Abstract
Cauliflower mosaic virus (CaMV) encodes a 520 aa polypeptide, P6, which participates in several essential activities in the virus life cycle including suppressing RNA silencing and salicylic acid-responsive defence signalling. We infected Arabidopsis with CaMV mutants containing short in-frame deletions within the P6 ORF. A deletion in the distal end of domain D-I (the N-terminal 112 aa) of P6 did not affect virus replication but compromised symptom development and curtailed the ability to restore GFP fluorescence in a GFP-silenced transgenic Arabidopsis line. A deletion in the minimum transactivator domain was defective in virus replication but retained the capacity to suppress RNA silencing locally. Symptom expression in CaMV-infected plants is apparently linked to the ability to suppress RNA silencing. When transiently co-expressed with tomato bushy stunt virus P19, an elicitor of programmed cell death in Nicotiana tabacum, WT P6 suppressed the hypersensitive response, but three mutants, two with deletions within the distal end of domain D-I and one involving the N-terminal nuclear export signal (NES), were unable to do so. Deleting the N-terminal 20 aa also abolished the suppression of pathogen-associated molecular pattern-dependent PR1a expression following agroinfiltration. However, the two other deletions in domain D-I retained this activity, evidence that the mechanisms underlying these functions are not identical. The D-I domain of P6 when expressed alone failed to suppress either cell death or PR1a expression and is therefore necessary but not sufficient for all three defence suppression activities. Consequently, concerns about the biosafety of genetically modified crops carrying truncated ORFVI sequences appear unfounded.
Introduction
Members of the family Caulimoviridae of pararetroviruses infect plants and replicate by reverse transcription of a circular dsDNA genome (Haas et al., 2002). The family contains six known genera of which the most extensively studied member is cauliflower mosaic virus (CaMV), the type member of the genus Caulimovirus. CaMV has a genome of ~8 kb comprising six major ORFs (I–VI). Five of the six major virus proteins are translated sequentially from a single polycistronic RNA, the 35S RNA (Ryabova et al., 2002, 2004). This unusual translational strategy is found in members of only two genera of viruses, Caulimovirus and the closely related Soymovirus (Ryabova et al., 2002, 2006).
P6, a 62 kDa polypeptide encoded by CaMV ORFVI, was initially identified as the major component of cytoplasmic inclusion bodies, which constitute the sites of virus assembly (Haas et al., 2002). P6, which is translated from its own monocistronic mRNA, plays an essential role in different aspects of virus replication. It functions in infected cells to facilitate translation of the downstream ORFs on the 35S RNA (Bonneville et al., 1989; Zijlstra & Hohn, 1992) via interaction with components of the translational machinery (Bureau et al., 2004; Leh et al., 2000; Park et al., 2001; Ryabova et al., 2004), a mechanism known as translation transactivation (TAV). P6 prevents ribosome detachment at the stop codon, enabling polypeptide synthesis to reinitiate at the next start codon (Ryabova et al., 2004).
At least four more roles for P6 have been identified. P6 interacts with at least two of the other CaMV proteins involved in aphid transmission, P2 and P3 (Lutz et al., 2012). It forms cytoplasmic inclusion bodies of various sizes; the smaller ones associate with microtubules and the endoplasmic reticulum and move dynamically along actin filaments (Harries et al., 2009). This movement is probably essential for intracellular virus trafficking and involves the interaction of P6 with the CaMV movement protein P1 (Hapiak et al., 2008) and CHUP1, which mediates association between chloroplasts and the cytoskeleton. These findings suggest that P6 subverts the mechanism responsible for chloroplast movement for intracellular trafficking of CaMV (Angel et al., 2013).
P6 is the major genetic determinant of virus pathogenicity (Baughman et al., 1988; Kobayashi & Hohn, 2004; Schoelz et al., 1986; Stratford & Covey, 1989) and expression from a transgene results in a symptom-like phenotype (Baughman et al., 1988; Cecchini et al., 1997; Zijlstra et al., 1996). P6 exhibits virus-encoded suppressor of RNA silencing (VSR) activity (Haas et al., 2008; Love et al., 2007), probably through its interaction with the dsRNA-binding protein DRB4 (Haas et al., 2008), a component of the Dicer4 complex.
Finally, ectopic expression of P6 in Arabidopsis and Nicotiana benthamiana profoundly affects signalling mediated by salicylic acid (SA), jasmonic acid, ethylene and auxin (Geri et al., 2004; Love et al., 2012; Smith, 2007). The ability of P6 to manipulate multiple components of the host defence suggests its central role as a pathogenicity determinant during virus infection, and the pleiotropic phenotypes that result from transgene-mediated expression in planta derive from its activity as a pathogenicity effector.
How does a single protein achieve such a diverse range of activities? Outwith closely related members of the family Caulimoviridae, P6 has no obvious homologues and its three-dimensional structure is unknown. Li & Leisner (2002) defined four domains based on self-association, and sequence analysis has revealed several structural motifs and functional domains (Fig. 1a). These include RNA binding, RNase H, a short N-terminal helical domain and several predicted nuclear localization signals (NLSs) (Cerritelli et al., 1998; De Tapia et al., 1993; Haas et al., 2008; Kobayashi & Hohn, 2003; Ryabova et al., 2004). De Tapia et al. (1993) identified aa 111–242 (domain D-II) as containing the minimum functional domain (miniTAV domain) able to facilitate TAV. The miniTAV domain overlaps an RNase H domain (Cerritelli et al., 1998) and contains the interaction motif for RL18; those for RL24, eIF4G and eIF2B are located within domain D-III (Ryabova et al., 2004). Domains D-II and D-IV are involved in the interaction with CHUP1 (and presumably therefore intracellular trafficking (Angel et al., 2013). P6 is a nucleocytoplasmic shuttle protein, and both nuclear localization and export functions are essential for infectivity. Mutation of a nuclear export signal (NES) at the N terminus abolishes infectivity and mutation of the predicted NLS within the C-terminal domains abolishes both VSR activity and infectivity (Haas et al., 2008; Kobayashi & Hohn, 2004).
Fig. 1.
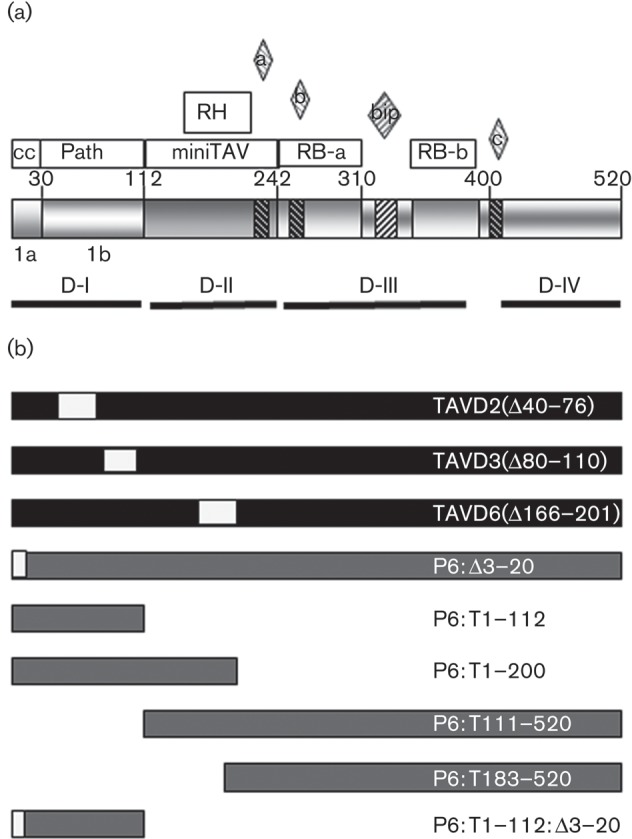
Map of the P6 domains and mutants used in this study. (a) Schematic representation of P6 domains: amino acid numbers at the boundaries of known domains are indicated. Open boxes show the coiled-coil (cc) α-helix, pathogenicity/host-range/avirulence (Path), minimum transactivator (miniTAV), RNase H (RH) and RNA binding (RB-a and RB-b). The bipartite nuclear localization signals (NLS; bip) and three non-conventional NLS (a, b and c) are indicated by diamonds above and cross-hatching. The self-association domains D-I to D-IV and subdomains 1a and 1b are indicated by solid lines. Data from Haas et al. (2005), Haas et al. (2008), Kobayashi & Hohn (2004) and Hapiak et al. (2008). (b) Deletions in P6 coding sequences in CaMV-TAV mutants and in the corresponding P6 expression constructs. Filled boxes indicate sequence from CaMV CM1841, shaded boxes indicate sequence derived from CaMV Cabb B-JI and open boxes indicate internal deletions.
