Abstract
Human pegivirus (HPgV), formerly ‘GB virus C’ or ‘hepatitis G virus’, is a member of the genus Flavivirus (Flaviviridae) that has garnered significant attention due to its inhibition of HIV, including slowing disease progression and prolonging survival in HIV-infected patients. Currently, there are six proposed HPgV genotypes that have roughly distinct geographical distributions. Genotypes 2 and 3 are the most comprehensively characterized, whereas those genotypes occurring on the African continent, where HPgV prevalence is highest, are less well studied. Using deep sequencing methods, we identified complete coding HPgV sequences in four of 28 patients (14.3 %) in rural Uganda, east Africa. One of these sequences corresponds to genotype 1 and is the first complete genome of this genotype from east Africa. The remaining three sequences correspond to genotype 5, a genotype that was previously considered exclusively South African. All four positive samples were collected within a geographical area of less than 25 km2, showing that multiple HPgV genotypes co-circulate in this area. Analysis of intra-host viral genetic diversity revealed that total single-nucleotide polymorphism frequency was approximately tenfold lower in HPgV than in hepatitis C virus. Finally, one patient was co-infected with HPgV and HIV, which, in combination with the high prevalence of HIV, suggests that this region would be a useful locale to study the interactions and co-evolution of these viruses.
Introduction
Human pegivirus (HPgV), formerly ‘GB virus C’ or ‘hepatitis G virus’, was discovered in 1995 and assigned to the Flaviviridae, a family containing several important human disease agents, including yellow fever virus, West Nile virus, Dengue fever virus, and hepatitis C virus (HCV; ICTV, 2012; Leary et al., 1996; Simons et al., 1995; Stapleton et al., 2011). Within the flaviviridae, HPgV has recently been classified as a member of the newly proposed genus Pegivirus, which infects non-human primates (Birkenmeyer et al., 1998), rodents (Drexler et al., 2013; Kapoor et al., 2013b), bats (Quan et al., 2013) and horses (Chandriani et al., 2013; Kapoor et al., 2013a). Like all flaviviruses, HPgV has a single stranded, positive sense RNA genome (Leary et al., 1996; Linnen et al., 1996b). The HPgV genome is 9.4 kb in length and contains a single long ORF flanked by 5′ and 3′ untranslated regions (Mohr & Stapleton, 2009). The closest relative of HPgV known to infect humans is HCV; however, unlike HCV, HPgV is lymphotropic, establishes subclinical infection, and is not associated with hepatitis (George et al., 2006; reviewed by Stapleton, 2003).
HPgV has a worldwide distribution, with studies in developed countries showing that between 1 % and 5 % of healthy blood donors are viraemic (Moaven et al., 1996; Mohr & Stapleton, 2009; Stapleton, 2003). Prevalence is higher in developing countries, with the highest blood donor infection rates (between 11.1 and 18.9 %) on the African continent (Mohr & Stapleton, 2009). HPgV is transmitted sexually, vertically, and through exposure to infected blood (Bhattarai & Stapleton, 2012). It is therefore highly prevalent in populations with sexually transmitted or blood borne infections. For example, previous studies have reported HPgV viraemia in 17–40 % of HIV-positive individuals (Heringlake et al., 1998b; Mohr & Stapleton, 2009; Stapleton et al., 2004).
Despite being non-pathogenic (Berzsenyi et al., 2005; Polgreen et al., 2003), HPgV has garnered significant research attention because of its interactions with HIV. Several clinical studies have shown that HPgV infection slows disease progression in individuals infected with HIV. Specifically, HPgV infection can prolong survival and reduce mortality in HIV-infected patients through reduction in HIV viral load, increased CD4+ T-cell counts, and improved response to antiretroviral therapy (Heringlake et al., 1998a; Tillmann et al., 2001; Williams et al., 2004; Xiang et al., 2001). The mechanisms by which HPgV modulates HIV infection may include direct interference with HIV entry and replication and indirect regulation of host factors that prevent or reduce disease progression (reviewed by Bhattarai & Stapleton, 2012).
Given the proposed use of HPgV as a bio-therapeutic agent in the treatment of AIDS (Gretch, 2012), significant attention has been placed on understanding the epidemiology, evolution and genetic diversity of this virus (Bagasra et al., 2012; Wu et al., 2012). HPgV sorts into six genotypes with roughly distinct geographical origins (Muerhoff et al., 1996, 2005; Saito et al., 1998; Smith et al., 2000). Initially, three genotypes were described as originating in Africa (genotype 1), Europe (genotype 2), and Japan (genotype 3). Later, viruses from south-east Asia (genotype 4), South Africa (genotype 5) and Indonesia (genotype 6) were described (Muerhoff et al., 2005, 2006; Naito et al., 1999; Sathar et al., 1999; Smith et al., 2000; Tucker et al., 1999). Most recently, three novel full-length genomes were sequenced from injecting drug users in south-western China; these have been proposed to represent a seventh genotype (Feng et al., 2011). Of the 46 full-length coding genomes now available in public databases, the majority are genotypes 2 and 3, likely representing focused sampling from the geographical areas corresponding to these genotypes. Only two full coding genomes exist from genotype 1, despite evidence from partial-genome sequencing suggesting that genotype 1 may be the most genetically diverse genotype (Parreira et al., 2012). Full coding genomes from the African continent include two from west Africa (Leary et al., 1996; Saito et al., 1999), one from east Africa (Erker et al., 1996), and one from South Africa (Muerhoff et al., 2005).
Here, we describe four new HPgV genome sequences from western Uganda. These sequences were collected from a region of east Africa that is heavily burdened by infectious diseases, including a high prevalence of HIV, reported at 16.1 % in individuals age 15–49 in 2008 (Rubaihayo et al., 2010). The sequences correspond to genotypes 1 and 5, the most poorly characterized groups in terms of availability of full-length viral genome sequences (Parreira et al., 2012). Our results expand the known diversity and geographical range of these genotypes and demonstrate natural co-circulation of HPgV and HIV in rural east Africa.
Results
As part of an effort to identify the causative agents of febrile illness, blood samples from a total of 28 subjects presenting with fevers of unknown origin at one of two health clinics in rural Uganda were deep sequenced to identify RNA viruses. From this cohort, four patients showed evidence of HPgV viraemia. De novo assembly of HPgV deep sequencing reads (total of 13 880 to 148 032) from these patients yielded near-complete HPgV genomes (9 281–9 339 bp), with an average coverage depth ranging from 686× to 6326×. Two of these patients were female (Hu5 and Hu6), two were male (Hu20 and Hu21), and each resided in one of three villages located within 25 km2 of each other. HPgV titres, assayed by quantitative reverse transcription polymerase chain reaction (RT-qPCR) and expressed as HPgV genome copies ml-1 serum, were high in all four infected individuals: Hu5, 8.0×107; Hu6, 5.1×107, Hu20, 1.4×106; and Hu21, 3.1×106.
