Summary
We used adaptive evolution to improve freeze tolerance of industrial baker's yeast. Our hypothesis was that adaptation to low temperature is accompanied by enhanced resistance of yeast to freezing. Based on this hypothesis, yeast was propagated in a flour‐free liquid dough model system, which contained sorbitol and NaCl, by successive batch refreshments maintained constantly at 12°C over at least 200 generations. Relative to the parental population, the maximal growth rate (µmax) under the restrictive conditions, increased gradually over the time course of the experiment. This increase was accompanied by enhanced freeze tolerance. However, these changes were not the consequence of genetic adaptation to low temperature, a fact that was confirmed by prolonged selection of yeast cells in YPD at 12°C. Instead, the experimental populations showed a progressive increase in NaCl tolerance. This phenotype was likely achieved at the expense of others traits, since evolved cells showed a ploidy reduction, a defect in the glucose derepression mechanism and a loss in their ability to utilize gluconeogenic carbon sources. We discuss the genetic flexibility of S. cerevisiae in terms of adaptation to the multiple constraints of the experimental design applied to drive adaptive evolution and the technologically advantageous phenotype of the evolved population.
Introduction
Adaptive evolution is a natural process by which living beings are able to survive in a constantly changing environment. Under these circumstances, the best‐fitting phenotypes are selected and the corresponding genotypes fixed in the population, so the species are able to perpetuate in the new environment. The molecular aspects of these phenomena have been studied in depth in microorganisms, because their short generation time enables follow‐up studies of many generations of continuous culture in a reasonable time period (Wright, 2004). In the case of the budding yeast Saccharomyces cerevisiae, studies on adaptive evolution have revealed that this organism displays a fast evolutionary rate when it is grown in a continuous nutrient‐limited environment, with an adaptive change occurring every 40 generations (Paquin and Adams, 1983). Adaptive changes involve significant improvements in cell properties, i.e. fitness, and are frequently associated with sequence alterations involving Ty elements (Adams and Oeller, 1986).
In parallel with basic studies, scientists have tried to take advantage of the adaptive potential of S. cerevisiae. Industrial S. cerevisiae strains have been adapted and selected (a process called ‘domestication’), for their better performance in baking, brewing and wine making (Randez‐Gil et al., 1999; Querol et al., 2003; Randez‐Gil et al., 2003), showing that adaptive evolution has tangible effects in different industrial environments. In combination with genetic engineering, recombinant strains have also been evolved to develop highly specialized yeasts in technologically important properties like pyruvate production (van Maris et al., 2004), efficient xylose (Kuyper et al., 2004) and lactose fermentation (Guimarães et al., 2008), or enhanced tolerance against toxins (Heer and Sauer, 2008). Nevertheless, the full potential of adaptive evolution in the development of industrial strains has hardly been explored (Patnaik, 2008). Moreover, the acquisition of new traits, as well as the flexibility of the adaptive process, could be constrained by intrinsic properties of yeasts (Dykhuizen, 1990), or even be counter to ‘biological design’, as has been proposed for certain kind of stresses (Attfield, 1997).
Stress tolerance of commercial Saccharomyces cerevisiae yeasts is, without doubt, a trait that needs improvements (Attfield, 1997; Dequin, 2001; Randez‐Gil et al., 2003). In the bakery sector, the huge growth in frozen dough manufacture, both for bread and pastries, has revealed that this organism has problems when withstanding temperatures below 0°C (Tanghe et al., 2003; Aguilera et al., 2007). Freezing and frozen storage of dough are stressful situations that cause cell injury, leading to longer proofing times and reduced product volume. Therefore, the development of freeze‐tolerant baker's yeast is of great economic interest and efforts have been made to understand the effects of freezing and define targets for strain selection and improvement (Randez‐Gil et al., 2003).
Freezing is a complex and multifaceted stress, in which different stressors, such as osmotic (Wolfe and Bryant, 1999), mechanical (Morris et al., 1988) or oxidative stress (Hermes‐Lima and Storey, 1993) appear to play important roles. Consequently, diverse strategies, including activation of cross‐protection mechanisms (Kronberg et al., 2007) or pre‐loading of yeast cells with freeze‐protective molecules such as trehalose (Hirasawa et al., 2001) or glycerol (Myers and Attfield, 1999), have been proposed to increase freeze resistance of yeast cells in baking applications. However, physiological conditioning approaches do not solve the lack of intrinsic freeze resistance of yeast cells and their effects are rapidly lost at the onset of fermentation (Tanghe et al., 2003). Alternatively, enhanced freeze tolerance has been achieved in industrial baker's yeast by homologous or heterologous expression of genes involved in water transport (Tanghe et al., 2002), membrane fluidity (Rodriguez‐Vargas et al., 2002; 2007) or anti‐freeze proteins (Panadero et al., 2005b). However, the use of such strains by the industry remains constrained due to consumer's reticence with respect to genetically modified organisms. As a result, there is still no industrial baker's yeast on the market that combines good fermentative capacity with an appropriate resistance to freezing.
Studies in different organisms, including bacteria (Murga et al., 2000) and fungi (Thammavongs et al., 2000), have established a correlation between cold acclimatization and acquired freeze tolerance. However, the existence of such a physiological link in S. cerevisiae remains controversial (Park et al., 1997; Diniz‐Mendez et al., 1999), although recent evidence emphasizes the parallelism between the genetic and molecular aspects of both freeze and cold‐shock responses (Aguilera et al., 2007). When cells are subjected to low (0–12°C) temperatures, protective molecules like trehalose or glycerol are synthesized. Although neither trehalose nor glycerol is needed for growth at 10–12°C (Tai et al., 2007), their accumulation provides freeze protection (Kandror et al., 2004; Panadero et al., 2006). Likewise, fluidization of membranes via synthesis of unsaturated fatty acids, which is a well‐known cellular adaptation to low temperatures (Aguilera et al., 2007), also has a positive effect on survival after incubation at −20°C (Rodriguez‐Vargas et al., 2007). These results suggest that strains that are well adapted to low temperatures (10 to 18°C) will also be more resistant to freezing stress. Based on this idea, here we report on the selection, by means of experimental evolution, of industrial yeast populations with increased growth rates in a liquid dough model system at 12°C. We also report the physiological and technological characterization of evolved clones isolated from the 200‐generation terminal population, and discuss the evolutionary driving force for several of the observed changes.
Results
Adaptive evolution in LD at 12°C leads to an enhanced growth rate
We explored the possibility of enhancing the freeze‐stress tolerance of industrial baker's yeast cells by generating adaptations to growth at 12°C. Since strains suited for commercial use should maintain other important traits, such as growth and maltose fermentation capacity, cells were cultured in a synthetic flour‐free liquid‐dough (LD) medium. Liquid‐dough model systems effectively mimic the nutritional status and environment encountered by baker's yeast cells in bread dough (Panadero et al., 2005a). Yeast was propagated by successive batch refreshments maintained constantly at 12°C during at least 200 generations. Indeed, the maximal growth rate (µmax) at 12°C of the parental strain (0.012 ± 0.001 h−1) increased gradually over the course of the experiment (Table 1). Thus, the doubling time decreased from around 58 h for the parental strain to 14 h for the 200‐generation evolved population, indicating that the original yeast population was capable of adapting to our experimental conditions in order to optimize growth.
Table 1.
