Abstract
Objectives
Despite improvements in the management of ovarian cancer patients over the last 30 years, there has been only a minimal improvement in overall survival. While targeted therapeutic approaches for the treatment of cancer have evolved, major challenges in ovarian cancer research persist, including the identification of predictive biomarkers with clinical relevance, so that empirical drug selection can be avoided. In this article, we review published genomic analysis studies including data generated in our laboratory and how they have been incorporated into modern clinical trials in a rational and effective way.
Methods
Multiple published genomic analysis studies were collected for review and discussion with emphasis on their potential clinical applicability.
Results
Genomic analysis has been shown to be a powerful tool to identify dysregulated genes, aberrantly activated pathways and to uncover uniqueness of subclasses of ovarian tumors. The application of this technology has provided a solid molecular basis for different clinical behaviors associated with tumor histology and grade. Genomic signatures have been obtained to predict clinical end points for patients with cancer, including response rates, progression-free survival, and overall survival. In addition, genomic analysis has provided opportunities to identify biomarkers, which either result in a modification of existing clinical management or to stratification of patients to novel therapeutic approaches designed as clinical trials.
Conclusions
Genomic analyses have accelerated the identification of relevant biomarkers and extended our understanding of the molecular biology of ovarian cancer. This in turn, will hopefully lead to a paradigm shift from empirical, uniform treatment to a more rational, personalized treatment of ovarian cancers. However, validation of potential biomarkers on both the statistical and biological levels is needed to confirm they are of clinical relevance, in order to increase the likelihood that the desired outcome can be predicted and achieved.
Keywords: ovarian, cancer, genomics, clinical trials
introduction
Ovarian cancer is the seventh leading cause of cancer-related death among women worldwide and has the highest case-fatality rate among gynecologic cancers. Globally, ovarian cancer claims ∼160 500 deaths in 2010, increased from 113 600 in 1990 and 140 200 in 2008 [1, 2]. Epithelial ovarian carcinoma (EOC) comprises 90%–95% of all cases, while sex cord-stromal tumors, malignant ovarian germ cell tumors and ovarian carcinosarcoma are uncommon [3, 4]. Over 70% of women with EOC present with advanced disease involving the upper abdomen (FIGO stage III/IV) due to the lack of early symptoms and effective screening strategies. The standard management for newly diagnosed epithelial ovarian cancer includes aggressive cytoreductive (debulking) surgery followed by the administration of platinum and taxane-based chemotherapy. Cytoreduction to <1 cm of residual disease after surgery is an independent favorable predictor for prognosis [5–7]. However, some argue that the ability to achieve this so-called optimal debulking reflects an intrinsically biologically more favorable and indolent cancer [8]. Despite the inherent resistance to chemotherapy (refractoriness) in some patients, ∼80% of patients achieve an initial clinical complete response [9]. Nevertheless, the majority of EOC patients will eventually relapse. Some patients relapse within 6 months and have a short survival due to platinum-resistance disease; other patients have late relapses with platinum-sensitive disease, and substantially longer survival. Currently, clinicians do not have good prognostic tools to gage which patients are destined to have platinum-resistant or sensitive disease.
The current standard of care is to apply a universal paradigm to all women with ovarian cancer (surgery followed by adjuvant chemotherapy), as mentioned above. Considering the highly diverse population and the variable response to currently standardized therapeutic regimens, a better understanding of the genetic and molecular mechanisms underlying the ovarian cancer pathogenesis and chemoresistance is increasingly needed to allow optimized and individualized patient care with the aim to improve patient outcome.
Due to insufficient power of clinicopathological features and traditional molecular predicators of outcome for ovarian cancer, [10, 11] high throughput genomic analyses such as comparative genomic hybridization (CGH) and gene expression profiling have been proposed to identify gene signatures or signaling pathways as clinically relevant diagnostic and prognostic biomarkers. These high throughput technologies enable the evaluation of multiple targets allowing the development of interrelated gene signature predicting for a clinical end point. In addition, expression changes and genomic alterations can be correlated on a global level to identify key genes or pathways that play causative roles in tumor progression. As a result, several novel therapeutic approaches have been designed or are under active investigation.
epithelial ovarian cancer, a heterogeneous disease
Epithelial ovarian cancer is a heterogeneous disease consisting of tumors with different histology and grade. The most common EOC types are the serous tumors followed by endometrioid, mucinous, and clear-cell cancers which represent 50%–60%, 25%, 4%, and 4% of all ovarian tumors, respectively [12]. Serous tumors are thought to arise from the fallopian tube. Mucinous tumors are cystic tumors with a smooth lining of mucin-secreting epithelial cells resembling either endocervical or colonic epithelium. Endometrioid and clear-cell lesions are thought to arise from dysregulated endometriosis.
There are notable differences in clinical behavior among these subtypes. Mucinous and clear-cell tumors present at an early stage with amenability of complete surgical resection and are often resistant to conventional chemotherapy (<30% response rate). In contrast, the more prevalent serous and endometrioid cancers are aggressive tumors that are often presented at an advanced stage with abdominal spreading yet usually very chemoresponsive (>70% response rate).
