Abstract
The mitochondrial DNA (mtDNA) 4977-bp deletion (ΔmtDNA4977 mutation) is one of the most frequently observed mtDNA mutations in human tissues, and may play a role in carcinogenesis. Only a few studies have evaluated ΔmtDNA4977 mutation in breast cancer tissue, and the findings have been inconsistent, which may be due to methodological differences. In this study, we developed a quantitative real-time PCR assay to assess the level of the ΔmtDNA4977 mutation in tumor tissue samples from 55 primary breast cancer patients and 21 patients with benign breast disease (BBD). The ΔmtDNA4977 mutation was detected in all of the samples with levels varying from 0.000149% to 7.0%. The ΔmtDNA4977 mutation levels were lower in tumor tissues than in adjacent normal tissues in both breast cancer and BBD subjects. The differences, however, were not statistically significant. No significant difference between breast cancer and BBD patients was found in the ΔmtDNA4977 mutation levels of tumor tissues and adjacent normal tissues. The ΔmtDNA4977 mutation levels were not significantly associated with clinicopathological characteristics (age, histology, tumor stage, and ER/PR status) in breast cancer or BBD patients. These results do not support the notion that the mitochondrial DNA 4977-bp deletion plays a major role in breast carcinogenesis.
Keywords: benign breast disease, breast cancer, mtDNA 4977 mutation, quantitative real-time PCR assay
INTRODUCTION
The development of cancer involves the accumulation of various genetic alterations, which are present both in the mitochondrial and nuclear genomes (1). Human mitochondrial DNA (mtDNA) is a circular molecule consisting of 16,571 bp encoding 2 rRNAs, 22 tRNAs, and 13 polypeptides (2, 3). Mitochondria play a critical role in energy production and oxidative phosphorylation. MtDNA is particularly susceptible to damage by environmental carcinogens because it contains no introns, has no protective histones or non-histone proteins, and is exposed continuously to endogenous reactive oxygen species (ROS) (4, 5). Therefore, mtDNA may serve as a potential sensor for cellular DNA damage and marker for cancer development. Moreover, susceptibility to mtDNA mutations may be particularly important in breast cancer, because the normal metabolism of estradiol through redox-cycling intermediates may generate ROS and lead to oxidative injury that facilitates neoplastic transformation in the breast (6).
The mitochondrial 4977-bp deletion (ΔmtDNA4977 mutation), also known as the common deletion, is one of the most frequently observed mtDNA mutations. This deletion occurs between nucleotides 8,470 and 13,447 of human mtDNA and spans five tRNA genes and seven genes encoding subunits of cytochrome c oxidase, complex I, and ATPases (4). The ΔmtDNA4977 mutation may contribute to respiratory defects in many types of human malignancy such as colorectal, liver, lung, and prostate cancers, and may be associated with tumor progression. To date, only few studies have evaluated the association between the ΔmtDNA4977 mutation and breast cancer risk, and the findings from these studies have been inconsistent (4, 7–9). The inconsistency in previous studies may be due, in part, to the use of non-quantitative detection methods and/or failure in quantitative PCR strategies to identify, and disqualify from use, pairs of primers which will also efficiently co-amplify nuclear DNA. Further, none of these studies have assayed the ΔmtDNA4977 mutation in benign breast disease (BBD).
In this study, we developed a new absolute quantitative PCR method, which is more sensitive than other methods previously used. A substantial benefit of using this mtDNA-specific assay design lies in significantly-decreased risk of quantitating genomic DNA normally present in total-DNA preparations, and without the need for exhaustive mtDNA-purification methods. We applied this method to quantify the level of the ΔmtDNA4977 mutation among 55 patients diagnosed with breast cancer and 21 patients diagnosed with BBD.
MATERIALS AND METHODS
Study subjects and control samples
This study examined surgically-resected tumor tissues and adjacent normal tissues of 55 patients with primary breast cancer and 21 patients with BBD, who are a subset of patients recruited as part of the Shanghai Breast Cancer Study (10). These patients were diagnosed with breast cancer or BBD between 1996 and 1998 and were identified through a network of major hospitals that treat over 80% of breast cancer patients in urban Shanghai. All patients provided written, informed consent to participate in the study, and the study protocols were approved by the Institutional Review Boards of all institutes involved in the study. Medical charts were reviewed using a standard protocol to obtain information on cancer treatment, clinical stage, and cancer characteristics, such as estrogen and progesterone receptor status. Two senior pathologists reviewed pathology slides to confirm the diagnosis for breast cancer or BBD. BBDs were classified based on published criteria developed by Page and colleagues (11). During surgery, tumor tissue samples were obtained from the tumor and the adjacent normal tissue samples were obtained from the distal edge of the resection. These samples were snap-frozen in liquid nitrogen as soon as possible, typically within 10 minutes. Samples were stored at −70°C until the relevant assays were performed.
