Summary
Interest in understanding prokaryotic biotransformation of high‐molecular‐weight polycyclic aromatic hydrocarbons (HMW PAHs) has continued to grow and the scientific literature shows that studies in this field are originating from research groups from many different locations throughout the world. In the last 10 years, research in regard to HMW PAH biodegradation by bacteria has been further advanced through the documentation of new isolates that represent diverse bacterial types that have been isolated from different environments and that possess different metabolic capabilities. This has occurred in addition to the continuation of in‐depth comprehensive characterizations of previously isolated organisms, such as Mycobacterium vanbaalenii PYR‐1. New metabolites derived from prokaryotic biodegradation of four‐ and five‐ring PAHs have been characterized, our knowledge of the enzymes involved in these transformations has been advanced and HMW PAH biodegradation pathways have been further developed, expanded upon and refined. At the same time, investigation of prokaryotic consortia has furthered our understanding of the capabilities of microorganisms functioning as communities during HMW PAH biodegradation.
Introduction
Research in regard to prokaryotic biodegradation of high‐molecular‐weight polycyclic aromatic hydrocarbons (HMW PAHs) has continued to advance worldwide. Although some of the first reports of HMW PAH biotransformations by bacteria were published in 1975 (Barnsley, 1975; Gibson et al., 1975) and 1984 (Jerina et al., 1984), it was from the late 1980s that research in this field began to proceed more quickly when studies that documented HMW PAH‐utilizing bacteria and ring fission products of bacterial HMW PAH biodegradation were reported (Heitkamp and Cerniglia, 1988; Heitkamp et al., 1988a,b; Mahaffey et al., 1988; Mueller et al., 1989). By the late 1990s, a diverse number of studies had been published that probed the field through different scientific approaches and that led to the isolation of HMW PAH‐degrading and HMW PAH‐utilizing bacterial strains, characterization of HMW PAH biodegradation metabolites and proposals for mechanisms of HMW PAH biodegradation. These studies were later the subject of reviews pertaining specifically to the field of HMW PAH biodegradation by bacteria (Kanaly and Harayama, 2000) or as part of excellent wider reviews that also covered HMW PAH biodegradation by eukaryotic organisms and biodegradation of other classes of hydrocarbon pollutants (Juhasz and Naidu, 2000; Van Hamme et al., 2003). Since that time, advances in molecular biology and more recently, high‐throughput technologies, such as whole‐genome sequencing, proteomics and metabolomics, are equipping the field with new tools that are facilitating even more rapid advancements. Research pertaining to bacterial biodegradation of HMW PAHs has indeed expanded worldwide and is the subject of this review.
HMW PAH properties and occurrence
The PAHs that possess more than three aromatic rings have been referred to as HMW PAHs in the environmental microbiology literature. The physical and chemical properties of HMW PAHs are such that they generally appear to be persistent in the environment and may pose risks to human and ecological health in parent molecule form or after biological and/or chemical transformations (Lundstedt et al., 2007). As is widely known, HMW PAHs are sparingly soluble in water, are electrochemically stable and may be acutely toxic, genotoxic, immunotoxic (Burchiel and Luster, 2001) or act as agents of hormone disruption (Van de Wiele et al., 2005) depending upon circumstances and the mode of exposure. Due to their elevated octanol–water partition coefficients (Kow), HMW PAHs may partition into organic phases, soil and sediment organic matter and membranes of living organisms, and are candidates for bioconcentration, bioaccumulation and sometimes biomagnification through trophic transfers in terrestrial and marine food webs (Neff, 2002; Meador, 2003). The chemical structures of 12 HMW PAHs whose biodegradation is discussed within this review are shown in Fig. 1.
Figure 1.
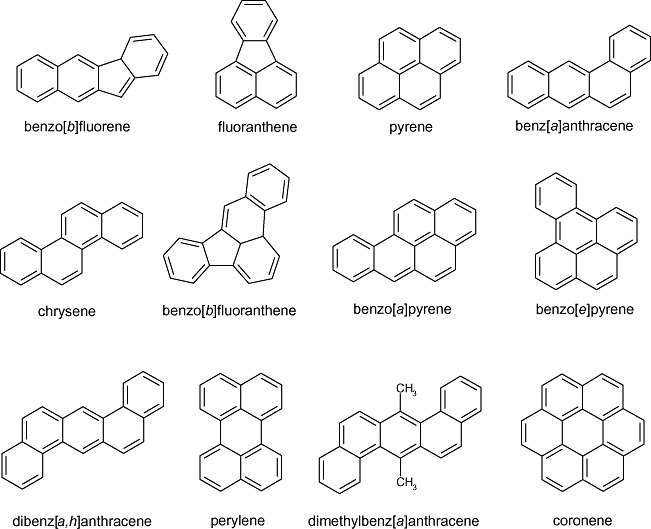
Structures of representative HMW PAH molecules discussed in this review.
HMW PAH molecular stability, hydrophobicity and low water solubility appear to be some of the main factors that contribute to their persistence in the environment and these factors may also be correlated with the size of the molecule or the total number of aromatic rings (Cerniglia, 1992). HMW PAHs are formed by pyrogenic processes during the incomplete combustion of organic material through pyrolysis and pyrosynthesis (Mastral and Callén, 2000), are generally present in relatively high concentrations in fossil fuels and in products of fossil fuel refining (petrogenic origin; Quann, 1998) and are produced biologically. In areas of concentrated HMW PAH pollution caused by anthropogenic releases they may co‐occur in complex non‐polar aqueous phase liquid (NAPL) mixtures, such as in the case of creosote, a coal tar distillate used as a wood preservative. However, HMW PAHs may originate from many different types of sources, both anthropogenic and non‐anthropogenic (Van Metre et al., 2000; Hylland, 2006; Johnsen and Karlson, 2007; Li et al., 2007; Achten and Hofmann, 2009) and their levels of occurrence in the environment are dependent upon numerous factors including the degree of industrial development and proximity to point and/or non‐point sources combined with the effects of regional and global transport phenomena. The environmental levels of HMW PAHs vary widely, they appear to occur just about everywhere and their occurrence has been studied in the atmosphere (Baek et al., 1991; Lang et al., 2008), in air (Finlayson‐Pitts and Pitts, 1997; Han and Naeher, 2006), in soil (Wilcke, 2000; Nam et al., 2009), in freshwater and marine sediments (Cornelissen et al., 1998; Zakaria et al., 2002), in ice cores (Kawamura and Suzuki, 1994), in the deep ocean (Ohkouchi et al., 1999) and in numerous other media ranging from vegetation to food (Wagrowski and Hites, 1997; Phillips, 1999; Howsam et al., 2000; Fismes et al., 2002). As a class, PAHs are currently considered to be the most abundant free organic molecules in space (Ehrenfreund and Charnley, 2000) and detection of HMW PAHs in interstellar regions indicates that their ubiquity transcends planet earth (Ehrenfreund and Sephton, 2006).
On earth there is interest to understand the environmental fates of these chemicals because such information may prove beneficial for controlling their concentrations in the environment and for limiting exposure. At the same time, demand for optically pure chemical products created by microorganisms is increasing and biological transformation of HMW PAHs may be advantageous for the production of regioselective and stereoselective organic chemicals in some settings.
A complete pyrene biotransformation pathway and Mycobacterium vanbaalenii PYR‐1
Research in regard to the biotransformation of the four‐ring peri‐condensed PAHs pyrene and fluoranthene by different genera of bacteria has been significantly advanced. Biodegradation pathways for these PAHs have been further developed since the first reports of metabolites and biodegradation pathways for these compounds were published for actinomycetes and Gram‐negative organisms (Heitkamp et al., 1988b; Mueller et al., 1990; Weissenfels et al., 1991; Cerniglia, 1992). In the last 10 years many new bacteria that possess HMW PAH biodegradation capabilities for pyrene and fluoranthene have been isolated from different environments throughout the world and the characteristics of these bacteria have been examined to various degrees and shall be discussed in this review. At the same time, comprehensive, rigorous investigations of previously discovered isolates, such as Mycobacterium sp. strain PYR‐1 (Heitkamp and Cerniglia, 1988), now known as M. vanbaalenii PYR‐1 (Khan et al., 2002), have continued and are significantly expanding our understanding of prokaryotic HMW PAH biodegradation.
Due to the enormous amount of high‐quality research that has been conducted on M. vanbaalenii PYR‐1 it most likely needs to be addressed as the subject of its own review; however, these authors will attempt to discuss some of the results pertaining to this organism herein. M. vanbaalenii PYR‐1 was originally isolated from estuarine sediments near an oil field (Heitkamp and Cerniglia, 1988) and was shown at that time to biodegrade various HMW PAHs, including the four‐ring PAHs pyrene and fluoranthene by fortuitous metabolism. Over 20 years later, this organism has come to be one of the most extensively studied organisms in the field of prokaryotic HMW PAH biodegradation research and PAH biodegradation research in general. Building upon many previous studies that have involved this organism, investigations have continued over the last 10 years, which have resulted in further development and refinement of the HMW PAH catabolic pathways for pyrene, fluoranthene, benz[a]anthracene, 7,12‐dimethylbenz[a]anthracene and even benzo[a]pyrene in addition to many other comprehensive studies.
The proposed pathways for pyrene biodegradation by M. vanbaalenii PYR‐1 have been continuously developed since its isolation and it was originally determined that oxidation by this organism occurred by both dioxygenation and monooxygenation (Heitkamp et al., 1988b). Within dioxygenation‐initiated pyrene metabolism, the predominant pathway was shown to occur via dioxygenation at the 4,5 positions of pyrene and resulted in cis‐4,5‐dihydroxy‐4,5‐dihydropyrene (pyrene cis‐4,5‐dihydrodiol), which after multiple transformations ultimately led to the tricarboxylic acid cycle (TCA) cycle. At least a second pathway that began with initial dioxygenation at the 1,2 positions to form O‐methylated derivatives of pyrene cis‐1,2‐dihydrodiol appeared to serve as a detoxification process. In 2007, Kim and colleagues (2007) constructed the first complete pathway for pyrene biotransformation by M. vanbaalenii PYR‐1 to TCA intermediates by utilizing a combination of metabolite identification, genomic and proteomic analyses that indicated that pyrene was metabolized through the β‐ketoadipate pathway. Twenty‐seven enzymes that included 14 responsible for the degradation of pyrene to phthalate, six responsible for the degradation of phthalate to protocatechuate and seven responsible for the lower pathway from protocatechuate to acetyl‐coenzyme A and succinyl coenzyme A were identified in addition to three enzymes responsible for the detoxification pathway of pyrene to 1,2‐dimethoxypyrene. It was reported that all genes involved in the pyrene metabolic pathways were identified through these analyses and that alternative routes of pyrene degradation were not detected based upon genome sequence screening. The proposed pathways for pyrene biodegradation by mycobacteria, including the complete pathway proposed for M. vanbaalenii PYR‐1 to the TCA cycle, are shown in Fig. 2, with further discussion in regard to pyrene biodegradation by other organisms discussed in later sections of this review.