Ovarian cancer also spans a broad spectrum of histologic grade [13, 14]. Tumors are graded from 1 to 3, with grade 3 being the least differentiated. A unique feature of ovarian cancer is the subdivision of ‘low malignant potential’ (LMP or borderline) tumors, which are defined as grade 0 tumors. LMP tumors display the atypical cellular features (presence of nuclear atypia and micropapillary morphology), but do not invade into the ovarian stroma. Even in advanced stage disease, patients with LMP have an excellent 5-year survival [15]. In direct contrast with high-grade (Grade 2,3) invasive ovarian cancers, low-grade (Grade 1) ovarian tumors are typically less aggressive, slow-growing tumors, resulting in much better 5-year survival (∼85%) [16].
genomic analysis of histological subtypes of ovarian cancer
It has been increasingly established that tumor heterogeneity in both histology and clinical phenotypes has a molecular basis. Identification of distinct patterns of signaling pathway disruption among different histologic subtypes should enable specific clinical approach to improve the treatment efficacy and patient outcome. Recently, genomic expression profiling studies on epithelial ovarian cancer of various histologies have elucidated distinguishable global gene expression profiles and signaling pathways which contribute to the biological and clinicopathological features seen in the four major subtypes, although some overlap exists between high-grade serous and endometroid carcinomas [17–24]. Unsupervised hierarchical clustering from two independent studies have shown that the mucinous and clear-cell tumors are separated groups, and the endometroid and serous tumors initially group together and then separate into two distinct branches, each dominated by either the endometrial or the serous tumors [19, 24].
Distinct genomic abnormities (gene amplification, deletion, and mutation) also exist among the various subtypes of ovarian cancer. For example there is a high prevalence of TP53 mutations among high-grade serous ovarian cancer (>90% frequency), as well as a high prevalence of mutations involving BRCA1 and BRCA2 [25]. In contrast, mucinous adenocarcinoma and low-grade serous carcinoma (LGSC) are characterized by aberrant Ras/MAPK signaling due to the prevalence of activating KRAS and BRAF mutations [26–28]. Mutations of CTNNB1/β-catenin and PTEN, resulting in hyperactivation of Wnt and PI3K/Akt signaling, are common in endometroid tumors but are rare in the other three major histotypes [23, 29, 30]. Mutations of PIK3CA and corresponding PI3 K/Akt hyperactivation are most frequently observed in clear-cell carcinomas [31–33]. Approximately 50% of clear-cell carcinoma cases also present loss-of-function mutation of ARID1A gene which functions as a tumor suppressor [34–36].
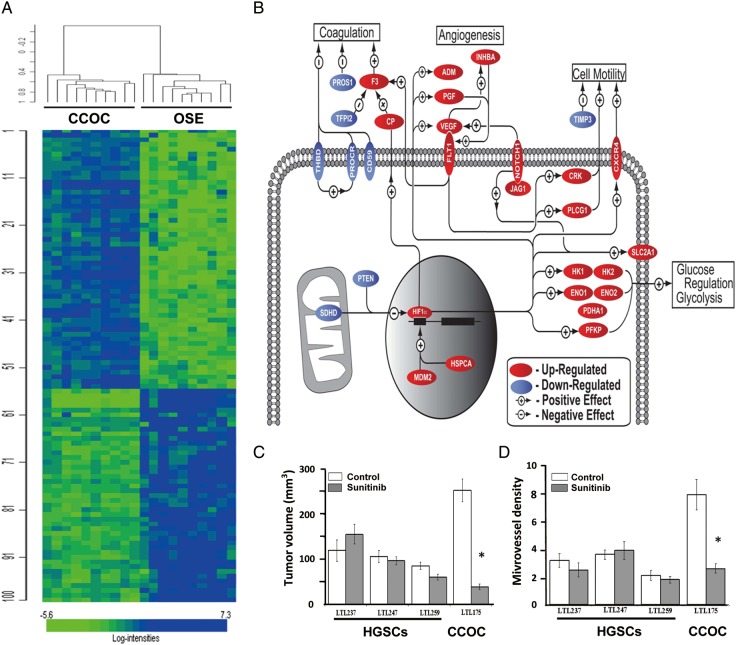
Several targeted therapeutic approaches have been proposed based on the distinct molecular signatures of ovarian clear-cell carcinoma which generally shows resistance to standard platinum–taxane-based regimes. For example a gene expression profile previously identified indicates signature pathway revolving enhanced response to hypoxia, oxidative stress, and cytokine signaling in ovarian clear-cell carcinomas (Figure 1A and B) [21, 37, 38]. Hypoxia inducible factor 1, alpha (HIF-1α), is overexpressed in ovarian clear-cell carcinoma when compared with other EOC histologic subtypes [21, 37]. In addition, major activated pathways in clear-cell carcinoma are not seen in other subtypes and involve HIF-1α and HIF-2α/EPAS1 centered angiogenesis, hypoxic cell growth, and glucose metabolism [21]. Enhanced IL6/STAT3 signaling has also been characterized to upregulate HIF-1α and HIF-2α, contributing to hypoxia response. These activated pathways suggest ovarian clear-cell tumor cells may have enhanced capacity to survive in an environment with limited nutrients and oxygen. This was also shown in preclinical studies; when compared with serous cell lines, clear-cell tumor cells lines were less likely to be affected by hypoxia and glucose deprivation. Knockdown of key genes in hypoxia pathways (HIF-1α and ENO-1) sensitized clear-cell ovarian cancer cell lines to hypoxia/glucose deprivation [21]. Therefore, disruption of the hypoxia response or the derivative angiogenic pathways may become an effective therapeutic regime against ovarian clear-cell carcinoma.
Figure 1.
Whole-genome expression profiling of clear-cell ovarian carcinomas. (A) Graphic representation of whole-genome expression profiling of the specimens from clear-cell ovarian carcinoma (CCOC) and ovarian surface epithelium (OSE). (B) Pathway analysis of differentially regulated genes identified by the transcriptome profiling. Genes included in the analysis were required to have a fold change ≥1.5 (over OSE). Multiple probe sets were averaged for each gene. (C) Effect of a 2-week treatment with sunitinib on growth of subrenal capsule xenografts in NOD-SCID mice (6 mice/group; 2 grafts per kidney) of transplantable high-grade serous (HGSC) and clear-cell ovarian carcinoma (CCOC) tissue lines derived from patients' cancers. Growth of the xenografts is expressed as tumor volume. Data are presented as means ± SEM. (D) Effect of sunitinib on microvessel density of subrenal capsule xenografts in NOD-SCID mice of serous (HGSC) and clear-cell (CCOC) ovarian carcinoma tissue lines. Data are presented as the average number of blood vessels per 400× microscopic field ± SEM. *P < 0.01 Adapted from Stany et al. [21].