Total DNA was extracted using TRIzol® Reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The concentration of DNA was measured with a TBS-380 Fluorometer (Turner Biosystems, Sunnyvale, CA) using the DNA-specific binding dye Hoechst 33258 (Sigma, St Louis, MO) to stain the DNA. Four reference mitochondrial DNA samples (SHE049i, SHE187i, SHE049v, and SHE145v), with known (ΔmtDNA4977 mutation):(total mtDNA) ratios were provided by Dr. Konrad Huppi (Cancer Prevention Studies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD). SRM 2392, a human mtDNA standard reference material, was obtained from the National Institute of Standards and Technology (Gaithersburg, MD) (12).
Plasmid constructions for the standard curve
To identify two target regions in the mtDNA genome (a conserved control region present in 100% of all normal mtDNA and a mutated sequence produced only by the ΔmtDNA4977 mutation), published sequences for mtDNA (GenBank Accession No. NC001807 and the MITOMAP database (http://www.mitomap.org)) were used. We conducted BLASTN searches against dbEST and nr (the nonredundant set of Genbank, EMBL and DDBJ database sequences) to confirm that the sequences chosen for the primers targeting both regions should be single-copy in the mtDNA genome and not conserved within human genomic DNA sequences recorded in those databases. The following primers were used to amplify DNA fragments representing the normal mtDNA (mtDNAnormal) or the ΔmtDNA4977 mutation respectively: mtDNAnormal forward primer 5′-acgccataaaactcttcaccaa -3′, reverse primer 5′-ggttcggttggtctcgtcta -3′; ΔmtDNA4977 mutation forward primer 5′-accactttcaccgctacacg -3′, reverse primer 5′-agggaggtagcgatgagagt -3′. The putative amplicon sequences for both the wild-type and mutated targets had been previously screened by BLASTn to eliminate any redundancies in human genome DNA and screened against both the NCBI SNP database and the Japanese Mitochondrial SNP Database (13, 14). The PCR products of mtDNAnormal and the ΔmtDNA4977 mutation yield 437 bp and 484 bp amplicons, respectively.
The standard PCR reaction was carried out in the following mixture: 1× PCR Buffer; 4 mmol/l MgCl2; 200 mmol/L dATP, dCTP, dGTP and dTTP; 200 nmol/L each primers; and 1 U of HotStar Taq DNA polymerase (Qiagen, Valencia, CA). Five ng of mtDNA (SRM 2392 used for the mtDNAnormal, SHE049i for the ΔmtDNA4977 mutation) in 2μl H2O were added to 18 μl of PCR mixture. Thermocycling conditions used were: initial denaturation and hot start at 95°C for 15 min; 40 cycles consisting of 30 s at 92°C, 30 s at 60°C and 30 s at 72°C, followed by a final extension at 72°C for 10 min. The PCR product was purified using the Wizard PCR Preps DNA Purification System (Promega, San Luis Obispo, CA) according to the manufacturer’s instructions. The purified PCR product was cloned into the pCR 2.1-TOPO plasmid (Invitrogen, Carlsbad, CA) in accordance with manufacturer’s instructions. Blue-white screening was performed before plasmids were isolated. Plasmids were analyzed by EcoRI restriction and further confirmed by BigDye Terminator Chemistry (Applied Biosystems, Foster, CA), performed by GenePASS, Inc. (Nashville, TN). Confirmed clones were renamed pCR437 for the mtDNAnormal and pCR484 for the ΔmtDNA4977 mutation. The recombinant plasmid DNA was isolated and purified using the QIAprep Plasmid Miniprep Kit (Qiagen, Valencia, CA). The concentration of plasmid was measured by Hoechst 33258 (Sigma, St Louis, MO) on a TBS-380 Fluorometer (Turner Biosystems, Sunnyvale, CA), and the copy number calculated according to the known molecular weight of the plasmid. Serial dilutions of each standard were made in the range of 101–107 copies per 2 μl for long-term use and storage (15, 16).