Although TAV activity and virus-trafficking functions have been mapped, the domain(s) responsible for VSR activity and SA signalling suppression remain(s) to be identified. Domain D-I plays a major role in pathogenicity and host range and acts as an avirulence domain in Arabidopsis and Solanaceous hosts (Agama et al., 2002; Baughman et al., 1988; Palanichelvam et al., 2000; Palanichelvam & Schoelz, 2002; Schoelz et al., 1986; Stratford & Covey, 1989). D-I has been divided into subdomain 1a, comprising the N-terminal 30 aa containing the NES (Haas et al., 2008; Haas et al., 2005), and subdomain 1b (aa 31–110) containing avirulence and pathogenicity functions. Mutants with deletions within 1b retain replication competence but exhibit delayed virus spread in turnip (Kobayashi & Hohn, 2004).
We carried out infection studies using CaMV mutants with deletions in P6 and found that at least one mutation within subdomain 1b abolished both VSR activity and symptom development without significantly reducing systemic virus titre. We transiently expressed WT and mutant P6 in Nicotiana benthamiana and Nicotiana tabacum and assayed the ability to suppress expression of an SA-responsive marker gene, PR1a, and cell death in response to a gene-for-gene elicitor. Deletions in subdomains 1a and 1b abolished VSR activity and also abolished the suppression of the cell-death response seen with WT P6. However, only the deletion in subdomain 1a eliminated suppression of PR1a expression. Domain D-I evidently plays an essential role in several pathogenicity effector activities. Suppression of RNA silencing and cell death may be functionally linked, but suppression of SA-responsive gene expression must involve an at least partially independent mechanism.
Results
Infectivity of WT and P6 deletion mutants of CaMV in Arabidopsis
CaMV mutants with in-frame deletions in subdomain 1b of P6 are replication competent in turnip but show delayed long-distance spread (Kobayashi & Hohn, 2004). We inoculated WT Arabidopsis (ecotype Col-0) with WT virus (CaMV-CW) and three mutants (Fig. 1b). CaMV-TAVD2 and CaMV-TAVD3 carry deletions in subdomain 1b, whilst CaMV-TAVD6 has a deletion in the miniTAV domain and cannot replicate in turnip.
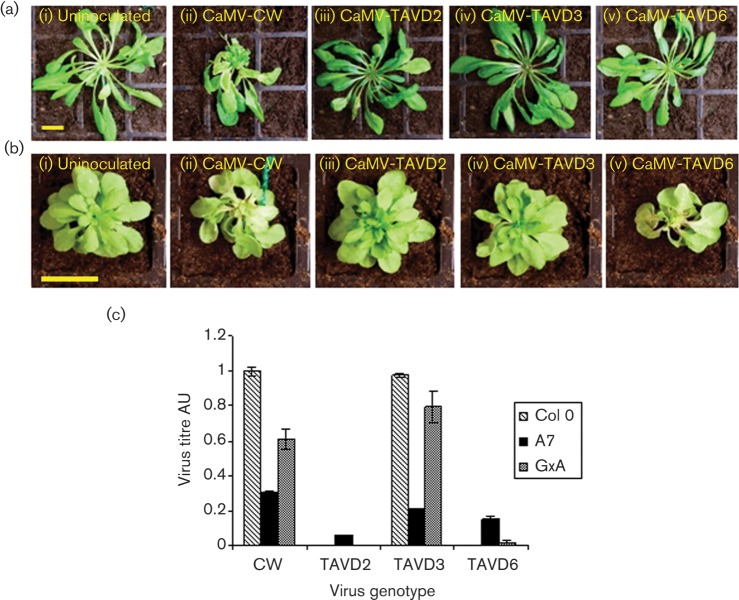
Agroinoculation with CaMV-CW was remarkably efficient, with symptoms appearing at ~13 days post-infection (p.i.) and essentially 100 % of plants developing obvious stunting, leaf distortion and mosaics by 28 days p.i. (Fig. 2a). Plants inoculated with CaMV-TAVD2 and CaMV-TAVD6 did not develop any symptoms (Fig. 2a). With CaMV-TAVD3 inoculation, plants were usually asymptomatic, although by 28 days p.i. the occasional leaf exhibited subtle vein clearing. We measured virus titres at 28 days p.i. using ELISA (Fig. 2c). Col-0 plants inoculated with CaMV-CW contained high titres of virus, but CaMV-TAVD2 and CaMV-TAVD6 were not detectable by ELISA. Surprisingly, despite the lack of symptoms, titres of CaMV-TAVD3 were consistently very similar to titres of CaMV-CW. Thus, this deletion did not appear to significantly reduce virus accumulation, at least under the conditions of our experiment, but profoundly affected symptom development.
Fig. 2.
Infectivity of CaMV (WT and mutants) on WT and P6 transgenic Arabidopsis. (a) Symptoms on Col-0 at 28 days p.i.: (i) uninfected, (ii) CaMV-CW, (iii–v) CaMV TAV mutants as indicated. Bar, 2 cm. (b) Symptoms on P6 transgenic plants (line A7) at 28 days p.i.: (i) uninfected, (ii) CaMV-CW, (iii–v) CaMV TAV mutants as indicated. Bar, 2 cm (note difference in scale between a and b). (c) Virus titres at 28 days p.i. in Col-0, A7 and GxA plants determined by ELISA. Bars shows mean titres (±sd) of three tissue samples each comprising three pooled plants. Titres in arbitrary units (AU) are normalized to the mean of Col-0 plants infected with CaMV-CW.
We next tested whether we could complement the mutations in ORFVI by providing P6 from the transgenic Arabidopsis line A7, which expresses P6 at levels similar to those in infected plants (Cecchini et al., 1997) (Fig. 2b). Plants started to exhibit subtle symptoms of leaf distortion at around 14 days p.i., and by 28 days p.i., stunting and leaf distortion were visible on all A7 plants inoculated with CaMV-CW. Plants inoculated with CaMV-TAVD6 also all developed symptoms similar to CaMV-CW and at around the same time, but those inoculated with CaMV-TAVD2 and CaMV-TAVD3 did not. Titres of CaMV-CW were approximately 30 % of those in Col-0 plants, consistent with our previous reports of reduced titres in P6 transgenics (Love et al., 2007, 2012). All three mutants also accumulated to significant titres (Fig. 2c). These results suggested that functional P6 provided from a transgene can act in trans to facilitate the replication of the mutants. ORFVI sequences provided from a transgene can under some circumstances recombine with defective CaMV genomes when infection proceeds over an extended period (Király et al., 1998). Although we cannot absolutely rule out the possibility of recombination in our complementation experiments, we believe that it is unlikely because virus titres for the WT and mutants were similar at 28 days p.i. and, in the case of CaMV-TAVD6, symptoms started to appear at a similar relatively early stage in infection. The absence of symptoms in A7 plants infected with CaMV-TAVD2 and CaMV-TAVD3 suggested that, even when WT P6 is provided from a transgene, symptom development is blocked in the presence of virus-encoded P6 containing deletions in subdomain 1b. We did not observe this with CaMV-TAVD6 in which the deletion affects the miniTAV domain.