HPgV polyprotein cleavage sites were predicted based on an alignment with members of the Pegivirus and Hepacivirus genera and through manual (Nielsen et al., 1997) and in silico (Petersen et al., 2011) signallase and NS3-NS4A protease cleavage site prediction. The genomic architecture of the four viral sequences was consistent with that of known HPgV sequences, containing a single ORF of 8 528 bp that putatively encodes two envelope proteins (E1 and E2), a 6.4 kDa ion channel protein (P7-like protein; as identified using the ExPASY server; Gasteiger et al., 2005) and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B; reviewed by Stapleton et al., 2011). For each of the new HPgVs, the first Met in-frame with the coding sequence aligned well with the experimentally determined translation initiation codon for HPgV (Simons et al., 1996). As observed previously in other pegiviruses, the sequence encoding a putative core (i.e. nucleocapsid) protein was absent or truncated in all four new genomes. The four viral nucleotide sequences were annotated and deposited in GenBank with accession numbers KC618398–KC698401.
Partial 5′UTR sequences of 532 to 534 nt were recovered by deep sequencing, representing ≥96 % of the 554 nt HPgV 5′UTR (Linnen et al., 1996a; Xiang et al., 2000). These sequences were 88 to 89 % similar to the 5′UTRs of HPgV reference strains (AF121950 and NC_001710) and 90 % to 99 % similar to each other. Partial 3′UTR sequences of 219 to 278 nt were recovered for the new HPgVs, representing 70 % to 89 % of the 312 nt HPgV 3′UTR (Linnen et al., 1996b; Xiang et al., 2000). As observed for the 5′UTR, the 3′UTRs of the novel HPgVs and HPgV reference strains (AF121950 and NC_001710) were well conserved; 3′UTR sequences acquired by deep sequencing were ≥97 % similar to aligned regions of the reference strains and ≥98 % similar to each other.
Deep sequencing also identified five HIV-positive individuals within the study cohort, containing 40 to 180 reads per sample. Four individuals were infected with HIV-1 subtype A1/D and one was infected with HIV-1 subtype A1. This infection frequency (17.9 %) is consistent with a previously reported HIV prevalence for this region of 16.1 % (Rubaihayo et al., 2010). One of the above patients, Hu20, was co-infected with HIV-1 subtype A1/D and HPgV. HPgV motifs previously proposed to be important for HIV antagonism were conserved in all new sequences described herein, including the co-infected sample (George et al., 2012; Jung et al., 2007; Koedel et al., 2011; Timmons et al., 2013; Xiang et al., 2006, 2008).
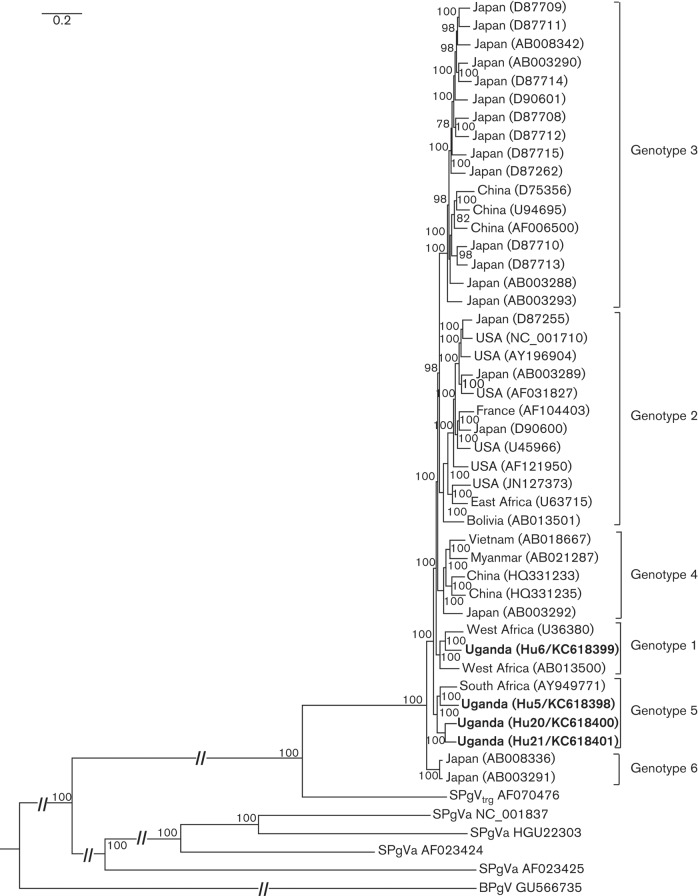
To compare our newly described sequences to published HPgV sequences, we reconstructed a Bayesian phylogenetic tree that included our sequences as well as publicly available full-length HPgV sequences. Our Bayesian phylogeny is largely consistent with recently published HPgV phylogenies (Feng et al., 2011; Parreira et al., 2012), showing 100 % posterior clade support for proposed genotypes 1–5 (Fig. 1). The phylogeny includes sequences identified as recombinant, as assessed using the recombination analysis software RDP4 (Martin et al., 2010): AB021287 (genotype 4; recombination event spanning E2, P7 and NS2), U75356 (genotype 3; recombination events in NS4B and NS5B), and D87715 (genotype 3; recombination event in NS5B). These sequences were retained because removal of these putative recombinants did not change the topology of the tree.
Fig. 1.
HPgV Bayesian phylogenetic tree produced from an alignment of 38 full coding genomes including the four new full coding sequences described herein. Putative recombinants AB021287, U75356, D87715 were included because their inclusion did not alter the topology of the tree. More distantly related pegiviruses, including simian pegivirus from chimpanzees (SPgVtrg; AF070476) and New World monkeys (AF023424, AF023425, HGU22303 and NC_001837), as well as bat pegivirus (BPgV; GU566735) were included as outgroups. Newly discovered sequences are in bold with sample identification numbers and GenBank accession numbers in parentheses. Posterior clade probability values are shown as percentages at nodes; scale bar indicates nucleotide substitutions per site.