Real-time quantitative PCR
Absolute DNA quantification was performed using the standard curve method (16). Initial efforts to validate a relative quantitation approach (ΔΔCt) for this assay, as previously described (17), were unsuccessful, due to inefficacy of a primer starvation strategy to adequately modulate differences in amplification reaction efficiency (data not shown). The primers and probes for the mtDNAnormal or the ΔmtDNA4977 mutation were designed by and obtained from Applied Biosystems (Foster, CA) (Table 1). Submitted sequences for both the wild-type and mutated targets had been previously screened by BLASTn to eliminate any redundancies in human genome DNA and were screened against both the NCBI SNP database and the Japanese Mitochondrial SNP Database (13, 14). Quantitative real-time PCR was performed in a 96-well optical plate on an ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems), and the data were analyzed using SDS (Ver. 2.2) software. A total reaction volume of 50 μl containing 10 ng DNA template, 25 μl 1x TaqMan Universal PCR Master Mix (without UNG), 5 μl 5 μM primers, 1 μl 10 μM probes. The thermal cycling conditions were as follows: 95°C for 10 minutes to activate the AmpliTaq Gold enzyme, followed by 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. Every sample was tested in triplicate. These positive controls were derived from Centre d’Etude du Polymorphisme Human (CEPH) 1347-02 DNA, extracted from a single human cell line, which was previously evaluated in this assay design at 2 ng, 4 ng, 6 ng, 8 ng, 10 ng, 12 ng and 14 ng per reaction. In that trial, DNA amount as low as 10 ng/reaction were found to be consistently detectable. Thus 10 ng DNA per reaction was used in the assay for all samples.
Table 1.
Primer and probe sequences for quantitative real time PCR
| Name | Primer sequence (5′-3′) | Probe sequence (5′-3′) |
|---|---|---|
| mtDNAnormal | Forward Seq: CCT CTC CAC CCT TAT CAC AAC AC Reverse Seq: TCA TAT TAT GGC CAA GGG TCA T |
FAM AGA ACA CCT CTG ATT ACT C MGBNFQ |
| ΔmtDNA4977 mutation | Forward Seq: CTA AAT ACT ACC GTA TGG CCC ACC Reverse Seq: CAG CTA ATG CTA GGC TGC CAA TGG |
FAM AAC ACA AAC TAC CAC CTA CC MGBNFQ |
In each PCR run, standard reference curves containing serial dilutions of the corresponding plasmid clones with known amounts of input copy number were included. Data generated by these standard plasmids allowed the generation of two standard curves showing the absolute number of copies of the mtDNAnormal or the ΔmtDNA4977 mutation. The threshold cycle (Ct) is the cycle at which sufficient new amplification product is detectable as ambient fluorescence from non-quenched probe. The higher the initial amount of available DNA template, the faster the detection in the PCR process and the lower the Ct value. The Ct values of the samples could then easily be converted to the number of DNA copies per reaction, by integrating Ct values for each sample well onto the reference dilution curve consisting of the respective standard reference plasmid at known concentrations (15, 16). CEPH 1347-02 total DNA at fixed concentration was used in every assay plate amplified, as a positive PCR quality control.
Statistical Analysis
The ΔmtDNA4977 mutation levels were calculated as the copy numbers of the ΔmtDNA4977 mutation molecules per total mtDNA molecules (ΔmtDNA4977 mutation/(mtDNAnormal + ΔmtDNA4977 mutation)). The average mutation levels were estimated with the medians (25th, 75th percentiles). The differences of the mutation levels between tumor tissues and adjacent non-tumor tissues from the same individuals were evaluated using the Wilcoxon signed rank test, and the difference of the mutation levels between breast cancer cases and benign breast disease controls, as well as the differences between different histology types/stages of breast cancer and benign breast disease were evaluated using the Wilcoxon rank sum test.