µmax of the experimental populations under different culture conditions.
| Culture conditions | µmax (h−1)a | |||
|---|---|---|---|---|
| Parental | 50G | 100G | 200G | |
| LD 12°C | 0.012 ± 0.001 | 0.017 ± 0.002 | 0.033 ± 0.001 | 0.049 ± 0.004 |
| YPD 12°C | 0.119 ± 0.004 | n.d. | n.d. | 0.108 ± 0.003 |
| YPD 30°C | 0.540 ± 0.009 | n.d. | n.d. | 0.497 ± 0.012 |
| YPD‐NaCl 30°C | 0.174 ± 0.003 | 0.226 ± 0.005 | 0.244 ± 0.005 | 0.271 ± 0.004 |
The maximal growth rate constant (µmax) of the parental, 50‐ (50G), 100‐ (100G) and 200‐generation (200G) evolved populations in LD at 12°C, was calculated from the slope of the line obtained after plotting ln X versus t, where X is the cell density of the culture, measured as OD600, at multiple time points (t) during exponential growth in LD, YPD or YPD containing 1 M NaCl, at the indicated temperature.
n.d., not determined.
The parental and evolved populations differ in freeze tolerance
We analysed whether the higher growth rate of the evolved populations in LD at 12°C correlates with enhanced freeze tolerance. Molasses‐grown yeast cells from the parental, 50‐, 100‐ and 200‐generation evolved populations were transferred to LD medium and total CO2 production at 30°C was measured before and after freezing and frozen storage for 7, 14 or 21 days. As expected, the fermentative capacity of yeast cells greatly decreased upon freezing/thawing stress. Indeed, values of CO2 production by the parental strain were only 19% of those of the corresponding unfrozen control after 21 days of storage at −20°C (Fig. 1). This decrease was less pronounced when yeast cells from the 12°C evolutionary experiment were tested. Under the same conditions, the 50‐generation evolved population showed CO2 values of approximately 41% with respect to those of its unfrozen control (Fig. 1). However, further selection in a LD environment at 12°C did not significantly increase the magnitude of this difference. Moreover, the evolutionary experiment had negative effects on the fermentative capacity of the parental strain before freezing. Indeed, CO2 production diminished approximately 20% in unfrozen control samples of the terminal population (200 generations) relative to the parental population (Fig. 1, time 0).
Figure 1.
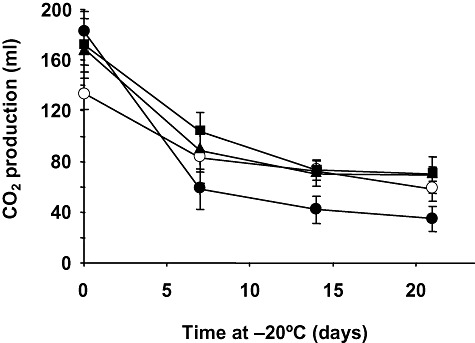
Freeze tolerance of cells harvested from the evolved populations. Molasses‐grown yeast cells from the parental (●), 50‐ ( ), 100‐ (
), 100‐ ( ) and 200‐generation (○) evolved populations were transferred to LD medium and total CO2 production at 30°C was measured before (control, time 0) and after freezing and frozen storage at −20°C for 7, 14 or 21 days. The amount of CO2 produced for 180 min at 30°C was recorded in a home‐made fermentometer. Frozen samples were thawed at 30°C for 30 min before measuring gassing power. Values are expressed as ml of CO2 per sample. In all cases, values represent the means of at least three independent experiments. The error associated with the points was calculated by using the formula:
) and 200‐generation (○) evolved populations were transferred to LD medium and total CO2 production at 30°C was measured before (control, time 0) and after freezing and frozen storage at −20°C for 7, 14 or 21 days. The amount of CO2 produced for 180 min at 30°C was recorded in a home‐made fermentometer. Frozen samples were thawed at 30°C for 30 min before measuring gassing power. Values are expressed as ml of CO2 per sample. In all cases, values represent the means of at least three independent experiments. The error associated with the points was calculated by using the formula: 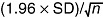 , where n is the number of measurements. Additional details are given in the Experimental procedures section.
, where n is the number of measurements. Additional details are given in the Experimental procedures section.
Selection of adaptive clones within the terminal population
Previous work has shown that polymorphisms can arise among microbial populations evolving under different conditions. Therefore, we tried to select adaptive clones within the experimental population with improved freeze tolerance. According to our initial hypothesis, we screened the terminal population in order to isolate adaptive clones with faster growth in LD at 12°C. As a first step, we cultured yeast cells on solid LD in Petri dishes at 12°C and selected those that formed the biggest colonies. After inspecting no less than 2000 colonies, we selected 24 clones, CR1 to CR24 (data not shown). Then, we analysed these selected clones in two different ways. First, we calculated the average colony size of each of the individual clones grown on solid LD at 12°C. Second, we determined, by means of an impedance analyser, the freeze‐provoked loss of CO2 production in LD samples stored for 14 days at −20°C. However, there was no correlation between these two parameters in the set of clones tested (see Fig. S1). As a result, we decided to randomly select two of these clones, CR19 and CR20, for further characterization. Because the results of all the subsequent analyses were found to be consistent for both clones, we only report in most cases those obtained from the CR20 strain.
The evolved clones show enhanced growth and freeze tolerance
As expected, the growth rate in LD at 12°C of the evolved strain CR20 was significantly higher, 0.055 ± 0.003 h−1, than that of the parental strain reported above (Table 1). However, there were no significant differences between this and the entire 200‐generation evolved population (Table 1). Similarly, the evolved strain demonstrated higher freeze tolerance than the parental population, but again differences in this parameter between the isolated clone and the 200‐generation heterogeneous population were scarce. CO2 production for the CR20 strain in 14‐day frozen samples was 101.4 ± 8.8 ml, around 58% of that found for the corresponding unfrozen control, while the parental and terminal population maintained for this time‐period at frozen‐storage, 23% and 54%, respectively, of their CO2 production capacity in the LD model system (Fig. 1). Similar results were observed for the CR19 strain (data not shown). Hence, the analysed evolved clones had similar properties to those of the evolved population from which they were derived.
CO2 production in flour‐based dough
We compared the gassing power of yeast cells from the parental and the CR19 and CR20 strains inoculated in bread dough after freezing and frozen storage. CO2 production by all the yeast strains analysed was strongly reduced following 21 days of storage at −20°C, as expected, but again there were significant differences between parental and evolved cells. Thus, survival of yeast cells, as measured by CO2 production, was higher for both of the evolved strains tested (Fig. 2). Hence, the enhanced freeze tolerance measured in the LD model system also applies in real dough samples.
Figure 2.
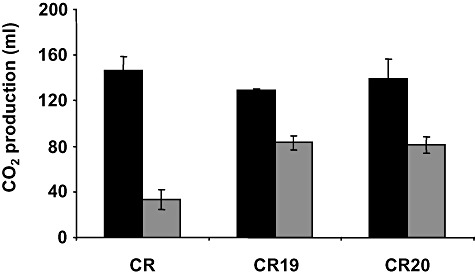
The evolved strains show increased freeze tolerance in flour‐based dough. Molasses‐plate‐grown cells of the parental CR and evolved CR19 and CR20 baker's yeast strains were used to prepare lean dough as described in the Experimental procedures section. Samples were quickly frozen at −80°C for 1 h and stored at −20°C for 21 days. Then, the frozen dough (grey bars) was left to thaw at 30°C for 30 min and CO2 production was recorded in a home‐made fermentometer (Chittick apparatus). Unfrozen samples (black bars) were used as control. Values are expressed as ml of CO2 produced after 180 min of dough fermentation and represent the means of at least three independent experiments. The error was calculated as described in Fig. 1.