Sunitinib, a pan-receptor tyrosine kinases inhibitor primarily targeting tumor microvasculature through the inhibition of angiogenic signals from VEGF, PDGF, and c-kit [39], was thus predicted as an effective therapeutic agent specifically for clear-cell carcinoma. The therapeutic potential of sunitinib was subsequently demonstrated by us using an orthotopic mouse model engrafted with patient-derived clear-cell and serous ovarian tumors. Sunitinib's effect on growth and viability of clear-cell tumor grafts, distinct from serous tumor grafts, highlights sunitinib's potential use as a targeted agent in the treatment of ovarian clear-cell carcinomas (Figure 1C) [21]. Combination therapy of sunitinib and 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine liposome encapsulated small interfering RNA against HIF-1α further shows synergistic activity, providing the rationale for targeted therapy in patients with ovarian clear-cell carcinoma [21].
The administration of sunitinib maleate, an FDA approved oral agent, as therapeutic reagent against chemotherapy-resistant ovarian clear-cell carcinoma patients has promising results in small clinical trials [37, 40]. The Gynecologic Oncology Group (GOG) has initiated GOG-254, a phase II trial of sunitinib in the treatment of persistent or recurrent ovarian clear-cell carcinoma.
In addition to transcription profile, distinct genomic abnormities have also been identified in ovarian clear-cell carcinoma. High frequency (estimated to be 40%) of phosphoinositide 3-kinase catalytic alpha (PIK3CA) mutations has been well documented for ovarian clear-cell carcinomas [31–33]. Activating mutations of PIK3CA results in activation of Akt-mediated signaling to mammalian target of rapamycin (mTOR) for protein synthesis and other downstream effectors for stress response (e.g. apoptosis evasion, chemoresistance, and hypoxia response). Higher level of activated phosphor-mTOR has been specifically observed in ovarian clear-cell carcinoma, suggesting that the PI3K–Akt–mTOR–HIF pathway may be a target with therapeutic potential [41]. Phase I/II trial data exist for mTOR inhibitory agents such as rapamycin, temsirolimus, everolimus, and deferolimus. In a small series of patients, three of six women with ovarian clear-cell carcinoma had a response to temsirolimus [42]. A phase II evaluation of temsirolimus in combination with carboplatin and paclitaxel followed by temsirolimus consolidation as first-line therapy in the treatment of ovarian clear-cell carcinoma is currently in practice as a GOG trial (GOG-268).
genomic analysis of low-grade serous ovarian cancer
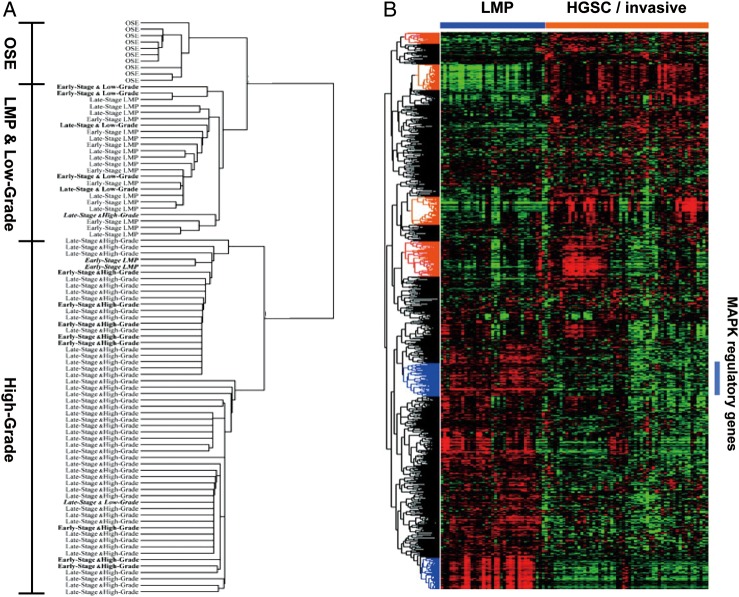
LGSC of the ovary differs from high-grade serous carcinoma (HGSC) in epidemiology, histopathology, associated molecular changes, and clinical course [43]. Equally compelling are data that suggest a link between serous tumors of LMP and LGSC and show differences between LMP tumors and HGSCs [17, 44]. We have previously demonstrated that the gene expression profile of LMP serous tumors and LGSCs are similar and very distinct from the expression profiles of HGSCs (Figure 2) [17, 45–48]. Serous LMP tumors and LGSCs have activating mutations in KRAS, BRAF, and ERBB2, suggesting MAPK hyperactivation. Mutations of either KRAS, BRAF, or the receptor tyrosine kinase ERBB2 lead to constitutive activation of MAPK/Erk pathway, which subsequently activates growth factor stimuli independent and enhanced cell proliferation and/or survival. Interrogation of expression profiles further delineate the distinct molecular features of LMP tumors and LGSCs contributed by the hyperactivation of MAPK/Erk pathway [46, 47].
Figure 2.