RESULTS
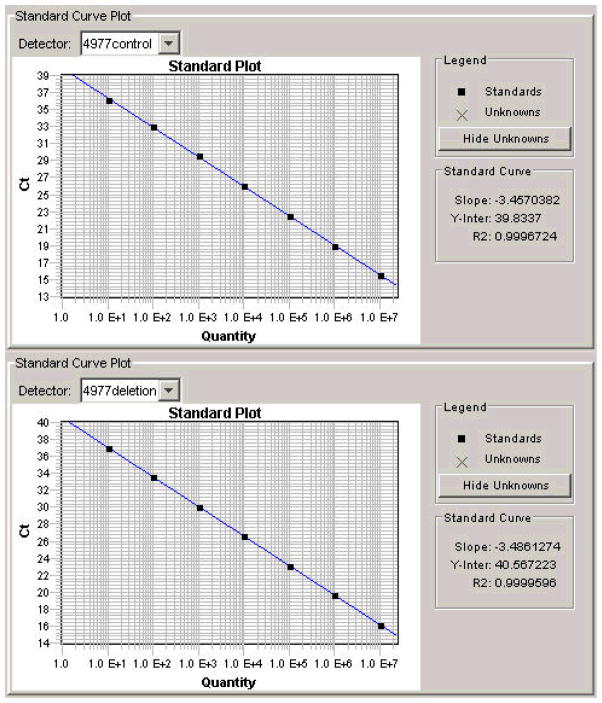
We first determined whether it was possible to reliably detect and quantify the percentage of copies of the mitochondrial genome harboring the ΔmtDNA4977 mutation. Calibration curves for the mtDNAnormal and the ΔmtDNA4977 mutation were constructed. There was excellent correlation between the cycle number and mtDNA copy number, across six log-orders of magnitude from 10 copies to 107 copies, with correlation coefficients of 0.9997 and 0.9999 for the mtDNAnormal and the ΔmtDNA4977 mutation, respectively (Figure 1). Similar results were observed using different amounts of CEPH1347-02 total DNA (from 10 to 14 ng DNA per reaction). To further validate the method, we prepared combinations of 4 different reference samples containing known amounts of ΔmtDNA4977 mutation to obtain 12 samples containing various proportions of mutant mtDNA. These 12 validation samples were then quantified for levels of ΔmtDNA4977 mutation using our quantitative PCR method. The data from this experiment were completely consistent with the predicted values. As few as 10 copies of the target mtDNA was detectable by this method.
Figure 1.
Sensitivity of real-time PCR to template copy number. Threshold cycle (i.e. Ct, vertical axis) at decreasing concentrations of template DNA (horizontal axis) for the mtDNAtotal (A) or the ΔmtDNA4977 mutation (B) are shown. There is a linear relationship between template concentration and the threshold cycle number (Ct) for both amplifications.
The demographic characteristics of the breast cancer cases and BBD controls are shown in Table 2. Compared to BBD subjects in this study, breast cancer cases were older at time of interview, more likely to be post-menopausal, and had a higher waist-to-hip ratio.
Table 2.
Selected demographic characteristics and known breast cancer risk factors in patients diagnosed with breast cancer or benign breast diseases (BBD)
| Breast cancer (n = 55) | BBD (n = 21) | p-valuea | |
|---|---|---|---|
| Age (mean) | 46.9 | 42.8 | 0.034 |
| Education>=high school (%) | 38.2 | 57.1 | 0.134 |
| Age at menarche (mean) | 14.2 | 14.7 | 0.184 |
| Post-menopause (%) | 32.7 | 14.3 | 0.108 |
| Body mass index (mean) | 24.4 | 22.4 | 0.009 |
| Waist-to-hip ratio (mean) | 0.79 | 0.79 | 0.951 |
| Regular physical activity (%) | 20.0 | 9.5 | 0.278 |
| Family history of BC among first degree relatives | 7.3 | 4.8 | 0.693 |
The p-values were derived from t-test for continuous variables and Chi-squared test for categorical variables.
The percentage of ΔmtDNA4977 mutation for samples in our study was detectable in a range from 0.000149% to 7% of all DNA present per sample. Table 3 compares the ΔmtDNA4977 mutation levels in tumor tissues and adjacent non-tumor tissues. In primary breast cancer subjects, the ΔmtDNA4977 mutation levels were lower in tumor tissue than in adjacent non-tumor tissue, however, the differences were not statistically significant (median 4.28 vs 7.60, p=0.093). This was also true for benign tumor tissues and their corresponding adjacent tissues (median 3.89 vs 5.07, p=0.448) (Table 3). No significant difference between breast cancer and BBD patients was found in the ΔmtDNA4977 mutation levels of tumor and adjacent normal tissues (Table 3).
Table 3.
