Abstract
Analysis of plasma microRNAs (miRNAs) by quantitative polymerase chain reaction (qPCR) provides a potential approach for cancer diagnosis. However, absolutely quantifying low abundant plasma miRNAs is challenging with qPCR. Digital PCR offers a unique means for assessment of nucleic acids presenting at low levels in plasma. This study aimed to evaluate the efficacy of digital PCR for quantification of plasma miRNAs and the potential utility of this technique for cancer diagnosis. We used digital PCR to quantify the copy number of plasma microRNA-21-5p (miR-21–5p) and microRNA-335–3p (miR-335–3p) in 36 lung cancer patients and 38 controls. Digital PCR showed a high degree of linearity and quantitative correlation with miRNAs in a dynamic range from 1 to 10,000 copies/μL of input, with high reproducibility. qPCR exhibited a dynamic range from 100 to 1×107 copies/μL of input. Digital PCR had a higher sensitivity to detect copy number of the miRNAs compared with qPCR. In plasma, digital PCR could detect copy number of both miR-21–5p and miR-335–3p, whereas qPCR was only able to assess miR-21–5p. Quantification of the plasma miRNAs by digital PCR provided 71.8% sensitivity and 80.6% specificity in distinguishing lung cancer patients from cancer-free subjects.
Keywords: digital PCR, miRNAs, plasma, diagnosis, lung cancer
Introduction
Non-small-cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer that is the number one cancer killer.1 The 5-year survival rate for stage IV NSCLC is only 10%, whereas it is nearly 80% for stage IA NSCLC.1 These statistics provide the primary rationale to improve the early detection of NSCLC.2 MicroRNAs (miRNAs) are small molecules that have important functions in diverse biological processes, including cell proliferation, differentiation, and apoptosis.3–5 miRNAs can transcriptionally regulate expressions of more than 30% of human protein-coding genes.3,4 Furthermore, dysfunction of some miRNAs contributes to the development and progression of human malignancies, including lung cancer.6 In addition, miRNA expression profiles offer molecular signatures for the classification, diagnosis, and progression of cancer, and thus could be developed as cancer biomarkers.5,7 Blood is one of the most easily and noninvasively accessible body fluids. The use of routine blood plasma samples for assessment of miRNAs originating from primary tumors either as a result of metastasizing cells or the leakage of sequences from the tumors into the peripheral circulation would provide a useful tool for lung cancer early detection.8–10
Quantitative polymerase chain reaction (qPCR) is one of the most commonly used techniques that can estimate expression levels of miRNAs in clinical specimens.9–12 However, qPCR has 2 major challenges for the assessment of plasma miRNAs.13,14 First, qPCR is an indirect and labor-consuming approach to analyzing miRNAs, as it relies on an increase in fluorescence signal that is proportional to the polymerase reaction product, and uses the cycle threshold (CT) as a metric. CT values for miRNA targets are referenced to endogenous small RNA controls across samples and used for normalization. This can become problematic, because expression levels of the endogenous controls and their transcripts may differ between samples.13,15 Furthermore, numerous endogenous genes have been evaluated for determination of target miRNAs, including U6, U6B, 18S rRNA, 5S RNA, RNU38B, and RNU43, yet none has been widely accepted as a standard control.9,10 These problems can be partially solved through the use of an exogenous “spike-in” control, which, however, does not account for any template-specific effect or bias introduced through primer design. Moreover, to estimate the absolute abundance of a given miRNA, data must be compared to a previously-generated standard curve from the same template with identical primers and conditions. However, the additional manipulations are labor intensive, and extreme care should be taken when measuring the reference samples and comparing the references and experimental standard curves.13 Second, the sensitivity of qPCR for the detection of a low copy number in genes is not high enough, as it only resolves ~1.5-fold changes of nucleic acids.14 Given that a proportion of the cancer-associated miRNAs is derived from primary tumor and could be ‘diluted’ in a background of normal miRNAs,8,16,17 the miRNAs presenting at low levels in plasma could be undetectable by qPCR.
Droplet digital PCR is a direct method for quantitatively measuring nucleic acids,18–25 as it depends on limiting partition of the PCR volume, where a positive result of a large number of microreactions indicates the presence of a single molecule in a given reaction. The number of positive reactions, together with Poisson’s distribution, can be used to produce a straight and high-confidence measurement of the original target concentration.23 Therefore, digital PCR does not require a reliance on rate-based measurements (CT values), endogenous controls, and the use of calibration curves. Furthermore, previous studies targeting low copy numbers in nucleic acids have demonstrated that digital PCR has a high degree of sensitivity and precision as compared to qPCR.26–28 However, limited data25,29 exist regarding the application of this new technology to quantify miRNAs in clinical specimens, particularly plasma, in the field of molecular oncology.
The objective of the present study was to evaluate the efficacy of digital PCR for the quantification of plasma miRNAs and its potential utility for cancer diagnosis. We first investigated the efficacy of using digital PCR for quantitative detection of 2 miRNAs (miRs-21–5p and 335–3p) in artificially-seeded samples, RNA of cancer cells, and clinical plasma samples. miRs-21–5p and 335–3p were chosen, because our previous studies9–12 showed that miR-21–5p displayed a high expression level, whereas miR-335–3p had an endogenously low level in plasma. We then used digital PCR to quantify the copy number of plasma miR-21–5p and miR-335–3p in 36 lung cancer patients and 38 controls. This study presents the earliest assessment of digital PCR as a potential tool for quantitative detection of miRNAs in clinical samples for cancer diagnosis.