Distinct expression profiles between low-grade/low malignant potential (LMP) and high-grade serous ovarian tumors. (A and B) Unsupervised hierarchical clustering from two independent studies Bonome et al. [17] (A) and Anglesio et al. [47] (B) are presented. The second study further suggests a gene expression matrix consisting of MAPK pathway regulated genes (as defined by ontology classification tools DAVID and Panther, highlighted by a bar) in LMP serous ovarian tumors.
Given the poor response of LGSCs to conventional platinum-taxane chemotherapy, therapeutic targeting of hyperactivated Ras/Raf/MEK/Erk may be a novel approach to this disease. A phase II clinical trial evaluating selumetinib (AZD6244), an oral non-ATP competitive small molecule inhibitor of MEK1/2 in patients with recurrent ovarian LGSC (n = 53) has recently been completed by the GOG (GOG-239) [49]. Selumetinib has demonstrated substantial activity in recurrent low-grade serous tumors with less toxicity, particularly when compared with that accompanied with genotoxic chemotherapy. In contrast to the minimal overall responsive rate of 3.7% with conventional cytotoxic agents, [50] selumetinib significantly increased the objective response rate to 15%, with another 65% of patients presenting stable disease. In addition, substantial improvement of progression-free interval was observed with selumetinib, from 29 weeks to 11 months. These findings suggest that inhibitors of the MAPK pathway warrant further investigation in patients with LMP tumor or LGSC.
genomic analysis of high-grade serous ovarian cancer
HGSC comprise the majority of advanced-stage serous ovarian tumors. Substantial efforts have been devoted to identify genomic abnormalities and expression profiles of high-grade serous tumors. The Cancer Genome Atlas (TCGA) Project analyzed ∼500 HGSC samples and demonstrated large amounts of genomic/epigenomic abnormalities [25]. Multiple areas of chromosomal gain or loss were detected with the amplification of more than 30 growth-stimulatory genes including MECOM, MYC and CCNE1, which are amplified in at least 10% of the cases. In contrast, few high-frequent mutations were detected except the inactivating mutations of TP53 (over 90%) and BRCA1/2, which functionally contribute to the genomic instability of HGSCs. In addition to somatic mutation, BRCA1/2 function is inhibited in HGSC through additional mechanisms including germline mutations, upstream mutations, and hypermethylation [25, 51]. These alterations result in deficiency of DNA double-strand break repair through homologous recombination (HR) in a subset of HGSCs. Preclinical work has supported the synthetic lethality of DNA single-strand break repair inhibition by poly(adenosine diphosphate–ribose) polymerases inhibitor olaparib (AZD2281) with impaired HR repairing, including in the setting of BRCA dysfunction [52–54]. In addition, the therapeutic efficacy of olaparib in combination with the anti-angiogenic cediranib (AZD2171) is currently under investigation in recurrent HGSC patient as a phase II clinical trial, based on the recently completed phase I assessment of the safety and clinical response (>60% clinical benefit rate) [55].
prognostic gene expression signature reveals underlying biology
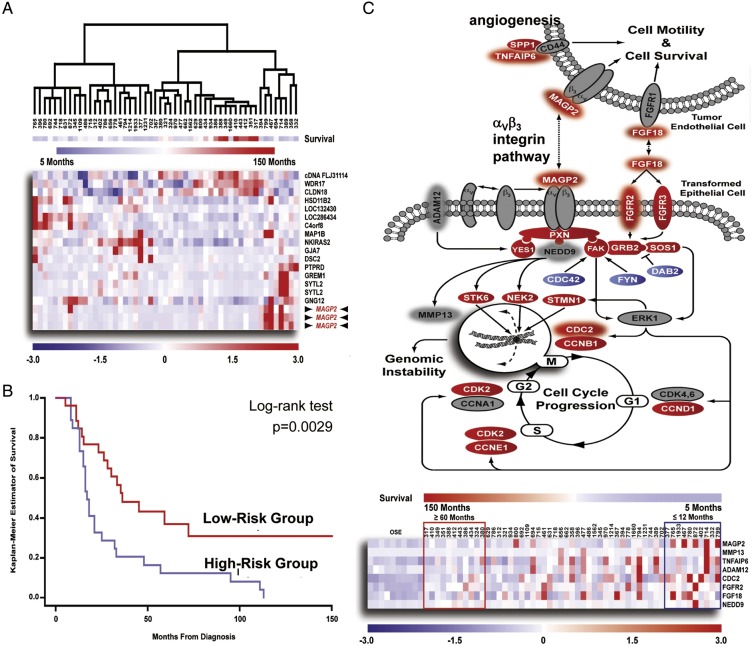
The development of genomic classifiers of patient survival using microarray based gene expression profiling has the potential to stratify patients according to prognosis and identify new opportunities for therapeutic intervention. Initial efforts have focused on discrete patient groups by supervised clustering analysis of dichotomized data of short- and long-term survivals [56, 57]. Yet, expression patterns identified in this manner may not adequately differentiate the majority of patients who will die at an intermediate end point. In addition, due to the heterogeneity of tumor tissue, directly evaluating raw tumor samples may introduce erroneous data attributable to varying amounts of intervening stroma and lymphocytic infiltrates. To limit the complexities of the tissue analysis, we have procured tumor specimens with laser captured microdissection to increase the accuracy of expression profiling [21, 58]. Considering survival as a continuous variable, we have identified and validated a survival signature in a cohort of 53 microdissected HGSCs using a two-step ‘semi-supervised’ approach with Cox regression analysis and leave-one-out cross-validation [58]. The performance of the prediction analysis was demonstrated by hierarchical clustering, which demonstrated the ability of the top scoring genes (Cox hazard ratio >10) to cluster the 53 specimens according to survival (Figure 3). The validity of the entire 200 probe set predictor was confirmed by Kaplan–Meier analysis with a significant difference in survival time. The identified prognosis signature implicates a group of genes that reflect hyperactivation of αVβ3 integrin pathway as a prominent survival associated event.
