancer odds ratio (OR) = 1.05 [95% confidence intervals (95% CI)] 1.03–1.07, Ptrend = 2 × 10−5; serous invasive ovarian cancer OR = 1.12 (95% CI 1.09–1.16), Ptrend = 3 × 10−9; serous LMP ovarian cancer OR = 1.45 (95% CI 1.36–1.55), Ptrend = 2 × 10−14; prostate cancer OR = 0.88 (95% CI 0.85–0.91), Ptrend = 2 × 10−18]. The only other significant association, allowing for multiple testing, was OBFC1 SNP rs2487999 with serous LMP non-invasive ovarian cancer risk Ptrend = 6 × 10−4, although the number of cases here (n = 979) was relatively small compared with the other case–control comparisons.
DISCUSSION
From our analyses, we find four loci that are associated with TL at genome-wide significance levels; the previously reported TERC (3q26.2), TERT (5p15.33) and OFBC1 (10q24.3) loci, and a novel locus—PXK on 3p14.1. We also find evidence for association at ZNF311 (6p22.1) and BCL2L1 (20q11.2), but these have not yet reached the accepted significance levels for genome-wide studies. The recent publication by Codd et al. (27) reported seven loci associated with TL and here we find genome-wide significance levels for three of these regions: TERC, TERT and OBFC1 and supportive evidence (Ptrend < 5.0 × 10−4) for a further three: ACYP2 (2p16.2), NAF1 (4q32.2), RTEL1 (20q13.3). However, at 2p16.2 we found the minor allele of surrogate SNP (rs10165485) to be associated with longer TL in contrast to the published rs11125529 association with shorter TL. We did not have any good surrogate for the reportedly associated SNP at 19p12 (ZNF208) and so could not test this. We could not confirm previously reported associations (26) at CTC1 (17p13.1) and ZNF676 (19p12) (data not shown). It should be noted that although the ZNF208 and ZNF676 loci are only ∼350 kb apart, their lead SNPs are uncorrelated [pairwise r2 = 0.002 (35)].
In our own GWAS, we observed 272 SNPs with Ptrend < 10−3 among healthy control subjects, significantly more than the expected number of 184. Eighty-six of these 272 variants became more significant upon addition of the data from cancer cases, demonstrating that despite having overall differences in TL, when compared with healthy controls, cancer cases continue to display consistent genetic associations with TL. In the entire set of 26 089 participants (analysis of both healthy controls and cancer cases), 595 SNPs displayed Ptrend < 10−3, a 3-fold excess over the 194 variants that would have been expected by chance. Of the 1558 SNPs nominated from the GWAS meta-analysis, three displayed Ptrend < 10−4 in the total 26 089 ‘Cases and Controls’ analysis, including two SNPs at the TERC locus (marked on Fig. 1), and a further seven SNPs displayed Ptrend < 10−3 (data not shown). We note that none of the SNPs most significantly associated with TL at any of the loci were identified from the initial TL GWAS meta-analysis. This is evidence that our initial study was underpowered, and that future larger studies will continue to confirm the existence of many more TL-associated loci.
Until fine-scale mapping has been completed, we cannot be certain of the functional gene at each locus. The most significant SNP at 3p14.1, rs6772228, is in intron 4 of the PXK gene. This codes for a serine/threonine kinase involved in the regulation of electrical excitability and synaptic transmission, a seemingly unlikely candidate for involvement in telomere biology. It should also be noted that, from the study-by-study results shown in Supplementary Material, Table S2, the PXK association seems driven by the SEARCH cases and controls. The top SNP at 6p22.1 is 13 kb upstream of ZNF311, a gene which codes for a zinc finger protein that may work as a transcription regulator. The most significant SNP at 20q11.2 is in intron 3 of the BCL2L1 gene, which belongs to the BCL-2 family of proteins, integral in the regulation of apoptosis. All six loci are being fine-mapped, with all SNPs correlated with the lead SNP in each region (r2 > 0.3) submitted for genotyping on another upcoming collaborative custom chip.