Phenotypic stability and genome size
We were interested in determining whether the observed effects of the long‐term cultivation experiment in LD at 12°C were the consequence of genetic changes or a mere acclimatization. First, we tested the stability of the evolved clone CR20. After 50 generations of exponential growth in YPD at 30°C, cells maintained the enhanced freeze tolerance observed in the initial CR20 population (data not shown). Then, we analysed the average ploidy level of the parental CR and evolved CR20 strains by flow cytometry. Cells of approximately pentaploid DNA content (∼ 4.8n) were observed for the parental strain (Fig. 3). This ploidy level is in good correspondence with the five copies of the TRP1 gene found in a likely isogenic commercial baker's yeast strain (Estruch and Prieto, 2003). However, cells of CR20 were approximately tetraploid (3.9n, Fig. 3). Hence, selection for 200 generations in the LD system at 12°C favoured a reduction in the genomic content of the parental CR strain.
Figure 3.
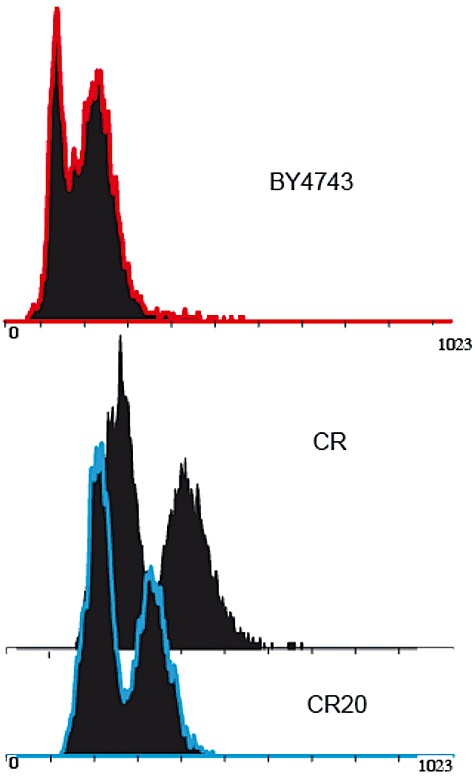
Flow cytometric analysis is used to estimate approximate genome size. Logarithmic YPD‐grown cells of the industrial (CR and CR20) and laboratory (BY4743) S. cerevisiae strains were stained with propidium iodine and the DNA content of 30 000 individuals cells was measured by flow citometry. The horizontal axes in the graphs are measures of dye fluorescence and are proxies for genome size. Diploid cells of the BY4743 strain were used as control. The first peak indicates the unreplicated DNA content of the population, G1 phase (2n for the BY4743 strain), while the second peak indicates the replicated DNA content, G2 phase (4n for the BY4743 strain). A representative experiment is shown.
Continuous batch culture at 12°C does not confer better fitness at low temperature
We examined whether selection at low temperature is the evolutionary force that drove the physiological response observed in the experimental populations. First, we tested growth under non‐restrictive conditions. To our surprise, evolved cells showed a slight growth defect as compared with parental cells on YPD at both 30°C (Fig. 4A and B) and 12°C (Fig. 4A). On the contrary, the evolved strains showed better growth than the parental strain on solid LD even at 30°C (Fig. 4B). Similar phenotypes were observed for the entire terminal population. Indeed, the parental CR strain grew slightly better than the 200‐generation evolved population in liquid YPD at both 12°C and 30°C (Table 1). Hence, yeast cells were unable to adapt to low temperature per se when cultivated in the LD system.
Figure 4.
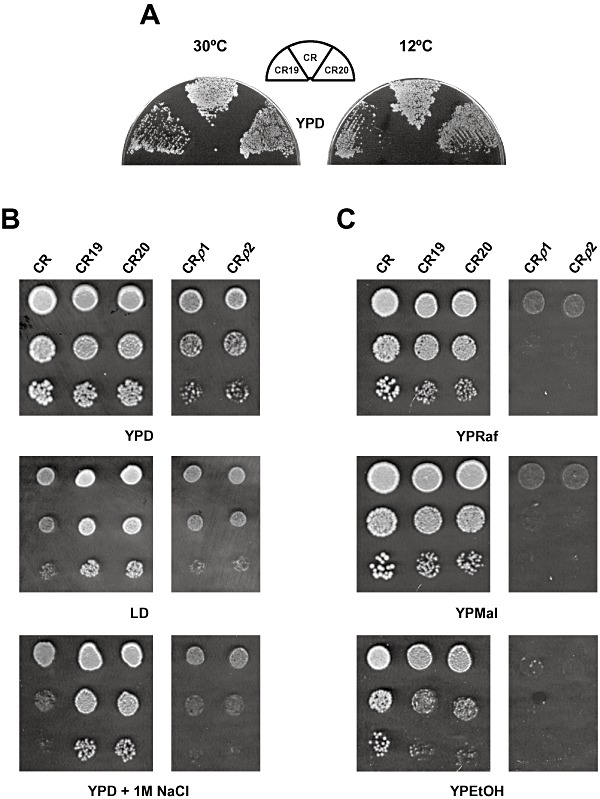
Phenotypic characterization of evolved clones and petite mutants. Cells of the parental CR and evolved CR19 and CR20 strains were assayed for growth on different culture media and/or conditions. (A) YPD at 30°C or 12°C. (B) YPD, LD or YPD containing 1 M NaCl at 30°C. (C) YP containing raffinose (YPRaf), maltose (YPMal) or ethanol (YPEtOH) as the sole carbon source at 30°C. In some cases, two petite yeast mutants of the CR strain, CRρ1 and CRρ2, were tested under the same conditions (panel B and C). YPD‐exponentially growing cultures (OD600 = 1.0) were diluted (1–10−3) and aliquots were extended (10 µl 10−3, A) or spotted (2.5 µl 1–10−2, B and C) on Petri dishes. Cells were inspected for growth after 2–4 (30°C) or 10 (12°C) days. Results of a representative experiment are shown.
At this point, we rationalized that those genetic changes that give cells better fitness at 12°C might be purged from cultures if they compromised growth under other stress conditions. To examine this possibility, yeast cells of the parental CR strain were cultured for 200 generations in liquid YPD medium at 12°C, and the parental and evolved cells were analysed for growth differences. Again, the estimated µmax in YPD at 12°C was again slightly higher for the parental, 0.116 ± 0.005 h−1, than for the terminal population, 0.103 ± 0.004 h−1. Similar results were observed when cells of another commercial baker's yeast strain, Plus Vital (PV), were selected under the same conditions. Indeed, there was a small advantage in the maximum growth rate at 12°C of the parental versus the evolved population, 0.105 ± 0.003 and 0.095 ± 0.004 h−1 respectively. No other apparent phenotypic characteristic was found to be altered in response to selection of baker's yeast cells in YPD at 12°C (data not shown).
NaCl resistance is the main target of evolution in the LD system at 12°C
The LD model system contains sorbitol and NaCl (Panadero et al., 2005a). Therefore, we analysed the contribution of high osmotic pressure on the selection process. Exposure to pure hyperosmolarity provided by 1–2 M sorbitol did not seem to exert any differential influence on growth, because all the strains analysed grew equally (data not shown). In contrast, cells of the evolved clones showed enhanced growth on 1 M NaCl‐containing plates (Fig. 4B), indicating a marked resistance to the toxic effects of this salt. To confirm this trait, the parental and the 50‐, 100‐ and 200‐generation evolved populations were grown at 30°C in liquid 1 M NaCl‐YPD and the µmax of growth was estimated (Table 1). As can be seen, the 50‐generation evolved population displayed a significant increase in its ability to grow in the presence of NaCl as compared with the parental population. Moreover, the magnitude of this difference was greater over the course of the evolutionary experiment (Table 1). Hence, NaCl tolerance seemed to be the main response to selection of yeast cells in LD at 12°C.