Materials and Methods
Determination of the dynamic range and sensitivity of miRNA quantification by using digital PCR and qPCR
Synthetic single-stranded RNA oligonucleotides corresponding to human mature miRNAs, miR-21–5p and miR-335–3p, were purchased from Integrated DNA Technologies (Integrated DNA Technologies, San Diego, CA). To generate standard curves for the miRNAs, the synthetic miRNAs were inputted into the reverse transcription (RT) reaction over an empirically-derived range of copies, as previously described.17,30 Briefly, the varying dilutions of each oligonucleotide were made in diethylpyrocarbonate (DEPC) H2O such that the final input into the RT reaction had a volume of 1.67 μL. Input RNA was reversely transcribed by a T100 thermal cycler (Applied Biosystems, Foster City, CA) using TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied BioSystems). Information about the oligo sequences of miR-21–5p and miR-335–3p used for the RT and digital/qPCR reactions can be found on the website of Life Technologies Corporation (Life Technologies Corporation, Frederick, MD) with Cat. # 4427975 and Cat. # 4427975. The thermocycler parameters were as follows: hold for 30 min at 16°C, for 30 min at 42°C, and 5 min at 85°C. For conventional qPCR analysis, 2.25 μL of cDNA was combined with 2.75 μL of PCR reagents to produce a PCR reaction in a total volume of 5.0 μL. qPCR was carried out on an Applied BioSystems 7900HT thermocycler (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 min. qPCR data was analyzed by using the RQ Manager software (Applied Biosystems) with an automatic Ct setting for assigning the baseline and threshold for Ct determination. A standard curve from each synthetic miRNA was established, as previously described.17,30 All tests were performed in triplicates.
Digital PCR was performed in parallel for the measurement of miRNAs in the serially-diluted oligonucleotides. 20 μL of the reaction mixture containing 5 μL of cDNA solution, 10 μL of digital PCRTM Supermix (Bio-Rad, Hercules, CA), and 1 μL of Taqman primer/probe mix (Applied Biosystems) and DEPC H2O was loaded into a plastic cartridge (Bio-Rad) with 70 μL of QX100 Droplet Generation oil (Bio-Rad) and then placed into the QX100 Droplet Generator (Bio-Rad). The droplets generated from each sample were transferred to a 96-well PCR plate (Eppendorf, Germany). PCR amplification was carried on a T100 thermal cycler (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles of 95°C for 30 seconds and 60°C for 1 min, then 1 cycle of 98°C for 10 min, ending at 4°C. The plate was then loaded on Droplet Reader (Bio-Rad) and read automatically. Absolute quantification of each miRNA was calculated from the number of positive counts per panel using the Poisson distribution, as previously described.13,27,29,31 The quantification of the target miRNAs was presented as the number of copies/μL of PCR mixture. All tests were performed in triplicates.
To further determine the dynamic range and sensitivity of miRNA quantification by using digital PCR, we isolated total RNA from a lung cancer cell line (SK-MES-1) (American Type Culture Collection, Manassas, VA), using the miRVana™ PARIS™ Kit (Ambion, Austin, TX). cDNA was reversely transcribed using miRNA-specific primers for miR-335–3p in 15 μl reaction mixture. The RT product was diluted in DEPC water by 7 orders of magnitude ranging from 1:1 to 1:4096. The diluted samples were run by using digital PCR and qPCR, respectively, for the quantification of miR-335–3p.
miRNA quantification by using digital PCR and qPCR in plasma
We recruited 15 cancer-free healthy nonsmokers from University of Maryland Medical Center under our Institutional Review Board (IRB)-approved research protocol. Written consent was obtained from the subjects of the study. RNA was isolated from plasma of 15 cancer-free nonsmokers using the miRVana™ PARIS™ Kit (Ambion).12 1 μL RNA was reversely transcribed to cDNA with miRNA-specific primers for miR-21–5p and miR-335–3p, respectively, in a 15 μL reaction mixture. 5 μL of the RT product was used for digital PCR and qPCR analyses as described above for miRNA quantification in a 20 μL reaction mixture. To quantify the miRNAs in plasma by qPCR, CT values were converted into absolute copy number per μL reaction mixture by using the standard curves generated above. To quantify the miRNAs by digital PCR, the copy number of each miRNA per μL PCR reaction mixture was directly determined using the Droplet Reader (Bio-Rad). The copy number of miRNA per μL of plasma was calculated based on a formula: copy number of each miRNA per μL of PCR reaction mixture is multiplied by the number of dilution folds in the plasma.
Determination of diagnostic values of miRNA quantification in plasma of lung cancer patients and cancer-free controls by digital PCR
We recruited 36 patients with histological types of stage I NSCLC and 38 cancer-free individuals, including heavy smokers, from University of Maryland Medical Center under our IRB protocol. Geographic and clinical characteristics of the cases and controls are shown in Table 1. 10 mL of peripheral blood was drawn from the subjects using standardized phlebotomy procedures in BD Vacutainer spray-coated K2EDTA Tubes (BD, Franklin Lakes, NJ). The blood samples from lung cancer patients were collected at the time of initial consultation, prior to definitive surgical management and/or adjuvant therapy. The specimens were processed within 2 hours of collection by centrifugation at 1,300 × g at 4°C for 10 minutes. Plasma was then transferred to a fresh tube and stored at −80°C until use.