Sequence Hu6 (KC618399) fell within the genotype 1 clade (100 % posterior probability), sharing 89.6 % and 88.8 % nucleotide identity with the other complete genotype 1 sequences, U36380 and AB013500, respectively (Fig. 1). The three remaining sequences, Hu5 (KC618398), Hu20 (KC618400) and Hu21 (KC618401) sorted with genotype 5 (Muerhoff et al., 2005) with 100 % posterior clade probability. Hu20 and Hu21 are sister taxa (92 % similarity), and are 88.8 % similar to Hu5 and published sequence AY949771. Hu5 and AY949771 share 89.2 % similarity with each other. Of note, our phylogeny supports the presence of a sixth genotype, represented by two complete sequences from Japan (AB008336 and AB003291). These sequences share 97.7 % nucleotide identity with each other and are 87.7 % and 87.8 % similar to genotype 1 and 5, respectively (Table 1). Simian pegivirus from chimpanzees (SPgVtro) was approximately 71 % similar to all human genotypes (Table 1).
Table 1. Percentage nucleotide identity between HPgV genotypes.
Percentage nucleotide identities below the diagonal and standard errors above the diagonal are shown. Simian pegivirus from chimpanzees (SPgVtro) was included for comparison.
| Genotype 1 | Genotype 2 | Genotype 3 | Genotype 4 | Genotype 5 | Genotype 6 | SPgVtro | |
| Genotype 1 | . | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.005 |
| Genotype 2 | 86.7 | . | 0.003 | 0.003 | 0.003 | 0.004 | 0.004 |
| Genotype 3 | 87.3 | 86.8 | . | 0.003 | 0.003 | 0.003 | 0.004 |
| Genotype 4 | 87.0 | 86.5 | 87.1 | . | 0.003 | 0.003 | 0.004 |
| Genotype 5 | 87.6 | 86.7 | 87.3 | 87.0 | . | 0.003 | 0.005 |
| Genotype 6 | 87.7 | 86.5 | 87.1 | 86.6 | 87.9 | . | 0.005 |
| SPgVtro | 70.7 | 70.5 | 70.7 | 70.8 | 70.9 | 70.7 | . |
The new genotype 1 and 5 sequences made possible the calculation of within-genotype average pairwise nucleotide similarity. For genotype 1, this value was 88.8 % (standard error, 0.003), and for genotype 5 this value was 89.3 % (standard error, 0.002). Among putative proteins, nucleotide sequences were most conserved in the truncated core (94.0 %) and NS5A (92.2 %) regions. The least conserved areas were located in the P7-like region (89.8 %) and in NS2 (89.7 %; Table 2).
Table 2. Average percentage nucleotide identity among sequences within HPgV by gene.
| Genotype | Overall | Core | E1 | E2 | P7-like | NS2 | NS3 | NS4A | NS4B | NS5A | NS5B |
| 1 | 88.8 | 95.2 | 88.4 | 88.3 | 89.9 | 87.3 | 87.7 | 88.8 | 87.7 | 90.6 | 89.9 |
| 2 | 89.7 | 94.3 | 89.1 | 89.0 | 87.2 | 88.9 | 89.0 | 90.0 | 89.0 | 90.6 | 91.0 |
| 3 | 90.5 | 92.5 | 89.7 | 91.0 | 91.3 | 89.7 | 89.5 | 92.5 | 89.5 | 91.9 | 91.2 |
| 4 | 89.0 | 96.2 | 89.7 | 88.3 | 86.5 | 87.7 | 87.7 | 91.8 | 88.5 | 91.5 | 89.4 |
| 5 | 89.3 | 92.9 | 89.6 | 88.3 | 86.5 | 87.5 | 88.1 | 91.0 | 89.2 | 81.5 | 90.6 |
| 6 | 97.7 | 92.9 | 97.5 | 97.6 | 98.1 | 87.0 | 98.7 | 96.4 | 97.2 | 97.3 | 97.5 |
| Average | 90.8 | 94.0 | 90.7 | 90.4 | 89.8 | 89.7 | 90.1 | 91.8 | 90.2 | 92.2 | 91.6 |
To characterize HPgV diversity within infected hosts, we quantified single-nucleotide polymorphisms (SNP) within each individual, which reflects the diversity of the viral ‘quasispecies’ (Lauring & Andino, 2010). SNP frequencies in HPgV were lower compared to HCV for both synonymous (silent) and non-synonymous (amino acid changing) mutations (Lauck et al., 2012a). HPgV from patient Hu5 contained the highest total number of SNP (123 synonymous and six non-synonymous), although Hu20 had a higher total number of non-synonymous SNP, at seven (Table 3). The majority of SNP occurred at frequencies of 5–10 %, suggesting that these SNP may reflect random nucleotide misincorporations. We also estimated nucleotide diversity as the average pairwise difference between sequences for all sites (π), synonymous sites (πS) and non-synonymous sites (πN). Hu5 displayed the highest overall nucleotide diversity throughout the genome (π of 0.00921 substitutions per site, Table 4). Comparing values of πS and πN for each protein-coding region, πS was consistently higher than πN, with the exception of the core protein region from patient Hu6 (Table 4). While no HPgV protein-coding gene was consistently more diverse than the other protein-coding genes, NS5B had the highest average πN value in three out of the four patients: Hu5, Hu20 and Hu21. Intra-host diversity estimates for patient Hu20, co-infected with HIV, did not differ appreciably from estimates for other patients infected with HPgV alone (Table 4).
Table 3. Intra-host diversity of HPgV.
Assessed as the number of synonymous (S) and non-synonymous (NS) mutations and dN/dS ratios (for whole coding genomes only) across the full coding viral genomes from four patients in western Uganda. –, not calculated.
| Frequency (%) | Hu5 | Hu6 | Hu20 | Hu21 | ||||||||
| S | NS | dN/dS | S | NS | dN/dS | S | NS | dN/dS | S | NS | dN/dS | |
| 5–10 | 66 | 4 | – | 17 | 2 | – | 36 | 5 | – | 14 | 1 | – |
| 10–30 | 31 | 1 | – | 24 | 2 | – | 16 | 2 | – | 11 | 1 | – |
| 30–50 | 26 | 1 | – | 1 | 0 | – | 6 | 0 | – | 5 | 0 | – |
| Total | 123 | 6 | 0.049 | 42 | 4 | 0.095 | 58 | 7 | 0.121 | 30 | 2 | 0.0667 |
Table 4. Nucleotide diversity.