Physiological characterization of the evolved strains
We assayed the growth of the parental, CR19 and CR20 strains in different culture media. Like on glucose (Fig. 4B, YPD), cells of the evolved strains showed a slight growth defect when maltose or raffinose, were supplied as the sole carbon source (Fig. 4C). This phenotype was more evident when cells were cultured on ethanol (Fig. 4C). Hence, increased fitness in LD at 12°C compromises growth performance under non‐limiting conditions.
Subsequently, we analysed how sugars were consumed during LD fermentation and the level of metabolic by‐products formed (Fig. 5). The level of residual glucose during 180 min of fermentation showed almost the same profiles for the parental and the evolved strain CR20, and the same trend was observed for maltose (Fig. 5A), for which consumption commenced when the glucose level was low (approximately 90 min). Similarly, there was no significant difference in the kinetics of ethanol and glycerol formation, the two main products of yeast fermentation (Fig. 5B). Only at a late stage did the ethanol level of the evolved strain appear to be a little higher than that found in the parental strain.
Figure 5.
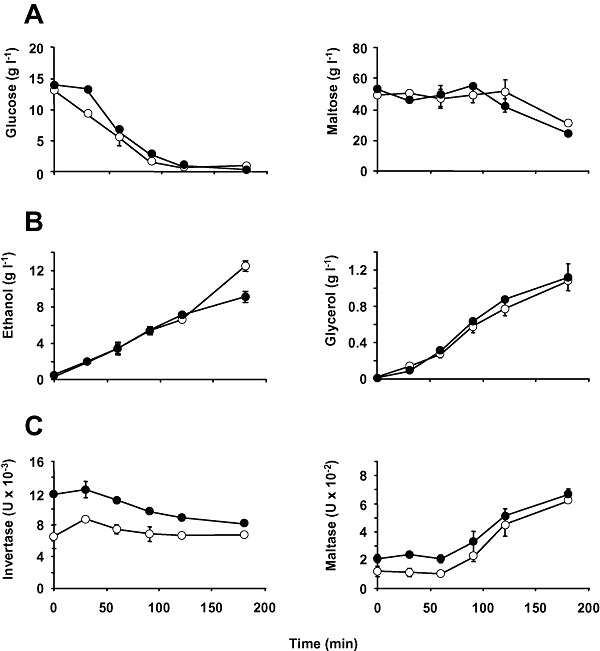
Enzymatic and metabolic profile. Molasses‐grown cells of the parental CR (●) and evolved CR20 (○) strains were transferred to LD medium and incubated at 30°C. At the indicated times, samples of the cultures were taken for further analysis. A. Residual glucose and maltose in the culture supernatant. B. Production of glycerol and ethanol. C. Invertase and maltase activity in cell‐free extracts. Experimental details are given in the Experimental procedures section. In all cases, values represent the means of at least three independent experiments. The error associated with the points was calculated as described in Fig. 1.
We also measured the invertase and maltase activities: the key enzymes for the metabolism of yeast cells during yeast propagation and fermentation processes. Quite remarkably, the initial levels of both enzyme activities were significantly lower, around twofold, in the evolved CR20 strain compared with the parental strain (Fig. 5C). Thereafter, enzyme activities in the two strains were concordant. The level of invertase decreased slightly over the time‐course of the fermentation experiment, whereas maltase activity increased in the presence of maltose in the LD system (Fig. 5C).
The evolved strains do not show a petite phenotype
We examined in more depth the phenotype of the evolved strains on ethanol. First, we tested whether the growth defect was due to a mitochondrial respiration deficiency (petite phenotype), by measuring the presence of functional cytochrome c oxidase activity. For this, we isolated two petite yeast mutants of the CR strain by growing cells on YPD plates in the presence of ethidium bromide and used them as controls. Our results showed that the activity of this mitochondrial electron transport chain enzyme was apparently unaffected by the evolutionary experiment (see Fig. S2). Then, we tested the growth behaviour of the petite mutants. As can be seen in Fig. 4, cells of the petite mutants exhibited a severe deficiency in utilizing raffinose or maltose as a carbon source, and a complete inability to grow on ethanol (Fig. 4C). As in the parental strain, they exhibited NaCl‐sensitivity (Fig. 4B), and thus, cultures from the petite mutants showed a similar behaviour to the parental strain on LD at 30°C (Fig. 4B). Altogether, our results appear to discount a mitochondrial functional deficiency as the cause of the growth phenotype on ethanol of the evolved population.
The catabolite derepression process is altered in the CR20 strain
We analysed whether the CR20 strain might be altered in the glucose derepression process as suggested by the invertase and maltase data shown above (Fig. 5C). Cells of the parental and evolved strain were grown on glucose, transferred to ethanol and assayed for the gluconeogenic activities fructose 1,6‐bisphosphatase (FbPase) and phosphoenolpyruvate carboxykinase (PEPCK), as well as for isocitrate lyase, an enzyme of the glyoxalate cycle (Table 2). These enzymes are targets of glucose repression, and are strictly required for growth on ethanol when it is the sole carbon source (Gancedo, 1998). As expected, the activity of all of these enzymes was almost undetectable in glucose‐grown cells of either the parental or the evolved CR20 strain (data not shown). Both strains were also sensitive to the absence of glucose. However, the level of FbPase, PEPCK and isocitrate lyase displayed significantly lower values in the evolved than in the parental strain after 4 h of ethanol culture (Table 2).
Table 2.
Derepression of fructose 1,6‐bisphosphatase (FbPase), phosphoenolpyruvate carboxykinase (PEPCK) and isocitrate lyase.
| Strain | Activity (nmol min−1 mg−1 of protein)a | ||
|---|---|---|---|
| FbPase | PEPCK | Isocitrate lyase | |
| CR | 10 ± 1 | 46 ± 5 | 17 ± 3 |
| CR20 | 4 ± 1 | 17 ± 2 | 10 ± 2 |
Enzymatic activities were measured in cell‐crude extracts after 4 h in YP‐Ethanol medium, as described in the text. Values are mean of at least three independent experiments.
Discussion
The aim of this work was to obtain evolved industrial strains with improved freeze tolerance. In agreement with this, we show here that a baker's yeast population that has evolved at 12°C in a dough‐like environment exhibits enhanced CO2 production capability after freezing and frozen storage. Moreover, the evolved population displays NaCl tolerance. To our knowledge, this is the first report of a NaCl‐resistant industrial baker's yeast strain, a property that has commercial interest. The addition of salt to bread dough has harmful effects on yeast performance (Hernandez‐Lopez et al., 2003) and consequently, NaCl resistance is considered important in the combination of traits that an industrial strain should exhibit (Randez‐Gil et al., 2003). Hence, the evolved strains reported in our study represent a clear example of the potential of targeted evolutionary approaches in the development of novel industrial strains.
The initial hypothesis of this study was that baker's yeast adapts to cold environments, and that improvements in this trait may have positive effects on freeze tolerance. However, the assumption of low temperature adaptation in S. cerevisiae is not supported by our experimental results. Baker's yeast cells that evolved in either LD or YPD at 12°C did not experience phenotypic adaptation to this temperature. On the contrary, both evolutionary experiments caused a negative effect on growth of the industrial strain under non‐restrictive conditions (YPD medium) at 12°C, which was accompanied by a similar loss of specific growth rate at 30°C. Hence, it seems that, at least under our experimental conditions, low temperature per se did not impose a selective pressure on industrial baker's yeast.