Table 1.
Demographic and clinical data of 36 lung cancer patients and 38 cancer-free controls.
| PARAMETER | LUNG CANCER PATIENTS | % | CANCER-FREE CONTROLS | % |
|---|---|---|---|---|
|
|
|
|||
| MEAN | MEAN | |||
| Age, years | 66.7 | 64.6 | ||
|
| ||||
| Gender | ||||
|
| ||||
| Men | 22 | 61.1 | 25 | 65.8 |
|
| ||||
| Women | 14 | 38.9 | 13 | 34.2 |
|
| ||||
| Race | ||||
|
| ||||
| White American | 27 | 75 | 28 | 73.7 |
|
| ||||
| African American | 9 | 25 | 10 | 26.3 |
|
| ||||
| Smoking, pack-years | 49.3 | 19.6 | ||
|
| ||||
| 28.6 | 12.7 | |||
|
| ||||
| Histology | ||||
|
| ||||
| Adenocarcinoma | 20 | 55.6 | ||
|
| ||||
| Squamous cell carcinoma | 16 | 44.4 | ||
|
| ||||
| All are stage I NSCLC | ||||
Abbreviation: NSCLC, non-small cell lung cancer.
Statistical analysis
A t-test was applied to determine significant differences of values between groups. Kappa statistics were used to evaluate agreement between the different individuals for quantification of miRNAs. Bland-Altman plots were used to analyze the correlation of the 2 methods. A Pearson’s correlation analysis was utilized to assess the relationship between plasma miRNA expressions and demographic characteristics of the patients and cancer-free controls. Clinicopathologic diagnoses were used as reference standards to decide the sensitivity and specificity of the miRNAs. Receiver-operator characteristic (ROC) curve and area under the curve (AUC) analyses were applied to determine the accuracy of each miRNA and cut-off values in a specimen with a given specificity rate. We used logistic regression models with constrained parameters as in least absolute shrinkage and selection operator (lasso) to determine the performance of the combined use of 2 genes. All P values shown were 2-sided, and a P-value of < 0.05 was considered statistically significant.
Results
The dynamic ranges and sensitivities of miRNA quantification by using digital PCR and qPCR
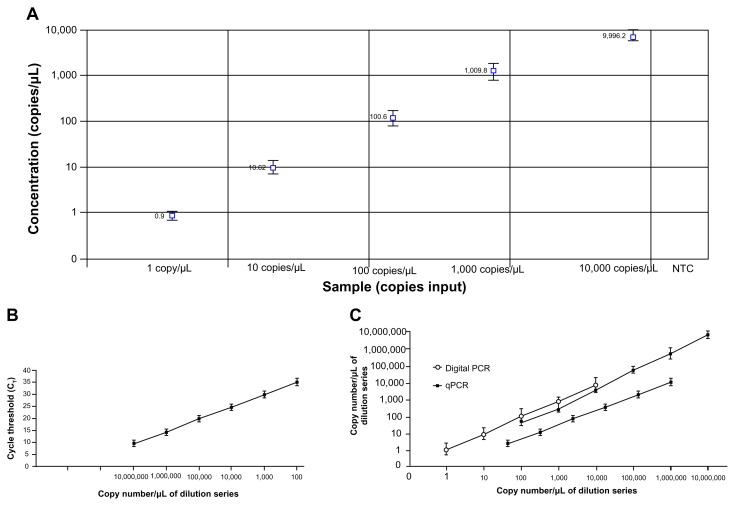
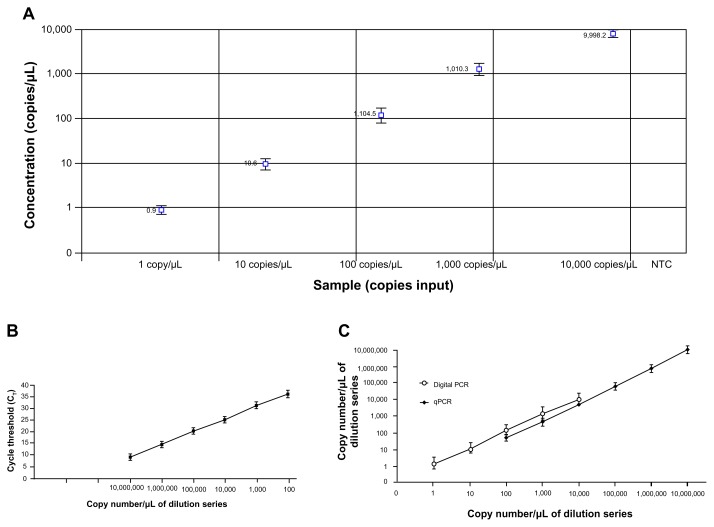
Synthetic single-stranded RNA oligonucleotides corresponding to human mature miR-335–3p and miR-21–5p were serially diluted and run with digital PCR and qPCR for measuring their copy number. Digital PCR analysis showed that both miR-335–3p and miR-21–5p exhibited excellent linearity between the target input and measured values in a dynamic range of 5 orders of magnitude from 1 to 10,000 copies/μL of input (Figs. 1 and 2). Furthermore, in this dynamic range, digital PCR displayed an estimated slope coefficient that was close to 1 (0.95–0.98) and R2 values that were close to 1 (0.96–0.99) for detection of the miRNAs. To determine reproducibility between digital PCR assays, the panel of dilutions of the synthetic RNA oligonucleotides was independently analyzed by 2 research staff members. There was a high agreement between the results produced from the independent digital PCR assays (the kappa statistic for concordance was 0.86, P < 0.01). Standard curves created by qPCR assays showed that the 2 miRNAs displayed excellent linearity between target input and CT values in a dynamic range from 100 to 1 × 107 copies per μL (Figs. 1 and 2). However, qPCR produced more than 35 Ct values for the samples that had less than 100 copies of miRNAs per μL of input, suggesting that there was no significant amplification signal in the samples. Overall, digital PCR had a narrower dynamic range (approximately 1 and 10,000 copies per μL of input) compared with qPCR (100 and 1 × 107 copies per μL of input) for quantification of the miRNAs (Figs. 1 and 2). However, the lowest copy number detected by digital PCR was significantly lower (1 copy per μL of input) than that by qPCR (approximately 100 copies per μL of input). Therefore, digital PCR might have a higher sensitivity compared with qPCR for quantitatively measuring copy number of the miRNAs tested in the present study.