VSR activity of CaMV deletion mutants
The transgenic Arabidopsis line GxA contains a 35S–GFP transgene whose expression is silenced by a second transgene, a potato virus X amplicon containing part of the GFP-coding sequence (Dalmay et al., 2000; Schwach et al., 2005). CaMV infection of GxA suppresses silencing of the GFP transgene, restoring strong fluorescence to infected tissue (Love et al., 2007). We used this assay to compare the VSR activities of WT and mutant virus. Virus levels in GxA were similar to those in Col-0, with CaMV-CW and CaMV-TAVD3 accumulating to high titres but CaMV-TAVD2 and CaMV-TAVD6 undetectable by ELISA (Fig. 2c). As with Col-0, only CaMV-CW induced symptoms in GxA.
We examined the upper leaves for GFP fluorescence using confocal microscopy (Fig. 3a). Uninoculated controls showed no detectable fluorescence, even at high gain, but tissue from CaMV-CW-infected plants consistently showed strong fluorescence. We did not observe any fluorescent cells in systemic leaves of GxA inoculated with any of the three mutants, despite the high titres of CaMV-TAVD3. To test for local silencing suppression (around the sites of inoculation), we examined inoculated leaves (Fig. 3b). With CaMV-CW, by 8–11 days p.i. we consistently observed groups of cells showing strong GFP fluorescence. Inoculated leaves recovered from the microscope slide and assayed by ELISA all contained moderate to high titres of virus (with some leaf-to-leaf variation). Titres of CaMV-TAVD3 in inoculated leaves were similar to those of CaMV-CW but we did not observe any GFP fluorescence. Leaves inoculated with CaMV-TAVD2 and CaMV-TAVD6 contained no detectable titres of virus. With CaMV-TAVD2, we did not observe any fluorescent cells, but with CaMV-TAVD6 we consistently observed fluorescence in groups of cells within every leaf we examined between 8 and 11 days p.i. (Fig. 3b), albeit at lower intensity than with CaMV-CW; by 14 days p.i. fluorescence had become undetectable. Kobayashi & Hohn (2003) showed using sensitive PCR that CaMV-TAVD6 is unable to replicate in single cells. However, agroinoculation could provide transient P6 expression through direct transcription of ORFVI (from its own 19S promoter) of the replication-incompetent CaMV-TAVD6 genome (Kobayashi & Hohn, 2003, 2004). This result demonstrates that deleting aa 166–201 does not abolish VSR activity.
Fig. 3.
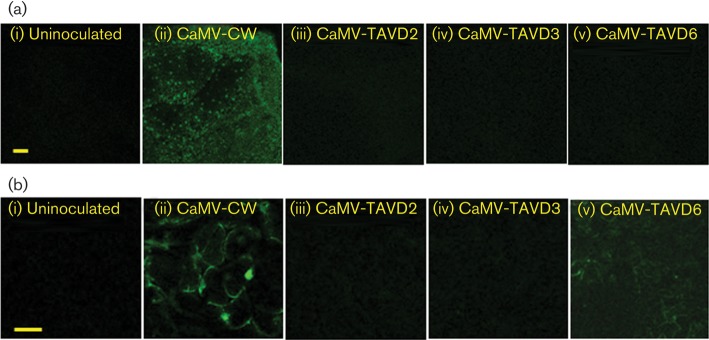
GFP fluorescence in leaves of GxA plants inoculated with CaMV WT and mutants. (a) Confocal microscope images of representative upper leaves of plants at 28 days p.i.: (i) uninoculated, (ii) CaMV-CW, (iii–v) CaMV TAV mutants. The panels show low-magnification images of GFP fluorescence. All panels were taken at the same microscope gain settings. (b) Confocal microscope images of representative inoculated leaves of plants at 28 days p.i.: (i) uninfected, (ii) CaMV-CW, (iii–v) CaMV TAV mutants. Note that the images in (b) are taken at a higher magnification than those in (a). All panels were taken with the same microscope gain settings. Bars, 100 µm.
Mutations in P6 affect the ability to suppress SA-responsive cell death
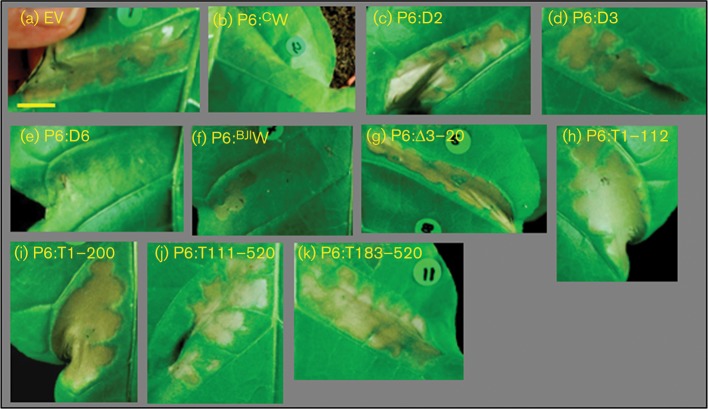
Expression of P6 from a transgene in Arabidopsis reduces and delays cell death following treatment with SA or inoculation with an avirulent pathogen (Love et al., 2012). To identify the domain(s) responsible for this activity, we exploited the ability of tomato bushy stunt virus (TBSV) P19 to elicit a gene-for-gene hypersensitive response (HR) in N. tabacum in an SA-dependent manner (Angel & Schoelz, 2013; Sansregret et al., 2013). Agrobacterium-mediated expression of P19 in N. tabacum gave a strong HR that was complete by 36 h but could be extended to 3–5 days by reducing the Agrobacterium titre to one-quarter the usual level. At the higher titre of P19, co-infiltration with a hypervirulent strain of Agrobacterium containing pGWB-P6BJIW or pGWB-P6CW delayed the onset of HR by approximately 24 h. At the reduced titre of P19, co-infiltration with either WT P6 construct substantially halted the progress of this HR compared with co-infiltration with empty vector (EV) (Fig. 4a, b, f). Neither WT nor mutant P6 elicited HR in the absence of P19.
Fig. 4.
Suppression of TBSV P19-dependent cell death by co-infiltration with WT and mutant variants of P6. Photographs of leaf patches 4 days after co-agroinfiltration with a construct expressing P19 plus EV control (a) or WT or mutant P6 as indicated (b–k). All images are shown at similar magnification. Bar, 1.0 cm.
We cloned the P6 coding sequences from the three CaMV mutants into Ti expression vectors and tested their ability to suppress HR (Fig. 4). In contrast to P6:CW (wild-type P6 from CaMV CM1841), neither of the two mutants with deletions in subdomain 1b, P6:D2 and P6:D3, was able to suppress the development of HR, but P6:D6, with a deletion in the miniTAV domain, suppressed cell death with an efficiency similar to WT (Fig. 4c–e). The N-terminal subdomain 1a contains the NES, and mutations that abolish nuclear export also abolish VSR activity (Haas et al., 2008). We therefore deleted 18 of the 20 aa at the N terminus to produce what we predicted would be a functionally equivalent construct, P6:Δ3–20 (Fig. 1b). When transiently co-expressed with P19, P6:Δ3–20 was unable to suppress HR (Fig. 4g).
To test whether the D-I domain was able to suppress P19-induced HR in the absence of the C-terminal domains, we produced a further series of expression constructs (for details, see Fig. 1b). Truncated polypeptides comprising the N-terminal 112 or 200 aa (P6:T1–112, P6:T1–200) did not suppress the HR elicited by P19 (Fig. 4h, i). Neither did the corresponding C-terminally truncated variants (P6:T111–520 and P6:T183–520) (Fig. 4j, k). The ability to suppress cell death thus broadly paralleled the ability to suppress RNA silencing, with deletions in both D-I subdomains, but not in the miniTAV domain, affecting this activity. However, the D-I domain expressed alone was not sufficient to suppress cell death; therefore other regions of P6 must also be required for this activity.