The above observations do not necessarily mean that S. cerevisiae is unable to achieve low temperature adaptation. Aneuploidy and/or polyploidy is a common feature among industrial yeasts that has been retained, or even promoted, by prolonged cultivation under nutrient‐limiting conditions, such as those used for biomass production (Higgins et al., 2001). Conversely, large‐scale transitions in genome size from tetraploid or triploid to diploid, the predominant vegetative state of S. cerevisiae, have been observed during long‐term evolution experiments in non‐limiting carbon conditions (Gerstein et al., 2006; 2008). At this point, a loss of ploidy, like that observed in the evolved CR20 strain in this study, might represent an evolutionary advantage for cells grown under our experimental conditions. However, this genomic adjustment might have masked subtle increases in relative fitness of the evolved population at 12°C. Hence, comparative experiments employing haploid strains should be conducted to provide evidence of the real adaptive flexibility of S. cerevisiae to low temperature.
Selection in LD medium at 12°C increased NaCl tolerance in baker's yeast cells. This is consistent with the presence of this stressor in the LD model system and suggests that high NaCl levels are perceived as a lethal stress, able to induce adaptive evolution. Growing evidence suggests that a variety of environmental stresses are able to induce a mutagenesis program, which promotes local concerted evolution (Heidenreich, 2007). This would appear not to be the case of high osmotic pressure, since cells of the parental and evolved populations grew equally in the pure high osmolarity provided by sorbitol. It also seems that genetic changes in the evolved strain affected critical processes specific for NaCl tolerance, not related to high osmolarity. Although the transcriptional responses to osmotic and NaCl stresses are remarkably similar to each other (Causton et al., 2001), the mechanisms of halo‐ and osmo‐tolerance appear to be rather different. Consistent with this view, key genes involved in salt tolerance do not seem to participate in osmotic adjustment. For instance, expression of HAL1, which is involved in K+ homeostasis (Gaxiola et al., 1992), and ENA1, encoding the main P‐type ATPase that mediates the active efflux of Na+ (Haro et al., 1991), is enhanced both by osmotic and saline stress (Marquez et al., 1998). However, overexpression of HAL1 confers NaCl, but not osmotic tolerance (Gaxiola et al., 1992). Similarly, mutants in the ENA1 gene are NaCl sensitive, but they do not show osmo‐sensitivity (Haro et al., 1991). Thus, the existence of a complete genetic association between salt‐ and osmo‐tolerance is uncertain.
We observed adaptation to NaCl and freeze resistance in the evolved population. This observation is consistent with an important role of ionic stress in cell injury during freezing, as has been previously suggested (Wolfe and Bryant, 1999). Recently, we have shown that overproduction of Crz1p, the calcineurin‐target transcription factor (Stathopoulos and Cyert, 1997), increases yeast tolerance to both NaCl and freeze stress (Panadero et al., 2007). The calcineurin/Crz1p pathway is the main pathway implicated in the NaCl response (Rusnak and Mertz, 2000; Cyert, 2003) and its activation induces the expression of most NaCl‐responsive genes (Yoshimoto et al., 2002).
As expected, improvements in NaCl tolerance appeared to be achieved at the expense of others traits. Indeed, evolved cells showed a clear deterioration in their ability to utilize gluconeogenic carbon sources. This inability probably reflects a defect in the glucose derepression mechanism, since the CR20 strain achieved twofold lower invertase‐ and maltase‐specific activity levels, two enzymes sensitive to catabolite repression (Gancedo, 1998), than the parental CR strain in molasses medium. The activity of key enzymes of the glyoxylate cycle and gluconeogenesis was also lower in ethanol cultures of the CR20 strain. Synthesis of these enzymes is essential for the assimilation of ethanol and other gluconeogenic carbon sources, and is tightly controlled by glucose repression (Gancedo, 1998). It also seems plausible that the growth defect on ethanol of the CR20 strain might reflect a pleiotropic side effect of the same genetic changes that produced adaptation to NaCl, as has been suggested in other evolution experiments (Cooper and Lenski, 2000). Thus, recent theoretical work suggests that most of the phenotypic change during an episode of adaptation can result from the selection of a few mutations with relatively large effects (Zeyl, 2005).
Overall, our study demonstrates the potential of adaptive evolution to achieve industrial yeast strains with better characteristics. It also highlights how the relationships between different stresses may influence the final results. This is of major importance for designing the conditions for targeted evolution experiments. In our case, it seems that the observed improvements in CO2 production after freezing could be a result of the genetic adaptation to NaCl stress by the yeast, and not to low temperatures per se. However, due to the complexity of the culture medium employed, it cannot be ruled out that the selection pressure acting on the final phenotype is due to synergistic effects or interactions between different stressors. Remarkably, we observed that increases in freeze tolerance were limited and did not vary in magnitude when cells were cultivated longer than 50 generations. Freezing is a complex and multifaceted stress, in which different stress responses appear to play important roles (Tanghe et al., 2003; Aguilera et al., 2007). In this scenario, a better knowledge of targets for resistance to freezing damage would be a valuable tool for the rational design of selective environments that could drive further improvements in the desired phenotype.
Experimental procedures
Yeast strains and culture media
Two commercial baker's yeast strains, Cinta Roja (CR) and PV, produced by Burns Philp Food S.A. (Cordoba, Spain) and CGL (Lesaffre Group, Valladolid, Spain), respectively, were used in the evolutionary experiments reported in this study. The diploid S. cerevisiae BY4743 laboratory strain was also used in different experiments. Cells were regularly maintained on solid YPD containing 20 g l−1 agar, 20 g l−1 peptone, 20 g l−1 glucose and 10 g l−1 yeast extract. For the evolution experiments yeast cells were grown in a flour‐free LD model system (Panadero et al., 2005a). Briefly, this consisted of 45 g l−1 maltose, 15 g l−1 glucose, 4.7 g l−1 (NH4)2HPO4 and 2.5 g l−1 yeast extract, buffered with 14.14 g l−1 sodium citrate at pH 5.5 (adjusted with citric acid). The medium also contained 60 g l−1 sorbitol (∼0.33 M) and 13.5 g l−1 NaCl (∼0.23 M), which reduce water activity to bread dough values (aw ∼ 0.97), and a mixture of essential nutrients, 2 g l−1 MgSO4·7H2O, 0.8 g l−1 KCl, 40 mg l−1 nicotinic acid, 4.0 mg l−1 thiamine and 4.0 mg l−1 pyridoxine.
Prolonged cultivation at low temperature
Evolution experiments were conducted using batch culture techniques in a similar manner than that described by Gerstein and colleagues (2006). Fifty millilitres of medium, LD or YPD, was inoculated at OD600 = 0.01 and cultured in 250 ml Erlenmeyer flasks in a cooling incubator set at 12°C and 200 r.p.m. When OD600 reached 1 (±0.2), the culture was transferred to a new 12°C pre‐cooled flask (500 µl culture into 50 ml medium) and cultivated in the same way until a minimum of 200 generations was attained. As each transfer allowed ∼6.64 mitotic divisions, a total of ∼35 transfers were carried out in each independent experiment. At 50, 100, 150 and 200 generations, samples from the experimental population were taken, maintained as frozen (−80°C) glycerol stocks and then rescued in YPD agar plates at 30°C for 24 h before further analysis. These conditions preserved the characteristics of the evolved populations and ensured that yeast cells were growing under the same conditions.
Growth measurements
Cells were grown in YPD at 30°C to mid‐exponential phase, collected by centrifugation (3000 g, 2 min, 4°C), washed, transferred to fresh LD, YPD or YPD containing 1 M NaCl, and incubated at 30°C or 12°C as indicated, after which growth was monitored. Doubling times (g) were calculated from the formula g = ln2/µ, where µ is the specific growth rate constant of the culture. µ was calculated from the slope of the line obtained after plotting ln X versus t, where X is the cell density of the culture, measured as OD600, at multiple time points (t) during logarithmic growth.