Figure 1.
The dynamic ranges and sensitivities of digital PCR and qPCR for miR-335–3p quantification. A. Linearity of miR-335–3p concentration measured by digital PCR in a dilution series of known input amounts of synthetic oligonucleotide corresponding to miR-355–3p and a negative template control (NTC) sample. The x-axis represented concentrations (copy number/μL) of the dilution series; y-axis represented measured concentrations by digital PCR in triplets. Digital PCR had a dynamic range of 5 orders of magnitude from 1 to 10,000 copies per μL. B. A standard curve was generated for miR-335–3p by qPCR in the serially-diluted samples. The x-axis displayed concentrations (copy number/μL) of the dilution series; the y-axis represented their Ct measured by qPCR. qPCR had a dynamic range of 6 orders of magnitude ranging from 100 to 1 × 107 copies per μL. C. Digital PCR had a narrower dynamic range (1 and 10,000 copies per μL of input) compared with qPCR (100 and 1 × 107 copies per μL of input) for quantification of miR-335–3p. The lowest copy number detected by digital PCR is significantly lower (one copy per μL of input) than that by qPCR (100 copies per μL of input). The experiments were performed 3 times.
Figure 2.
The dynamic ranges and sensitivities of digital PCR and qPCR for miR-21–5p quantification. A. Linearity of miR-21–5p concentration measured by digital PCR in a dilution series of known input amounts of synthetic oligonucleotide corresponding to miR-21–5p and a negative template control (NTC) sample. Digital PCR had a dynamic range of 5 orders of magnitude from one to 10,000 copies per μL. B. qPCR had a dynamic range of six orders of magnitude ranging from 100 to 1 × 107 copies per μL. C. Digital PCR had a narrower dynamic range compared with qPCR for quantification of miR-21–5p. The lowest copy number detected by digital PCR is significantly lower (one copy per μL of input) than that by qPCR (100 copies per μL of input). The experiments were performed 3 times.
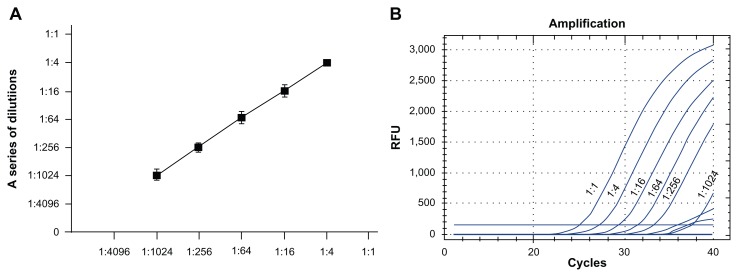
To further determine the sensitivity of miRNA quantification by using digital PCR, total RNA was isolated from a lung cancer cell line, SK-MES–1. The generated cDNA was diluted over 1:4 series by 7 orders of magnitude ranging from 1:1 to 1:4096, and then run with digital PCR and qPCR, respectively. The values generated by digital PCR were associated well with the RNA input (R2 = 0.95), at over 5 orders of magnitude ranging from 1:4 to 1:1024 (Fig. 3A). The lowest concentration of miR-335–3p measured by digital PCR was 0.63 copies/μL of reaction in 1:1024 dilutions. qPCR analysis showed that the CT values correlated well with the RNA input (R2 = 0.94), at over 5 orders of magnitude ranging from 1:1 to 1:256 (Fig. 3B) in serially-diluted samples. However, qPCR could not produce amplification signal in the samples that had a low amount of RNA input (1:1024 dilutions; Fig. 3B; Supplementary Fig. 1). These observations confirm that digital PCR might have a higher sensitivity than does qPCR assay for quantitative assessment of the 2 miRNAs.
Figure 3.