Mutations in P6 affect the ability to suppress expression of an SA-responsive marker gene
Agroinfiltration of N. benthamiana elicits pathogen-associated molecular pattern (PAMP)-responsive expression of PR1a, a reliable marker of SA-responsive gene expression (Volko et al., 1998); this response is strongly suppressed by transient expression of P6 (Love et al., 2012). P6:CW gave the expected reduction in PR1a transcripts to ~30 % that with EV (Fig. 5a). We anticipated that P6 mutants with deletions in subdomain 1b might also fail to suppress PR1a expression. However, P6:D2 and P6:D3 as well as P6:D6 all reduced PR1a transcripts to a broadly similar level to that of P6:CW (Fig. 5a). All three mutants evidently retained the ability to suppress SA-responsive gene expression. In contrast, infiltration with P6:Δ3–20 resulted in levels of PR1a expression similar to EV (Fig. 5b). Therefore, sequences required for suppression of SA-responsive gene expression are present in subdomain 1a but apparently not 1b.
Fig. 5.
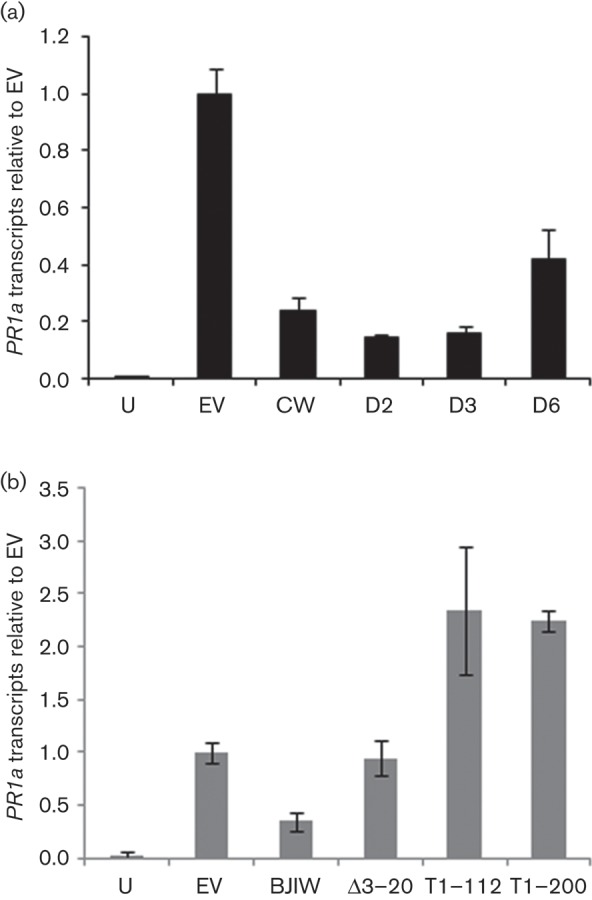
Quantification of PR1a expression in N. benthamiana leaves following transient expression of WT and mutant P6 by agroinfiltration. (a) PR-1a transcripts, determined by quantitative PCR, in N. benthamiana leaves harvested 48 h after agroinfiltration. Samples were uninfiltrated leaves (U), and leaves infiltrated with Agrobacterium carrying the following vectors: pGWB17 (EV), P6:CW (CW), P6:D2 (D2), P6:D3 (D3) and P6:D6 (D6). (b) PR-1a transcripts, determined as above. Samples were uninfiltrated leaves (U), and leaves infiltrated with Agrobacterium carrying the following vectors pGWB17 (EV), P6BJIW (BJIW), P6:Δ3–20 (Δ3–20), P6:T1–112 (T1–112) and P6:T1–200 (T1–200). Bars show means±sd (in arbitrary units) of three independent biological samples each comprising three pooled infiltrated leaf sections. Values were normalized to values for EV.
We next investigated whether expressing the N-terminal domain alone was sufficient to suppress PR1a expression. P6:T1–112 and P6:T1–200 not only failed to reduce PR1a transcript levels but also produced a consistent increase of more than twofold over and above EV controls (Fig. 5b). The D-I domain is therefore not sufficient for suppression of SA-dependent gene expression but may play some role in this activity because expression on its own promoted elevated expression of PR1a.
Intracellular localization of mutant P6
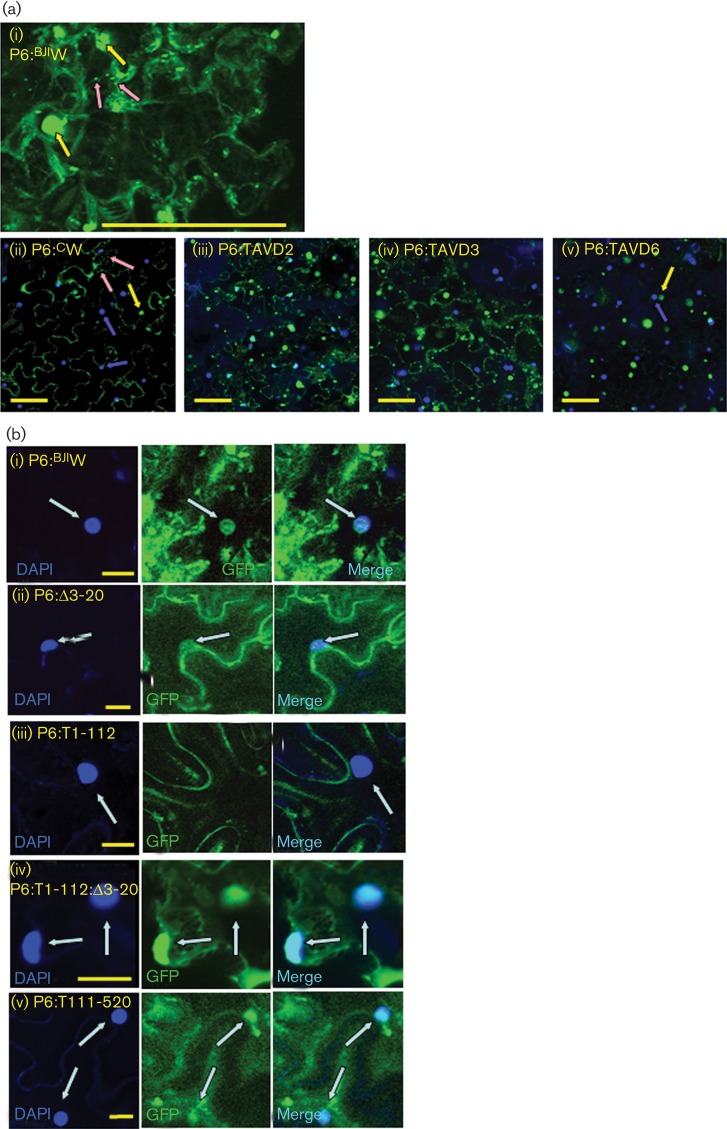
To test whether loss of defence suppression activity in some mutants might be attributable to mislocalization and to confirm appropriate expression of P6, we analysed its intracellular localization after transiently expressing the mutant forms of P6 as C-terminal GFP fusions in N. benthamiana (Fig. 6). GFP-tagged P6BJIW (wild-type P6 from CaMV Cabb B-JI) suppressed PR1a expression with an efficiency similar to a myc-tagged construct (see Methods), indicating that this activity was unaffected by the GFP tag (data not shown). Epidermal cells expressing WT P6 from the two isolates CM1841 and Cabb B-JI showed identical patterns of intracellular fluorescence, reminiscent of those reported by Harries et al. (2009) (Fig. 6a, i and ii). Cells contained cytoplasmic inclusion bodies that were highly variable in size. Large numbers of small inclusion bodies were present, some of which appeared to be associated with cytoplasmic strands. Large inclusion bodies were often clustered around the nucleus, but we observed only weak GFP fluorescence co-localizing with DAPI (Fig. 6b, i), consistent with the findings of Haas et al. (2005) who reported rapid nuclear export of P6.
Fig. 6.