ΔmtDNA4977 mutation levels in tumor and tumor-adjacent normal tissue from patients diagnosed with breast cancer or benign breast diseases (BBD)
| Median (25th, 75th percentile)a | p-valueb | ||
|---|---|---|---|
|
| |||
| Tumor tissue | Adjacent normal tissue | ||
| Breast cancer patients (n = 55) | 4.28 (2.05, 25.50) | 7.60 (1.43, 47.10) | 0.093 |
| BBD patients (n = 21) | 3.89 (1.01, 21.10) | 5.07 (1.57, 33.70) | 0.448 |
| p-valuec | 0.486 | 0.844 | |
The ratios of ΔmtDNA4977/mtDNATotal have been multiplied by 100,000
p-value derived from Wilcoxon signed rank test for the comparison between tumor tissues and adjacent normal tissues.
p-value derived from Wilcoxon rank sum test for the comparison between breast cancer and BBD.
Table 4 compares the ΔmtDNA4977 mutation levels of tumor and adjacent normal tissue across different clinicopathological characteristics (histology, tumor stage) of breast cancer or BBD patients. Compared with breast cancer cases with early stage disease, breast cancer cases with advanced stage disease had lower ΔmtDNA4977 mutation levels in both tumor and adjacent normal tissues. The difference, however, was not statistically significant. In BBD patients, on the other hand, ΔmtDNA4977 mutation levels in both tumor and adjacent tissues were higher in those who were non-proliferative than those who were proliferative. However, the differences did not reach statistical significance. In addition, we found no association of the ΔmtDNA4977 mutation with age at diagnosis or ER/PR status (data not shown)
Table 4.
Histology, tumor stage and the ΔmtDNA4977 mutation (Median, 25th and 75th percentile) of tumor and adjacent tissue in patients with breast cancer and benign breast disease
| Tumor tissue
|
Adjacent tissue
|
||
|---|---|---|---|
| No. of patients | Median (25th, 75th percentile) | Median (25th, 75th percentile) | |
| Benign breast diseases | |||
| Non-proliferative lesions | 9 | 6.7 (2.0, 7.7) | 14.6 (5.1, 67.4) |
| Proliferative lesions | 9 | 1.3 (0.9, 21.1) | 2.9 (1.1, 15.9) |
| p-valuea | 0.723 | 0.185 | |
| Breast cancer (by TNM stage) | |||
| I, IIa | 30 | 5.1(1.9, 28.4) | 12.3(2.0, 77.1) |
| IIb, III | 25 | 4.0(2.1 13.7) | 4.6(1.2, 33.5) |
| p-value a | 0.966 | 0.343 | |
p-value derived from Wilcoxon rank sum test
DISCUSSION
In this paper, we describe a novel, sensitive, and convenient real-time PCR assay capable of identifying the ΔmtDNA4977 mutation. The wide detection range (from 10 to 107 copies) and continuous scale of quantification of this method suggests that this is a sensitive, specific, reproducible, rapid, and convenient technique for the detection of the ΔmtDNA4977 mutation without post-PCR procedures. Such an assay could be a valuable tool for performing rapid assessment of the mtDNA4977 mutation. Several methods have been used previously to quantify the level of mtDNA4977 mutation. Long range PCR, primer-shift PCR and semi-quantitative PCR are time-consuming and the results are sometimes not reproducible (18–20). Furthermore, these PCR methods use an endpoint determination that may not be truly quantitative because of plateau effects (21). There have been some reports demonstrating the advantages of using the real-time PCR method and its potential clinical value for the ΔmtDNA4977 mutation (8, 18–20, 22). However, only two of these reports used the standard curve method, which is believed to be more reliable and sensitive (18, 19). Compared to these previously described real-time PCR methods, the target primer sequence regions we identified are single-copy in the mtDNA genome and not homologous to any genomic DNA sequences. Therefore, this method has less potential for PCR contamination during performance, without amplification of any sequence repeated within genomic DNA.