Quantification of miR-335–3p by digital PCR and qPCR using RNA isolated from a lung cancer cell line, SK-MES-1. The generated cDNA was diluted over 1:4 series by 7 orders of magnitude ranging from 1:1 to 1:4096. A. The values produced by digital PCR were related to the RNA input over 5 orders of magnitude ranging from 1:4 to 1:2014. The x-axis represented input concentrations of the dilution series; y-axis represented measured concentrations by digital PCR. B. qPCR analysis showed that the CT values associated to the RNA input over 5 orders of the magnitude ranging from 1:1 to 1:256. However, qPCR was not able to reliably detect the sample that had low amount RNA input (1:1024 dilutions). This experiment was performed 3 times. Figure 3B only shows 1 well for the amplification at each dilution.
The sensitivity of digital PCR for quantitatively detecting miRNAs in plasma
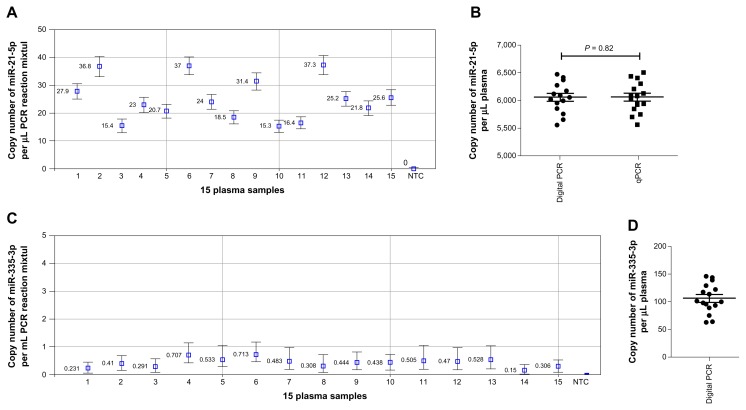
To investigate the sensitivity of digital PCR for assaying copy number of miRNAs in plasma, RNA was isolated from 15 healthy nonsmokers, and then run with digital PCR and qPCR in parallel for the quantification of miR-21–5p and miR-335–3p. For qPCR assay, standard curves generated from the above experiments in the above serially-diluted samples were used to determine copy number of the miRNAs. For digital PCR, the copy number of each miRNA was determined by using the Droplet Reader. Each well of the plasma samples contained at least 10,000 droplets. Therefore, the samples were successfully “read” by a fluorescence detector. The copy number of miR-21–5p/μL PCR reaction mixture assessed by digital PCR was 25.09 ± 1.93 (Fig. 4A). The copy number of miR-21–5p/μL reaction determined by qPCR was 24.69 ± 2.2. Consequently, the copy number of miR-21–5p/μL plasma determined by digital PCR and qPCR was 6,021.6 ± 463.2 and 5,925.6 ± 480, respectively (Fig. 4B). There was no significant difference of copy number of miR-21–5p assessed by the different techniques (P = 0.82). Furthermore, there was close agreement between the 2 methods for measuring miR-21–5p in this set of specimens as determined by Bland-Altman Analysis (Supplementary Fig. 2 and Supplementary Table 1). Consistently, a Pearson’s correlation analysis showed a high correlation between the 2 methods (R2 = 0.96). The copy number of miR-335–3p was also successfully assessed in the plasma samples by digital PCR. As shown in Figure 4C, the copy number of miR-335–3p per μL PCR reaction mixture assessed by digital PCR was 0.44 ± 0.13. Subsequently, the copy number of miR-335–3p per μL plasma evaluated by digital PCR was 106.2 ± 7.0 (Fig. 4D). The Ct value of qPCR for miR-335–3p in the plasma samples was more than 40, and thus amplification curves for the miRNAs were not generated. The absence of amplification for miR-335–3p indicated that qPCR could not efficiently detect the miRNA in plasma. Therefore, digital PCR rather than qPCR might reliably and sensitively measure the copy number of miR-335–3p that has endogenous low-level expression in plasma.
Figure 4.
Quantification of miRNAs in plasma of 15 healthy nonsmokers by digital PCR and qPCR. A. Concentration represented by copy number of miR-21–5p per μL PCR reaction by digital PCR assay. B. The graph indicated copy number of miR-21–5p per μL plasma measured in the 15 plasma samples by digital PCR and qPCR, respectively. C. Concentration represented by copy number of miR-335–3p per μL PCR reaction by digital PCR. D. Copy number of miR-335–3p per μL plasma measured in the 15 samples by digital PCR.
Diagnostic potential of miRNA quantification in plasma by digital PCR
The absolute copy number of 2 lung cancer-associated miRNAs, miR-21–5p and miR-335–3p, was evaluated in plasma obtained from a cohort of 36 NSCLC patients and 38 cancer-free subjects. The copy number for miR-21–5p/μL plasma in the specimens of cancer patients and cancer-free individuals was 23,458 ± 5,249 and 6,384 ± 796.7, respectively (P = 0.0013). The copy number for miR-335–3p/μL plasma of cancer patients and cancer-free individuals was 278.4 ± 70.49 and 91.19 ± 16.64, respectively (P = 0.009) (Supplementary Fig. 3). Overall, both miR-21–5p and miR-335–3p had a significantly higher copy number in the plasma of lung cancer patients compared with cancer-free controls.