Figure 2.
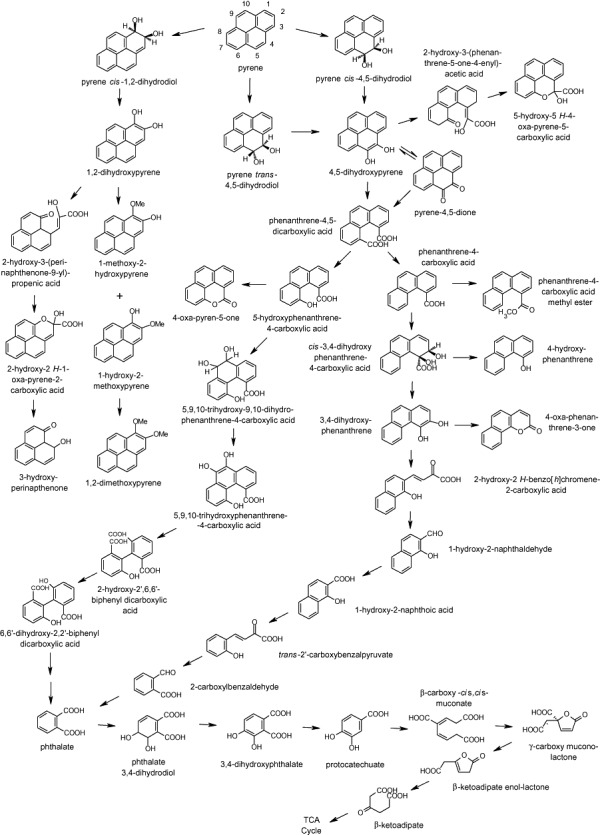
Pathways proposed for the biodegradation of pyrene by mycobacteria, including the complete pathway proposed for M. vanbaalenii PYR‐1 to the TCA cycle (Heitkamp et al., 1988b; Walter et al., 1991; Cerniglia, 1992; Dean‐Ross and Cerniglia, 1996; Schneider et al., 1996; Rehmann et al., 1998; Vila et al., 2001; Krivobok et al., 2003; Habe et al., 2004; Kim et al., 2005b; 2007; Liang et al., 2006; Zhong et al., 2006).
Genes relevant to HMW PAH metabolism in M. vanbaalenii PYR‐1
To construct the complete pyrene pathway for M. vanbaalenii PYR‐1 by such a biosystems approach (and for the fluoranthene biodegradation pathways discussed below), results were required from earlier studies where the enzymatic functions of various genes involved in PAH biodegradation processes were determined from M. vanbaalenii PYR‐1 (Khan et al., 2001; Kim et al., 2004b; Stingley et al., 2004a), Mycobacterium sp. strain 6PY1 (Krivobok et al., 2003) and from phenanthrene biodegradation by Nocardioides sp. strain KP7 (Iwabuchi and Harayama, 1998a,b; Saito et al., 2000) for example. In 2001, Khan et al. was the first to characterize the genes encoding a polycyclic aromatic ring dioxygenase in M. vanbaalenii PYR‐1 whereby an aldehyde dehydrogenase (nidD), the dioxygenase small (β‐)subunit (nidB) and the dioxygenase large (α‐)subunit (nidA) were cloned and sequenced and found to be arranged in a sequence different from genes found in other bacterial dioxygenase systems. These genes were later shown by Brezna and colleagues (2003) to have conserved homologues in four other distantly related Mycobacterium species through comparisons of Mycobacterium sp. strain RJGII‐135, (Schneider et al., 1996), M. flavescens PYR‐GCK, (Dean‐Ross and Cerniglia, 1996) M. frederiksbergense FAn9T (Willumsen et al., 2001) and M. gilvum BB1 (Boldrin et al., 1993; Böttger et al., 1997). M. austroafricanum ATCC 33464, which did not show high PAH‐degrading activity, did not possess the nidA gene even though it was reported to be almost identical to M. vanbaalenii PYR‐1 at the 16S rDNA level.
Results from previous studies on PAH biodegradation by M. vanbaalenii PYR‐1 and the three‐ring compounds anthracene and phenanthrene suggested that this organism might possess multiple copies of different dioxygenase genes or at least possess relaxed dioxygenase specificity because the organism was capable of biotransforming a broad range of aromatic substrates (Moody et al., 2001). Indeed, in 2004, Stingley and colleagues (2004a) focused on characterizing additional genes in the nidDBA region of M. vanbaalenii PYR‐1 and identified and characterized a putative phthalate operon in a Mycobacterium species for the first time that was located approximately 12–19 kb upstream of the nidA large subunit gene. They showed that a putative regulatory protein (phtR) was encoded divergently with five tandem genes: phthalate dioxygenase large subunit (phtAa), small subunit (phtAb), phthalate dihydrodiol dehydrogenase (phtB), phthalate dioxygenase ferredoxin subunit (phtAc) and phthalate dioxygenase ferredoxin reductase (phtAd). When cloned into Escherichia coli, the authors concluded that phthalate was converted to 3,4‐dihydroxyphthalate based upon the fragmentation pattern of the trimethylchlorosilylated metabolite derivative recovered by GC‐MS analyses. Using PCR and Southern hybridization analyses, it was shown that homologues to the pht operon region were present in PAH‐degrading Mycobacterium spp. that were assayed previously for the nidA gene by Brezna and colleagues (2003), with the exception of Mycobacterium sp. strain RJGII‐135. M. austroafricanum GTI‐23 (Bogan et al., 2003) and a Rhodococcus sp. (Dean‐Ross et al., 2001) were also assayed and found to possess the pht operon.
Further investigations with M. vanbaalenii PYR‐1 resulted in the characterization of ring‐hydroxylating dioxygenase genes Orf25 and Orf26 that encoded putative aromatic oxygenase large and small subunits (Stingley et al., 2004b). This was the first time that genes in the phthalate pathway were identified in a Mycobacterium sp. and confirmed previous reports by Moody and colleagues (2001) in their biodegradation studies with phenanthrene. Depending upon one's perspective, the main (4,5) pathway for the biodegradation of pyrene enters into the phenanthrene pathway through the formation of 3,4‐dihydroxyphenanthrene during biodegradation to phthalate and the TCA cycle. By using a proteomics approach that consisted of protein profiling by two‐dimensional electrophoresis analyses (2‐DE) combined with liquid chromatography electrospray ionization tandem mass spectrometry, Kim and colleagues (2004a) documented the upregulation of multiple proteins in M. vanbaalenii PYR‐1 cultures that were induced with PAH compounds. These included a catalase‐peroxidase, putative monooxygenase, dioxygenase small subunit, a naphthalene‐inducible dioxygenase small subunit and an aldehyde dehydrogenase. The dioxygenase small subunit (β‐subunit) was a 20 kDa protein that showed a sevenfold increase when M. vanbaalenii PYR‐1 was exposed to fluoranthene compared with other induction substrates. Further investigation by PCR with degenerate primers that were designed based upon de novo peptide sequencing combined with a series of plaque hybridizations were carried out to screen the genomic library of M. vanbaalenii PYR‐1 and the genes nidA3B3 were cloned and sequenced (Kim et al., 2006). In cloned E. coli cells, nidA3B3 converted the HMW PAHs fluoranthene, pyrene and benz[a]anthracene to cis‐dihydrodiols with the aid of ferredoxin (PhdC) and ferredoxin reductase (PhdD) genes. nidA3B3 represented the fourth copy of a terminal dioxygenase gene identified from M. vanbaalenii PYR‐1.
In addition to the terminal dioxygenase genes documented in M. vanbaalenii PYR‐1, a catalase‐peroxidase gene, the katG gene was cloned, expressed and characterized when it was induced by pyrene. It was the first gene characterized in M. vanbaalenii PYR‐1 and it was postulated that the enzyme served to protect dioxygenases from oxidative inactivation by exogenous oxidants or that the enzyme was induced to remove endogenous hydrogen peroxide that may have been generated as an intermediate during PAH metabolism (Wang et al., 2000). The cytochrome P450 genes cyp151 (pipA), cyp 150 and cyp 51 were characterized in M. vanbaalenii PYR‐1 by Brezna and colleagues (2006) and represented the first evidence of functional cytochrome P450 genes coexisting with aromatic ring‐hydroxylating dioxygenase in a HMW PAH‐utilizing organism. These data corroborated data from previous work by Moody and colleagues (2001; 2003; 2004; 2005) that indicated monooxygenase attacks on phenanthrene, benz[a]anthracene, 7,12‐dimethylbenz[a]anthracene and at the 11,12 position of benzo[a]pyrene for example (see below). In the case of pyrene, M. vanbaalenii PYR‐1 monooxygenation was discussed as early as 1988 (Heitkamp et al., 1988b). After gene cloning, pipA and cyp 150 were partially expressed in E. coli and resulted in the production of mono‐oxygenated products of the HMW PAHs pyrene and 7‐methylbenz[a]anthracene. In the case of pyrene, 1‐, 2‐ and 4‐hydroxypyrene were produced and it was noted that due to the symmetry of pyrene these were the only possible isomers. Further screening of 13 mycobacterium isolates for pipA, cyp 150 and cyp 51 indicated that there was little correlation between PAH‐degrading ability and the presence of these genes and this was in contrast to assays for nidA and nidB carried out in this study (Brezna et al., 2006) and in a previous study (Brezna et al., 2003) discussed above.