Proposed mechanisms for the role of the ΔmtDNA4977 mutation in carcinogenesis remain unclear. The ΔmtDNA4977 mutation affects genes encoding 7 polypeptide components of the mitochondrial respiratory chain and 5 of the 22 tRNAs necessary for mitochondrial protein synthesis (23). Thus, the mutated ΔmtDNA4977 may represent a strong functional disadvantage, possibly thereby repressing the growth of cancer cells harboring deleted mtDNA. It was recently demonstrated in transmitochondrial cybrid cells that the common mtDNA deletion sensitized the cells to apoptosis at low heteroplasmy levels (8, 24). A significant increased proportion of the ΔmtDNA4977 mutation has been reported in various type of cancer, including nasopharyngeal, esophageal cancer, stomach, thyroid, salivary glands, hepatocellular and oral cancers (5, 18, 25–28). However, in more recent studies, a decreased proportion of the ΔmtDNA4977 mutation in tumor tissue as compared with corresponding non-tumorous tissue has been observed in gastric, hepatocellular, oral, and skin cancers (29–32). This phenomenon has been explained as being either the result of a dilution effect due to rapid cytoplasmic division or from the result of a selective effect via an unknown mechanism by which cancer cells harboring mtDNA with the large-scale deletion are eliminated because of apoptosis or selective purification during growth (30).
The role of ΔmtDNA4977 mutation has not been adequately investigated in relation to breast cancer risk, and existing studies have reported conflicting results. The ΔmtDNA4977 mutation was first reported as occurring in one out of seven breast cancer samples (7). Later, three studies reported a different pattern of ΔmtDNA4977 mutation in tumors compared to normal tissue. In a study conducted using samples from 39 breast cancer patients and 23 women without breast cancer, the ΔmtDNA4977 mutation was not specific to breast cancer and was present in 17% of normal breast tissues, 31% of adjacent normal tissues from a cancerous breast and 44% of breast cancers (9). Two more recent studies found lower levels of the ΔmtDNA4977 mutation in breast tumor tissues compared to normal control tissue. In a study conducted among 17 Brazilian women, lower levels of ΔmtDNA4977 mutation were observed in breast cancer tissue than in either the normal control tissue or adjacent normal tissue of breast cancer patients, suggesting that cancer cells are essentially free of this mutation (4). In the second study, a hospital-based study of 60 Taiwanese women with breast cancer, the ΔmtDNA4977 mutation was detected in 28 non-tumorous breast tissue samples (47%) and only three of the breast cancer samples (5%) (8). In our study, we found the ΔmtDNA4977 mutation to be ubiquitous in both tumor and non-tumor tissues. In primary breast cancer subjects, the ΔmtDNA4977 mutation levels were lower in tumor tissues than in adjacent non-tumor tissues, however, the differences were not statistically significant. This pattern of mutation levels was also found in benign tumor tissues and their corresponding adjacent tissues. No significant difference was found between breast cancer and BBD patients for the ΔmtDNA4977 mutation levels of tumor and adjacent normal tissues. We believe the higher detection rate for the ΔmtDNA4977 mutation in our study is a result of the increased sensitivity and specificity of the quantitative assay we employed, in contrast to the methods used in previous studies (long PCR or semi-quantitative PCR methods). Our finding is also biologically plausible, due to the fact that cancer cells are metabolically adapted for rapid growth and proliferation under conditions of low pH and oxygen tension, solid cancer cells generate energy by glycolysis in strong preference to oxidative phosphorylation (33–35). Furthermore, previous in vitro studies have suggested that mitochondrial protein synthesis is defective and the activity of cytochrome oxidase decreases when the proportion of the ΔmtDNA4977 mutation was greater than 60% in the cancer cell (23). The amount of the ΔmtDNA4977 mutation that accumulates may not be sufficient, by itself, to cause a significant defect or dysfunction in breast cancer cells.
The current study has several strengths. We included both tumor tissue and adjacent normal tissue samples from patients diagnosed with breast cancer or BBD, which enabled a systematic evaluation of the ΔmtDNA4977 mutation based on the types of tissue and disease. To our knowledge, no study has examined the ΔmtDNA4977 mutation content in breast tumor tissue or adjacent normal tissue from BBD patients. Since it is difficult to obtain breast tissue from women without any breast disease, we were unable to evaluate ΔmtDNA4977 mutation in normal individuals. Because of a relatively small sample size, we are unable to analyze the relationship of the ΔmtDNA4977 mutation with outcomes of breast cancer.
In summary, this study describes the development of a rapid, sensitive, and practical real-time PCR method to quantify the ΔmtDNA4977 mutation in tissue samples. Our results show that the ΔmtDNA4977 mutation was ubiquitous in both tumor and non-tumor tissues of both breast cancer and BBD subjects. Our results do not support the notion that the ΔmtDNA4977 mutation plays a major role in breast carcinogenesis.