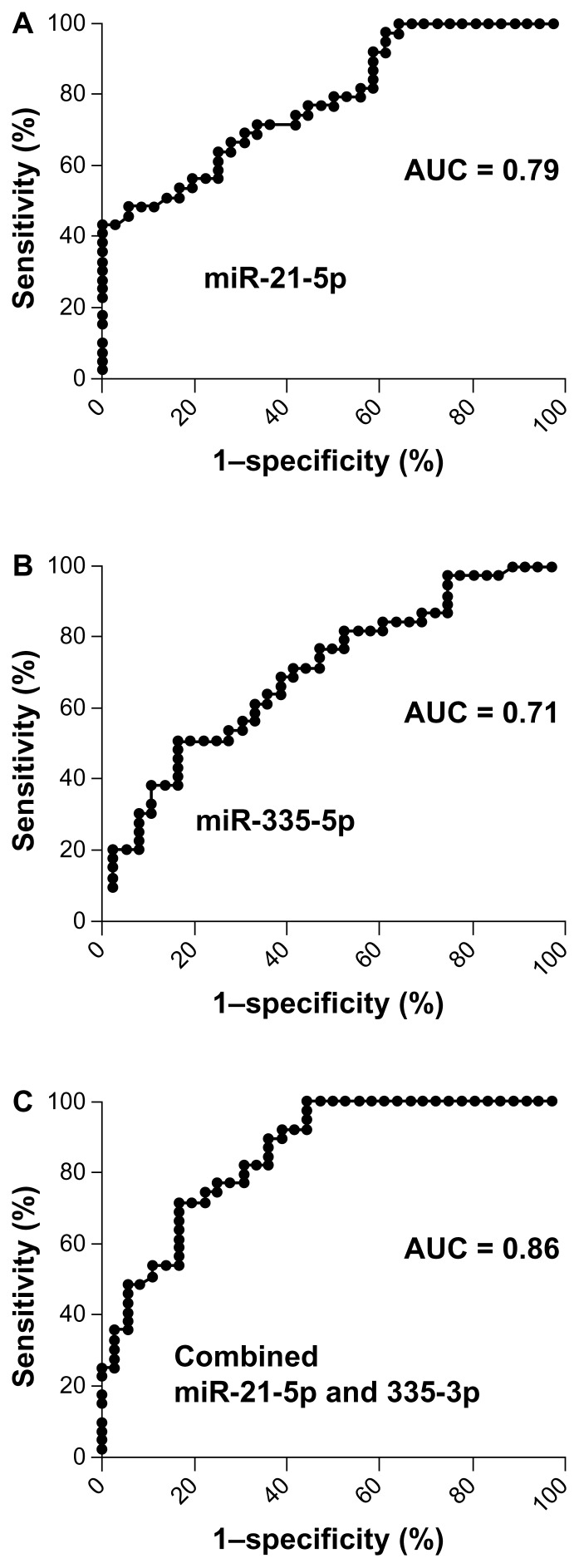
The 2 miRNAs exhibited AUC values of 0.79 and 0.71, respectively, in distinguishing NSCLC patients from the controls (Fig. 5A–B). Furthermore, combined quantification of the 2 miRNAs by digital PCR produced 0.86 AUC, which was statistically higher than that of individual ones used alone (Fig. 5C; all P < 0.05). Given a specificity of 80.6%, the 2 miRNAs used in conjunction revealed a sensitivity of 71.8% in differentiating the NSCLC patients from the cancer-free subjects. Given a sensitivity of 100%, the 2 miRNAs used in combination produced a specificity of 52.6% in the set of cases and controls. Furthermore, the estimated correlation between the miRNAs was low (all R’s < 0.50, all P’s > 0.05), implying that the copy number of the 2 miRNAs in plasma could be complementary to each other.
Figure 5.
Diagnostic values of miRNA quantification by digital PCR in plasma. The ROC and AUC were used to determine sensitivity and specificity of each miRNA. AUC for each miRNA conveyed its accuracy in differentiating lung cancer patients from the control subjects in terms of sensitivity and specificity. A. ROC curve analysis of copy numbers of the miR-21–5p in plasma of the 36 lung cancer patients and 38 cancer-free individuals produced 0.79 AUC value. B. miR-335–3p created 0.71 AUC value. C. Combined quantification of miR-21–5p and miR-335–3p by digital PCR produced 0.86 AUC that was statistically higher than that of individual one used alone. Analysis of the two miRNAs generated a sensitivity of 71.8% and a specificity of 80.6% in differentiating NSCLC patients from the cancer-free subjects.
Discussion
Quantitative determination of low amounts of miRNA templates is challenging with qPCR. Our head-to-head comparison of digital PCR and qPCR for measuring miRNAs in both artificially-seeded samples and cancer cells demonstrates that digital PCR exhibits a higher sensitivity than qPCR to quantitatively detect the copy number of miRNAs. Furthermore, digital PCR displays a high reproducibility in analysis of miRNAs. In addition, digital PCR rather than qPCR not only determines the copy number of miR-21–5p, which is highly abundant in plasma, but also of miR-335–3p, which is lowly abundant. Moreover, the performance characteristic of digital PCR for quantitative assessment of the miRNAs is validated in clinical plasma specimens of lung cancer cases and controls. The observations imply that digital PCR might overcome the weakness of the conventional PCR technique for quantitative detection of the low abundant miRNAs in the circulating body fluid. Therefore, digital PCR could potentially provide a useful means for quantification of low abundant plasma miRNAs, whose changes are the hallmark of lung cancer.