Gas production assays
From an overnight YPD batch culture, yeasts were centrifuged, washed with H2O and extended onto 140‐mm‐diameter plates (10 units of OD600 per plate) containing molasses agar [5.0 g l−1 beet molasses (49% sucrose), 0.5 g l−1 (NH4)2HPO4, 26.0 g l−1 agar and 20 mg l−1 biotin; adjusted to a final pH of 5.0]. Plates were incubated for 22–24 h at 30°C. Cells were then recovered by washing the plate surface with 2 × 3 ml of distilled water, and saving the yeast suspension. Collections from several plates were mixed, centrifuged for 5 min at 4500 r.p.m. and resuspended in distilled water (4°C) containing 27 g l−1 of NaCl, vortexed, and the OD600 of the resulting suspension measured. The final yeast concentration was adjusted to 30 mg (dry weight) ml−1 (OD600 = 1 equals 0.35 mg (cells dry weight) ml−1; Panadero et al., 2005a). Fifteen millilitres of the yeast mixture was poured into a 100‐ ml screw cap graduated bottle, placed in a 30°C water bath and gently shaken (80 r.p.m.). After 15 min, 15 ml of 30°C pre‐warmed 2× LD solution lacking NaCl was added and the amount of CO2 produced recorded in a home‐made fermentometer (Chittick) as previously described (Panadero et al., 2005a). For flour‐based dough experiments, 15 ml of the yeast suspension was poured into a 30°C pre‐warmed 250 ml screw cap graduated bottle containing 25 g of wheat flour. The ingredients were mixed gently with a glass rod, and the resulting homogenous dough was incubated at 30°C in a water bath, connected to the Chittick apparatus and pre‐incubated for 10 min before CO2 production was recorded. Samples for freezing were kept at −80°C for 1 h and then stored at −20°C. At different times, they were thawed at 30°C for 30 min before measuring gassing power. In all cases, CO2 production was recorded for 180 min. Values are expressed as ml of CO2 per sample.
Alternatively, CO2 production was estimated by impedance techniques. For this purpose, cells were grown in Petri dishes containing molasses agar, recovered and washed as described above. A volume equivalent to 4.9 mg of biomass (dry weight) was centrifuged, resuspended in 4 ml of saline solution, and mixed with an equal volume of 2× LD without NaCl. Production of CO2 was indirectly estimated in a BacTrac 4300 Microbiological Analyzer (SY‐LAB Geräte GmbH, Austria), by measuring the evolution of the impedance of a solution of 2 g l−1 KOH at 30°C. Strains with higher gas production were those able to reduce impedance to 50% in shorter time.
Colony size analysis
For selection of individuals with bigger colony size from the terminal population, cells were grown overnight in YPD and then centrifuged, washed and resuspended in H2O to a final OD600 = 1. The suspension was diluted to 10−4 and extended on Petri dishes (100 µl per plate, corresponding to 80–100 cfu) containing LD agar medium. Plates were incubated at 12°C for 10 days. At least 2000 colonies were visually inspected, and bigger colonies were rescued.
Average colony size was measured as follows: cells were grown, diluted, plated and incubated as described (1 plate per clone). Plates were then scanned, and the image transformed to a black and white image. Colony area was measured with the Image Gauge software (Fujifilm, Kanagawa, Japan), and the average colony size was calculated for each plate, corresponding to each of the selected individuals.
Genome size
Flow cytometric analysis (Hutter and Eipel, 1979) was used to estimate the approximate genome size of the industrial yeast strains. Cells were grown in liquid YPD to exponential‐phase, harvested, washed and fixed in 70% ethanol at 4°C for 5 min. Then, cells were collected by centrifugation and resuspended in 10 mM PBS buffer (pH 7.2), containing 400 µl of RNase (10 mg ml−1). After incubation at 37°C for 30 min, cells were harvested by centrifugation and resuspended in 1 ml of the same buffer containing 2.5 mg l−1 of propidium iodine. Samples were analysed using a flow cytometer FACScan analyser (Becton Dickinson, USA). Ploidy determinations were done by comparing with the diploid BY4743 strain.
Phenotype analysis
For plate growth analysis, cells were grown in YPD and then washed, adjusted to OD600 = 1 and diluted to 10−3. Ten microlitres of the yeast suspension was extended on Petri dishes containing LD agar, YPD agar, YPD agar plus 1 M NaCl or 1 M sorbitol, YP agar plus 20 g l−1 maltose, 20 g l−1 sucrose, 20 g l−1 raffinose or 30 g l−1 ethanol. More‐detailed sensitivity assays were performed by spotting (2.5 µl) 10‐fold serial dilutions of the cell culture. Unless otherwise indicated, colony growth was inspected after 2–5 days of incubation at 30°C.
Preparation of cell‐free extracts
Cells (approximately 15 units of OD600) from ethanol‐grown cultures were harvested by centrifugation (3000 g, 2 min, 4°C), washed twice with chilled distilled water, resuspended in 20 mM imidazole‐HCl buffer, pH 7.0, and transferred into a tube containing 1.0 g of glass beads (acid‐washed, 0.4 mm diameter). The mixture was vortexed at maximum speed for 3 min, then centrifuged at 12 000 g for 10 min (4°C), and the supernatant was used for the determination of FbPase, PEPCK and isocitrate lyase activity. For maltase and invertase measurements, cells grown on molasses plates were recovered, washed, resuspended in distilled water (4°C) containing 27 g l−1 of NaCl, and the yeast suspension mixed with an equal volume of 2× LD solution lacking NaCl. Cells were incubated at 30°C and samples of 2 ml were taken at different times. Cell‐free extracts were obtained as above except that 0.1 M potassium phosphate adjusted to pH 6.5 was used as buffer (KPB).
Enzyme activity determinations
Maltase (Okada and Halvorson, 1964), invertase (Niederacher and Entian, 1987), FbPase (Gancedo and Gancedo, 1971), PEPCK (Hansen et al., 1976) and isocitrate lyase (Dixon and Kornberg, 1959) were determined spectrophotometrically. All assays were performed at 30°C. One unit is defined as the amount of enzyme transforming 1 nmol of substrate per minute under the assay conditions. Total protein was determined using the commercial Bio‐Rad protein assay kit and rabbit IgG as standard.
Metabolite analysis
Glucose was measured colorimetrically by the glucose oxidase and peroxidase method in the presence of 2,2′‐azino‐bis(3‐ethylbenzothiazoline‐6‐sulfonic acid) diammonium salt (ABTS, Sigma), following manufacturer's instructions. Maltose and sucrose were measured by hydrolysis with maltase (α‐glucosidase) and invertase, respectively, as previously described (Hernandez‐Lopez et al., 2003), and subsequent determination of the quantity of released glucose. Glycerol and ethanol were measured with commercial kits (Roche) following manufacturer's instructions.
Induction and analysis of petite yeast mutants
Petite yeast mutants of the CR strain were obtained by growing cells onto YPD plates in the presence of ethidium bromide and used as control. Mutants were detected following the protocol described by McEwen and colleagues (1985), using tetramethyl‐p‐phenylenediamine as a cytochrome c oxidase activity stain for yeast colonies. Colonies containing cytochrome c oxidase were stained blue, whereas cytochrome c oxidase‐deficient mutants remained white or were stained light blue.