Intracellular localization of WT and mutant P6 tagged with GFP. Confocal microscope images of tissue from N. benthamiana leaves 3 days after agroinfiltration with WT and mutant variants of P6 fused at the C terminus to GFP. GFP fluorescence is green and DAPI fluorescence (nuclear staining) is blue. (a) Intracellular distribution of WT and TAVD mutant P6: (i) P6BJIW in a single epidermal cell. Representative large inclusion bodies are indicated by yellow arrows and small inclusion bodies by pink arrows. (ii) P6CW. Yellow and pink arrows are as in (i), whilst blue arrows indicate nuclei (DAPI staining). (iii–v) P6:D2, P6:D3 and P6:D6. Bars, 100 µm. (b) High-magnification images showing nuclear localization of WT and truncated forms of P6: (i) P6BJIW, (ii) P6:Δ3–20, (iii) P6:T1–112, (iv) P6:T1–112:Δ3–20 and (v) P6:T111–520. Panels from left to right: DAPI, GFP, merge. Nuclear fluorescence is indicated by arrows. Bars, 20 µm.
Localization of P6:D2-GFP and P6:D3-GFP was indistinguishable from that of the WT (Fig. 6a, ii–iv). These deletions did not cause obvious changes in intracellular distribution. With P6:D6-GFP, we observed very few small inclusion bodies, although the large ones were still abundant (Fig. 6a, v). Domain D-II, which contains the deletion in P6:D6 (aa 166–201), has been identified as interacting with CHUP1 in connection with intracellular virus trafficking (Angel et al., 2013). The region may be required for cytoskeletal association and for the formation of small inclusion bodies.
The deletion in P6:Δ3–20–GFP included three residues identified as essential for nuclear export (Haas et al., 2008), so we anticipated that it would show enhanced co-localization with DAPI. Unexpectedly, the nuclear localization was similar to that of WT (Fig. 6b, ii). Haas et al. (2005) expressed the N-terminal 110 aa alone and compared localization with the same polypeptide with the N-terminal α-helix deleted. We produced equivalent constructs (although with a C-terminal GFP tag), P6:T1–112–GFP and P6:T1–112:Δ3–20–GFP. Whereas P6:T1–112–GFP showed no nuclear localization whatsoever, P6:T1–112:Δ3–20–GFP co-localized strongly with DAPI (Fig. 6b, iii, iv). P6:T111–520–GFP, which lacks the entire D-I domain (including the NES), also showed enhanced nuclear localization (Fig. 6, v). Our results are therefore consistent with deletion of the N-terminal 20 aa affecting nuclear export. Haas et al. (2005) fused GFP to the N terminus; our use of a C-terminal tag might account for the differences for full-length P6.
Sequence conservation of domain D-I across members of the Caulimoviridae
We used the programs Jalview (Waterhouse et al., 2009) and JPred3 (Cole et al., 2008) to align the sequences of P6 from CaMV with ten other members of the genus Caulimovirus and four members of the genus Soymoviruses. P6 from all ten caulimoviruses showed significant homology with CaMV over almost the entire sequence, but none of the soymoviruses showed significant similarity to the caulimovirus D-I domain. (Figs 7 and S1, available in JGV Online). Within domain D-I, homology varied between different members of the genus Caulimovirus (Fig. 7), but there was notable sequence conservation between aa 67 and 88, in particular the GK(D/E)X(S/T)NPLXXXXLXK motif (aa 74–88) conserved in 10 of 11 sequences. Interestingly, this motif extends across the junction between the TAVD2 and TAVD3 deletions.
Fig. 7.

Alignment of sequences of the N-terminal amino acids of P6 from caulimoviruses. The sequences from CaMV, horseradish latent virus (HLV), lamium leaf distortion associated virus (LLDAV), carnation etched ring virus (CERV), dahlia mosaic virus-Holland (DaMV-Holl), eupatorium vein clearing virus (EVCV), mirabilis mosaic virus (MMV), dahlia mosaic virus (DaMV), figwort mosaic virus (FMV), cestrum yellow leaf curling virus (CmYLCV) and strawberry vein banding virus (SVBV) that precede the RNaseH domain (aa 140 in CaMV) are aligned, with the consensus sequence shown below in logo form. Residues are coloured according to the clustal_x colouring scheme and the Uniprot accession numbers indicated.
The N-terminal sequences of the soymoviruses are shorter than those of the caulimoviruses and are rather diverse (Fig. S1). Possibly, members of the genus Soymovirus lack a functional D-I domain.
Discussion
We have shown that the N-terminal domain of P6 contains sequences essential for its activities as a suppressor of RNA silencing and of SA-dependent defence responses. These appear to be distinct from its TAV and virus-trafficking functions. Deleting the distal end of subdomain 1b (aa 80–110) abolished VSR activity within the context of an infectious virus clone. The same deletion also abolished the ability to suppress one aspect of SA-dependent signalling, cell death triggered by the elicitor TBSV P19, but not another, PAMP-driven PR1a expression. The mechanisms underlying these three activities may therefore overlap but are clearly not identical. However, the N-terminal subdomain 1a, which includes the NES, is essential for all three.
CaMV-TAVD6, with a deletion within the miniTAV domain, consistently produced a transient silencing suppression in inoculated leaves, evidence that this mutant retains VSR activity. As CaMV-TAVD6 is completely unable to replicate in protoplasts (Kobayashi & Hohn, 2003), we assume that P6 mRNA is transcribed directly from CaMV-TAVD6 genomes introduced by agroinoculation. Although we were unable to detect CaMV-TAVD2 accumulation in inoculated leaves using ELISA, we might have expected similar limited P6 expression by direct transcription of the T-DNA following agroinoculation. If so, the failure of CaMV-TAVD2 to stimulate similar transient GFP expression in inoculated leaves suggests that it too may be deficient in VSR activity.
Suppression of RNA silencing by VSR is a major contributor to symptom induction (Burgyán & Havelda, 2011). Despite titres similar to WT virus, CaMV-TAVD3 was essentially asymptomatic on Col-0 plants, suggesting that the symptoms of CaMV infection (at least in Arabidopsis) are probably linked to the VSR activity of P6. Complementation by transgene-derived P6 allowed all three mutants to replicate, but, whereas CaMV-CW and CaMV-TAVD6 caused obvious stunting and leaf distortion in A7, CaMV-TAVD3 and CaMV-TAVD2 were both asymptomatic. As A7 produces high levels of WT P6 (Cecchini et al., 1997), the absence of symptoms in CaMV-TAVD2- and CaMV-TAVD3-infected plants must be a dominant-negative effect, presumably linked to the loss of VSR activity, further evidence that CaMV-TAVD2 is also defective in this respect.
A role for domain D-I in defence suppression is consistent with its identification as the major genetic determinant of virus pathogenicity, host range and avirulence (Kobayashi & Hohn, 2003; Palanichelvam & Schoelz, 2002; Schoelz & Shepherd, 1988; Stratford & Covey, 1989). Both subdomains are involved. Subdomain 1a clearly plays an essential role in pathogenicity as deleting it eliminated both the suppression of cell death and PAMP-responsive gene expression. Correct localization of P6 may be required for these activities. Deletions within subdomain 1b abolished the ability to suppress cell death in our assay but not the ability to suppress PAMP-triggered expression of PR1a. The apparent discrepancy between the effects of deletions in subdomain 1b on these two different SA-dependent responses may be explained by the recent report that extreme resistance to TBSV in N. tabacum is elicited by a complex of P19 plus small interfering RNAs (siRNAs) (Sansregret et al., 2013). Interaction with DRB4, a component of the Dicer4 complex (Haas et al., 2008), provides a probable mechanism for the VSR activity of P6. As Dicer4 is involved in generating siRNAs, both defence suppression activities of P6 (on RNA silencing and SA signalling) could potentially play a role in inhibiting the HR elicited by P19.