From our data on the lead SNPs in these six most significant loci, the TL-altering alleles have additive effects in healthy control individuals: those carrying 10 ‘shorter telomere’ alleles have TL on average 731 bp shorter than individuals with one or fewer ‘shorter telomere’ alleles. Since TL decreases by ∼20 bp per year of age (11,28), this is equivalent in magnitude to an age difference of 37 years respectively for these few individuals at the two extremes of the distribution (Fig. 2). Correspondingly, there is a mean 410 bp difference in TL between the controls (9%) with three shorter telomere alleles and those (4%) with eight shorter telomere alleles—equivalent to a >20 year age difference.
The first of our study aims—to find genetic variants that are strongly associated with TL—has been successful, and we have therefore been able to continue to the second aim—to examine whether any of these TL loci also have additional effects on cancer risk in the hormone-related cancer data available to us. When examined individually, only one of the top SNPs (TERT SNP rs7726159) displays highly significant associations with cancer risk (Table 2). This complex locus has already been the in-depth subject of two fine-scale mapping studies (28,40), and both these studies demonstrated that most of the TERT variants associated with TL and those associated with the strongest cancer risks are quite distinct. Although SNP rs7726159 may have a direct functional effect on telomere maintenance, its association with cancer risk is more probably due to its linkage disequilibrium (correlation) with two other SNPs (rs2242652 and rs10069690), which do have likely directly functional effects on cancer risk (28,40). The only other reported TL loci displaying significant evidence of association with hormone-related cancers are OBFC1 SNP rs2487999 with serous LMP ovarian cancer [OR = 1.29 (95% CI 1.14–1.43); Ptrend = 6 × 10−4; Table 2], and RTEL1 SNP rs2738783 with prostate cancer risk, [OR = 0.94 (95% CI 0.90–0.97); Ptrend = 3 × 10−4; Table 3]. Further investigation may be merited in these two cases. The remaining TL-associated SNPs have negligible effects on cancer risk and thus support the findings from prospective studies on TL and future cancer risk (13,18).
In conclusion, this study confirms the genetic control of TL by common SNPs in at least six loci. None of the SNPs at these loci are likely to have substantial direct effects on risk of hormone-related cancer. This is further evidence refuting the hypothesis that shorter mean TL in blood cells is a major causal factor in breast, ovarian or prostate cancer development.
MATERIALS AND METHODS
GWAS and SNP selection
Three individual GWAS were performed to look for variants associated with TL in three studies of disease-free individuals from the East Anglian region of Britain; men from the ProtecT prostate cancer case–control study (34) (n = 1148), women from the Sisters in Breast Screening (SIBS) study (41) (http://ccge.medschl.cam.ac.uk/research/local/sibs-study/) (n = 796) and participants in the European Prospective Investigation into Cancer (EPIC-Norfolk) (http://www.srl.cam.ac.uk/epic/) (n = 296). All the three studies had mean TL determined by the same quantitative real-time PCR (13) and genome-wide SNP genotypes determined using varying platforms (see Supplementary Material). The ProtecT GWAS was carried out using a set of 522 098 SNPs (after quality control (QC)) genotyped on the Illumina InfiniumTM platform. The SIBS GWAS was performed using a set of 255 051 SNPs (after QC), genotyped on the Illumina HumanCytoSNP-12™ chip. The EPIC GWAS genotyping was performed using a custom high-density array by Perlegen Sciences Inc. and after QC, 187 508 SNPs were included in further analyses. From all these datasets, the genotypes of 2 448 093 SNPs with minor allele frequency (MAF) >0.01 were imputed using MaCH in all 2240 participants and a meta-analysis of the three studies was carried out using ProbABEL (see Supplementary Material). From this, we identified and selected 1629 potentially TL-associated variants (Ptrend < 10−4) for inclusion on the iCOGs chip. Of these, 1558 passed QC and were included in the final analyses.