Evidence for constitutive catechol‐O‐methyl transferase and PAH‐quinone reductase was also provided from studies with M. vanbaalenii PYR‐1. It was discussed that catechol‐O‐methyl transferase may play an important role in detoxifying non‐K‐ and non‐bay‐region PAH catechols that are produced inside the cell during PAH metabolism (Kim et al., 2004b). These compounds are not considered to serve as carbon and energy sources for M. vanbaalenii PYR‐1 and it has been postulated that quinone reductases might be necessary for protecting the cell against cytotoxicity and to provide a PAH catechol supply during metabolism. One year before, Kim and colleagues (2003b) also reported on the recovery of two o‐quinone reductases from the cell extracts of German soil isolate Mycobacterium sp. strain PYR100, which was isolated on pyrene as a sole source of carbon and energy and which also degraded fluoranthene and phenanthrene. Comparative analysis of the 16S rDNA and 16S‐23S intergenic spacer sequences of Mycobacterium sp. strain PYR100 and M. vanbaalenii PYR‐1 later revealed that strain PYR100 was an intergenic spacer sequence sequevar variant of M. vanbaalenii PYR‐1 (Kim et al., 2005a). Furthermore, the activity of M. vanbaalenii PYR‐1 to metabolize pyrene under two different pH conditions was reported in 2005 in addition to a then updated version of the pyrene biodegradation pathway by this organism (Kim et al., 2005b). Interestingly, a decrease in one pH unit, from 7.5 to 6.5, resulted in a fourfold faster degradation rate of 0.5 mM pyrene by a resting cell suspension of M. vanbaalenii PYR‐1. Higher levels of pyrene were detected in cytosolic fractions and it was concluded that the acidic pH facilitated increased cell permeability to PAHs. However, M. vanbaalenii PYR‐1 produced relatively higher levels of O‐methylated derivatives of non‐bay‐region and non‐K‐region pyrene‐diols in the cytosol at pH 6.5 compared with the presence of metabolic intermediates in the culture fluid at pH 7.5, and the authors concluded that acidic pH may become a significant burden to cells when toxic metabolites accumulate in the cytosol.
Mycobacterial biodegradation of pyrene and fluoranthene
Many new pyrene‐ and fluoranthene‐degrading mycobacteria have been isolated throughout the world in the last 10 years. In 1998, pyrene utilization as a sole source of carbon and energy and metabolite production by Mycobacterium sp. strain KR2, isolated from a German gas works soil, was documented by Rehmann and colleagues (1998). When the organism was fed 500 mg l−1 pyrene, approximately 60% was metabolized in 8 days. Eight metabolites were recovered and contributed to our understanding of the pyrene catabolic pathways, including 2‐carboxybenzaldehyde, 1‐hydroxy‐2‐naphthoic acid and cis‐3,4‐phenanthrene dihydrodiol‐4‐carboxylic acid that were recovered for the first time (Fig. 2). The identity of cis‐3,4‐phenanthrene dihydrodiol‐4‐carboxylic acid was postulated from the identification of 4‐phenanthrol in acidified media. Mycobacterium sp. strain KR2 continued to biodegrade pyrene even though the organism was maintained on nutrient agar slants without pyrene addition and the authors discussed that this was supportive evidence for chromosomal location of the degradative genes and further noted that ortho‐cleavage pathways generally appeared to be chromosomally encoded (van der Meer et al., 1992). Attempts to detect plasmids in strain KR2 were unsuccessful. In 1999, pyrene‐utilizing Mycobacterium sp. strain MR‐1 was documented after isolation from PAH‐contaminated sediments from the Buffalo River, New York, USA by Molina and colleagues (1999). Cross‐induction of pyrene mineralization by phenanthrene was demonstrated for strain MR‐1 and an enrichment culture from the same sediments whereby it was shown that new protein synthesis was not required for [4,5,9,10‐14C]pyrene mineralization after pre‐acclimation on phenanthrene.
Mycobacterium sp. strain AP1 was isolated from crude oil‐contaminated sand in Spain on enrichment cultures that consisted of 1 g l−1 pyrene in mineral salts medium (Vila et al., 2001). The organism also utilized fluoranthene, phenanthrene and hexadecane but not anthracene for example, as sole sources of carbon and energy. Investigation of the biodegradation mechanisms of Mycobacterium sp. strain AP1 during growth on pyrene showed that initiation of attack occurred by either mono‐ or dioxygenase activity at the 4,5 positions that resulted in either trans‐ or cis‐4,5‐dihydroxy‐4,5‐dihydropyrene metabolites in addition to other metabolites that further confirmed the pathways proposed for pyrene biodegradation. Vila and colleagues (2001) also identified the novel metabolite, 6,6′‐dihydroxy‐2,2′‐biphenyl dicarboxylic acid whose presence indicated that a new branch of the proposed pathways for pyrene occurred by two sequential cleavages of both central K‐region rings. This branch of the pathway is also indicated in Fig. 2. They reported that the new metabolite was formed after dioxygenation at the 4,5 and 9,10 positions with subsequent cleavage of both central rings and that meta‐ and ortho‐cleavage reactions followed by one‐ or two‐carbon excisions might be possible. The metabolite was also found to increase in the culture medium and it was suggested that it might therefore be a dead‐end‐product. Interestingly, the UV‐visible spectrum for 6,6′‐dihydroxy‐2,2′‐biphenyl dicarboxylic acid was similar to an unidentified metabolite (metabolite IV) reported by Rehmann and colleagues (1998) during growth of Mycobacterium sp. strain KR2 on pyrene.
In 2003, Krivobok and colleagues (2003) identified two pyrene‐induced proteins from Mycobacterium sp. strain 6PY1, originally isolated from PAH‐contaminated soil that grew on pyrene as a sole source of carbon and energy. In 14C[PAH] assays, cells that were grown on pyrene or phenanthrene exhibited high levels of catabolic activity, but cells that were grown on acetate did not and this situation was exploited by examining protein extracts via 2‐DE combined with in vivo protein‐labelling assays. Pyrene‐induced proteins were tentatively identified by peptide sequence analyses and the genes encoding the terminal components of the two enzymes were successfully cloned. These two new terminal dioxygenase components, Pdo1 and Pdo2, were reported to be similar in other actinomycetes, including M. vanbaalenii PYR‐1 and Nocardioides strain KP7, and it was concluded that they were regulated differentially. Mycobacterium sp. strain 6PY1 was also utilized in pyrene biodegradation investigations in freshwater sediments whereby 15 mg of pyrene and radiolabelled pyrene was applied in 300 g of wet sediment (Jouanneau et al., 2005). It was concluded that [4,5,9,10‐14C]pyrene mineralization was enhanced by sediment inoculation, 8.8% 14CO2 in 61 days, and that inoculation plus planting with the reed Phragmites australis resulted in twofold less enhancement, 4.4% 14CO2 over the same time period. The authors also reported that they recovered the water‐soluble metabolites, 4,5‐phenanthroic acid and 4‐phenanthroic acid from their microcosms and isolated two pyrene‐degrading strains most closely related to M. austroafricanum and Stenotrophomonas maltophila. Mycobacterium sp. strain 6PY1 was also employed in experiments designed to investigate the effects of organic and inorganic materials on pyrene removal (Cottin and Merlin, 2008).
New Mycobacterium spp. strains JLS, KMS and MCS were isolated from creosote‐ and pentachlorophenol‐contaminated soil from Montana, USA that was undergoing bioremediation through a prepared‐bed land treatment unit process. All three isolates mineralized pyrene as a sole source of carbon and energy in radiolabel studies and were shown to possess dioxygenase nid genes, nidA and nidB (Miller et al., 2004). They were employed in a comparative study with M. gilvum PYR‐GCK and M. vanbaalenii PYR‐1 whereby various biochemical and genetic differences among the strains were documented (Miller et al., 2007). Mycobacterium sp. strain JLS was the subject of synchrotron radiation‐based Fourier transform infrared spectromicroscopy investigations where it was shown that the presence of humic acids significantly shortened the lag time for pyrene biodegradation by this strain from 168 to 2 h when it was studied on a magnetite chip (Holman et al., 2002). These strains were also investigated in relation to their abilities to associate with plant roots when inoculated onto germinating barley seeds whereby strain KMS in the presence of diluted root wash (Child et al., 2007a) or barley root colonization by strain KMS resulted in enhanced pyrene mineralization by radiotracer analyses (Child et al., 2007b).
In other studies that involved Mycobacterium sp. strain KMS, the effects of varying soil humic acid concentrations on pyrene mineralization in liquid culture were investigated (Liang et al., 2007) in addition to pyrene metabolite humification studies where it was incubated with [4‐13C]‐ or [4,5,9,10‐14C]pyrene with and without a soil humic acid standard to characterize the nature of the produced residues (Nieman et al., 2007). Results of the [13C]/[14C]pyrene investigations indicated that most of the pyrene carbon was incorporated into cellular material and that direct coupling of extracellular pyrene metabolites to soil organic matter was not a primary fate mechanism under the experimental conditions. These results were in agreement with previous [4,9‐13C]pyrene‐sediment investigations using flash‐pyrolysis combined with 13C NMR where sequestered pyrene was found to be non‐covalently associated with sediment organic matter and was not transformed (Guthrie et al., 1999).
In 2006, Mycobacterium sp. strain KMS was the subject of a proteomics study (Liang et al., 2006) whereby three metabolites, including pyrene‐4,5‐dione, were identified. Initial attack by Mycobacterium sp. strain KMS on pyrene was reported to occur by mono‐ or dioxygenation at the 4,5 positions based upon metabolite and protein identification and a pathway for this organism was proposed that confirmed previous biodegradation pathways proposed for other mycobacteria (Fig. 2). Although the metabolite trans‐4,5‐pyrene dihydrodiol was not recovered in this study, the gene coding an epoxide hydrolase was identified and, therefore, monooxygenation was included in the proposed pathway for strain KMS. It was noted that this was the first study to show transformation of pyrene to pyrene‐4,5‐dione by a Mycobacterium species. Interestingly, strain KMS metabolized pyrene‐4,5‐dione when it was added directly to culture media, indicating that it may have unique abilities to deal with pyrene‐4,5‐dione toxicity. The accumulation of pyrene‐4,5‐dione had been previously shown by Kazunga and Aitken (2000) in M. vanbaalenii PYR‐1 when it was incubated with cis‐4,5‐pyrene dihydrodiol and as a pyrene metabolite of Sphingomonas yanoikuyae R1 (isolated from a gas manufacturing plant; Aitken et al., 1998). In whole‐sediment incubations, the presence of pyrene‐4,5‐dione had been shown by Guthrie‐Nichols and colleagues (2003) for the first time during studies designed to examine the effects of ageing on pyrene transformation in sediments. cis‐4,5‐Pyrene dihydrodiol had been documented in sediments in 1996 by Li and colleagues (1996) Additionally, diones of fluoranthene, fluoranthene‐2,3‐ and ‐1,5‐dione were documented for the first time from bacteria by Kazunga and colleagues (2001) by studying four strains that produced these compounds during fluoranthene metabolism. Fluoranthene‐2,3‐dione inhibited the mineralization of other HMW PAHs by some of the strains and, although the inhibition mechanism appeared to occur through cytotoxicity, results indicated that other less straightforward mechanisms also appeared to be possible.