Estimated as average pairwise distances between sequences for non-synonymous sites (πN), synonymous sites (πS), and all sites (π) by gene and for the entire coding region (ORF).
| Core | E1 | E2 | P7-like | NS2 | NS3 | NS4A | NS4B | NS5A | NS5B | ORF | ||
| Hu5 | Mean πS | 0.0151 | 0.0189 | 0.0239 | 0.0430 | 0.0296 | 0.0277 | 0.0151 | 0.0268 | 0.0216 | 0.0256 | 0.0253 |
| Mean πN | 0.0022 | 0.0024 | 0.0021 | 0.0013 | 0.0025 | 0.0016 | 0.0016 | 0.0018 | 0.0017 | 0.0031 | 0.0021 | |
| Mean π | 0.0067 | 0.0073 | 0.0088 | 0.0144 | 0.0109 | 0.0096 | 0.0057 | 0.0097 | 0.0075 | 0.0099 | 0.0092 | |
| Hu6 | Mean πS | 0.0072 | 0.0077 | 0.0092 | 0.0093 | 0.0076 | 0.0069 | 0.0042 | 0.0099 | 0.0064 | 0.0081 | 0.0078 |
| Mean πN | 0.0075 | 0.0014 | 0.0012 | 0.0010 | 0.0017 | 0.0014 | 0.0010 | 0.0014 | 0.0017 | 0.0024 | 0.0017 | |
| Mean π | 0.0074 | 0.0034 | 0.0036 | 0.0038 | 0.0035 | 0.0031 | 0.0020 | 0.0040 | 0.0031 | 0.0041 | 0.0035 | |
| Hu20 | Mean πS | 0.0277 | 0.0200 | 0.0172 | 0.0183 | 0.0131 | 0.0168 | 0.0189 | 0.0155 | 0.0129 | 0.0200 | 0.0168 |
| Mean πN | 0.0012 | 0.0019 | 0.0018 | 0.0024 | 0.0027 | 0.0033 | 0.0024 | 0.0015 | 0.0018 | 0.0039 | 0.0026 | |
| Mean π | 0.0132 | 0.0075 | 0.0065 | 0.0073 | 0.0059 | 0.0075 | 0.0074 | 0.0060 | 0.0051 | 0.0087 | 0.0069 | |
| Hu21 | Mean πS | 0.0034 | 0.0090 | 0.0144 | 0.0144 | 0.0103 | 0.0118 | 0.0130 | 0.0121 | 0.0102 | 0.0135 | 0.0119 |
| Mean πN | 0.0027 | 0.0016 | 0.0014 | 0.0014 | 0.0024 | 0.0014 | 0.0017 | 0.0017 | 0.0015 | 0.0032 | 0.0019 | |
| Mean π | 0.0029 | 0.0039 | 0.0054 | 0.0054 | 0.0048 | 0.0046 | 0.0051 | 0.0049 | 0.0041 | 0.0063 | 0.0050 | |
Discussion
This study examined HPgV infection in western Uganda, where 4 out of 28 (14.3 %) of patients screened were identified as HPgV-positive. This result is consistent with previous findings of high HPgV prevalence in developing countries, ranging from 5 % to 18.9 % (Mohr & Stapleton, 2009). This study identified two genotypes, 1 and 5, co-circulating in the area. To our knowledge, the only complete genome previously reported from east Africa sorted with genotype 2, a lineage containing a number of European sequences, which is suggested to reflect early human migrations from Africa to Europe (Smith et al., 2000). Genotype 1 has previously been identified as an Africa-wide clade, originating from west and central Africa, (Leary et al., 1996; Smith et al., 1997; Tanaka et al., 1998). Our discovery of a genotype 1 sequence in east Africa confirms that this genotype is widespread throughout the African continent. Genotype 5 has previously been characterized as a South African clade, with a single complete coding sequence and several partial sequences reported from this country (Muerhoff et al., 2005; Sathar & York, 2001; Tucker et al., 1999; Tucker & Smuts, 2000). Our discovery of three genotype 5 sequences in Uganda demonstrates that the geographical range of this viral genotype is more extensive than previously documented.
The four complete HPgV coding sequences described herein provide new support for the phylogenetic positions of previously undersampled genotypes. While genotype 2 and 3 have been well characterized phylogenetically, previous studies have had limited success resolving HPgV evolutionary history because of few genotype 1, 4 and 5 complete coding sequences (Parreira et al., 2012). The additional complete coding sequences described here demonstrate consistency in the phylogenetic relationships previously established for genotypes 1 and 5 despite expansion of their known geographical range.
Our sequence analyses and Bayesian phylogeny suggest the existence of six HPgV genotypes based on six strongly supported clades. The proposed sixth genotype comprises two Japanese sequences (Suzuki et al., 1999; Tanaka et al., 1998) previously considered members of genotypes 1 and 4, respectively (Feng et al., 2011; Muerhoff et al., 2005), but also shown to be genetically distinct from other genotypes by principal coordinate analysis (Parreira et al., 2012). Our results also suggest that another previously proposed genotype, represented by a single sequence (Muerhoff et al., 2006), as well as other sequences proposed to form a seventh genotype (Feng et al., 2011), should be considered part of genotype 4, with which they cluster with 100 % posterior clade support according to our analyses. Interestingly, one of our putative genotype 6 sequences, (GenBank accession number AB003291), clusters with 15 partial African NS3 and NS5a ‘indel types’ from central Africa (Tanaka et al., 1998). The 12 amino acid indel that typifies this group may be an ancestral trait that was lost in other HPgV lineages, perhaps indicating that this is an ancestral viral lineage with a geographical origin in Africa (Pavesi, 2001; Tanaka et al., 1998). Our results demonstrating multiple, diverse HPgV genotypes co-circulating within a very small geographical area (<25 km2, with genotype 1 from Hu6 and genotype 5 from Hu20 originating from the same village) further supports the idea of a centre of HPgV endemism and diversity in sub-Saharan Africa.
Our study also confirms that HIV-1 and HPgV co-circulate in sub-Saharan Africa on a local scale. Previous studies have identified motifs within the HPgV genome that are associated with the inhibition of HIV replication. Particularly, the HPgV non-structural phosphoprotein NS5A inhibits replication by reducing surface expression of the HIV co-receptor CXCR4 and increasing the release of its ligand, SDF-1 into supernatant (Xiang et al., 2006, 2008). The NS5A motif (position 152–165) responsible for this activity is conserved in all known sequences, including the four new sequences identified here. Similarly, HPgV E2 envelope protein has been identified as preventing HIV viral entry by modifying HIV gp41 fusion peptide (Jung et al., 2007; Koedel et al., 2011; Timmons et al., 2013). E2 (269–286) is also conserved among the new sequences, suggesting that these new sequences also have HIV-inhibitory properties.