Figure 3.
Identification of a prognostic gene expression signature correlating with survival in microdissected advanced-stage HGSCs. (A) Hierarchical clustering of 53 advanced-stage HGSCs using expression values for genes possessing a Cox score >10. (B) Kaplan–Meier analysis of the predictor demonstrated a significant difference in survival time (P = 0.0029). (C) Assessment of putative signaling events contributing to patient survival through pathway analysis. Differentially regulated genes identified in the 53 microdissected serous tumors, when compared with 10 normal OSE brushings were labeled with white fonts. Genes predictive of poor prognosis is cycled with halo. A heat map is shown below to demonstrate association between survival signature genes identified in the pathway and overall patient survival. Adapted from Mok et al. [58].
The gene possessing the highest hazard ratio, MAGP2, activates αVβ3 integrin signaling through its N-terminal RGD domain [59]. Mutation of the RGD motif or αVβ3 integrin neutralizing antibody blocks MAGP2 mediated adhesion, migration, invasion and chemoresistance of HGSC cells. Considering αVβ3 integrin is expressed on endothelial cells and has been associated with tumor angiogenesis, [60] secreted MAGP2 was predicted to modulate the biology of surrounding endothelial cells to promote neovascularization. MAGP2 knock-down significantly decreases the tumor burden of HGSC cell line xenografts in vivo, as well as the tumor microvessel density. Validation using independent patient samples confirmed the prognostic value of MAGP2 through both qRT-PCR and IHC, while increased MAGP2 expression correlated with microvessel density further confirmed the association between MAGP2 and angiogenesis.
The biological activity of MAGP2 emphasized the importance of angiogenesis in the pathogenesis of ovarian cancer. Recently, targeting tumor vasculature becomes a promising approach for ovarian cancer treatment [61]. Bevacizumab is an antiangiogenic monoclonal antibody that inhibits VEGF activity. Two independent large-scale phase III studies conducted in the US (GOG-218) and Europe (the International Collaboration for Ovarian Neoplasms [ICON]-7 trial) tested the incorporation of bevacizumab as a first-line therapy with concomitant standard (3-weekly carboplatin and paclitaxel) chemotherapy in ovarian cancer patients. Maintenance treatment with bevacizumab extended the length of progression-free survival (both studies) as well as overall survival (ICON-7) in a subset of patients [62, 63].
integrative genomic analysis identifies genes critical for clinicopathological feature of HGSC
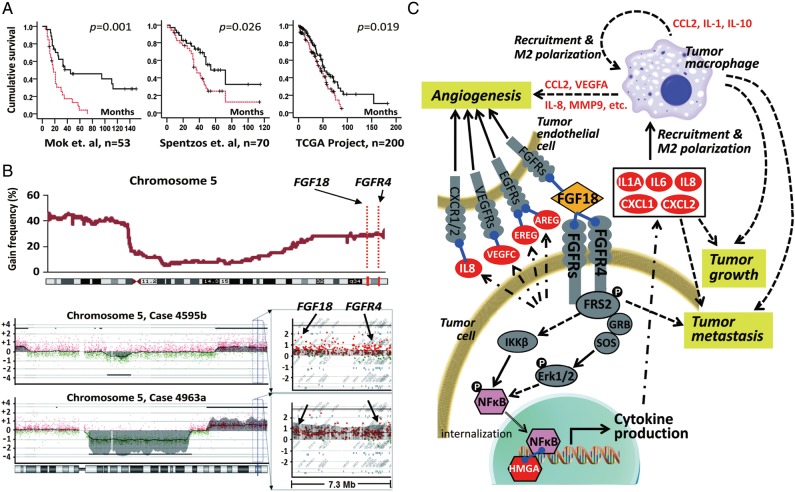
Considering the extensive genomic instability in HGSCs, integrated analysis of DNA copy number difference and gene expression profiles becomes another important way for biomarker discovery. In our previous study, oligonucleotide array CGH analysis on microdissected HGSC samples demonstrated the amplification of chromosome segment 5q31 to 5q35.3 as one of the most significant copy number abnormalities associated with poor overall survival [64]. To identify candidate genes that drive tumorigenesis in this aberrantly amplified chromosome segment, genes located between 5q31 and 5q35.3 were compared with a prognostic gene expression signature generated with microdissected samples [58], and fibroblast growth factor 18 (FGF18, located on chromosome 5q35.1) was found to be the gene possessing the strongest prognostic value in segment 5q31–5q35.3 (Figure 4A and B) [65]. Subsequent validation with two independent profiling studies [25, 66] and an independent cohort for IHC staining confirmed the FGF18 as an independent predictive marker for poor clinical outcome. Functional validation studies have demonstrated that FGF18 promotes migration, invasion and tumorigenicity of ovarian cancer cells in vitro and in vivo. In addition, FGF18 activates NF-κB to increase the production of oncogenic cytokines and chemokines. This in turn, results in a more malignant tumor microenvironment characterized by enhanced angiogenesis and augmented tumor-associated macrophage infiltration and M2 polarization [67, 68]. In summary, we propose that the aberrant upregulation of FGF18 in ovarian cancer cells may initiate reciprocal heterotypic signaling interactions and cascades in neoplastic cells and non-neoplastic cells within the tumor microenvironment and eventually accelerates the malignant progression to convey a poor outcome in serous ovarian cancer (Figure 4C). We further demonstrated that the FGF18 signaling in HGSC cells is at least partially conveyed by FGFR4 (located on chromosome 5q35.2). The genomic proximity of FGF18 and FGFR4 implicates co-amplification, which was observed in the initial study [64] and confirmed by qPCR using an independent cohort [69]. The co-amplification of both the ligand (FGF18) and the receptor (FGFR4) contributes to the hyperactivation of the FGF signal, mediating the 5q31–5q31.3 amplicon-related poor patient survival. Taken together, this study has revealed the hyperactivation of FGF signaling as one of the potential mechanisms defining poor prognosis in a subset of HGSCs. Therefore, blocking FGF signaling becomes a rational therapeutic approach for HGSC patients, especially for those with 5q31–5q35.3 amplification. So far, several therapeutic approaches against FGF signaling have been developed, including receptor tyrosine kinase inhibitors, receptor neutralizing antibodies and FGF ligand traps [70–72]. A phase II evaluation of the triple VEGFR/PDGFR/FGFR inhibitor BIBF-1120 has recently been completed. BIBF-1120 was well tolerated and was associated with an improvement in progression-free survival, which justifies further study within a large phase III trial [73]. An alternative inhibition approach is using a decoy FGF receptor to sequester FGF ligands, which largely avoids toxicity from small molecule inhibitors [74]. One of the FGF ligand trap proteins, FP-1039 has completed phase I testing and entered phase II trials for endometrial cancer [75]. Subsequent work will be needed to confirm the prognostic value of FGF18 in larger groups of patients and identify individuals suitable for FGF-targeted therapy.