Further isolation of pyrene and fluoranthene‐degrading strains of mycobacteria was accomplished by different research groups and in 2003, Bogan and colleagues (2003) reported on the characterization of M. austroafricanum strain GTI‐23, isolated from a manufactured gas plant site soil in Iowa, USA. Strain GTI‐23 grew on pyrene and fluoranthene as sole sources of carbon and energy and also degraded benzo[a]pyrene. Inhibition was shown to occur when mixed with phenanthrene or pyrene. Interestingly, GTI‐23 grew on both HMW PAHs and the aliphatics dodecane and hexadecane. As more studies document such occurrences, it appears that aromatic and aliphatic hydrocarbon degradative capacities within a single strain may not be as unusual as previously thought. In pyrene‐contaminated soil inoculation studies that employed M. austroafricanum strain GTI‐23, increased pyrene contact time resulted in increased sequestration and reduced biodegradation (Bogan and Sullivan, 2003). Zhong and colleagues (2006) have reported on the degradation of pyrene and PAH mixtures by Mycobacterium sp. strain A1‐PYR that was isolated from mangrove sediments. It utilized pyrene as a sole source of carbon and energy and some pyrene metabolites were identified. Pyrene‐ and fluoranthene‐degrading Mycobacterium sp. strains HH1 through HH3 were also isolated from mangrove sediments in the Ho Chung basin in Hong Kong (Zhou et al., 2006; 2008) and the authors reported on fluoranthene biodegradation by a Terrabacter sp. strain. Terrabacter species were previously shown to metabolize low‐molecular‐weight PAHs but not HMW PAHs (Habe et al., 2005). In 1999, Mycobacterium sp. strain CH1 was isolated from PAH‐contaminated freshwater sediments in the USA by Churchill and colleagues (1999) and was shown to mineralize pyrene and fluoranthene with pyrene as a sole carbon and energy source. Strain CH1 was also capable of utilizing branched‐ and n‐alkanes as sole carbon and energy sources. Alkaliphilic Mycobacterium sp. strain MHP‐1 was isolated from soil in Chiba, Japan (Habe et al., 2004) and utilized pyrene as a sole source of carbon and energy via the 4,5 biodegradation pathway. Optimum growth pH was 9 and the strain was found to possess the aromatic ring hydroxylase gene nidAB.
The characterization of the new species Mycobacterium pyrenivorans was documented in 2004 by Derz and colleagues (2004) following isolation from PAH‐contaminated soil at a former coking plant site in Übach‐Palenberg, Germany. Mycobacterium pyrenivorans was found to grow on the HMW PAHs pyrene and fluoranthene as a sole source of carbon and energy but not benzo[a]pyrene. Sho and colleagues (2004) identified two pyrene degradation gene clusters in Mycobacterium sp. strain S65 which was isolated from a jet fuel‐contaminated site in Quebec, Canada. Using Southern hybridization, nidA homologues were shown to occur in two separate loci in the strain. The organism used pyrene, fluoranthene and phenanthrene as sole carbon and energy sources and pyrene mineralization was enhanced by the addition of less water‐soluble HMW PAHs, such as benzo[a]pyrene or benz[a]anthracene. M. frederiksbergense FAn9T was isolated from a coal tar‐contaminated former gas works site in Frederiksberg, Denmark and it was reported to use the HMW PAH fluoranthene and pyrene as sole sources of carbon and energy (Willumsen et al., 2001). In 2000, Bastiaens and colleagues (2000) compared two different procedures for the isolation of PAH‐utilizing bacteria and showed clear differences in the types of organisms isolated by each mode of isolation. They reported that liquid enrichment selected for Sphingomonas species and a membrane method selected for Mycobacterium species. A pyrene‐utilizing strain LB208 was determined to be a Mycobacterium species that was 100% similar to pyrene‐degrading strain Mycobacterium gilvum strain BB1 (Boldrin et al., 1993; Böttger et al., 1997). M. frederiksbergense strain LB501T was recovered in this study and although it has not been shown to biodegrade HMW PAHs, it has been the subject of various comprehensive investigations that have contributed to our understanding of PAH biodegradation (Wick et al., 2001; 2002; 2003a,b; van Herwijnen et al., 2003a).
Mycobacterium gilvum strain VF1 and Gordonia‐like strain BP9 – the etymologically correct Gordonia was proposed in 1997 to replace Gordona (Stackebrandt et al., 1997; Arenskötter et al., 2004) – was originally isolated from hydrocarbon‐contaminated soil and was capable of utilizing fluoranthene and pyrene as sole carbon and energy sources (Kästner et al., 1994). It was demonstrated to grow on pyrene microcrystals formed by sonication with and without silicone oil (Mutnuri et al., 2005) and under these conditions biodegradation was enhanced. Pyrene and fluoranthene biodegradation by strain B9 were also assessed in soil under different conditions including variable pH (Kästner et al., 1998).
Mycobacterium sp. strain LP1 was isolated from enrichment culture produced from a control agricultural soil, Uppsala, Sweden, which was part of a previous study (Pizzul et al., 2006) and was shown to grow on pyrene resulting in greater than 90% degradation in 2 weeks when incubated at a concentration of 50 mg l−1 (Pizzul et al., 2007). Incubation with a mixture of PAHs resulted in less than 25% pyrene biodegradation. After growth of Mycobacterium sp. strain SNP11, isolated from a grassland soil near Metz, France, on pyrene, fluoranthene and other PAHs, metabolite screening for acute, chronic and genotoxic effects was carried out and indicated significant reductions in all cases (Pagnout et al., 2006). Genes encoding the α‐subunit of two ring‐hydroxylating dioxygenases Nid and Pdo2 were detected in this organism (Pagnout et al., 2007) and the pdoA2 gene region (called phdA in their study) was cloned and sequenced. Expression of PAH‐degrading genes in Mycobacterium smegmatis mc2155 was carried out for the first time and broad ring‐hydroxylating substrate specificity was revealed.
Biotransformation pathways of fluoranthene by actinomycetes
Although there have been numerous reports of both Gram‐negative and Gram‐positive bacteria specifically capable of degrading fluoranthene, with approximately 50 bacterial isolates reported by Rehmann and colleagues (2001), characterization of fluoranthene metabolites has occurred with less frequency. In the last 10 years however, various key studies that employed both classical and new approaches have resulted in the documentation of new metabolites and new proposals for pathways of fluoranthene biodegradation by actinomycetes. The currently proposed pathways for the biodegradation of fluoranthene by actinomycetes are given in Fig. 3 and are discussed below.
Figure 3.
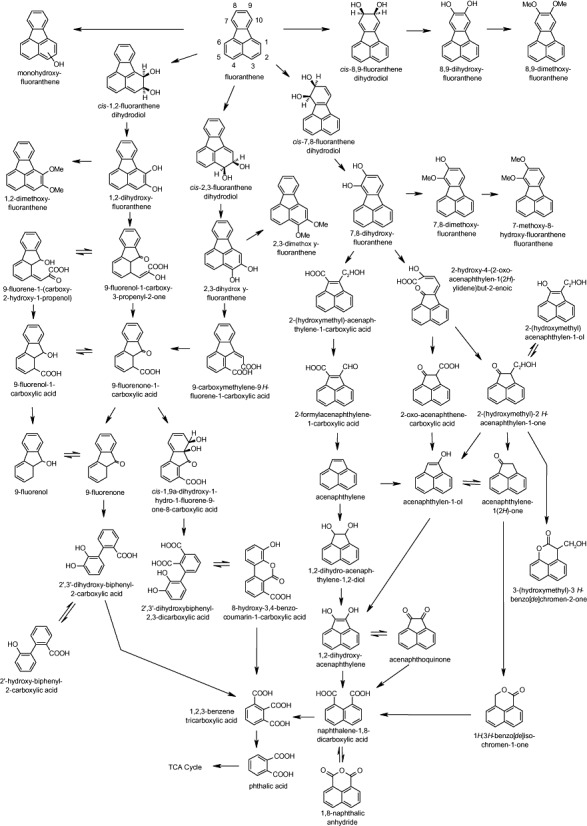
Pathways proposed for the biodegradation of fluoranthene by bacteria (Kelley et al., 1991; 1993; Weissenfels et al., 1991; Cerniglia, 1992; Sepic et al., 1998; Rehmann et al., 2001; Dean‐Ross et al., 2002; López et al., 2005; 2006; Kweon et al., 2007; Lee et al., 2007). Metabolites detected from fluoranthene, acenaphthene and acenaphthylene transformations are shown.
In 2001, an alternative pathway for fluoranthene biodegradation through carbon positions 2,3 by Mycobacterium sp. strain KR20 was proposed by Rehmann and colleagues (2001). This new information complemented the existing proposed pathways for fluoranthene biodegradation by Alcaligenes denitrificans (Weissenfels et al., 1991), M. vanbaalenii PYR‐1 (Kelley et al., 1991; Cerniglia, 1992; Kelley et al., 1993) and Pasteurella sp. strain IFA (Sepic et al., 1998) that showed 1,2 position‐ and 7,8 position‐initiating attacks on fluoranthene but for which the corresponding cis‐1,2‐fluoranthene and cis‐7,8‐fluoranthene dihydrodiols had yet to be recovered. Mycobacterium sp. strain KR20, isolated from a PAH‐contaminated German gas works site, was grown on 500 mg l−1 fluoranthene as a sole source of carbon and energy, and seven metabolites were documented. Five metabolites, cis‐2,3‐fluoranthene dihydrodiol, Z‐9‐carboxymethylene‐fluorene‐1‐carboxylic acid, 4‐hydroxybenzochromene‐6‐one‐7‐carboxylic acid (indicated as 9‐carboxymethylene‐9H‐fluorene‐1‐carboxylic acid and 8‐hydroxy‐3,4‐benzocoumarin‐1‐carboxylic acid respectively in Fig. 3), cis‐1,9a‐dihydroxy‐1‐hydro‐fluorene‐9‐one‐8‐carboxylic acid and benzene‐1,2,3‐tricarboxylic acid, were identified by NMR and MS spectroscopy and a new pathway was proposed. At that time, the possibility of initial dioxygenation at the 2,3 positions for M. vanbaalenii PYR‐1 and Pasteurella sp. strain IFA was also discussed even though comparison of downstream fluoranthene metabolites among the strains indicated that pathway divergenence was likely. This was later confirmed in M. vanbaalenii PYR‐1 in 2007 when Kim and colleagues (2007) published a comprehensive investigation of fluoranthene metabolites for this organism and explained that the 2,3 pathway was most likely the main mechanism of fluoranthene biodegradation.