Finally, this study expands our understanding of HPgV intra-host viral genetic diversity. It is has been suggested that HPgV can exist as a ‘quasispecies’ with different tissue tropisms in a single individual (Fogeda et al., 2000; Ruiz et al., 2010). However, intra-host genetic diversity of HPgV has, to our knowledge, remained previously undescribed. We found that the total number of SNPs detected in HPgV was approximately tenfold lower than in HCV, measured using nearly identical methods (Lauck et al., 2012a). Although the reasons for this difference are unclear, we speculate that the non-pathogenic nature of HPgV reflects its stable co-evolution with human hosts and immunological tolerance, while pathogenic viruses like HCV must diversify to evade host immunity. Additionally, the majority of HPgV intra-host mutations occurred at a frequency of less than 10 % but were not randomly distributed across the genome. Of particular interest, we found the greatest number of synonymous and non-synonymous intra-host mutations in NS5B. This result was surprising, given that NS5B contains conserved stem–loop structures that may constrain its evolution (Cuceanu et al., 2001). Both our intra-host diversity data and nucleotide diversity estimates demonstrated a tendency toward synonymous mutations over non-synonymous mutation, which indicates purifying selection (i.e. selection against potentially deleterious non-synonymous mutations). These results corroborate previous findings of low rates of non-synonymous-to-synonymous substitutions, averaging 0.04 across all coding regions (Romano et al., 2008).
Overall, our results expand the known geographical range of HPgV genotypes 1 and 5 and demonstrate that multiple HPgV lineages can co-circulate in the same area. We also show that levels and patterns of intra-host HPgV diversity do not mirror levels or patterns of inter-host HPgV variability. Furthermore, the presence of diverse HPgV lineages in an HIV-endemic region, and documented co-infection with both viruses, suggests that this region of sub-Saharan Africa may be a useful locale for investigating immunological and evolutionary interactions between HPgV and HIV, including their clinical effects in co-infected patients. As more data are generated on the distribution and prevalence of the HPgV genotypes and co-infection with HIV, it should be possible to investigate lineage-specific HPgV–HIV interactions. Understanding these natural dynamics of infection and co-infection may, in turn, lead to novel therapeutic approaches for patients infected with HIV.
Methods
Ethics.
All samples and data were collected with informed consent in accordance with World Health Organization guidelines. Permission to conduct this research was granted by the Uganda National Council of Science and Technology, McGill University, and the University of Wisconsin–Madison. Samples were shipped internationally following IATA guidelines and regulations.
Patients and sample collection.
Individuals visiting one of two health clinics outside of Fort Portal (one of the largest towns in western Uganda) who presented with a fever over 100.4 °F were asked to participate in a larger study whose aim was to identify aetiologies of fever. Demographic information including gender, age, and village were recorded, and blood was drawn from the median cubital vein into an evacuated plasma collection tube (Becton Dickinson) by staff nurses at the clinic. Blood was kept cool until processing (<4 h), then was separated using centrifugation in a field laboratory and frozen immediately in liquid nitrogen for storage and transport to the USA. Samples were shipped to North America in an IATA-approved dry shipper.
Amplification of viral RNA.
RNA was prepared from blood plasma and directly sequenced on an Illumina MiSeq instrument as previously described (Lauck et al., 2011; Lauck et al., 2013). Sequence data were processed using CLC Genomics Workbench (version 5.5; CLC Bio); sequences were stripped of adaptor sequences, quality/length trimmed (minimum of q30, ≥100 bp) and assembled de novo. Contiguous sequences were characterized using blastn (Altschul et al., 1990); all sequences were mapped to a reference viral database acquired from GenBank. Novel and reference HPgV ORF sequences were aligned using TranslatorX (Abascal et al., 2010). HPgV protein products were identified by comparison to published sequences and annotated using CLC Genomics Workbench. The four complete sequences generated were deposited in GenBank under accession numbers KC618398–KC618401.
Analysis.
Phylogenetic relationships within the Pegivirus genus were estimated from nucleotide sequences of full-length published coding genome sequences. If two or more genomes were greater than 99 % identical, one sequence was chosen to represent both. All genomes with minimum sequence ambiguity were selected (AB003288, AB003289, AB003290, AB003291, AB003292, AB003293, AB008336, AB008342, AB013500, AB013501, AB018667, AB021287, AF006500, AF031827, AF121950, AY196904, AY949771, D87255, D87262, D87708, D87709, D87710, D87711, D87712, D87713, D87714, D87715, D90600, D90601, HQ331233, HQ331235, JN127373, NC_001710, U36380, U45966, U63715, U75356, U94695). Putatively recombinant forms were identified using rdp4, version 4.16 (Martin et al., 2010). Sequences were aligned using the MAAFT algorithm in TranslatorX (Abascal et al., 2010), and regions of ambiguous alignment were removed using the embedded Gblocks algorithm under default settings (Castresana, 2000). More distantly related pegiviruses, including simian pegivirus (SPgV) from chimpanzees (SPgVtro; AF070476), New World monkeys (AF023425, AF023424, NC_001837, HGU22303), and bat pegivirus (BPgV; GU566735) were used as outgroups. Bayesian trees were reconstructed using MrBayes, version 3.2.1 (Ronquist & Huelsenbeck, 2003) with a substitution model of the form GTR+ , selected using jModelTest and the Akaike information criterion (AIC), with ΔAIC to second-best model GTR+I+
, selected using jModelTest and the Akaike information criterion (AIC), with ΔAIC to second-best model GTR+I+ = 70.1 (Posada, 2008), model parameters estimated from the data under default priors, and Markov chains run for 2.5 million generations, with the first 25 % of sampled trees discarded as burn-in.
= 70.1 (Posada, 2008), model parameters estimated from the data under default priors, and Markov chains run for 2.5 million generations, with the first 25 % of sampled trees discarded as burn-in.
To estimate inter-host viral genetic diversity, nucleotide-level percentage sequence identity among sequences was calculated as the pairwise proportion of nucleotide differences (p-distance) in mega5.05 (Tamura et al., 2011). To estimate intra-host viral genetic diversity, single-nucleotide polymorphism (SNP) analysis was performed using CLC’s SNP analysis tool, with the following parameters: window length, 7; maximum gap and mismatch count, 2; minimum central quality base, 30; minimum average quality for window bases, 25; minimum coverage, 100×; and minimum variant frequency, 5 %. This method has been used previously for intra-host viral genetic diversity analysis of HCV to ensure that only high quality and high coverage areas are considered in SNP calling (Lauck et al., 2012a). To achieve fine genetic resolution of each viral population, estimates of nucleotide diversity were calculated using all available deep sequences, including those sequencing with SNPs below our highly conservative threshold. Nucleotide diversity estimates (π) were calculated using the PoPoolation software version 1.2.2 (Kofler et al., 2011) with disabled corrections and a transversion penalty of six.