Acknowledgments
We thank A. Blasco and B. Esteve‐Zarzoso for technical assistance. This research was jointly funded by the Spanish Ministry of Science and Technology (CICYT projects, AGL2004‐00462 and AGL2007‐65498‐C02‐01) and the EU's Sixth Framework Program (Marie Curie Reintegration Grants). J.A. was the recipient of a post‐doctoral contract within the ‘I3P’ Program from CSIC.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Correlation between growth in LD at 12°C and freeze tolerance of evolved clones. Twenty‐four individuals sampled from the 200‐generation terminal population of our evolutionary experiment were analysed for growth in plates of LD agar at 12°C and freeze tolerance. Growth was assumed to be proportional to the colony size and was estimated by measuring the average colony area of at least 80–100 colonies corresponding to each of the selected individuals. Relative freeze tolerance was estimated by measuring the loss of CO2 production in an impedance analyser for LD samples stored for 14 days at −20°C. Unfrozen samples were used as control. Data points are the mean of at least two independent experiments. The correlation coefficient (r2) for the linear regression of relative freeze tolerance against average colony size was 0.046.
The strains evolved in LD at 12°C are not deficient in respiration. Cells of the CR and CR20 strains, and two independent petite mutants from the parental CR strain (notated as CRρ1 and CRρ2) were spotted onto YPD agar plates and cultured for 2–3 days. Colonies were transferred to a filter paper and assayed for cytochrome c oxidase by the addition of the electron donor tetramethyl‐p‐phenylenediamine (TMPD). Colonies oxidizing TMPD immediately acquired a dark blue colour, whereas cells lacking the oxidase activity remained white.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adams J., Oeller P.W. Structure of evolving populations of saccharomyces cerevisiae: adaptive changes are frequently associated with sequence alterations involving mobile elements belonging to the Ty family. Proc Natl Acad Sci USA. 1986;83:7124–7127. doi: 10.1073/pnas.83.18.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera J., Randez‐Gil F., Prieto J.A. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol Rev. 2007;31:327–341. doi: 10.1111/j.1574-6976.2007.00066.x. [DOI] [PubMed] [Google Scholar]
- Attfield P.V. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat Biotechnol. 1997;15:1351–1357. doi: 10.1038/nbt1297-1351. [DOI] [PubMed] [Google Scholar]
- Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper V.S., Lenski R.E. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Cyert M.S. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun. 2003;311:1143–1150. doi: 10.1016/s0006-291x(03)01552-3. [DOI] [PubMed] [Google Scholar]
- Dequin S. The potential of genetic engineering for improving brewing, wine‐making and baking yeasts. Appl Microbiol Biotechnol. 2001;56:577–588. doi: 10.1007/s002530100700. [DOI] [PubMed] [Google Scholar]
- Diniz‐Mendez L., Bernardes E., De Araujo P.S., Panek A.D., Paschoalin V.M.F. Preservation of frozen yeast cells by trehalose. Biotechnol Bioeng. 1999;65:572–578. doi: 10.1002/(sici)1097-0290(19991205)65:5<572::aid-bit10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Dixon G.H., Kornberg H.L. Assay methods for key enzymes of the glyoxylate cycle. Biochem J. 1959;72:3p. [Google Scholar]
- Dykhuizen D.E. Mountaineering with microbes. Nature. 1990;346:15–16. doi: 10.1038/346015a0. [DOI] [PubMed] [Google Scholar]
- Estruch F., Prieto J.A. Construction of a Trp‐ commercial baker's yeast strain by using food‐safe‐grade dominant drug resistance cassettes. FEMS Yeast Res. 2003;4:329–338. doi: 10.1016/S1567-1356(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Gancedo J.M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J.M., Gancedo C. Fructose‐1,6‐diphosphatase, phosphofructokinase and glucose‐6‐phosphate dehydrogenase from fermenting and non fermenting yeasts. Arch Microbiol. 1971;76:132–138. doi: 10.1007/BF00411787. [DOI] [PubMed] [Google Scholar]
- Gaxiola R., De Larrinoa I.F., Villalba J.M., Serrano R. A novel and conserved salt‐induced protein is an important determinant of salt tolerance in yeast. EMBO J. 1992;11:3157–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A.C., Chun H.J., Grant A., Otto S.P. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A.C., McBride R.M., Otto S.P. Ploidy reduction in Saccharomyces cerevisiae. Biol Lett. 2008;4:91–94. doi: 10.1098/rsbl.2007.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães P.M., François J., Parrou J.L., Teixeira J.A., Domingues L. Adaptive evolution of a lactose‐consuming Saccharomyces cerevisiae recombinant. Appl Environ Microbiol. 2008;74:1748–1756. doi: 10.1128/AEM.00186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R.J., Hinze H., Holzer H. Assay of phosphoenolpyruvate carboxykinase in crude yeast extracts. Anal Biochem. 1976;74:576–584. doi: 10.1016/0003-2697(76)90240-2. [DOI] [PubMed] [Google Scholar]
- Haro R., Garciadeblas B., Rodriguez‐Navarro A. A novel P‐type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- Heer D., Sauer U. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microbial Biotechnol. 2008;1:497–506. doi: 10.1111/j.1751-7915.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich E. Adaptive mutation in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 2007;42:285–311. doi: 10.1080/10409230701507773. [DOI] [PubMed] [Google Scholar]
- Hermes‐Lima M., Storey K.B. Antioxidant defences in the tolerance of freezing and anoxia by garter snakes. Am J Physiol. 1993;265:R646–R652. doi: 10.1152/ajpregu.1993.265.3.R646. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Lopez M.J., Prieto J.A., Randez‐Gil F. Osmotolerance and leavening ability in sweet and frozen sweet dough. Comparative analysis between Torulaspora delbrueckii and Saccharomyces cerevisiae baker's yeast strains. Antonie Van Leeuwenhoek. 2003;84:125–134. doi: 10.1023/a:1025413520192. [DOI] [PubMed] [Google Scholar]
- Higgins V.J., Bell P.J., Dawes I.W., Attfield P.V. Generation of a novel Saccharomyces cerevisiae strain that exhibits strong maltose utilization and hyperosmotic resistance using nonrecombinant techniques. Appl Environ Microbiol. 2001;67:4346–4348. doi: 10.1128/AEM.67.9.4346-4348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R., Yokoigawa K., Isobe Y., Kawai H. Improving the freeze tolerance of bakers’ yeast by loading with trehalose. Biosci Biotechnol Biochem. 2001;65:522–526. doi: 10.1271/bbb.65.522. [DOI] [PubMed] [Google Scholar]
- Hutter K.J., Eipel H. Microbial determinations by flow cytometry. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Kandror O., Bretschneider N., Kreydin E., Cavalieri D., Goldberg A.L. Yeast adapt to near‐freezing temperatures by STRE/Msn2,4‐dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- Kronberg M.F., Nikel P.I., Cerrutti P., Galvagno M.A. Modelling the freezing response of baker's yeast prestressed cells: a statistical approach. J Appl Microbiol. 2007;104:716–727. doi: 10.1111/j.1365-2672.2007.03588.x. [DOI] [PubMed] [Google Scholar]