Mycobacterium holderi was isolated from PAH‐contaminated soil and was reported to grow on fluoranthene and co‐oxidize pyrene in the presence of fluoranthene. It produced 29 metabolites during fluoranthene biodegradation and the cis‐2,3‐fluoranthene dihydrodiol was the only metabolite reported to be identified; however, evidence was not provided (Kleespies et al., 1996). In 2002, Dean‐Ross and colleagues. (2002) documented biodegradation of fluoranthene via fortuitous metabolism by a M. flavescens strain and a Rhodococcus sp. strain. Both strains acted on fluoranthene through the 1,2 pathway and meta cleavage; however, it was also proposed that the Rhodococcus sp. strain produced 9‐(carboxymethylene)fluorene‐1‐carboxylic acid (9‐carboxymethylene‐9H‐fluorene‐1‐carboxylic acid) by ortho‐cleavage of 2,3‐dihydroxy‐fluoranthene similar to Mycobacterium sp. strain KR20.
Metabolites of fluoranthene biodegradation by various mycobacteria were characterized by López et al. in two reports from 2005 and 2006 whereby Mycobacterium sp. strains CP1, CP2, Cft2 and Cft6 and Mycobacterium sp. strain AP1, which was described above in the context of pyrene biodegradation (Vila et al., 2001), were investigated under conditions where fluoranthene was administered as the sole source of carbon and energy (López et al., 2005; 2006). In the 2005 report, strains CP1, CP2, Cft2 and Cft6, isolated from creosote‐contaminated soil in Andújar, Spain were all found to oxidize fluoranthene by dioxygenation at the 1,2 positions followed by meta cleavage and a two‐carbon excision to produce 9‐fluorenone‐1‐carboxylic acid and by dioxygenation at the 2,3 positions followed by ortho‐cleavage to yield 9‐carboxymethylene‐9H‐fluorene‐1‐carboxylic acid. Additionally, strains CP1 and CP2 metabolized fluoranthene by dioxygenation at the 7,8 positions followed by meta cleavage and eventual transformation to acenaphthone. These three routes of fluoranthene biodegradation were also shown to occur in Mycobacterium sp. strain AP1 when it was grown on fluoranthene as a sole source of carbon and energy (López et al., 2006).
In 2007, through the use of genomic, metabolomic and proteomic approaches, Kweon and colleagues (2007) reported that there were 53 potential enzymes likely to be involved in the biodegradation of fluoranthene by M. vanbaalenii PYR‐1. They proposed a comprehensive pathway that included four possible metabolic routes that could be initiated by either mono‐ or dioxygenation reactions. They explained that most of the enzymes used in the pyrene degradation pathway were identified in fluoranthene‐induced cultures and that similar to pyrene degradation in M. vanbaalenii PYR‐1, all of the routes channelled through phthalate and the β‐ketoadipate pathway with the exception of detoxification pathways. Also in 2007, fluoranthene metabolism was characterized by proteomic methods using Mycobacterium sp. strain JS14, which was isolated from PAH‐contaminated soil in Hilo, Hawaii, USA along with other strains, including Mycobacterium sp. strain JS19b1, which grew on fluoranthene and pyrene (Lee et al., 2007; Seo et al., 2007). Sixteen metabolites of fluoranthene biodegradation generated by strain JS14 were detected following 2 weeks of incubation with 200 mg l−1 fluoranthene. Comparison of glucose‐fed and fluoranthene‐fed cultures by 1‐D PAGE or 2‐DE and nano‐LC‐MS/MS revealed upregulation of 25 proteins related to fluoranthene biodegradation including catalase and superoxide dismutase. In strain JS14, initial dioxygenation at the 1,2‐, 2,3‐, 7,8‐ and 8,9‐positions were revealed and pathways for fluoranthene biodegradation by strain JS14 were proposed. Strain JS14 was also shown to transform the organophosphorous pesticides chlorfenvinphos and diazinon (Seo et al., 2007).
Biotransformation pathways of benz[a]anthracene by Mycobacterium spp.
Reports that document the biodegradation of kata‐annelated HMW PAHs occur with less frequency in the biodegradation literature, even though reports of their biodegradation were published as early as 1947 when it was shown that a marine consortium biodegraded 47% and 13% of 25 mg each of benz[a]anthracene and dibenz[a,h]anthracene through the detection of carbon dioxide (Sisler and ZoBell, 1947). Since the 1980s, reports that documented the biodegradation of the four‐ring benz[a]anthracene by bacterial isolates representing the genera Sphingobium (Jerina et al., 1984; Mahaffey et al., 1988; Ye et al., 1996; Boyd et al., 2006), Sphingomonas (Demanèche et al., 2004; Jouanneau et al., 2006), Stenotrophomonas (Juhasz et al., 1996; 1997; 2000b), Alcaligenes (Weissenfels et al., 1991), Pseudomonas (Caldini et al., 1995; Chen and Aitken, 1999) and Mycobacterium (Schneider et al., 1996) were published and documentations of metabolites under various circumstances were published for Sphingobium yanoikuyae strains B8/36 and B1 (Jerina et al., 1984; Mahaffey et al., 1988; Boyd et al., 2006), Sphingomonas sp. strain CHY‐1 (Demanèche et al., 2004; Jouanneau and Meyer, 2006; Jouanneau et al., 2006) and Mycobacterium sp. strain RJGII‐135 (Schneider et al., 1996). In 2005, Moody and colleagues (2005) characterized multiple metabolites from cultures of M. vanbaalenii PYR‐1 exposed to 120 mg l−1 benz[a]anthracene applied in dimethylformamide. HPLC, UV‐visible absorption, GC‐MS and NMR analyses revealed that M. vanbaalenii PYR‐1 attacked benz[a]anthracene at the 1,2‐, 5,6‐, 7,12‐, and 10,11‐positions and it was concluded that the 10,11‐pathway was the predominant route of metabolism. ortho‐ and meta‐ring cleavages resulted in various benz[a]anthracene biodegradation metabolites and multiple pathways for biodegradation by M. vanbaalenii PYR‐1 were proposed (Fig. 4). Previously, only the cis‐5,6‐ and 10,11‐dihydrodiols had been documented from mycobacteria (Schneider et al., 1996).
Figure 4.
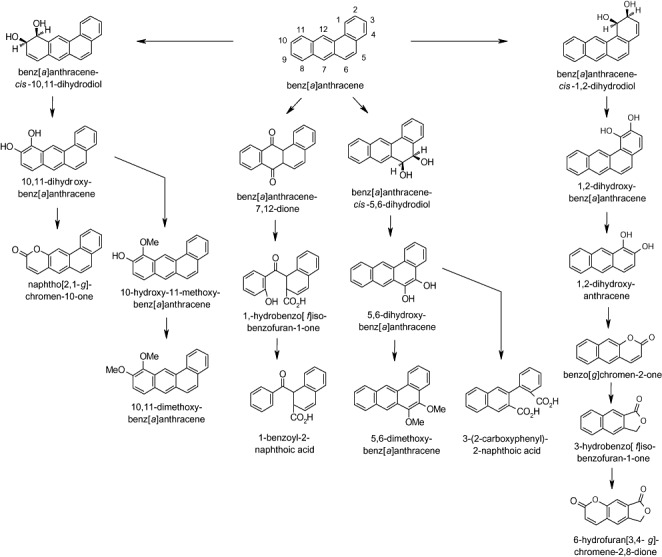
Pathways proposed for the biodegradation of benz[a]anthracene by mycobacteria (Schneider et al., 1996; Moody et al., 2005).
Regio‐ and stereo‐selective biodegradation of 7,12‐dimethylbenz[a]anthracene was also documented in M. vanbaalenii PYR‐1 whereby mono‐ and dioxygenation reactions at the 5,6 positions and monooxygenation of the C‐7 methyl group were shown to occur (Moody et al., 2003).
Biotransformation pathways of benzo[a]pyrene by Mycobacterium spp.
Benzo[a]pyrene is currently not known to be utilized as a sole source of carbon and energy by any bacteria and significant biodegradation appears to require bacterial cooperation although the mechanisms are still unclear. At the same time, some isolates have been shown to biotransform benzo[a]pyrene, metabolites have been characterized and our understanding of biodegradation pathways has been advanced. In 2004, Moody and colleagues (2004) showed that M. vanbaalenii PYR‐1 metabolized benzo[a]pyrene delivered in dimethylformamide, 0.48 mM final benzo[a]pyrene concentration, in cultures that consisted of 500 ml of basal salts medium supplemented with 0.38 g ml−1 each of peptone, yeast extract and soluble starch. Phenanthrene in dimethylformamide, 100 µl of a 70 mM solution, was also added for enzyme induction. After 96 h, a total of nine metabolites were identified by UV‐visible, mass, NMR and circular dichroism spectral analyses: cis‐4,5‐dihydro‐4,5‐dihydroxybenzo[a]pyrene (benzo[a]pyrene cis‐4,5‐dihydrodiol), cis‐11,12‐dihydro‐11,12‐dihydroxybenzo[a]pyrene (benzo[a]pyrene cis‐11,12‐dihydrodiol), trans‐11,12‐dihydro‐11,12‐dihydroxybenzo[a]pyrene (benzo[a]pyrene trans‐11,12‐dihydrodiol), 10‐oxabenzo‐[def]chrysen‐9‐one, and dimethoxy and hydroxymethoxy derivatives of benzo[a]pyrene. Included in their findings were three ortho‐ring fission metabolites that originated from cultures that were fed benzo[a]pyrene cis‐4,5‐dihydrodiol: 4‐formylchrysene‐5‐carboxylic acid, 4,5‐chrysene‐dicarboxylic acid and a monocarboxylated chrysene product. Initial dioxygenation of benzo[a]pyrene by M. vanbaalenii PYR‐1 at the 4,5 positions, K region, was consistent with findings from other studies on mycobacteria that degrade pyrene such as M. flavescens (Dean‐Ross and Cerniglia, 1996), Mycobacterium sp. strain KR2 (Rehmann et al., 1998) and Mycobacterium sp. strain AP1 (Vila et al., 2001).