Figure 4.
Identification of FGF18 as a prognostic gene in high-grade advanced-stage papillary serous ovarian tumors. (A) Kaplan–Meier analysis of FGF18 expression in patients in three independent sets of serous ovarian cancer samples. Analysis was done by median cut with the P-value of log-rank test presented for each set. Solid lines: samples with low FGF18, broken lines: samples with high FGF18, ‘+’: censored samples. (B) CGH analysis of 72 serous ovarian tumors showed an increased DNA copy number for chromosome segment 5q31.3–5q35.3 in ∼25% of the samples (upper panel). Chromosome 5 profiles of two representative tumors with the detail of a 7.3-Mb locus (chromosome 5 distance: 170.3–177.6 Mb) containing FGF18 and FGFR4 (lower panel). Copy number was presented by log2 − 1 (value of 0 mean diploid). FGF18 and FGFR4 are amplified to at least four copies among the 5615 probes in the x-axis. (C) The effect of FGF18 on the pathogenesis of serous ovarian cancer. Gene expression profiling was carried out to compare the transcriptome of ovarian cancer cell lines with ectopic overexpression of FGF18 or RFP as control. Genes directly induced by FGF18 are labeled in white font. Arrows indicate potential interactions that contribute to FGF18-mediated ovarian tumorigenesis. Adapted from Wei et al.
conclusion
Despite the evolution of surgical techniques and meticulously designed chemotherapy regimens, refractoriness and relapse remains major obstacles in improving the prognosis of patients with ovarian cancer. Therapeutic innovations in the management of ovarian cancer rely on translating basic science research into clinical trials to investigate targeted and personalized therapies. There is an important need to understand the molecular characteristics to identify prognostic and predictive biomarkers which biologically drive the pathogenesis and chemoresistance of ovarian tumors. Large-scale integrative genomic profiling technologies provided opportunities for biomarker discovery. However, comprehensive functional validation studies on both biological and clinical levels are needed to understand the mechanistic basis for these biomarkers and comprehend their full clinical significance and application. With the aid of genomic profiling technologies, the time of personalized medicine for patients with ovarian cancer is becoming more of a reality and by which patients with this deadly, yet heterogeneous disease, might have a higher chance to benefit more from the available therapies.
funding
The financial support of research grants is gratefully acknowledged: NIH grant RC4CA156551, NIH grant 1R01CA142832, NIH grant R01CA169200, and The Julie Fund.
disclosure
The authors have declared no conflicts of interest.
references
- 1.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chornokur G, Amankwah EK, Schildkraut JM, et al. Global ovarian cancer health disparities. Gynecol Oncol. 2013;129:258–264. doi: 10.1016/j.ygyno.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo N, Peiretti M, Castiglione M. Non-epithelial ovarian cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):24–26. doi: 10.1093/annonc/mdp118. [DOI] [PubMed] [Google Scholar]
- 4.del Carmen MG, Birrer M, Schorge JO. Carcinosarcoma of the ovary: a review of the literature. Gynecol Oncol. 2012;125:271–277. doi: 10.1016/j.ygyno.2011.12.418. [DOI] [PubMed] [Google Scholar]
- 5.Bristow RE, Tomacruz RS, Armstrong DK, et al. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20:1248–1259. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 6.Galaal K, Naik R, Bristow RE, et al. Cytoreductive surgery plus chemotherapy versus chemotherapy alone for recurrent epithelial ovarian cancer. Cochrane Database Syst Rev. 2010 doi: 10.1002/14651858.CD007822.pub2. CD007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang SJ, Hodeib M, Chang J, et al. Survival impact of complete cytoreduction to no gross residual disease for advanced-stage ovarian cancer: a meta-analysis. Gynecol Oncol. 2013;130:493–498. doi: 10.1016/j.ygyno.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 8.Borley J, Wilhelm-Benartzi C, Brown R, et al. Does tumour biology determine surgical success in the treatment of epithelial ovarian cancer? A systematic literature review. Br J Cancer. 2012;107:1069–1074. doi: 10.1038/bjc.2012.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 10.Darcy KM, Brady WE, McBroom JW, et al. Associations between p53 overexpression and multiple measures of clinical outcome in high-risk, early stage or suboptimally-resected, advanced stage epithelial ovarian cancers A Gynecologic Oncology Group study. Gynecol Oncol. 2008;111:487–495. doi: 10.1016/j.ygyno.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farley J, Fuchiuji S, Darcy KM, et al. Associations between ERBB2 amplification and progression-free survival and overall survival in advanced stage, suboptimally-resected epithelial ovarian cancers: a Gynecologic Oncology Group Study. Gynecol Oncol. 2009;113:341–347. doi: 10.1016/j.ygyno.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38:87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 13.Malpica A. Grading of ovarian cancer: a histotype-specific approach. Int J Gynecol Pathol. 2008;27:175–181. doi: 10.1097/PGP.0b013e31816085e0. [DOI] [PubMed] [Google Scholar]