Titres of HPgV were determined using TaqMan RT-qPCR. RNA was extracted from 200 µl of blood plasma using a Qiagen QIAamp MinElute virus spin kit (Qiagen). HPgV quantification was performed on a Lightcycler 480 real-time instrument (Roche) using a Superscript III platinum one-step RT-qPCR kit (Invitrogen) and the following original primers and probe: forward 5′-TACGACGACTGCCCITACAC-3′; reverse 5′-TTTGCCCAGCTIACATCAGG-3′; probe 5′-FAM-CGCAGCCGTCGCTGCTGACAT-BHQ-3′. Standard Lightcycler RT-qPCR conditions were used, incorporating primers at 500 nM and the TaqMan probe at 100 nM. A standard curve was prepared from viral RNA (from patient Hu5) amplified using RT-qPCR with the following primers: forward: 5′-GCACAGGGAGAGGAAGGTC-3′ and reverse: 5′-CCCAGTCTGTCACCACCAC-3′. The resulting 588 bp amplicon was (i) cloned (Zero Blunt® PCR Cloning kit; Life Technologies), (ii) linearized (HindIII digestion; NEB), (iii) transcribed (MEGAscript T7 kit; Ambion) and (iv) quantified fluorometrically (Qubit 2.0, Invitrogen). The fidelity of the RNA standard was verified by Sanger sequencing.
Acknowledgements
We gratefully acknowledge the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, and Makerere University Biological Field Station for granting permission to conduct this research. We also thank D. Hyeroba, M. Thurber, A. Tumukunde, and G. Weny for assistance in the field, D. Natamba and L. Kabajase for assistance with sample collection and follow-up, and L. Kilby for assistance with permitting and logistics. This work was funded by the joint NIH-NSF Ecology of Infectious Disease program (grant number TW009237) and the UK Economic and Social Research Council, and by the Wisconsin Partnership Program through the Wisconsin Center for Infectious Disease (WisCID).
References
- Abascal F., Zardoya R., Telford M. J. (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38 (Web Server issue), W7-13 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Bagasra O., Sheraz M., Pace D. G. (2012). Hepatitis G virus or GBV-C: a natural anti-HIV interfering virus. In Viruses: Essential Agents of Life, pp. 363–388 Edited by Witzany G. New York, NY: Springer; 10.1007/978-94-007-4899-6_18 [DOI] [Google Scholar]
- Berzsenyi M. D., Bowden D. S., Roberts S. K. (2005). GB virus C: insights into co-infection. J Clin Virol 33, 257–266 10.1016/j.jcv.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Bhattarai N., Stapleton J. T. (2012). GB virus C: the good boy virus? Trends Microbiol 20, 124–130 10.1016/j.tim.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeyer L. G., Desai S. M., Muerhoff A. S., Leary T. P., Simons J. N., Montes C. C., Mushahwar I. K. (1998). Isolation of a GB virus-related genome from a chimpanzee. J Med Virol 56, 44–51 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chandriani S., Skewes-Cox P., Weidong Z., Ganem D. E., Divers T. J. (2013). Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proceedings of the National Academy of Science. USA 110, 5733–5734 10.1073/pnas.1219217110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuceanu N. M., Tuplin A., Simmonds P. (2001). Evolutionarily conserved RNA secondary structures in coding and non-coding sequences at the 3′ end of the hepatitis G virus/GB-virus C genome. J Gen Virol 82, 713–722 [DOI] [PubMed] [Google Scholar]
- Drexler J. F., Corman V. M., Müller M. A., Lukashev A. N., Gmyl A., Coutard B., Adam A., Ritz D., Leijten L. M. & other authors (2013). Evidence for novel hepaciviruses in rodents. PLoS Pathog 9, e1003438 10.1371/journal.ppat.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker J. C., Simons J. N., Muerhoff A. S., Leary T. P., Chalmers M. L., Desai S. M., Mushahwar I. K. (1996). Molecular cloning and characterization of a GB virus C isolate from a patient with non-A-E hepatitis. J Gen Virol 77, 2713–2720 10.1099/0022-1317-77-11-2713 [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhao W., Feng Y., Dai J., Li Z., Zhang X., Liu L., Bai J., Zhang H. & other authors (2011). A novel genotype of GB virus C: its identification and predominance among injecting drug users in Yunnan, China. PLoS ONE 6, e21151 10.1371/journal.pone.0021151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeda M., López-Alcorocho J. M., Bartolomé J., Arocena C., Martín M. A., Carreño V. (2000). Existence of distinct GB virus C/hepatitis G virus variants with different tropism. J Virol 74, 7936–7942 10.1128/JVI.74.17.7936-7942.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005). Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, pp. 571–607 Edited by Walker J. M. Totowa, NJ: Humana Press; 10.1385/1-59259-890-0:571 [DOI] [Google Scholar]
- George S. L., Varmaz D., Stapleton J. T. (2006). GB virus C replicates in primary T and B lymphocytes. J Infect Dis 193, 451–454 10.1086/499435 [DOI] [PubMed] [Google Scholar]
- George S. L., Varmaz D., Tavis J. E., Chowdhury A. (2012). The GB virus C (GBV-C) NS3 serine protease inhibits HIV-1 replication in a CD4+ T lymphocyte cell line without decreasing HIV receptor expression. PLoS ONE 7, e30653 10.1371/journal.pone.0030653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretch D. (2012). Editorial commentary: advocating the concept of GB virus C biotherapy against AIDS. Clin Infect Dis 55, 1020–1021 10.1093/cid/cis591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heringlake S., Ockenga J., Tillmann H. L., Trautwein C., Meissner D., Stoll M., Hunt J., Jou C., Solomon N. & other authors (1998a). GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J Infect Dis 177, 1723–1726 10.1086/517431 [DOI] [PubMed] [Google Scholar]
- Heringlake S., Ockenga J., Tillmann H. L., Trautwein C., Meissner D., Stoll M., Hunt J., Jou C., Solomon N. & other authors (1998b). GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J Infect Dis 177, 1723–1726 10.1086/517431 [DOI] [PubMed] [Google Scholar]
- ICTV (2012). ICTV Master Species List 2012 v1, February 14, 2013 edn. Leuven, Belgium: Internation Committee on Taxonomy of Viruses [Google Scholar]