Associations with TL
Among the participating COGS studies, the TL had been determined in cases and controls from the UK Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) study (13,28) (http://ccge.medschl.cam.ac.uk/search/) controls from the Danish Copenhagen City Heart study (28,42,43) (CCHS) and breast cancer cases from the Copenhagen General Population Study (18,44) (CGPS). TL was measured using a real-time PCR methodology as described elsewhere (11–13,45,46) and a composite variable, putting the SEARCH, CCHS and CGPS data onto the same scale, was used for all analyses (28) (see Supplementary Material). All 187 647 SNPs with MAF >0.02 on the iCOGS chip that passed QC criteria were tested for association with TL in these subjects. Initially, analysis was confined to the healthy control subjects within each study (SEARCH females, n = 6766; CCHS females, n = 4537E and CCHS male, n = 3762). A total of 272 SNPs were associated with TL, at Ptrend < 10−3, in these control subjects (Supplementary Material, Table S1). All associations are consistent with a log-additive model.
In an exploratory attempt to increase the available sample size and thus the study power of this study, the above analysis was repeated using breast cancer cases as well as the healthy controls from these cancer studies. Therefore, SEARCH cases (n = 8210) and CGPS cases (n = 2814) were also included in the analysis. Of the 272 variants with per-allele Ptrend < 10−3 in the control-only analysis, 86 variants increased in significance upon inclusion of TL data from these breast cancer cases in our analysis (Supplementary Material, Table S1). Among these were SNPs within all the loci previously and independently reported to be associated with TL; therefore, we feel confident that the inclusion of cancer cases is a valid way of increasing the study power.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing of and/or approved the manuscript. Conceived and designed the experiment: K.A.P., S.E.B., D.F.E., A.M.D., B.G.N. SNP selection: K.A.P., S.E.B., A.A.A.O., Z.K.-J., E.D., R.Y., A.R., J.S., J.C.-C., B.B., G.C.-T., P.D.P.P., A.B., R.A.E., D.F.E., A.M.D. iCOGS genotyping, calling and QC: K.A.P., S.E.B., S.F.N., A.A.A.O., C.B., J.D., G.C.-T., P.D.P.P., D.F.E., A.M.D.. Imputation: D.T., A.A.A.O., D.F.E. TL determination and analysis: K.A.P., S.E.B., M.W., S.F.N., D.F.E., A.M.D., B.G.N. GWAS sample and information provision: D.T., A.A.A.O., J.B., T.A., R.L., K.-T.K., D.E.N., F.C.H., J.L.D., D.F.E. Statistical analyses and programming: K.A.P., S.E.B., D.T., A.A.A.O., K.M., J.P.T., Z.K.-J., D.F.E. COGS coordination: G.C.-T., P.D.P.P., D.F.E., A.M.D. BCAC coordination: G.C.-T., P.D.P.P., D.F.E., A.M.D. BCAC data management: M.S., M.K.B., Q.W., J.D., E.D. OCAC coordination: G.C.-T., P.D.P.P., A.B. OCAC data management: J.T., P.D.P.P. PRACTICAL coordination: R.A.E., D.F.E. PRACTICAL data management: A.A.A.O., S.B., Z.K.-J.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement No. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (No. 1 U19 CA 148537—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation and the Ovarian Cancer Research Fund. TL measurement and analysis were funded by CR-UK project grant C1287/A9540 and Chief Physician Johan Boserup and Lise Boserup's Fund. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK grant C8197/A10123.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. This study would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F. Easton (BCAC), Andrew Berchuck (OCAC), Rosalind A. Eeles, Douglas F. Easton, Ali Amin Al Olama, Zsofia Kote-Jarai (PRACTICAL), Georgia Chenevix-Trench, Antonis Antoniou, Fergus Couch and Ken Offit (CIMBA), Joe Dennis, Alison M. Dunning, Andrew Lee and Ed Dicks (Cambridge), Javier Benitez, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffret Meyer and the staff of Mayo Clinic Genotyping Core Facility.