A standard curve is essential for qPCR assay. Yet the development of multiple standard curves for multiple miRNAs is labor-intensive and time-consuming.25 Digital PCR does not require a standard curve. Using digital PCR, one could reproducibly quantify multiple RNAs without the time-consuming generation of multiple standard curves. Therefore, another significant advantage of digital PCR would be its convenience for efficient quantification of multiple miRNAs on numerous samples. Furthermore, Hayden et al.24 has shown that digital PCR can be used at nearly the same cost as qPCR, because most reagents used are identical in the 2 systems, and have scalable, rapid throughput. In addition, the digital nature of the results means that data handling is relatively straightforward. Therefore, digital PCR would be practically useful in laboratory settings for efficient quantification of multiple miRNAs in large numbers of samples.
However, a potential weakness of the current digital PCR technique should be emphasized. Accurate quantitation of a transcript by digital PCR relies on a proper distribution of positive (cDNA from the target miRNA present) to negative (no cDNA) droplets. If the concentration of target cDNA is too high, there might be many droplets with 2 or more templates, obscuring the potential for digital identification. Statistical analysis would become less reliable.25 Indeed, our present study shows that the highest copy number per μL of input detected by digital PCR is 10,000 in the serially-diluted samples, which is significantly lower than that of qPCR. To analyze the miRNAs that might have high concentrations in plasma, appropriately diluting cDNA samples to have suitable cDNA concentrations in PCR reaction would be required before digital PCR is performed. Furthermore, continuous improvement of this digital PCR technique to expand its capability is necessary. In addition, combed use of qPCR for quantification of higher abundant plasma miRNAs and digital PCR for lower amount miRNAs would provide a synergetic approach for cancer diagnosis.
Dysregulation of both miR-21–5p and miR-335–3p has been suggested to involve in the development and progression of a variety of human cancers.32–37 For instance, upregulation of miR-21–5p has important function in cell proliferation, migration, invasion and survival.38 Knock-down of miR-21–5p could induce apoptosis and repress cell proliferation and invasion. miR-21–5p was also found to be elevated in many cancers such as lymphoma, prostate cancer, colorectal cancer and breast cancer.32–35 We previously found that analysis of miR-21–5p in sputum could potentially assist diagnosis of lung cancer.39 Abnormal expression of miR-335-3p was associated with metastatic processes in breast cancer.36 Recently, an elevated expression level of miR-335-3p was found in multiple myeloma.37 The results created from our present study are consistent with these previous findings, and provide further evidence in support of using the miRNAs as potential biomarkers for cancer diagnosis.
There are limitations to the present study. First, the sample size is comparably small, so further evaluation of the miRNAs in large cases and controls cohorts is clearly required. Second, the sensitivity (72.2%) and specificity (82.5%) of using the only 2 miRNAs for diagnosis of NSCLC are not efficient in routine clinical application. Furthermore, dysregulation of the 2 miRNAs is associated with other types of cancers,32–37 not specific to lung cancer. We are extensively evaluating more lung tumor-specific miRNAs defined from our and other groups7,10–12 to identify additional plasma-based miRNAs that can be added to the current ones so that the diagnostic efficacy of digital PCR could be improved. We are also continually determining the performance characteristics of digital PCR for miRNA quantification by diluting the total RNA of differently treated samples/different clinical samples/cancer cell lines.
Taken together, this study demonstrates that digital PCR might absolutely quantify the copy number of low abundant miRNAs and provide a potential tool for the quantification of plasma miRNAs whose changes are associated with lung cancer. Nevertheless, the continued development of this technology in many clinical scenarios, and further work to explore its value for routine use in diagnostic testing for cancer is required.
Supplementary Materials
Quantification of miR-335–3p by qRT-PCR using RNA isolated from a lung cancer cell line, SK-MES-1. The generated cDNA was diluted over 1:4 series by seven orders of magnitude ranging from 1:1 to 1:4096. The values produced by qPCR were related to the RNA input over five orders of magnitude ranging from 1:1 to 1:256. However, qPCR was not able to detect the sample that had low amount RNA input (1:1024 dilutions). This experiment was performed three times.
A Bland-Altman plot was used to determine the agreement between copy number of miR-21–5p detected by digital PCR and qRT-PCR assays. The two methods had very similar results on average, and the bias (difference between the means) is 3.5. The 95% limits of agreement were between −546.3 and 553.4.
Comparison of plasma miRNA expressions in 36 lung cancer patients and 38 cancer-free controls. Horizontal lines indicate mean values. A. Copy number for miR-21–5p/μl plasma of cancer patients and cancer-free individuals was 23,458 ± 5,249 and 6,384 ± 796.7, respectively (P = 0.0013). B. Copy number for miR-335–3p/μl plasma of cancer patients and cancer-free individuals was 278.4 ± 70.49 and 91.19 ± 16.64, respectively (P = 0.009).