- Jung S., Eichenmüller M., Donhauser N., Neipel F., Engel A. M., Hess G., Fleckenstein B., Reil H. (2007). HIV entry inhibition by the envelope 2 glycoprotein of GB virus C. AIDS 21, 645–647 10.1097/QAD.0b013e32803277c7 [DOI] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Cullen J. M., Scheel T. K. H., Medina J. L., Giannitti F., Nishiuchi E., Brock K. V., Burbelo P. D. & other authors (2013a). Identification of a pegivirus (GB virus-like virus) that infects horses. J Virol 87, 7185–7190 10.1128/JVI.00324-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Scheel T. K. H., Hjelle B., Cullen J. M., Burbelo P. D., Chauhan L. V., Duraisamy R., Sanchez Leon M. & other authors (2013b). Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio 4, e00216–e13 10.1128/mBio.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedel Y., Eissmann K., Wend H., Fleckenstein B., Reil H. (2011). Peptides derived from a distinct region of GB virus C glycoprotein E2 mediate strain-specific HIV-1 entry inhibition. J Virol 85, 7037–7047 10.1128/JVI.02366-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Orozco-terWengel P., De Maio N., Pandey R. V., Nolte V., Futschik A., Kosiol C., Schlötterer C. (2011). PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS ONE 6, e15925 10.1371/journal.pone.0015925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Hyeroba D., Tumukunde A., Weny G., Lank S. M., Chapman C. A., O’Connor D. H., Friedrich T. C., Goldberg T. L. (2011). Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS ONE 6, e19056 10.1371/journal.pone.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Alvarado-Mora M. V., Becker E. A., Bhattacharya D., Striker R., Hughes A. L., Carrilho F. J., O’Connor D. H., Pinho J. R. (2012a). Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. J Virol 86, 3952–3960 10.1128/JVI.06627-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Sibley S. D., Hyeroba D., Tumukunde A., Weny G., Chapman C. A., Ting N., Switzer W. M., Kuhn J. H. & other authors (2013). Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol 87, 688–691 10.1128/JVI.02433-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A. S., Andino R. (2010). Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 6, e1001005 10.1371/journal.ppat.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary T. P., Muerhoff A. S., Simons J. N., Pilot-Matias T. J., Erker J. C., Chalmers M. L., Schlauder G. G., Dawson G. J., Desai S. M., Mushahwar I. K. (1996). Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol 48, 60–67 [DOI] [PubMed] [Google Scholar]
- Linnen J., Wages J., Jr, Zhang-Keck Z.-Y., Fry K. E., Krawczynski K. Z., Alter H., Koonin E., Gallagher M., Alter M. & other authors (1996a). Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271, 505–508 10.1126/science.271.5248.505 [DOI] [PubMed] [Google Scholar]
- Linnen J., Wages J., Jr, Zhang-Keck Z. Y., Fry K. E., Krawczynski K. Z., Alter H., Koonin E., Gallagher M., Alter M. & other authors (1996b). Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271, 505–508 10.1126/science.271.5248.505 [DOI] [PubMed] [Google Scholar]
- Martin D. P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. (2010). RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463 10.1093/bioinformatics/btq467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moaven L. D., Hyland C. A., Young I. F., Bowden D. S., McCaw R., Mison L., Locarnini S. A. (1996). Prevalence of hepatitis G virus in Queensland blood donors. Med J Aust 165, 369–371 [DOI] [PubMed] [Google Scholar]
- Mohr E. L., Stapleton J. T. (2009). GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat 16, 757–768 10.1111/j.1365-2893.2009.01194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerhoff A. S., Simons J. N., Leary T. P., Erker J. C., Chalmers M. L., Pilot-Matias T. J., Dawson G. J., Desai S. M., Mushahwar I. K. (1996). Sequence heterogeneity within the 5′-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol 25, 379–384 10.1016/S0168-8278(96)80125-5 [DOI] [PubMed] [Google Scholar]
- Muerhoff A. S., Leary T. P., Sathar M. A., Dawson G. J., Desai S. M. (2005). African origin of GB virus C determined by phylogenetic analysis of a complete genotype 5 genome from South Africa. J Gen Virol 86, 1729–1735 10.1099/vir.0.80854-0 [DOI] [PubMed] [Google Scholar]
- Muerhoff A. S., Dawson G. J., Desai S. M. (2006). A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5′-untranslated region sequences. J Med Virol 78, 105–111 10.1002/jmv.20510 [DOI] [PubMed] [Google Scholar]
- Naito H., Win K. M., Abe K. (1999). Identification of a novel genotype of hepatitis G virus in Southeast Asia. J Clin Microbiol 37, 1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10, 1–6 10.1093/protein/10.1.1 [DOI] [PubMed] [Google Scholar]
- Parreira R., Branco C., Piedade J., Esteves A. (2012). GB virus C (GBV-C) evolutionary patterns revealed by analyses of reference genomes, E2 and NS5B sequences amplified from viral strains circulating in the Lisbon area (Portugal). Infect Genet Evol 12, 86–93 10.1016/j.meegid.2011.10.011 [DOI] [PubMed] [Google Scholar]
- Pavesi A. (2001). Origin and evolution of GBV-C/hepatitis G virus and relationships with ancient human migrations. J Mol Evol 53, 104–113 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Polgreen P. M., Xiang J., Chang Q., Stapleton J. T. (2003). GB virus type C/hepatitis G virus: a non-pathogenic flavivirus associated with prolonged survival in HIV-infected individuals. Microbes Infect 5, 1255–1261 10.1016/j.micinf.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: Phylogenetic Model Averaging. Mol Biol Evol 25, 1253–1256 [DOI] [PubMed] [Google Scholar]