Acknowledgments
Financial Support: This research was supported by U.S. Department of Defense grant DAMD17-02-1-0603 and National Cancer Institute grant R01 CA064277.
We thank Dr. Konrad Huppi (Cancer Prevention Studies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD) for kindly providing mitochondrial DNA samples, Bethanie Hull and Brandy Venuti for technical assistance in the preparation of this article, and all of the study participants and research staff of the Shanghai Breast Cancer Study for their support.
Reference List
- 1.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–152. doi: 10.1093/carcin/bgh282. [DOI] [PubMed] [Google Scholar]
- 2.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 3.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 4.Dani MA, Dani SU, Lima SP, Martinez A, Rossi BM, Soares F, Zago MA, Simpson AJ. Less DeltamtDNA4977 than normal in various types of tumors suggests that cancer cells are essentially free of this mutation. Genet Mol Res. 2004;3:395–409. [PubMed] [Google Scholar]
- 5.Tan DJ, Chang J, Liu LL, Bai RK, Wang YF, Yeh KT, Wong LJ. Significance of somatic mutations and content alteration of mitochondrial DNA in esophageal cancer, BMC. Cancer. 2006;6:93. doi: 10.1186/1471-2407-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy D, Liehr JG. Estrogen. DNA damage and mutations. Mutat Res. 1999;424:107–115. doi: 10.1016/s0027-5107(99)00012-3. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi MS, Bianchi NO, Bailliet G. Mitochondrial DNA mutations in normal and tumor tissues from breast cancer patients. Cytogenet Cell Genet. 1995;71:99–103. doi: 10.1159/000134072. [DOI] [PubMed] [Google Scholar]
- 8.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer, Genes Chromosomes. Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W, Qin W, Sauter ER. Large-scale mitochondrial DNA deletion mutations and nuclear genome instability in human breast cancer. Cancer Detect Prev. 2004;28:119–126. doi: 10.1016/j.cdp.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Gao YT, Shu XO, Dai Q, Potter JD, Brinton LA, Wen W, Sellers TA, Kushi LH, Ruan Z, Bostick RM, Jin F, Zheng W. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Schnitt S, Connolly J. Pathology of benign breast disorders. In: Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Diseases of the breast. Lippincott Williams & Wilkins; Philadelphia: 1999. pp. 75–93. [Google Scholar]
- 12.Levin BC, Holland KA, Hancock DK, Coble M, Parsons TJ, Kienker LJ, Williams DW, Jones M, Richie KL. Comparison of the complete mtDNA genome sequences of human cell lines--HL-60 and GM10742A--from individuals with pro-myelocytic leukemia and leber hereditary optic neuropathy respectively, and the inclusion of HL-60 in the NIST human mitochondrial DNA standard reference material--SRM 2392-I. Mitochondrion. 2003;2:387–400. doi: 10.1016/S1567-7249(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 13.Hirakawa M, Tanaka T, Hashimoto Y, Kuroda M, Takagi T, Nakamura Y. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30:158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pushnova EA, Geier M, Zhu YS. An easy and accurate agarose gel assay for quantitation of bacterial plasmid copy numbers. Anal Biochem. 2000;284:70–76. doi: 10.1006/abio.2000.4668. [DOI] [PubMed] [Google Scholar]
- 16.Chuanzhong Y, Ming G, Fanglin Z, Haijiao C, Zhen L, Shiping C, YongKang Z. Real-time quantitative reverse transcription-PCR assay for renal cell carcinoma-associated antigen G250. Clin Chim Acta. 2002;318:33–40. doi: 10.1016/s0009-8981(01)00799-9. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Shieh DB, Chou WP, Wei YH, Wong TY, Jin YT. Mitochondrial DNA 4,977-bp deletion in paired oral cancer and precancerous lesions revealed by laser microdissection and real-time quantitative PCR. Ann N Y Acad Sci. 2004;1011:154–167. doi: 10.1007/978-3-662-41088-2_16. [DOI] [PubMed] [Google Scholar]