- 14.Bodurka DC, Deavers MT, Tian C, et al. Reclassification of serous ovarian carcinoma by a 2-tier system: a Gynecologic Oncology Group Study. Cancer. 2012;118:3087–3094. doi: 10.1002/cncr.26618. [DOI] [PubMed] [Google Scholar]
- 15.Fukumoto M, Nakayama K. Ovarian epithelial tumors of low malignant potential: are they precursors of ovarian carcinoma? Pathol Int. 2006;56:233–239. doi: 10.1111/j.1440-1827.2006.01960.x. [DOI] [PubMed] [Google Scholar]
- 16.Lachance JA, Shutter J, Atkins KA, et al. Utilization of a uniform grading system for interpreting serous ovarian cancer. Am J Obstet Gynecol. 2008;199:189.e1–189.e6. doi: 10.1016/j.ajog.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Bonome T, Lee JY, Park DC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 18.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, et al. A distinct molecular profile associated with mucinous epithelial ovarian cancer. Br J Cancer. 2006;94:904–913. doi: 10.1038/sj.bjc.6603003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz DR, Kardia SL, Shedden KA, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 20.Shedden KA, Kshirsagar MP, Schwartz DR, et al. Histologic type, organ of origin, and Wnt pathway status: effect on gene expression in ovarian and uterine carcinomas. Clin Cancer Res. 2005;11:2123–2131. doi: 10.1158/1078-0432.CCR-04-2061. [DOI] [PubMed] [Google Scholar]
- 21.Stany MP, Vathipadiekal V, Ozbun L, et al. Identification of novel therapeutic targets in microdissected clear cell ovarian cancers. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021121. e21121- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wamunyokoli FW, Bonome T, Lee JY, et al. Expression profiling of mucinous tumors of the ovary identifies genes of clinicopathologic importance. Clin Cancer Res. 2006;12:690–700. doi: 10.1158/1078-0432.CCR-05-1110. [DOI] [PubMed] [Google Scholar]
- 23.Wu R, Hendrix-Lucas N, Kuick R, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3 K/Pten signaling pathways. Cancer Cell. 2007;11:321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Zorn KK, Bonome T, Gangi L, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11:6422–6430. doi: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enomoto T, Weghorst CM, Inoue M, et al. K-ras activation occurs frequently in mucinous adenocarcinomas and rarely in other common epithelial tumors of the human ovary. Am J Pathol. 1991;139:777–785. [PMC free article] [PubMed] [Google Scholar]
- 27.Gemignani ML, Schlaerth AC, Bogomolniy F, et al. Role of KRAS and BRAF gene mutations in mucinous ovarian carcinoma. Gynecol Oncol. 2003;90:378–381. doi: 10.1016/s0090-8258(03)00264-6. [DOI] [PubMed] [Google Scholar]
- 28.Singer G, Oldt R, III, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 29.Wright K, Wilson P, Morland S, et al. beta-catenin mutation and expression analysis in ovarian cancer: exon 3 mutations and nuclear translocation in 16% of endometrioid tumours. Int J Cancer. 1999;82:625–629. doi: 10.1002/(sici)1097-0215(19990827)82:5<625::aid-ijc1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Obata K, Morland SJ, Watson RH, et al. Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors. Cancer Res. 1998;58:2095–2097. [PubMed] [Google Scholar]
- 31.Kuo KT, Mao TL, Jones S, et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–1601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 33.Anglesio MS, Carey MS, Kobel M, et al. Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol. 2011;121:407–415. doi: 10.1016/j.ygyno.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Jones S, Wang TL, Shih I, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 37.Anglesio MS, George J, Kulbe H, et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res. 2011;17:2538–2548. doi: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 38.Mandai M, Matsumura N, Baba T, et al. Ovarian clear cell carcinoma as a stress-responsive cancer: influence of the microenvironment on the carcinogenesis and cancer phenotype. Cancer Lett. 2011;310:129–133. doi: 10.1016/j.canlet.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 39.Le TC, Raymond E, Faivre S. Sunitinib: a novel tyrosine kinase inhibitor. A brief review of its therapeutic potential in the treatment of renal carcinoma and gastrointestinal stromal tumors (GIST) Ther Clin Risk Manag. 2007;3:341–348. doi: 10.2147/tcrm.2007.3.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauh-Hain JA, Penson RT. Potential benefit of Sunitinib in recurrent and refractory ovarian clear cell adenocarcinoma. Int J Gynecol Cancer. 2008;18:934–936. doi: 10.1111/j.1525-1438.2007.01156.x. [DOI] [PubMed] [Google Scholar]
- 41.Miyazawa M, Yasuda M, Fujita M, et al. Therapeutic strategy targeting the mTOR-HIF-1alpha-VEGF pathway in ovarian clear cell adenocarcinoma. Pathol Int. 2009;59:19–27. doi: 10.1111/j.1440-1827.2008.02320.x. [DOI] [PubMed] [Google Scholar]
- 42.Takano M, Kikuchi Y, Kudoh K, et al. Weekly administration of temsirolimus for heavily pretreated patients with clear cell carcinoma of the ovary: a report of six cases. Int J Clin Oncol. 2011;16:605–609. doi: 10.1007/s10147-010-0177-z. [DOI] [PubMed] [Google Scholar]
- 43.Daz-Padilla I, Malpica AL, Minig L, et al. Ovarian low-grade serous carcinoma: a comprehensive update. Gynecol Oncol. 2012;126:279–285. doi: 10.1016/j.ygyno.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 44.Gershenson DM, Sun CC, Lu KH, et al. Clinical behavior of stage II-IV low-grade serous carcinoma of the ovary. Obstet Gynecol. 2006;108:361–368. doi: 10.1097/01.AOG.0000227787.24587.d1. [DOI] [PubMed] [Google Scholar]