- Quan P.-L., Firth C., Conte J. M., Williams S. H., Zambrana-Torrelio C. M., Anthony S. J., Ellison J. A., Gilbert A. T., Kuzmin I. V. & other authors (2013). Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A 110, 8194–8199 10.1073/pnas.1303037110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C. M., Zanotto P. M., Holmes E. C. (2008). Bayesian coalescent analysis reveals a high rate of molecular evolution in GB virus C. J Mol Evol 66, 292–297 10.1007/s00239-008-9087-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rubaihayo J., Surat A., Ezekiel M., Andrew A. (2010). High HIV prevalence and associated factors in a remote community in the Rwenzori region of Western Uganda. Infectious Disease Reports 2, e13 10.4081/idr.2010.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz V., Giordano M., Rivero C. W., Minassian M. L., Cuestas M. L., Trinks J., Mathet V. L., Oubiña J. R. (2010). GB virus C quasispecies detected in plasma and lymphocyte subsets in a natural human infection. J Gen Virol 91, 1687–1692 10.1099/vir.0.019877-0 [DOI] [PubMed] [Google Scholar]
- Saito T., Shiino T., Arakawa Y., Hayashi S., Abe K. (1998). Geographical characterization of hepatitis G virus genome: Evidence for HGV genotypes based on phylogenetic analysis. Hepatol Res 10, 121–130 10.1016/S1386-6346(97)00115-0 [DOI] [Google Scholar]
- Saito T., Ishikawa K.-i., Osei-Kwasi M., Kaneko T., Brandful J. A. M., Nuvor V., Aidoo S., Ampofo W., Apeagyei F. A. & other authors (1999). Prevalence of hepatitis G virus and characterization of viral genome in Ghana. Hepatol Res 13, 221–231 10.1016/S1386-6346(98)00095-3 [DOI] [Google Scholar]
- Sathar M. A., York D. F. (2001). Group 5: GBV-C/HGV isolates from South Africa. J Med Virol 65, 121–122 10.1002/jmv.2010 [DOI] [PubMed] [Google Scholar]
- Sathar M. A., Soni P. N., Naicker S., Conradie J., Lockhat F., Gouws E. (1999). GB virus C/hepatitis G virus infection in KwaZulu Natal, South Africa. J Med Virol 59, 38–44 [DOI] [PubMed] [Google Scholar]
- Simons J. N., Leary T. P., Dawson G. J., Pilot-Matias T. J., Muerhoff A. S., Schlauder G. G., Desai S. M., Mushahwar I. K. (1995). Isolation of novel virus-like sequences associated with human hepatitis. Nat Med 1, 564–569 10.1038/nm0695-564 [DOI] [PubMed] [Google Scholar]
- Simons J. N., Desai S. M., Schultz D. E., Lemon S. M., Mushahwar I. K. (1996). Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genome organization. J Virol 70, 6126–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Cuceanu N., Davidson F., Jarvis L. M., Mokili J. L., Hamid S., Ludlam C. A., Simmonds P. (1997). Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5' non-coding region. J Gen Virol 78, 1533–1542 [DOI] [PubMed] [Google Scholar]
- Smith D. B., Basaras M., Frost S., Haydon D., Cuceanu N., Prescott L., Kamenka C., Millband D., Sathar M. A., Simmonds P. (2000). Phylogenetic analysis of GBV-C/hepatitis G virus. J Gen Virol 81, 769–780 [DOI] [PubMed] [Google Scholar]
- Stapleton J. T. (2003). GB virus type C/Hepatitis G virus. Semin Liver Dis 23, 137–148 10.1055/s-2003-39943 [DOI] [PubMed] [Google Scholar]
- Stapleton J. T., Williams C. F., Xiang J. (2004). GB virus type C: a beneficial infection? J Clin Microbiol 42, 3915–3919 10.1128/JCM.42.9.3915-3919.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J. T., Foung S., Muerhoff A. S., Bukh J., Simmonds P. (2011). The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92, 233–246 10.1099/vir.0.027490-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Katayama K., Fukushi S., Kageyama T., Oya A., Okamura H., Tanaka Y., Mizokami M., Gojobori T. (1999). Slow evolutionary rate of GB virus C/hepatitis G virus. J Mol Evol 48, 383–389 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Mizokami M., Orito E., Ohba K.-i., Kato T., Kondo Y., Mboudjeka I., Zekeng L., Kaptue L. & other authors (1998). African origin of GB virus C/hepatitis G virus. FEBS Lett 423, 143–148 10.1016/S0014-5793(98)00083-0 [DOI] [PubMed] [Google Scholar]
- Tillmann H. L., Heiken H., Knapik-Botor A., Heringlake S., Ockenga J., Wilber J. C., Goergen B., Detmer J., McMorrow M. & other authors (2001). Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 345, 715–724 10.1056/NEJMoa010398 [DOI] [PubMed] [Google Scholar]
- Timmons C. L., Shao Q., Wang C., Liu L., Liu H., Dong X., Liu B. (2013). GB virus type C E2 protein inhibits human immunodeficiency virus type 1 assembly through interference with HIV-1 gag plasma membrane targeting. J Infect Dis 207, 1171–1180 10.1093/infdis/jit001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker T. J., Smuts H. E. (2000). GBV-C/HGV genotypes: proposed nomenclature for genotypes 1-5. J Med Virol 62, 82–83 [DOI] [PubMed] [Google Scholar]
- Tucker T. J., Smuts H., Eickhaus P., Robson S. C., Kirsch R. E. (1999). Molecular characterization of the 5′ non-coding region of South African GBV-C/HGV isolates: major deletion and evidence for a fourth genotype. J Med Virol 59, 52–59 [DOI] [PubMed] [Google Scholar]
- Williams C. F., Klinzman D., Yamashita T. E., Xiang J., Polgreen P. M., Rinaldo C., Liu C., Phair J., Margolick J. B. & other authors (2004). Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 350, 981–990 10.1056/NEJMoa030107 [DOI] [PubMed] [Google Scholar]
- Wu H., Padhi A., Xu J., Gong X., Tien P. (2012). Intra-host diversity and emergence of unique GBV-C viral lineages in HIV infected subjects in central China. PLoS ONE 7, e48417 10.1371/journal.pone.0048417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Wünschmann S., Schmidt W., Shao J., Stapleton J. T. (2000). Full-length GB virus C (Hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J Virol 74, 9125–9133 10.1128/JVI.74.19.9125-9133.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Wünschmann S., Diekema D. J., Klinzman D., Patrick K. D., George S. L., Stapleton J. T. (2001). Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med 345, 707–714 10.1056/NEJMoa003364 [DOI] [PubMed] [Google Scholar]
- Xiang J., McLinden J. H., Chang Q., Kaufman T. M., Stapleton J. T. (2006). An 85-aa segment of the GB virus type C NS5A phosphoprotein inhibits HIV-1 replication in CD4+ Jurkat T cells. Proc Natl Acad Sci U S A 103, 15570–15575 10.1073/pnas.0604728103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., McLinden J. H., Chang Q., Jordan E. L., Stapleton J. T. (2008). Characterization of a peptide domain within the GB virus C NS5A phosphoprotein that inhibits HIV replication. PLoS ONE 3, e2580 10.1371/journal.pone.0002580 [DOI] [PMC free article] [PubMed] [Google Scholar]