The truncated proteins P6 : 1–112 and P6 : 1–200 elicited elevated levels of PR1a transcripts compared with EV controls. The C-terminal region of P6, which is absent from these constructs, contains four predicted NLSs (Fig. 1a). Nuclear localization is required for VSR activity (Haas et al., 2008), and our results are consistent with it also being essential for cell death and suppression of SA-dependent gene expression. We did not observe obvious differences in nuclear localization between WT and truncated proteins (Fig. 6). However, because the N-terminal NES promotes very efficient re-export of P6 from the nucleus, even WT P6, which is actively imported into the nucleus (Haas et al., 2005), gave only weak GFP fluorescence within nuclei.
The effects of mutations in subdomain 1b on VSR activity and the suppression of the HR elicited by P19 imply that it must play a key role in these functions. The motif GK(D/E)X(S/T)NPLXXXXLXK, which spans the ends of the TAVD2 and TAVD3 deletions, is very highly conserved across 10/11 members of the genus Caulimovirus. Such a degree of sequence homology provides additional support for the importance of this region of P6 to members of this genus.
The pleiotropic phenotype(s) of P6-transgenic Arabidopsis (Geri et al., 2004; Love et al., 2012; Smith, 2007) would be most elegantly accounted for by a common underlying mechanism, perhaps all involving RNA silencing, rather than by diverse direct interactions with multiple signalling intermediates. However, because deletion mutants of subdomain 1b suppressed PAMP-responsive PR1a expression with a similar efficiency to WT P6, VSR activity must not be essential for this activity.
Transgene-mediated expression of VSRs elicits pleiotropic effects on jasmonic acid and other phytohormone responses (Endres et al., 2010; Lewsey et al., 2010; Lozano-Durán et al., 2011; Yang et al., 2008), and CaMV infection is accompanied by profound changes in microRNA (miRNA) and trans-acting siRNA (tasiRNA) populations (Blevins et al., 2006; Moissiard & Voinnet, 2006; Shivaprasad et al., 2008). The 5′ leader sequence of the CaMV 35S RNA is a target for all four Arabidopsis Dicer complexes, producing siRNAs that appear to target host transcripts (Blevins et al., 2006; Moissiard & Voinnet, 2006; Shivaprasad et al., 2008), evidence of a complex interaction mediated at least partially by RNA silencing. miRNAs and tasiRNAs regulate signalling pathways involving auxin (Rubio-Somoza & Weigel, 2011; Rubio-Somoza et al., 2009), ethylene (Pei et al., 2013) and jasmonic acid (Lewsey et al., 2010; Schommer et al., 2008; Zhang et al., 2012), and evidence is emerging that they also regulate immune responses and cell death in Arabidopsis (Alonso-Peral et al., 2010; Li et al., 2012).
We previously identified NPR1, a central regulator of defence, as a target for P6. NPR1 acts through complex mechanisms entailing activation by SA, nuclear localization, modification (phosphorylation and S-nitrosylation) and targeted proteolysis (Mukhtar et al., 2009). Recent reports identify three SA receptors, NPR1 itself (Wu et al., 2012) plus two E3 ligases, NPR3 and NPR4. These suppress or activate both programmed cell death and PR gene expression by regulating NPR1 levels in response to changes in intracellular SA (Fu et al., 2012). Expression of P6 alters intracellular localization and enhances accumulation of NPR1 (Love et al., 2012). The possibility that this might be achieved through miRNAs or tasiRNAs is intriguing. All four Arabidopsis Dicers are believed to participate in the biogenesis of siRNAs from the CaMV leader (Blevins et al., 2006; Moissiard & Voinnet, 2006), but it is DCL1 that is primarily responsible for the generation of miRNAs from host-encoded precursors (Vazquez et al., 2010). It would be interesting to investigate whether P6 and HYL1 (the DRB4 homologue in Dicer1) also interact. Although the details of the mechanisms remain unknown, our results suggest that RNA silencing regulates at least one response involving SA signalling (cell death), and that CaMV targets multiple defence responses via the VSR activity of P6.
Fifty-four transgenic events commercialized in the USA contain up to 528 bp of the coding region of ORFVI (Podevin & du Jardin, 2012). The potential expression of a C-terminal P6 polypeptide with defence-suppressing properties has been identified as a possible hazard in genetically modified crops (Latham & Wilson, 2013). The essential role for the N-terminal region of P6 in these activities demonstrates that these concerns are unfounded.
Methods
Virus infection.
Arabidopsis plants were grown under short days as described previously (Cecchini et al., 1998). Details of the P6-transgenic line A7 have been published (Cecchini et al., 1997). For assaying VSR activity, we infected transgenic line GxA in which expression of GFP is silenced by a potato virus X amplicon (Dalmay et al., 2000; Love et al., 2007; Schwach et al., 2005).
Virus infection was achieved using agroinfectible constructs derived from WT CaMV isolate CM1841 (pFastWt) and its ORFVI mutants, pFastTavD2, pFastTavD3 and pFastTavD6 (Kobayashi & Hohn, 2003, 2004; Tsuge et al., 1994), which were designated in this study as CaMV-CW, CaMV-TAVD2, CaMV-TAVD3 and CaMV-TAVD6, respectively. Full details of the construction of the agroinfectible clones are given in Fig. S2.
For virus infection, the hypervirulent Agrobacterium strain AGL1+virG (Vain et al., 2004) containing the appropriate construct was grown overnight at 28° in Luria–Bertani medium containing kanamycin (50 µg ml−1), rifampicin (50 µg ml−1) and gentamicin (50 µg ml−1). Bacteria were resuspended at OD600 = 0.2 in 10 mM MgCl2 and incubated for 2 h with 200 µM acetosyringone at room temperature. Celite was added, and plants at the eight-leaf stage were inoculated by pipetting 2 µl bacterial suspension onto one lower leaf and rubbing with a sterile inoculating loop.
Virus titres were measured using DAS-ELISA kits (Bioreba, Lynchwood Diagnostics, UK). The entire above-ground parts of three infected plants were combined, ground in 10 vols of Extraction Buffer (Bioreba), clarified in a bench-top centrifuge and the supernatant assayed according to the manufacturer’s instructions. Samples with high virus titres were diluted a further 10-fold before assay.
Transient expression of WT and mutant variants of P6.
Transient expression was carried out by agroinfiltration in N. benthamiana as described previously (Bazzini et al., 2007; Love et al., 2012). Vectors for expressing WT or mutant P6 were constructed using the Gateway cloning system (Invitrogen). Sequences were amplified by PCR using the primer combinations listed in Table S1, inserted into pENTR-DTopo and transferred to Gateway binary vectors pGWB17 (giving a C-terminal 4×myc tag) or pGWB5 (giving a C-terminal GFP fusion). Details of the deletions and truncations are shown in Fig. 1(b). Constructs were derived from CaMV isolate CM1841 (GenBank accession no. V001440) or the closely related Cabb B-JI (GenBank accession no. DQ211685). Details of pGWB-P6BJIW have been described in Love et al. (2012; referred to as pGWB-P6myc). p35S-P19 for expression of TBSV P19 (Voinnet et al., 2003) was a kind gift from Professor David Baulcombe (Cambridge, UK).
For the cell death suppression assay, TBSV P19 and P6 were co-expressed in leaves of N. tabacum (cv. Petite Havana SR1). Overnight cultures of p35S-P19 in Agrobacterium GV3101, were resuspended at OD600 = 0.1. Agrobacterium AGL1+virG containing the appropriate P6 expression construct (or as control pGWB17) were resuspended at OD600 = 0.4 and mixed with an equal volume of the P19 culture for infiltration. The development of necrosis was assessed visually over 3–5 days. To allow for potential leaf-to-leaf differences in the development of HR, we always included one WT P6 as a positive control and one EV as a negative control on each leaf.
Quantification of transcripts by quantitative PCR (qPCR).
NbPR1a transcripts were quantified by real-time reverse transcription qPCR using a Stratagene MX4000 or MX3000 thermocycler as described previously (Love et al., 2005, 2012). The reference gene was NbEF1α. Each biological sample comprised RNA extracted from ~50 mg tissue taken from the infiltrated area of a single N. benthamiana leaf. The primers are given in Table S1(B).