These results were significant because they expanded our understanding of the enzymatic capabilities of bacteria to biodegrade benzo[a]pyrene and built upon a previous study whereby enzymatic attack on benzo[a]pyrene by Mycobacterium sp. strain RJGII‐135 at the 4,5‐, 7,8‐ and 9,10‐ positions was proposed based upon the identification of four metabolites and included the first documentation of ring fission products of benzo[a]pyrene metabolism by a bacterium (Schneider et al., 1996). At that time, fluorescence and mass analyses were used to identify benzo[a]pyrene cis‐7,8‐dihydrodiol plus the downstream products of either 7,8‐ or 9,10‐dioxygenase attack, cis‐4‐(8‐hydroxypyren‐7‐yl)‐2‐oxobut‐3‐enoic acid or cis‐4‐(7‐hydroxypyren‐8‐yl)‐2‐oxobut‐3‐enoic acid and 7,8‐dihydro‐pyrene‐7‐carboxylic acid or 7,8‐dihydro‐pyrene‐8‐carboxylic acid. Because the meta fission products through the 7,8‐bond or the 9,10‐bond were not distinguishable, there were two possible identities for each of the two of the metabolites and it was noted that direct evidence for 9,10 initial dioxygenation was lacking. In 2004, 10‐oxabenzo‐[def]chrysen‐9‐one was identified by Moody and colleagues (2004) and this has provided further evidence to the 9,10 pathway. The cis‐9,10‐dihydrodiol (and the cis‐7,8 dihydrodiol) had been documented previously only from S. yanoikuyae strain B8/36 discussed below.
In addition to expanding our understanding of the 4,5 and 9,10 pathways, Moody and colleagues (2004) also revealed two 11,12 pathways that occured by both a monooxygenase and dioxygenase attack on benzo[a]pyrene that resulted in trans‐11, 12‐ and cis‐11,12‐dihydrodiols respectively. For the first time, the absolute configurations of the benzo[a]pyrene cis‐ and trans‐dihydrodiols were documented whereby the regio‐ and stereo‐selective activities of the mono‐ and dioxygenases of M. vanbaalenii PYR‐1 were shown through the production of benzo[a]pyrene‐cis‐11,12‐dihydrodiol (11S, 12R, 100% optically pure), ‐trans‐11,12‐dihydrodiol (equal mixture of 11S, 12S and 11R, 12R compounds) and the ‐cis‐4,5‐dihydrodiol (30% 4S, 5R and 70% 4R, 5S absolute stereochemistry). It was discussed that monooxygenation of benzo[a]pyrene was most likely carried out by cytochrome P450 enzymes and as discussed above, three cytochrome P450 genes were previously characterized from M. vanbaalenii PYR‐1 (Brezna et al., 2006). Hydroxymethoxy and dimethoxy metabolites of benzo[a]pyrene were also identified and corroborated a report that demonstrated a catechol‐O‐methyltransferase located in the soluble fraction of M. vanbaalenii PYR‐1 cell extracts (Kim et al. 2004b). The pathways proposed for benzo[a]pyrene biodegradation by mycobacteria are given in Fig. 5.
Figure 5.
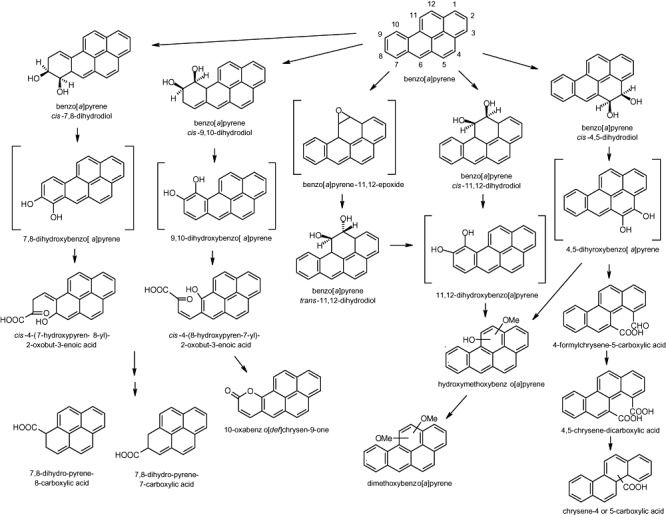
Pathways proposed for the biodegradation of benzo[a]pyrene by mycobacteria (Schneider et al., 1996; Moody et al., 2004). Hypothetical intermediates are indicated by brackets.
Further study on the effects of benzo[a]pyrene biodegradation in the presence of co‐occurring PAHs by Mycobacterium sp. strain RJGII‐135 revealed that biodegradation was inhibited by anthracene, benz[a]anthracene and pyrene, but were stimulated by phenanthrene under the conditions tested (McLellan et al., 2002). Mycobacterium sp. strain LP1, described above in the context of pyrene biodegradation, was also reported to mineralize approximately 10% of [7,10‐14C]benzo[a]pyrene in liquid culture, 50 mg l−1, when incubated with a mixture of PAHs for 2 weeks (Pizzul et al., 2006).
Sphingobium yanoikuyae strains B1 & B8/36, Sphingobium sp. strain EPA505 and HMW PAH biodegradation
Reports of HMW PAH biodegradation by non‐actinomycete bacteria appear to have occurred with less frequency when compared with the mycobacteria; however, since the late 1990s more numerous reporting has resulted in the documentation of isolates representing a range of non‐actinomycete organisms in regard to the biodegradation of the four‐ring PAHs, pyrene, fluoranthene, chrysene and benz[a]anthracene, the five‐ring benzo[a]pyrene, perylene, dibenz[a,h]anthracene and the seven‐ring coronene. During the same time, in‐depth research in regard to previously isolated strains, such as S. yanoikuyae strains B1 and B8/36, and Sphingobium sp. strain EPA505, has been advanced.
The term sphingomonad is used in the proceeding sections as described by Stolz (2009) (sensu latu) in a new review that details the sphingomonads in relation to their xenobiotic‐degrading capabilities. Indeed, member organisms of the sphingomonads are well known for their abilities to degrade a wide range of natural and xenobiotic compounds, including HMW PAHs in some cases, and the diverse catabolic capabilities of some sphingomonad isolates have been aptly demonstrated (Balkwill et al., 2006; Stolz, 2009). Among these, a Beijerinckia species, strain B1, was first isolated in 1973 by Gibson and colleagues (1973) and, in 1975 a mutant strain, Beijerinckia species strain B8/36, was reported to biotransform benz[a]anthracene and benzo[a]pyrene (Gibson et al., 1975; Jerina et al., 1984). These archetypal xenobiotic‐degrading organisms were reclassified in 1996 as Sphingomonas yanoikuyae strain B1; B8/36 (Khan et al., 1996; Gibson, 1999) and more recently reclassified as Sphingobium yanoikuyae strain B1; B8/36 following the proposal of the new genus Sphingobium (Takeuchi et al., 2001).
In rigorous chemical investigations of chrysene biotransformation, resting cells of S. yanoikuyae strain B8/36 were shown to oxidize chrysene to (+)‐cis‐(3S,4R)‐dihydroxy‐3,4‐dihydrochrysene (Boyd et al., 1997) with further investigations revealing that strain B8/36 produced (+)‐(3S,4R,9S,10R)‐3,4,9,10‐tetrahydroxy‐3,4,9,10‐tetrahydrochrysene, a bis‐cis‐dihydrodiol that represents a new class of metabolites derived from prokaryotic biotransformation of PAHs (Boyd et al., 1999). Most recently, Boyd and colleagues (2006) characterized enantiopure bis‐cis‐dihydrodiols from the biotransformation of the HMW PAH benz[a]anthracene (and anthracene) by strain B8/36, thus adding two more members to this new family of tetraol metabolites. Other recent studies with S. yanoikuyae strain B1 that involved HMW PAHs have resulted in the purification and characterization of the three protein components of a biphenyl 2,3 dioxygenase system, encoded by the genes bphA4, bphA3 and bphA1f,A2f. This system exhibited broad substrate specificity including dioxygenation of benzo[a]pyrene to benzo[a]pyrene cis‐7,8‐ and benzo[a]pyrene cis‐9,10‐dihydrodiols following expression in an E. coli recombinant (Ní Chadhain et al., 2007; Yu et al., 2007). The large subunit BphA1f was found to share 90% identity with and was most closely related to the ring hydroxylating dioxygenase from Sphingomonas sp. strain CHY‐1 discussed below (Demanèche et al., 2004). The crystal structures of the ferredoxin and terminal oxygenase components of the biphenyl 2,3 dioxygenase were documented by Ferraro and colleagues (2007) and this was the first report of a structure of a Rieske oxygenase that oxidized substrates with five aromatic rings. It was determined that the ability to catalyse the oxidation of such large substrates was made possible by both a larger active site entrance combined with the ability of the active site to accommodate larger substrates.
S. yanoikuyae strain B1 and other sphingomonads including Sphingobium sp. strain EPA505 were employed in autecological investigations by Cunliffe and Kertesz whereby it was concluded that phenotypic characteristics that may allow for sphingomonads to dominate in PAH‐contaminated soils did not appear to be necessary for their survival and propagation (Cunliffe and Kertesz, 2006a,b).
Examination of the versatile Sphingobium sp. strain EPA505 has also continued. Strain EPA505 was originally isolated from a creosote‐contaminated soil in Pensacola, Florida, USA (Mueller et al., 1989; 1990) and was shown to utilize or biotransform many HMW PAHs, including fluoranthene, benzo[b]fluorene, benz[a]anthracene, chrysene, pyrene, benzo[a]pyrene, benzo[b]fluoranthene and dibenz[a,h]anthracene under different circumstances (Ye et al., 1996). It was used as a model organism in surfactant (Triton X‐100, Tween 80), bioemulsifier (alasan) and sphingan biodegradation studies of the HMW PAHs fluoranthene and pyrene (Willumsen and Karlson, 1998; Barkay et al., 1999; Willumsen and Arvin, 1999; Luning Prak and Pritchard, 2002; Johnsen and Karlson, 2004), including an investigation with phenanthrene that documented the preferential utilization of Tween surfactant hydrophobic fractions as a carbon source by this strain (Kim and Weber, 2003) and the documentation of horizontal transfer of alasan to its cell surface (Osterreicher‐Ravid et al., 2000).
In 2001, Story and colleagues (2001) documented a biodegradation pathway for fluoranthene (and other PAHs) by Sphingobium sp. strain EPA505 using three classes of Tn5 mutant derivative strains that were defective for PAH degradation (Story et al., 2000). Biochemical evidence suggested that more than one operon was involved in aromatic catabolism by strain EPA505 and that the biodegradation pathway for fluoranthene diverged from the naphthalene, phenanthrene and anthracene pathways for this organism. Fluoranthene biodegradation by fluoranthene‐grown cultures of the wild‐type strain was proposed to occur through oxidative attack at the 7,8 position of fluoranthene and proceed through acenaphthoquinone and 1,8‐naphthalic anhydride similarly to the 7,8‐pathway in mycobacterium species (see Fig. 3). Further investigation of the biodegradation capability range of strain EPA505 was carried out whereby it was confirmed that the strain did not grow on chrysene, pyrene or benzo[b]fluoranthene (Story et al., 2004), but that oxidation products of pyrene (4,5‐dihydroxypyrene, phenanthrene‐4,5‐dicarboxylic acid and 5‐hydroxyphenanthrene‐4‐carboxylic acid; Fig. 3) and benzo[b]fluoranthene were detected by the wild‐type strain and mutants respectively. The authors noted that based upon these oxidation results that they also expected initial oxidation of chrysene and it was discussed that the lack of chrysene biotransformation may have been attributable to strict dioxygenase substrate specificity in strain EPA505. Metabolites from fortuitous metabolism of pyrene by strain EPA505 had been previously documented by Ho and colleagues (2000) as part of a wider study aimed at characterizing fluoranthene‐ and pyrene‐degrading bacteria from soils.