- Kuyper M., Winkler A.A., Van Dijken J.P., Pronk J.T. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 2004;4:655–664. doi: 10.1016/j.femsyr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- McEwen J.E., Cameron V.L., Poyton R.O. Rapid method for isolation and screening of cytochrome c oxidase‐deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1985;161:831–835. doi: 10.1128/jb.161.3.831-835.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maris A.J., Geertman J.M., Vermeulen A., Groothuizen M.K., Winkler A.A., Piper M.D. Directed evolution of pyruvate decarboxylase‐negative Saccharomyces cerevisiae, yielding a C2‐independent, glucose‐tolerant, and pyruvate‐hyperproducing yeast. Appl Environ Microbiol. 2004;70:159–166. doi: 10.1128/AEM.70.1.159-166.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez J.A., Pascual‐Ahuir A., Proft M., Serrano R. The Ssn6‐Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG‐dependent and –independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.J., Coulson G.E., Clarke K.J. Freezing injury in Saccharomyces cerevisiae. The effects of growth conditions. Cryobiology. 1988;25:471–472. [Google Scholar]
- Murga M.L., Cabrera G.M., De Valdez G.F., Disalvo A., Seldes A.M. Influence of growth temperature on cryotolerance and lipid composition of Lactobacillus acidophilus. J Appl Microbiol. 2000;88:342–348. doi: 10.1046/j.1365-2672.2000.00967.x. [DOI] [PubMed] [Google Scholar]
- Myers D.K., Attfield P.V. Intracellular concentration of exogenous glycerol in Saccharomyces cerevisiae provides for improved leavening of frozen sweet doughs. Food Microbiol. 1999;16:45–51. [Google Scholar]
- Niederacher D., Entian K.‐D. Isolation and caracterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987;206:505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- Okada H., Halvorson H.O. Uptake of alpha‐thioethyl‐glucopyranoside by Saccharomyces cerevisiae. 1. The genetic control of facilitated diffusion and active transport. Biochim Biophys Acta. 1964;82:538–542. doi: 10.1016/0304-4165(64)90445-3. [DOI] [PubMed] [Google Scholar]
- Panadero J., Randez‐Gil F., Prieto J.A. Validation of a flour‐free model dough system for throughput studies of baker's yeast. Appl Environ Microbiol. 2005a;71:1142–1147. doi: 10.1128/AEM.71.3.1142-1147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panadero J., Randez‐Gil F., Prieto J.A. Heterologous expression of type I antifreeze peptide GS‐5 in baker's yeast increases freeze tolerance and provides enhanced gas production in frozen dough. J Agric Food Chem. 2005b;53:9966–9970. doi: 10.1021/jf0515577. [DOI] [PubMed] [Google Scholar]
- Panadero J., Pallotti C., Rodríguez‐Vargas S., Randez‐Gil F., Prieto J.A. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J Biol Chem. 2006;281:4638–4645. doi: 10.1074/jbc.M512736200. [DOI] [PubMed] [Google Scholar]
- Panadero J., Hernández‐López M.J., Prieto J.A., Randez‐Gil F. Overexpression of the calcineurin target CRZ1 provides freeze tolerance and enhances the fermentative capacity of baker's yeast. Appl Environ Microbiol. 2007;73:4824–4831. doi: 10.1128/AEM.02651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C., Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- Park J.I., Grant C.M., Attfield P.V., Dawes I.W. The freeze‐thaw stress response of the yeast Saccharomyces cerevisiae is growth phase specific and is controlled by nutritional state via the RAS‐cyclic AMP signal transduction pathway. Appl Environ Microbiol. 1997;63:3818–3824. doi: 10.1128/aem.63.10.3818-3824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik R. Engineering complex phenotypes in industrial strains. Biotechnol Prog. 2008;24:38–47. doi: 10.1021/bp0701214. [DOI] [PubMed] [Google Scholar]
- Querol A., Fernández‐Espinar M.T., Del Olmo M., Barrio E. Adaptive evolution of wine yeast. Int J Food Microbiol. 2003;86:3–10. doi: 10.1016/s0168-1605(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Randez‐Gil F., Sanz P., Prieto J.A. Engineering baker's yeast: room for improvement. Trends Biotechnol. 1999;17:237–244. doi: 10.1016/s0167-7799(99)01318-9. [DOI] [PubMed] [Google Scholar]
- Randez‐Gil F., Aguilera J., Codón A., Rincón A.M., Estruch F., Prieto J.A. Baker's yeast: challenges and future prospects. In: De Winde J.H., editor. Springer‐Verlag; 2003. pp. 57–97. , and . In Functional Genetics of Industrial Yeasts (ed.). Heidelberg, Germany: , pp. [Google Scholar]
- Rodriguez‐Vargas S., Estruch F., Randez‐Gil F. Gene expression analysis of cold and freeze stress in baker's yeast. Appl Environ Microbiol. 2002;68:3024–3030. doi: 10.1128/AEM.68.6.3024-3030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Vargas S., Sánchez‐García A., Martínez‐Rivas J.M., Prieto J.A., Randez‐Gil F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol. 2007;73:110–116. doi: 10.1128/AEM.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak F., Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A.M., Cyert M.S. Calcineurin acts through the CRZ1/TCN1‐encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai S.L., Daran‐Lapujade P., Walsh M.C., Pronk J.T., Daran J.M. Acclimation of Saccharomyces cerevisiae to low temperature: a chemostat‐based transcriptome analysis. Mol Biol Cell. 2007;18:5100–5112. doi: 10.1091/mbc.E07-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanghe A., Van Dijck P., Dumortier F., Teunissen A., Hohmann S., Thevelein J.M. Aquaporin expression correlates with freeze tolerance in baker's yeast, and overexpression improves freeze tolerance in industrial strains. Appl Environ Microbiol. 2002;68:5981–5989. doi: 10.1128/AEM.68.12.5981-5989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanghe A., Van Dijck P., Thevelein J.M. Determinants of freeze tolerance in microorganisms, physiological importance, and biotechnological applications. Adv Appl Microbiol. 2003;53:129–176. doi: 10.1016/s0065-2164(03)53004-0. [DOI] [PubMed] [Google Scholar]
- Thammavongs B., Panoff J.M., Guéguen M. Phenotypic adaptation to freeze‐thaw stress of the yeast‐like fungus Geotrichum candidum. Int J Food Microbiol. 2000;60:99–105. doi: 10.1016/s0168-1605(00)00374-3. [DOI] [PubMed] [Google Scholar]
- Wolfe J., Bryant G. Freezing, crying, and/or vitrification of membrane‐solute‐water systems. Cryobiology. 1999;39:103–129. doi: 10.1006/cryo.1999.2195. [DOI] [PubMed] [Google Scholar]
- Wright B.E. Stress‐directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman K., Gasch A.P., Li H.X., Ogawa N., Botstein D. Genome‐wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. et al. [DOI] [PubMed] [Google Scholar]
- Zeyl C. The number of mutations selected during adaptation in a laboratory population of Saccharomyces cerevisiae. Genet. 2005;169:1825–1831. doi: 10.1534/genetics.104.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between growth in LD at 12°C and freeze tolerance of evolved clones. Twenty‐four individuals sampled from the 200‐generation terminal population of our evolutionary experiment were analysed for growth in plates of LD agar at 12°C and freeze tolerance. Growth was assumed to be proportional to the colony size and was estimated by measuring the average colony area of at least 80–100 colonies corresponding to each of the selected individuals. Relative freeze tolerance was estimated by measuring the loss of CO2 production in an impedance analyser for LD samples stored for 14 days at −20°C. Unfrozen samples were used as control. Data points are the mean of at least two independent experiments. The correlation coefficient (r2) for the linear regression of relative freeze tolerance against average colony size was 0.046.
The strains evolved in LD at 12°C are not deficient in respiration. Cells of the CR and CR20 strains, and two independent petite mutants from the parental CR strain (notated as CRρ1 and CRρ2) were spotted onto YPD agar plates and cultured for 2–3 days. Colonies were transferred to a filter paper and assayed for cytochrome c oxidase by the addition of the electron donor tetramethyl‐p‐phenylenediamine (TMPD). Colonies oxidizing TMPD immediately acquired a dark blue colour, whereas cells lacking the oxidase activity remained white.