Fluorescence microscopy.
GFP fluorescence in leaves of GxA and localization of P6–GFP in N. benthamiana leaves were followed using a Zeiss LSM510 confocal microscope essentially as described previously (Love et al., 2007, 2012). Nuclei were stained with DAPI (Molecular Probes, Life Technologies).
Protein sequence alignments.
The Jpred3 server was searched with the CaMV sequence (NCBI Protein no. Q3HM11) to obtain an alignment of diverse P6 sequences with redundancy removed. The selected sequences were retrieved intact from the Uniprot database and the alignment rebuilt with muscle (Edgar, 2004) and curated manually in Jalview.
Acknowledgements
This work was supported in part by funding from Glasgow University School of Life Sciences and by a grant from BBSRC (BB/D017319) to J. J. M. and A. S. We thank Professor David Baulcombe for the gift of p35S-P19.
Footnotes
Two supplementary figures and one table are available with the online version of this paper.
References
- Agama K., Beach S., Schoelz J., Leisner S. M. (2002). The 5' third of Cauliflower mosaic virus gene VI conditions resistance breakage in Arabidopsis ecotype Tsu-0. Phytopathology 92, 190–196 10.1094/PHYTO.2002.92.2.190 [DOI] [PubMed] [Google Scholar]
- Alonso-Peral M. M., Li J., Li Y., Allen R. S., Schnippenkoetter W., Ohms S., White R. G., Millar A. A. (2010). The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol 154, 757–771 10.1104/pp.110.160630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel C. A., Schoelz J. E. (2013). A survey of resistance to Tomato bushy stunt virus in the genus Nicotiana reveals that the hypersensitive response is triggered by one of three different viral proteins. Mol Plant Microbe Interact 26, 240–248 10.1094/MPMI-06-12-0157-R [DOI] [PubMed] [Google Scholar]
- Angel C. A., Lutz L., Yang X., Rodriguez A., Adair A., Zhang Y., Leisner S. M., Nelson R. S., Schoelz J. E. (2013). The P6 protein of Cauliflower mosaic virus interacts with CHUP1, a plant protein which moves chloroplasts on actin microfilaments. Virology 443, 363–374 10.1016/j.virol.2013.05.028 [DOI] [PubMed] [Google Scholar]
- Baughman G. A., Jacobs J. D., Howell S. H. (1988). Cauliflower mosaic virus gene VI produces a symptomatic phenotype in transgenic tobacco plants. Proc Natl Acad Sci U S A 85, 733–737 10.1073/pnas.85.3.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A. A., Mongelli V. C., Hopp H. E., del Vas M., Asurmendi S. (2007). A practical approach to the understanding and teaching of RNA silencing in plants. Electron J Biotechnol 10, 178–190 10.2225/vol10-issue2-fulltext-11 [DOI] [Google Scholar]
- Blevins T., Rajeswaran R., Shivaprasad P. V., Beknazariants D., Si-Ammour A., Park H. S., Vazquez F., Robertson D., Meins F., Jr & other authors (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34, 6233–6246 10.1093/nar/gkl886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville J. M., Sanfaçon H., Fütterer J., Hohn T. (1989). Posttranscriptional trans-activation in cauliflower mosaic virus. Cell 59, 1135–1143 10.1016/0092-8674(89)90769-1 [DOI] [PubMed] [Google Scholar]
- Bureau M., Leh V., Haas M., Geldreich A., Ryabova L., Yot P., Keller M. (2004). P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J Gen Virol 85, 3765–3775 10.1099/vir.0.80242-0 [DOI] [PubMed] [Google Scholar]
- Burgyán J., Havelda Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci 16, 265–272 10.1016/j.tplants.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Cecchini E., Gong Z. H., Geri C., Covey S. N., Milner J. J. (1997). Transgenic Arabidopsis lines expressing gene VI from cauliflower mosaic virus variants exhibit a range of symptom-like phenotypes and accumulate inclusion bodies. Mol Plant Microbe Interact 10, 1094–1101 10.1094/MPMI.1997.10.9.1094 [DOI] [PubMed] [Google Scholar]
- Cecchini E., Al Kaff N. S., Bannister A., Giannakou M. E., McCallum D. G., Maule A. J., Milner J. J., Covey S. N. (1998). Pathogenic interactions between variants of cauliflower mosaic virus and Arabidopsis thaliana. J Exp Bot 49, 731–737 [Google Scholar]
- Cerritelli S. M., Fedoroff O. Y., Reid B. R., Crouch R. J. (1998). A common 40 amino acid motif in eukaryotic RNases H1 and caulimovirus ORF VI proteins binds to duplex RNAs. Nucleic Acids Res 26, 1834–1840 10.1093/nar/26.7.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36 (Web Server issue), W197–W201 10.1093/nar/gkn238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D. C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553 10.1016/S0092-8674(00)80864-8 [DOI] [PubMed] [Google Scholar]
- De Tapia M., Himmelbach A., Hohn T. (1993). Molecular dissection of the cauliflower mosaic virus translation transactivator. EMBO J 12, 3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres M. W., Gregory B. D., Gao Z. H., Foreman A. W., Mlotshwa S., Ge X., Pruss G. J., Ecker J. R., Bowman L. H., Vance V. (2010). Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog 6, e1000729 10.1371/journal.ppat.1000729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S. H., Tada Y. & other authors (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geri C., Love A. J., Cecchini E., Barrett S. J., Laird J., Covey S. N., Milner J. J. (2004). Arabidopsis mutants that suppress the phenotype induced by transgene-mediated expression of cauliflower mosaic virus (CaMV) gene VI are less susceptible to CaMV-infection and show reduced ethylene sensitivity. Plant Mol Biol 56, 111–124 10.1007/s11103-004-2649-x [DOI] [PubMed] [Google Scholar]
- Haas M., Bureau M., Geldreich A., Yot P., Keller M. (2002). Cauliflower mosaic virus: still in the news. Mol Plant Pathol 3, 419–429 10.1046/j.1364-3703.2002.00136.x [DOI] [PubMed] [Google Scholar]
- Haas M., Geldreich A., Bureau M., Dupuis L., Leh V., Vetter G., Kobayashi K., Hohn T., Ryabova L. & other authors (2005). The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms. Plant Cell 17, 927–943 10.1105/tpc.104.029017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G., Azevedo J., Moissiard G., Geldreich A., Himber C., Bureau M., Fukuhara T., Keller M., Voinnet O. (2008). Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J 27, 2102–2112 10.1038/emboj.2008.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapiak M., Li Y. Z., Agama K., Swade S., Okenka G., Falk J., Khandekar S., Raikhy G., Anderson A. & other authors (2008). Cauliflower mosaic virus gene VI product N-terminus contains regions involved in resistance-breakage, self-association and interactions with movement protein. Virus Res 138, 119–129 10.1016/j.virusres.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P. A., Palanichelvam K., Yu W., Schoelz J. E., Nelson R. S. (2009). The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol 149, 1005–1016 10.1104/pp.108.131755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király L., Bourque J. E., Schoelz J. E. (1998). Temporal and spatial appearance of recombinant viruses formed between cauliflower mosaic virus (CaMV) and CaMV sequences present in transgenic Nicotiana bigelovii. Mol Plant Microbe Interact 11, 309–316 10.1094/MPMI.1998.11.4.309 [DOI] [Google Scholar]
- Kobayashi K., Hohn T. (2003). Dissection of cauliflower mosaic virus transactivator/viroplasmin reveals distinct essential functions in basic virus replication. J Virol 77, 8577–8583 10.1128/JVI.77.15.8577-8583.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Hohn T. (2004). The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol Plant Microbe Interact 17, 475–483 10.1094/MPMI.2004.17.5.475 [DOI] [PubMed] [Google Scholar]
- Latham J., Wilson A. (2013). Potentially dangerous virus gene hidden in commercial GM crops. Institute of Science in Society (Report). http://www.i-sis.org.uk/Hazardous_Virus_Gene_Discovered_in_GM_Crops.php [Google Scholar]
- Leh V., Yot P., Keller M. (2000). The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 266, 1–7 10.1006/viro.1999.0073 [DOI] [PubMed] [Google Scholar]