REFERENCES
- 1.Baird D.M. Telomeres. Exp. Gerontol. 2006;41:1223–1227. doi: 10.1016/j.exger.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan S.R., Blackburn E.H. Telomeres and telomerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Njajou O.T., Cawthon R.M., Damcott C.M., Wu S.H., Ott S., Garant M.J., Blackburn E.H., Mitchell B.D., Shuldiner A.R., Hsueh W.C. Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl Acad. Sci. USA. 2007;104:12135–12139. doi: 10.1073/pnas.0702703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slagboom P.E., Droog S., Boomsma D.I. Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 6.Andrew T., Aviv A., Falchi M., Surdulescu G.L., Gardner J.P., Lu X., Kimura M., Kato B.S., Valdes A.M., Spector T.D. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am. J. Hum. Genet. 2006;78:480–486. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 8.Harley C.B. Telomere loss: mitotic clock or genetic time bomb? Mutat. Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 9.Allsopp R.C., Vaziri H., Patterson C., Goldstein S., Younglai E.V., Futcher A.B., Greider C.W., Harley C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy M.Z., Allsopp R.C., Futcher A.B., Greider C.W., Harley C.B. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 11.Weischer M., Bojesen S.E., Cawthon R.M., Freiberg J.J., Tybjaerg-Hansen A., Nordestgaard B.G. Short telomere length, myocardial infarction, ischemic heart disease, and early death. Arterioscler. Thromb. Vasc. Biol. 2012;32:822–829. doi: 10.1161/ATVBAHA.111.237271. [DOI] [PubMed] [Google Scholar]
- 12.McGrath M., Wong J.Y., Michaud D., Hunter D.J., De Vivo I. Telomere length, cigarette smoking, and bladder cancer risk in men and women. Cancer Epidemiol. Biomarkers Prev. 2007;16:815–819. doi: 10.1158/1055-9965.EPI-06-0961. [DOI] [PubMed] [Google Scholar]
- 13.Pooley K.A., Sandhu M.S., Tyrer J., Shah M., Driver K.E., Luben R.N., Bingham S.A., Ponder B.A., Pharoah P.D., Khaw K.T., et al. Telomere length in prospective and retrospective cancer case–control studies. Cancer Res. 2010;70:3170–3176. doi: 10.1158/0008-5472.CAN-09-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J., Gammon M.D., Terry M.B., Wang Q., Bradshaw P., Teitelbaum S.L., Neugut A.I., Santella R.M. Telomere length, oxidative damage, antioxidants and breast cancer risk. Int. J. Cancer. 2009;124:1637–1643. doi: 10.1002/ijc.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wentzensen I.M., Mirabello L., Pfeiffer R.M., Savage S.A. The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2011;20:1238–1250. doi: 10.1158/1055-9965.EPI-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vivo I., Prescott J., Wong J.Y., Kraft P., Hankinson S.E., Hunter D.J. A prospective study of relative telomere length and postmenopausal breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2009;18:1152–1156. doi: 10.1158/1055-9965.EPI-08-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zee R.Y., Castonguay A.J., Barton N.S., Buring J.E. Mean telomere length and risk of incident colorectal carcinoma: a prospective, nested case-control approach. Cancer Epidemiol. Biomarkers Prev. 2009;18:2280–2282. doi: 10.1158/1055-9965.EPI-09-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weischer M., Nordestgaard B.G., Cawthon R.M., Freiberg J.J., Tybjaerg-Hansen A., Bojesen S.E. Short telomere length, cancer survival, and cancer risk in 47102 individuals. J. Natl Cancer Inst. 2013;105:459–468. doi: 10.1093/jnci/djt016. [DOI] [PubMed] [Google Scholar]
- 19.Allin K.H., Nordestgaard B.G., Zacho J., Tybjaerg-Hansen A., Bojesen S.E. C-reactive protein and the risk of cancer: a Mendelian randomization study. J. Natl. Cancer Inst. 2010;102:202–206. doi: 10.1093/jnci/djp459. [DOI] [PubMed] [Google Scholar]
- 20.Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 21.Benn M., Tybjaerg-Hansen A., Stender S., Frikke-Schmidt R., Nordestgaard B.G. Low-density lipoprotein cholesterol and the risk of cancer: a Mendelian randomization study. J. Natl Cancer Inst. 2011;103:508–519. doi: 10.1093/jnci/djr008. [DOI] [PubMed] [Google Scholar]