A new 16S rRNA gene‐based primer set was developed for the detection and monitoring of strain EPA505 and related strains by Leys and colleagues (2005b) in PAH‐contaminated soils (Leys et al., 2004) and detection of strain EPA505 in four out of five contaminated soils but not in five uncontaminated soils was documented and provided further evidence for the importance of this strain in PAH pollutant biodegradation in nature. Examination of fluoranthene and pyrene biodegradation in soil slurries under low‐nutrient conditions by strain EPA505 and Mycobacterium species strain VM552 (100% similar to M. gilvum by 16S rDNA sequence; Bastiaens et al., 2000), respectively, were also carried out by Leys and colleagues (2005a) and among other findings indicated that PAH degradation was unaffected by such conditions by these organisms.
With the purpose of developing pollutant biodegradation models, biodegradability parameters were reported for 20 out of 22 PAHs tested for Sphingobium sp. strain EPA505 (Dimitriou‐Christidis et al., 2007) and investigations into the biodegradation kinetics of 2‐methylphenanthrene, fluoranthene and pyrene in binary and ternary mixtures were performed (Dimitriou‐Christidis and Autenrieth, 2007). Strain EPA505, when combined with Sphingomonas aromaticivorans B0695 (Frederickson et al., 1995; Balkwill et al., 1997; now classified as a Novosphingobium, Takeuchi et al., 2001) was reported to degrade HMW PAHs in mixtures when the PAHs were dissolved in dodecane or silicone oil in two‐phase partitioning bioreactors (Daugulis and McCracken, 2003; Vandermeer and Daugulis, 2007). In UV‐irradiation studies, little effect on the biodegradaton of HMW PAH degradation in mixtures by strain EPA505 and S. yanoikuyae B1 were reported (Lehto et al., 2003).
Biodegradation of HMW PAHs by newly isolated sphingomonads
New HMW PAH‐degrading sphingomonad isolates have been reported to varying degrees in the literature since the late 1990s. Notably, Willison (2004) documented Sphingomonas species strain CHY‐1, isolated from PAH‐contaminated soil at a coal‐gasification site in the Rhône‐Alpes region of France that grew on chrysene as a sole source of carbon and energy and this organism has been the subject of many comprehensive investigations. When chrysene was administered at an initial concentration of 500 mg l−1 in silicone oil and an organic/aqueous phase ratio of 1:4, 50% of added chrysene was biodegraded in 5 days and greater than 97% after 35 days. [5,6,11,12‐14C]Chrysene radiotracer studies showed that 40–60% mineralization occurred and that addition of naphthalene increased the rate of mineralization. Further investigation with strain CHY‐1 through direct peptide analysis of PAH‐induced proteins, followed by cloning and functional analyses of genes encoding target proteins revealed that a single ring‐hydroxylating dioxygenase component, PhnI, catalysed the oxidation of a wide range of PAHs including the HMW PAHs chrysene and benz[a]anthracene and furthermore that PhnI was required for PAH catabolism by strain CHY‐1 (Demanèche et al., 2004). Benz[a]anthracene was reported to be converted into three compounds that most likely represented three isomeric dihydrodiol derivatives.
Jouanneau and colleagues (2006) separately purified and characterized the components of the CHY‐1 dioxygenase and documented its broad substrate specificity, initiating attack on PAHs ranging from two to five rings. Component ht‐PhnI was shown to convert chrysene and benz[a]anthracene to doubly dihydroxylated derivatives, the bis‐cis‐dihydrodiols that were documented by Boyd et al. in 1999 and 2006 for chrysene and benz[a]anthracene, respectively (Boyd et al., 1999; 2006). The crystal structure of PhnI oxygenase was determined and was reported to possess a substantially larger hydrophobic substrate binding pocket and residue flexibility at the pocket entrance (Jakoncic et al., 2007b), including that molecular modelling studies revealed that the pocket was large enough to accommodate benzo[a]pyrene (Jakoncic et al., 2007a). The authors concluded that the broad substrate specificity of strain CHY‐1 oxygenase was influenced by these factors. A versatile cis‐dihydrodiol dehydrogenase (PDDH) was also characterized from strain CHY‐1 whereby gene bphB was cloned and overexpressed in E. coli and was shown to oxidize two‐ to five‐ring PAH dihydrodiols (Jouanneau and Meyer, 2006). 3,4‐Dihydroxy‐3,4‐dihydrochrysene, 2,3‐dihydroxy‐2,3‐dihydrofluoranthene, 9,10‐dihydroxy‐9,10‐dihydrobenzo[a]pyrene and the 1,2‐, 8,9‐ and 10,11‐dihydrodiol isomers of benz[a]anthracene were all converted to their corresponding catechols by PDDH. Interestingly, 4,5‐dihydroxy‐4,5‐dihydropyrene was also converted by PDDH even though pyrene was not a substrate for PhnI. A three‐component salicylate 1‐hydroxylase was also characterized from strain CHY‐1 (Jouanneau et al., 2007).
Sphingomonas sp. strain LB126 isolated from PAH‐contaminated soil (Bastiaens et al., 2000) was shown to biodegrade fluoranthene by fortuitous metabolism and a pathway was proposed. 9‐Fluorenone‐1‐carboxylic was identified and indicated that 1,2 or 2,3 initial dioxygenation followed by either meta‐ or ortho‐ring cleavage was possible (van Herwijnen et al., 2003b). A novel angular dioxygenase complex, FlnA1–FlnA2, was also characterized from strain LB126 and was shown to transform fluoranthene to monohydroxyfluoranthene (Shuler et al., 2008). Sphingomonas sp. strain PheB4 was isolated from surface mangrove sediments in Shenzhen, China and was shown to biodegrade fluoranthene by fortuitous metabolism when phenanthrene (10 mg l−1) or a peptone/beef extract/NaCl/glucose mixture was added as co‐substrate (Zhong et al., 2007). When incubated together, phenanthrene biodegradation by strain PheB4 was found to be significantly inhibited by fluoranthene. Sphingomonas sp. strain VKM B‐2434 was isolated from contaminated sediment at a coke wastewater plant in Mariupol, Ukraine and was shown to grow on fluoranthene as a sole source of carbon and energy, but was also capable of biodegrading many other PAHs, including the HMW PAHs pyrene, benz[a]anthracene, chrysene and benzo[a]pyrene. Various metabolites were reported for many of the PAH substrates and indicated that strain VKM B‐2434 possessed broad substrate specificity for a range of compounds (Baboshin et al., 2008). Using a soil isolate that was 99.2% similar to S. yanoikuyae isolates based upon 862 sequenced bases, Rentz and colleagues (2008) reported that they identified cis‐7,8 dihydrodiol, pyrene‐8‐hydroxy‐7‐carboxylic acid and pyrene‐7‐hydroxy‐8‐carboxylic acid by LC‐MS analyses from benzo[a]pyrene biotransformation by this organism after induction with salicylate. In a previous study using the same organism, removal of approximately 1 mg l−1 or less of benzo[a]pyrene was reported to occur in cultures that were supplemented with plant root extracts (Rentz et al., 2005).
Among a group of sphingomonad strains isolated from the deep Atlantic coastal plain (Frederickson et al., 1995), Novosphingobium aromaticivorans strain B0695 was shown to biodegrade various PAHs including fluoranthene in the presence of Tween 80 (Shi et al., 2001). Isolated from an oil‐refinery waste‐contaminated sandpit, Sphingomonas sp. strain 107 was reported to grow on pyrene, produce a bioemulsifier and it was determined that its PAH degradation genes were plasmid‐encoded (Dagher et al., 1997). Sphingomonas sp. strain VTI was documented to mineralize low concentrations (below aqueous solubility) of various HMW PAHs (Aitken et al., 1998) and Sphingomonas paucimobilis strain TNE12 was shown to utilize fluoranthene as a sole source of carbon and energy (Shuttleworth et al., 2000). Using PAH‐degrading Sphingomonas xenophaga strain AJ1 (Iwabuchi et al., 1995), Navarro and colleagues (2008) reported on the removal of pyrene and benzo[a]pyrene from aqueous DNA solutions following extraction from PAH‐contaminated soil (Navarro et al., 2007).
Reports of HMW PAH biodegradation by bacteria other than mycobacteria and sphingomonads
Generally, the sphingomonads (α‐Proteobacteria) and actinomycetes, with specific emphasis on the mycobacteria, appear to be the most versatile groups of bacteria in the degradation of HMW PAHs; however, this may also be a result of enrichment and isolation methods and further research will be required (Bastiaens et al., 2000; Daane et al., 2001; Stach and Burns, 2002; Gaskin and Bentham, 2005). Whatever the case may be, new HMW PAH‐metabolizing organisms representing diverse genera have been documented in the last 10 years and among the Gram‐negative organisms, members of the γ‐Proteobacteria and β‐Proteobacteria have also been documented for their HMW PAH biodegradation capabilities. Members of the genus Pseudomonas, such as Pseudomonas saccharophila P15, isolated from creosote‐contaminated soil, was shown to exhibit enhanced removal rates of fluoranthene, pyrene, benz[a]anthracene, chrysene and benzo[a]pyrene when salicylate was used as an inducer of PAH dioxygenase activity (Chen and Aitken, 1999). The authors concluded that P. saccharophila P15 expressed low‐level constitutive metabolism that was inducible by phenanthrene to bring about HMW PAH metabolism. Pseudomonas alcaligenes PA‐10, isolated from a PAH‐contaminated bioreactor soil, was shown to biodegrade fluoranthene through fortuitous metabolism, four metabolites were identified (Gordon and Dobson, 2001) and it utilized fluoranthene in the presence of surfactants (Hickey et al., 2007). Pseudomonas strains, P. putida strain KBM‐1 (from sediments with background PAH levels) and P. stutzeri strain SAG‐R (from creosote‐contaminated soil) were shown to biodegrade pyrene both aerobically and anaerobically under nitrate‐reducing conditions and also represented the first documentation of HMW PAH degradation under such conditions (McNally et al., 1998; 1999). Kazunga and Aitken (2000) also documented pyrene transformation by P. stutzeri strain P16.