- 45.Meinhold-Heerlein I, Bauerschlag D, Hilpert F, et al. Molecular and prognostic distinction between serous ovarian carcinomas of varying grade and malignant potential. Oncogene. 2005;24:1053–1065. doi: 10.1038/sj.onc.1208298. [DOI] [PubMed] [Google Scholar]
- 46.May T, Virtanen C, Sharma M, et al. Low malignant potential tumors with micropapillary features are molecularly similar to low-grade serous carcinoma of the ovary. Gynecol Oncol. 2010;117:9–17. doi: 10.1016/j.ygyno.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Anglesio MS, Arnold JM, George J, et al. Mutation of ERBB2 provides a novel alternative mechanism for the ubiquitous activation of RAS-MAPK in ovarian serous low malignant potential tumors. Mol Cancer Res. 2008;6:1678–1690. doi: 10.1158/1541-7786.MCR-08-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 49.Farley J, Brady WE, Vathipadiekal V, et al. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol. 2013;14:134–140. doi: 10.1016/S1470-2045(12)70572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmeler KM, Sun CC, Bodurka DC, et al. Neoadjuvant chemotherapy for low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol. 2008;108:510–514. doi: 10.1016/j.ygyno.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Rigakos G, Razis E. BRCAness: finding the Achilles heel in ovarian cancer. Oncologist. 2012;17:956–962. doi: 10.1634/theoncologist.2012-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 53.Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee S, Kaye S. PARP inhibitors in BRCA gene-mutated ovarian cancer and beyond. Curr Oncol Rep. 2011;13:442–449. doi: 10.1007/s11912-011-0193-9. [DOI] [PubMed] [Google Scholar]
- 55.Liu JF, Tolaney SM, Birrer M, et al. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 2013 doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lancaster JM, Dressman HK, Whitaker RS, et al. Gene expression patterns that characterize advanced stage serous ovarian cancers. J Soc Gynecol Investig. 2004;11:51–59. doi: 10.1016/j.jsgi.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Berchuck A, Iversen ES, Lancaster JM, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 58.Mok SC, Bonome T, Vathipadiekal V, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson MA, Leavesley DI, Ashman LK. Microfibril-associated glycoprotein-2 specifically interacts with a range of bovine and human cell types via alphaVbeta3 integrin. J Biol Chem. 1999;274:13060–13065. doi: 10.1074/jbc.274.19.13060. [DOI] [PubMed] [Google Scholar]
- 60.Ellis PD, Metcalfe JC, Hyvonen M, et al. Adhesion of endothelial cells to NOV is mediated by the integrins alphavbeta3 and alpha5beta1. J Vasc Res. 2003;40:234–243. doi: 10.1159/000071887. [DOI] [PubMed] [Google Scholar]
- 61.Hall M, Gourley C, McNeish I, et al. Targeted anti-vascular therapies for ovarian cancer: current evidence. Br J Cancer. 2013;108:250–258. doi: 10.1038/bjc.2012.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 63.Perren TJ, Swart AM, Pfisterer J, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 64.Birrer MJ, Johnson ME, Hao K, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 65.Wei, et al. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J Clin Invest. 2013;123(10):4435–4448. doi: 10.1172/JCI70625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spentzos D, Levine DA, Ramoni MF, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4700–4710. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 67.Robinson-Smith TM, Isaacsohn I, Mercer CA, et al. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res. 2007;67:5708–5716. doi: 10.1158/0008-5472.CAN-06-4375. [DOI] [PubMed] [Google Scholar]
- 68.Thompson MS, Mok SC. Immunopathogenesis of ovarian cancer. Minerva Med. 2009;100:357–370. [PubMed] [Google Scholar]
- 69.Engler DA, Gupta S, Growdon WB, et al. Genome wide DNA copy number analysis of serous type ovarian carcinomas identifies genetic markers predictive of clinical outcome. PLoS One. 2012;7:e30996. doi: 10.1371/journal.pone.0030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 71.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med. 2011;17:283–292. doi: 10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harding TC, Long L, Palencia S, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5:178ra39. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 73.Ledermann JA, Hackshaw A, Kaye S, et al. Randomized phase II placebo-controlled trial of maintenance therapy using the oral triple angiokinase inhibitor BIBF 1120 after chemotherapy for relapsed ovarian cancer. J Clin Oncol. 2011;29:3798–3804. doi: 10.1200/JCO.2010.33.5208. [DOI] [PubMed] [Google Scholar]
- 74.Zhang H, Lorianne M, Baker K, et al. FP-1039 (FGFR1:Fc), a soluble FGFR1 receptor antagonist, inhibits tumor growth and angiogenesis. NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics; 22–26 October; San Francisco, CA. 2007. Abst B55. [Google Scholar]
- 75.Harding T, Palencia S, Long L, et al. Preclinical efficacy of FP-1039 (FGFR1:Fc) in endometrial carcinoma models with activating mutations in FGFR2. American Association for Cancer Research (AACR) 101st Annual Meeting 2010; 17–21 April 2010; Washington, DC. [Google Scholar]