- Lewsey M. G., Murphy A. M., Maclean D., Dalchau N., Westwood J. H., Macaulay K., Bennett M. H., Moulin M., Hanke D. E. & other authors (2010). Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol Plant Microbe Interact 23, 835–845 10.1094/MPMI-23-7-0835 [DOI] [PubMed] [Google Scholar]
- Li Y. Z., Leisner S. M. (2002). Multiple domains within the Cauliflower mosaic virus gene VI product interact with the full-length protein. Mol Plant Microbe Interact 15, 1050–1057 10.1094/MPMI.2002.15.10.1050 [DOI] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J. O., Cohn M. M., Tung J., Sun H. Y., Kumar P., Baker B. (2012). MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A 109, 1790–1795 10.1073/pnas.1118282109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A. J., Yun B. W., Laval V., Loake G. J., Milner J. J. (2005). Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol 139, 935–948 10.1104/pp.105.066803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A. J., Laird J., Holt J., Hamilton A. J., Sadanandom A., Milner J. J. (2007). Cauliflower mosaic virus protein P6 is a suppressor of RNA silencing. J Gen Virol 88, 3439–3444 10.1099/vir.0.83090-0 [DOI] [PubMed] [Google Scholar]
- Love A. J., Geri C., Laird J., Carr C., Yun B. W., Loake G. J., Tada Y., Sadanandom A., Milner J. J. (2012). Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS ONE 7, e47535 10.1371/journal.pone.0047535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Rosas-Díaz T., Gusmaroli G., Luna A. P., Taconnat L., Deng X. W., Bejarano E. R. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23, 1014–1032 10.1105/tpc.110.080267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz L., Raikhy G., Leisner S. M. (2012). Cauliflower mosaic virus major inclusion body protein interacts with the aphid transmission factor, the virion-associated protein, and gene VII product. Virus Res 170, 150–153 10.1016/j.virusres.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G., Voinnet O. (2006). RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci U S A 103, 19593–19598 10.1073/pnas.0604627103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mukhtar M. S., Nishimura M. T., Dangl J. (2009). NPR1 in plant defense: it's not over ’til it’s turned over. Cell 137, 804–806 10.1016/j.cell.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Palanichelvam K., Schoelz J. E. (2002). A comparative analysis of the avirulence and translational transactivator functions of gene VI of Cauliflower mosaic virus. Virology 293, 225–233 10.1006/viro.2001.1293 [DOI] [PubMed] [Google Scholar]
- Palanichelvam K., Cole A. B., Shababi M., Schoelz J. E. (2000). Agroinfiltration of Cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana species. Mol Plant Microbe Interact 13, 1275–1279 10.1094/MPMI.2000.13.11.1275 [DOI] [PubMed] [Google Scholar]
- Park H. S., Himmelbach A., Browning K. S., Hohn T., Ryabova L. A. (2001). A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106, 723–733 10.1016/S0092-8674(01)00487-1 [DOI] [PubMed] [Google Scholar]
- Pei H., Ma N., Chen J., Zheng Y., Tian J., Li J., Zhang S., Fei Z., Gao J. (2013). Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS ONE 8, e64290 10.1371/journal.pone.0064290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevin N., du Jardin P. (2012). Possible consequences of the overlap between the CaMV 35S promoter regions in plant transformation vectors used and the viral gene VI in transgenic plants. GM Crops Food 3, 296–300 10.4161/gmcr.21406 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2011). MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16, 258–264 10.1016/j.tplants.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Cuperus J. T., Weigel D., Carrington J. C. (2009). Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol 12, 622–627 10.1016/j.pbi.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Ryabova L. A., Pooggin M. M., Hohn T. (2002). Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog Nucleic Acid Res Mol Biol 72, 1–39 10.1016/S0079-6603(02)72066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L., Park H. S., Hohn T. (2004). Control of translation reinitiation on the cauliflower mosaic virus (CaMV) polycistronic RNA. Biochem Soc Trans 32, 592–596 10.1042/BST0320592 [DOI] [PubMed] [Google Scholar]
- Ryabova L. A., Pooggin M. M., Hohn T. (2006). Translation reinitiation and leaky scanning in plant viruses. Virus Res 119, 52–62 10.1016/j.virusres.2005.10.017 [DOI] [PubMed] [Google Scholar]
- Sansregret R., Dufour V., Langlois M., Daayf F., Dunoyer P., Voinnet O., Bouarab K. (2013). Extreme resistance as a host counter-counter defense against viral suppression of RNA silencing. PLoS Pathog 9, e1003435 10.1371/journal.ppat.1003435 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schoelz J. E., Shepherd R. J. (1988). Host range control of cauliflower mosaic virus. Virology 162, 30–37 10.1016/0042-6822(88)90391-1 [DOI] [PubMed] [Google Scholar]
- Schoelz J., Shepherd R. J., Daubert S. (1986). Region VI of cauliflower mosaic virus encodes a host range determinant. Mol Cell Biol 6, 2632–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6, e230 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F., Vaistij F. E., Jones L., Baulcombe D. C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138, 1842–1852 10.1104/pp.105.063537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P. V., Rajeswaran R., Blevins T., Schoelz J., Meins F., Jr, Hohn T., Pooggin M. M. (2008). The CaMV transactivator/viroplasmin interferes with RDR6-dependent trans-acting and secondary siRNA pathways in Arabidopsis. Nucleic Acids Res 36, 5896–5909 10.1093/nar/gkn590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. A. (2007). Interactions between cauliflower mosaic virus and auxin signalling. PhD Thesis, University of Glasgow, UK [Google Scholar]
- Stratford R., Covey S. N. (1989). Segregation of cauliflower mosaic virus symptom genetic determinants. Virology 172, 451–459 10.1016/0042-6822(89)90187-6 [DOI] [PubMed] [Google Scholar]
- Tsuge S., Kobayashi K., Nakayashiki H., Okuno T., Furusawa I. (1994). Replication of cauliflower mosaic virus ORF 1 mutants in turnip protoplasts. Ann Phytopathological Soc Jpn 60, 27–35 10.3186/jjphytopath.60.27 [DOI] [Google Scholar]
- Vain P., Harvey A., Worland B., Ross S., Snape J. W., Lonsdale D. (2004). The effect of additional virulence genes on transformation efficiency, transgene integration and expression in rice plants using the pGreen/pSoup dual binary vector system. Transgenic Res 13, 593–603 10.1007/s11248-004-2808-5 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Legrand S., Windels D. (2010). The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci 15, 337–345 10.1016/j.tplants.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33, 949–956 10.1046/j.1365-313X.2003.01676.x [DOI] [PubMed] [Google Scholar]
- Volko S. M., Boller T., Ausubel F. M. (1998). Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics 149, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009). Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., De Luca V., Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1, 639–647 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yang J. Y., Iwasaki M., Machida C., Machida Y., Zhou X., Chua N. H. (2008). βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22, 2564–2577 10.1101/gad.1682208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Jin Z., Xie D. (2012). Global analysis of non-coding small RNAs in Arabidopsis in response to jasmonate treatment by deep sequencing technology. J Integr Plant Biol 54, 73–86 10.1111/j.1744-7909.2012.01098.x [DOI] [PubMed] [Google Scholar]
- Zijlstra C., Hohn T. (1992). Cauliflower mosaic virus gene VI controls translation from dicistronic expression units in transgenic Arabidopsis plants. Plant Cell 4, 1471–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra C., Schärer-Hernández N., Gal S., Hohn T. (1996). Arabidopsis thaliana expressing the cauliflower mosaic virus ORF VI transgene has a late flowering phenotype. Virus Genes 13, 5–17 10.1007/BF00576974 [DOI] [PubMed] [Google Scholar]