Table 1.
| BIAS | 3.533 |
| SD of bias | 280.5 |
| 95% Limits of Agreement | |
| From | −546.3 |
| To | 553.4 |
Acknowledgments
This work was supported in part by National Cancer Institute grant R01CA161837, VA merit Award I01 CX000512, LUNGevity Foundation Early Detection Award, and University of Maryland Cancer Epidemiology Alliance Seed Grant (F. J.).
Footnotes
COMPETING INTERESTS: Author(s) disclose no potential conflicts of interest.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
FUNDING: This work was supported in part by National Cancer Institute grant R01CA161837, VA merit Award I01 CX000512, LUNGevity Foundation Early Detection Award, and University of Maryland Cancer Epidemiology Alliance Seed Grant (FJ).
Author Contributions
JM and NL performed molecular and analysis. MG collected specimens and reviewed clinical information. FJ conceived the study project, organized the whole study process, provided financial support, and finalized the manuscript.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Aberle DR, Adams AM, Berg CD, et al. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 4.Galasso M, Sana ME, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy? Genome Med. 2010;2(2):12. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33(6):1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5(3):492–7. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Jiang F. Applications of MicroRNAs in the Diagnosis and Prognosis of Lung Cancer. Expert Opin Med Diagn. 2012;6(3):197–207. doi: 10.1517/17530059.2012.672970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329(2):125–36. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91(4):579–87. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd DW, Gagnon KT, Corey DR. Digital quantitation of potential therapeutic target RNAs. Nucleic Acid Ther. 2013;23(3):188–94. doi: 10.1089/nat.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whale AS, Huggett JF, Cowen S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40(11):e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103(47):17807–12. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhat S, Herrmann J, Armishaw P, Corbisier P, Emslie KR. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem. 2009;394(2):457–67. doi: 10.1007/s00216-009-2729-5. [DOI] [PubMed] [Google Scholar]
- 19.Kiss MM, Ortoleva-Donnelly L, Beer NR, et al. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal Chem. 2008;80(23):8975–81. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreutz JE, Munson T, Huynh T, Shen F, Du W, Ismagilov RF. Theoretical design and analysis of multivolume digital assays with wide dynamic range validated experimentally with microfluidic digital PCR. Anal Chem. 2011;83(21):8158–68. doi: 10.1021/ac201658s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–11. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohl G, Shih IeM. PCR Principle and applications of digital. Expert Rev Mol Diagn. 2004;4(1):41–7. doi: 10.1586/14737159.4.1.41. [DOI] [PubMed] [Google Scholar]
- 23.Vogelstein B, Kinzler KW. PCR Digital. Proc Natl Acad Sci U S A. 1999;96(16):9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayden RT, Gu Z, Ingersoll J, et al. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J Clin Microbiol. 2013;51(2):540–6. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods. 2013;59(1):101–7. doi: 10.1016/j.ymeth.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Diehl F, Diaz LA. Digital quantification of mutant DNA in cancer patients. Curr Opin Oncol. 2007;19(1):36–42. doi: 10.1097/CCO.0b013e328011a8e7. [DOI] [PubMed] [Google Scholar]
- 27.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83(17):6474–84. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 29.Witwer KW, McAlexander MA, Queen SE, Adams RJ. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol. 2013;10(7):1080–86. doi: 10.4161/rna.25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heredia NJ, Belgrader P, Wang S, et al. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59(1):S20–S3. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Lawrie CH. MicroRNA expression in lymphoma. Expert Opin Biol Ther. 2007;7(9):1363–74. doi: 10.1517/14712598.7.9.1363. [DOI] [PubMed] [Google Scholar]
- 33.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121(5):1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 34.Siva AC, Nelson LJ, Fleischer CL, et al. Molecular assays for the detection of microRNAs in prostate cancer. Mol Cancer. 2009;8:17. doi: 10.1186/1476-4598-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian B, Katsaros D, Lu L, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117(1):131–40. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 36.Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronchetti D, Lionetti M, Mosca L, et al. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of miR-335–3p by qRT-PCR using RNA isolated from a lung cancer cell line, SK-MES-1. The generated cDNA was diluted over 1:4 series by seven orders of magnitude ranging from 1:1 to 1:4096. The values produced by qPCR were related to the RNA input over five orders of magnitude ranging from 1:1 to 1:256. However, qPCR was not able to detect the sample that had low amount RNA input (1:1024 dilutions). This experiment was performed three times.
A Bland-Altman plot was used to determine the agreement between copy number of miR-21–5p detected by digital PCR and qRT-PCR assays. The two methods had very similar results on average, and the bias (difference between the means) is 3.5. The 95% limits of agreement were between −546.3 and 553.4.
Comparison of plasma miRNA expressions in 36 lung cancer patients and 38 cancer-free controls. Horizontal lines indicate mean values. A. Copy number for miR-21–5p/μl plasma of cancer patients and cancer-free individuals was 23,458 ± 5,249 and 6,384 ± 796.7, respectively (P = 0.0013). B. Copy number for miR-335–3p/μl plasma of cancer patients and cancer-free individuals was 278.4 ± 70.49 and 91.19 ± 16.64, respectively (P = 0.009).
Table 1.
| BIAS | 3.533 |
| SD of bias | 280.5 |
| 95% Limits of Agreement | |
| From | −546.3 |
| To | 553.4 |