- 19.Pogozelski WK, Hamel CJ, Woeller CF, Jackson WE, Zullo SJ, Fischel-Ghodsian N, Blakely WF. Quantification of total mitochondrial DNA and the 4977-bp common deletion in Pearson’s syndrome lymphoblasts using a fluorogenic 5′-nuclease (TaqMan) real-time polymerase chain reaction assay and plasmid external calibration standards. Mitochondrion. 2003;2:415–427. doi: 10.1016/S1567-7249(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 20.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 2002;30:e68. doi: 10.1093/nar/gnf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicklas JA, Brooks EM, Hunter TC, Single R, Branda RF. Development of a quantitative PCR (TaqMan) assay for relative mitochondrial DNA copy number and the common mitochondrial DNA deletion in the rat. Environ Mol Mutagen. 2004;44:313–320. doi: 10.1002/em.20050. [DOI] [PubMed] [Google Scholar]
- 22.Kakiuchi C, Ishiwata M, Kametani M, Nelson C, Iwamoto K, Kato T. Quantitative analysis of mitochondrial DNA deletions in the brains of patients with bipolar disorder and schizophrenia. Int J Neuropsychopharmacol. 2005;8:515–522. doi: 10.1017/S1461145705005213. [DOI] [PubMed] [Google Scholar]
- 23.Porteous WK, James AM, Sheard PW, Porteous CM, Packer MA, Hyslop SJ, Melton JV, Pang CY, Wei YH, Murphy MP. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. Eur J Biochem. 1998;257:192–201. doi: 10.1046/j.1432-1327.1998.2570192.x. [DOI] [PubMed] [Google Scholar]
- 24.Schoeler S, Szibor R, Gellerich FN, Wartmann T, Mawrin C, Dietzmann K, Kirches E. Mitochondrial DNA deletions sensitize cells to apoptosis at low heteroplasmy levels. Biochem Biophys Res Commun. 2005;332:43–49. doi: 10.1016/j.bbrc.2005.04.086. [DOI] [PubMed] [Google Scholar]
- 25.Shao JY, Gao HY, Li YH, Zhang Y, Lu YY, Zeng YX. Quantitative detection of common deletion of mitochondrial DNA in hepatocellular carcinoma and hepatocellular nodular hyperplasia. World J Gastroenterol. 2004;10:1560–1564. doi: 10.3748/wjg.v10.i11.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao JY, Li YH, Gao HY, Mai HQ, Zhang Y, Guo X, Zeng YX. High frequency of common deletion (4981 bp) in mitochondrial DNA in nasopharyngeal carcinoma and its correlation with patient age and clinical stages. Cancer Biol Ther. 2004;3:1270–1274. doi: 10.4161/cbt.3.12.1243. [DOI] [PubMed] [Google Scholar]
- 27.Rogounovitch TI, Saenko VA, Shimizu-Yoshida Y, Abrosimov AY, Lushnikov EF, Roumiantsev PO, Ohtsuru A, Namba H, Tsyb AF, Yamashita S. Large deletions in mitochondrial DNA in radiation-associated human thyroid tumors. Cancer Res. 2002;62:7031–7041. [PubMed] [Google Scholar]
- 28.Lewis PD, Baxter P, Paul GA, Parry JM, Skibinski DO. Detection of damage to the mitochondrial genome in the oncocytic cells of Warthin’s tumour. J Pathol. 2000;191:274–281. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH634>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, Wei YH, Liu TY, Chi CW. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90:2390–2396. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer, Genes Chromosomes. Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 31.Lee HC, Yin PH, Yu TN, Chang YD, Hsu WC, Kao SY, Chi CW, Liu TY, Wei YH. Accumulation of mitochondrial DNA deletions in human oral tissues -- effects of betel quid chewing and oral cancer. Mutat Res. 2001;493:67–74. doi: 10.1016/s1383-5718(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 32.Yang JH, Lee HC, Chung JG, Wei YH. Mitochondrial DNA mutations in light-associated skin tumors. Anticancer Res. 2004;24:1753–1758. [PubMed] [Google Scholar]
- 33.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 34.Jia L, Liu KZ, Newland AC, Mantsch HH, Kelsey SM. Pgp-positive leukaemic cells have increased mtDNA but no increased rate of proliferation. Br J Haematol. 1999;107:861–869. doi: 10.1046/j.1365-2141.1999.01771.x. [DOI] [PubMed] [Google Scholar]
- 35.Han YC, Kong WJ, Zhang S, Wang YJ, Wang Y, Chen X. Mutation of mitochondrial DNA 4977 bp deletion in laryngeal squamous cell cancer. Ai Zheng. 2004;23:1297–1301. [PubMed] [Google Scholar]