- 22.Nordestgaard B.G., Palmer T.M., Benn M., Zacho J., Tybjaerg-Hansen A., Davey S.G., Timpson N.J. The effect of elevated body mass index on ischemic heart disease risk: causal estimates from a Mendelian randomisation approach. PLoS Med. 2012;9:e1001212. doi: 10.1371/journal.pmed.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Codd V., Mangino M., van der Harst P., Braund P.S., Kaiser M., Beveridge A.J., Rafelt S., Moore J., Nelson C., Soranzo N., et al. Common variants near TERC are associated with mean telomere length. Nat. Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy D., Neuhausen S.L., Hunt S.C., Kimura M., Hwang S.J., Chen W., Bis J.C., Fitzpatrick A.L., Smith E., Johnson A.D., et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl Acad. Sci. USA. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prescott J., Kraft P., Chasman D.I., Savage S.A., Mirabello L., Berndt S.I., Weissfeld J.L., Han J., Hayes R.B., Chanock S.J., et al. Genome-wide association study of relative telomere length. PLoS One. 2011;6:e19635. doi: 10.1371/journal.pone.0019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangino M., Hwang S.J., Spector T.D., Hunt S.C., Kimura M., Fitzpatrick A.L., Christiansen L., Petersen I., Elbers C.C., Harris T., et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012;21:5385–5394. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codd V., Nelson C.P., Albrecht E., Mangino M., Deelen J., Buxton J.L., Hottenga J.J., Fischer K., Esko T., Surakka I., et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 2013;45:422–427. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pharoah P.D., Tsai Y.Y., Ramus S.J., Phelan C.M., Goode E.L., Lawrenson K., Buckley M., Fridley B.L., Tyrer J.P., Shen H., et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013;45:362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaudet M.M., Kuchenbaecker K.B., Vijai J., Klein R.J., Kirchhoff T., McGuffog L., Barrowdale D., Dunning A.M., Lee A., Dennis J., et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J., et al. Genome-Wide Association Study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S., et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donovan J., Mills N., Smith M., Brindle L., Jacoby A., Peters T., Frankel S., Neal D., Hamdy F. Quality improvement report: improving design and conduct of randomised trials by embedding them in qualitative research: ProtecT (prostate testing for cancer and treatment) study. Commentary: presenting unbiased information to patients can be difficult. BMJ. 2002;325:766–770. doi: 10.1136/bmj.325.7367.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock J.H., Wright J.H., Yogo M. A survey of weak instruments and weak identification in generalized method of moments. J. Bus. Econ. Stat. 2002;20:518. http://ccge.medschl.cam.ac.uk/search/ [Google Scholar]
- 37.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., Sigurdsson A., Gudbjartsson D.F., Sigurgeirsson B., Benediktsdottir K.R., et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbull C., Rapley E.A., Seal S., Pernet D., Renwick A., Hughes D., Ricketts M., Linger R., Nsengimana J., Deloukas P., et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kote-Jarai Z., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Dadaev T., Jugurnauth-Little S., Ross-Adams H., Al Olama A.A., Benlloch S., Halim S., et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum. Mol. Genet. 2013;22:2520–2528. doi: 10.1093/hmg/ddt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pooley K.A., Tyrer J., Shah M., Driver K.E., Leyland J., Brown J., Audley T., McGuffog L., Ponder B.A., Pharoah P.D., et al. No association between TERT-CLPTM1L single nucleotide polymorphism rs401681 and mean telomere length or cancer risk. Cancer Epidemiol. Biomarkers Prev. 2010;19:1862–1865. doi: 10.1158/1055-9965.EPI-10-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bojesen S.E., Tybjaerg-Hansen A., Axelsson C.K., Nordestgaard B.G. No association of breast cancer risk with integrin beta3 (ITGB3) Leu33Pro genotype. Br. J. Cancer. 2005;93:167–171. doi: 10.1038/sj.bjc.6602674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allin K.H., Bojesen S.E., Nordestgaard B.G. Baseline C-reactive protein is associated with incident cancer and survival in patients with cancer. J. Clin. Oncol. 2009;27:2217–2224. doi: 10.1200/JCO.2008.19.8440. [DOI] [PubMed] [Google Scholar]
- 44.Zacho J., Tybjaerg-Hansen A., Jensen J.S., Grande P., Sillesen H., Nordestgaard B.G. Genetically elevated C-reactive protein and ischemic vascular disease. N. Engl. J. Med. 2008;359:1897–1908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]
- 45.Cawthon R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cawthon R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.