Pyrene biodegradation in the presence of biosurfactant‐producing strain Pseudomonas fluorescens strain 29L was demonstrated by Husain (2008) and in earlier studies, P. fluorescens strain P2a was reported to utilize chrysene and benz[a]anthracene as sole sources of carbon and energy (Caldini et al., 1995; Cenci and Caldini, 1997). Recently, Das and Mukherjee (2006) documented two biosurfactant‐producing strains of Pseudomonas aeruginosa isolated from a crude oil‐contaminated soil in northeast India that grew on pyrene and enhanced its apparent solubility five‐ to sevenfold and Obayori and colleagues (2008) documented the isolation of pyrene‐degrading P. aeruginosa strains LP5 and LP6 and Pseudomonas sp. strain LP1 that were isolated from differently polluted soils in Lagos, Nigeria.
Stenotrophomonas spp., also members of the γ‐Proteobacteria, have been shown to biodegrade HMW PAHs in different studies. The S. maltophila strain VUN 10,003 was reported to remove various HMW PAHs from solution by fortuitous metabolism under differing growth conditions and documentation for the removal of fluoranthene, benz[a]anthracene, benzo[a]pyrene, dibenz[a]anthracene and coronene was demonstrated in different studies (Juhasz et al., 1996; 1997; 2000b; 2002). Strain VUN 10,003 (previously characterized as Burkholderia cepacia) was originally isolated from soil on an abandoned gas works site in Victoria, Australia and grew on phenanthrene and pyrene as sole sources of carbon and energy (Juhasz et al., 1997). When incubated with pyrene as a co‐substrate, the strain was shown to biodegrade benzo[a]pyrene and dibenz[a,h]anthracene, 10–15 mg l−1, over 56 days concomitant with near complete pyrene degradation (250 mg l−1) by 3 weeks. Pyrene refeeding did not stimulate much additional biodegradation; however, various metabolites were recovered from the culture media and the authors concluded that metabolite repression may have occurred (Juhasz et al., 2002). Studies that documented decreased mutagenicity of PAHs after metabolism by S. maltophila strain VUN 10,003 (Juhasz et al., 2000b) and biodegradation of five‐ring PAHs during growth on phenanthrene by this strain and similar strains from the same site were also reported (Juhasz et al., 1997). Another S. maltophila strain, VUN 10,010 was reported to biodegrade a range of HMW PAHs through surfactant enhancement (Boonchan et al., 1998) and in fungal‐bacterial co‐cultures (Boonchan et al., 2000), including benzo[a]pyrene. In 2004, a second organism, Mycobacterium sp. strain B1 was also found to be present in this culture and was shown to biodegrade pyrene and fluoranthene but not benzo[a]pyrene, and it was discussed that the roles of these two organisms in the biodegradation of benzo[a]pyrene were still unclear (Dandie et al., 2004). Mentioned previously, S. maltophila sp. strain C‐7, isolated from freshwater sediments by Jouanneau and colleagues (2005), was capable of pyrene biodegradation and a Pasteurella strain (also γ‐Proteobacteria) biodegraded fluoranthene (Sepic et al., 1997) and was employed in toxicity studies (Sepic et al., 2003).
β‐Proteobacteria isolates, B. cepacia strain 2A‐12 (Kim et al., 2003a) and Burkholderia sp. strain VUN10013 (Somtrakoon et al., 2008) were both documented to biodegrade pyrene by fortuitous metabolism with yeast extract or phenanthrene, respectively, and a strain of Alcaligenes faecalis was recently reported to grow on fluoranthene (Toledo et al., 2006). The first documentation of a Paracoccus (α‐Proteobacteria), strain Ophe1, was reported to have been isolated from polluted soil in Attica, Greece and utilized pyrene as a sole source of carbon and energy in addition to other HMW PAHs (Zhang et al., 2004). Bacillus isolates, Bacillus cereus strain P21 (Kazunga and Aitken, 2000), B. cereus strain Py5, B. megaterium strain Py6 (both from Lin and Cai, 2008), B. subtilis strain DM‐04 (Das and Mukherjee, 2006) and three B. pumilis strains (Toledo et al., 2006) were recently reported to transform or grow on pyrene and the protein engineering of B. megaterium CYP102 for the purpose of accelerating PAH oxidation was also described (Harford‐Cross et al., 2000; Carmichael and Wong, 2001). Paenibacillus sp. strain PR‐P1 and Arthrobacter sp. strain PR‐P3, although unable to utilize pyrene in pure culture, were shown to facilitate pyrene degradation in sediment slurry microcosms (Daane et al., 2001).
Based upon the notion that higher transfer rates, PAH solubility and therefore bioavailablity may increase at higher temperatures, Feitkenhauer and colleagues (2003) examined the biodegradation of HMW PAHs under extreme thermophilic conditions (60–70°C) by Bacillus spp. and a Thermus sp. and documented the biodegradation of fluoranthene, pyrene and benzo[e]pyrene. At moderately thermophilic temperature, thermophilic Nocardia sp. strain TSH1 was shown to grow on pyrene at 50°C as a sole source of carbon and energy (Zeinali et al., 2007). An organism belonging to a genus that represents enteric bacteria, Leclercia adecarboxylata strain PS4040, was isolated from an oily sludge‐contaminated soil at the Digboi oil refinery in northeastern India and was shown to grow on pyrene resulting in approximately 60% biodegradation of 200 mg l−1 pyrene over 20 days (Sarma et al., 2004). The actinomycete, Saccharothrix xinjiangensis sp. strain PYX‐6, solated from Tianchi Lake, Xinjiang Uygur Autonomous Region, China, was reported to grow on pyrene as a sole source of carbon and energy and represented the first HMW PAH‐degrading organism from this genus (Hu et al., 2004).
Investigations in marine environments have led to the discovery of previously unreported HMW PAH‐degrading organisms as well. Geiselbrecht and colleagues (1998) recovered Cycloclasticus sp. strains from marine sediments that were shown to biodegrade low levels of pyrene or fluoranthene by fortuitous metabolism when phenanthrene was present. Recently, Wang and colleagues (2008) reported on the isolation of a HMW PAH‐utilizing organism from the deep sea for the first time. Cycloclasticus spirillensus strain P1, which grew on pyrene as a sole source of carbon and energy was sampled from a depth of 2682 m from Pacific deep sea sediments approximately 300 km from the northeast coast of the Philippines and currently represents the only member of Cycloclasticus capable of growth on a HMW PAH. Two new Gram‐negative marine strains, Novosphingobium pentaromativorans US6‐1 (Sohn et al., 2004) isolated from estuarine sediments in Ulsan Bay and Yeosauna aromativorans strain GW1‐1 (Kwon et al., 2006) isolated from estuarine sediments of the Korean Strait, Republic of Korea were described by the authors as HMW PAH‐degrading organisms that were capable of metabolizing a range of PAHs including benzo[a]pyrene.
Additionally, comprehensive environmental screening surveys that have resulted in the isolation and identification of numerous HMW PAH‐degrading bacteria representing different genera from diverse locations throughout the world have been conducted (Mueller et al., 1997; Aitken et al., 1998; Ho et al., 2000; Zhou et al., 2006; Johnsen et al., 2007; Seo et al., 2007; Hilyard et al., 2008; Launen et al., 2008; Zhou et al., 2008).
Prokaryotic consortia and HMW PAH biodegradation
Investigations of the catabolic capabilities of HMW PAH‐degrading bacterial consortia have increased in the last 10 years and research aimed at understanding consortia‐mediated biodegradation of HMW PAHs has been further enabled by advances in molecular biology and isolation‐independent techniques, such as denaturing gradient gel electrophoresis (DGGE). In an early demonstration of DGGE application to investigate HMW PAH‐biodegradation, Kanaly and colleagues (2000) compared the NAPL‐mediated mineralization of [7‐14C]benzo[a]pyrene by a bacterial consortium recovered from cattle pasture soil (Kanaly et al., 1997) to changes in consortium populations over time. Members of known HMW PAH‐degrading genera Sphingomonas, Mycobacterium, Alcaligenes and Burkholderia were documented by sequencing the V3 region of their 16S rDNA genes and eight of the 10 sequence types corresponded to Proteobacteria. Later isolation of bacterial strains from the consortium combined with sequence analyses of their 16s rDNA genes confirmed results obtained by DGGE, and one isolate, γ‐Proteobacteria member, Rhodanobacter sp. strain BPC1, was monitored in the consortium during NAPL and benzo[a]pyrene biodegradation by competitive PCR (Kanaly et al., 2002). Related studies of this consortium revealed the necessity of hydrocarbon NAPLs to facilitate the mineralization of benzo[a]pyrene (Kanaly et al., 2001; 2006) and the NAPL‐enhanced mineralization of chrysene and 3‐ring PAHs but not pyrene (Kanaly and Watanabe, 2004). Lafortune and colleagues (2009) used DGGE, construction of a 16S rDNA library and strain isolation to document the diversity of a consortium recovered from a creosote‐contaminated soil that degraded HMW PAHs in aqueous‐silicone oil two‐liquid phase (TLP) culturing systems. Results indicated that members of the α‐ and β‐Proteobacteria were most dominant in the consortium and that the biodegradative abilities of the isolates were limited. In support of previous investigations (e.g. Bouchez et al., 1999; Kanaly et al., 2002; Viñas et al., 2005a), their data indicated that the functionality of the consortium as a whole was more effective to perform the task of HMW PAH biodegradation when compared with organisms that were isolated from the community. In earlier studies using the same consortium in TLP systems, biodegradation of HMW PAHs including benzo[a]pyrene were documented (Marcoux et al., 2000; Villemur et al., 2000) and the activities of various isolates obtained from the consortium were also investigated (Gauthier et al., 2003). TLP systems were shown to enhance pyrene degradation by M. vanbaalenii PYR‐1 (MacLeod and Daugulis, 2003) through association with the organic phase (MacLeod and Daugulis, 2005).
Comparisons of consortia that were cultivated on PAHs including HMW PAHs in chemostat cultures were carried out by van Herwijnen and colleagues (2006) using DGGE and fluorescent in situ hybridization. Bioaugmentation was shown to have little effect on the PAH degradation‐capable community and the addition of PAHs resulted in increases in sphingomonads. Derz and colleagues (2006) recovered a consortium from creosote‐contaminated soil discussed above that contained M. pyrenivorans (Derz et al., 2004) and demonstrated extensive mineralization of [4,5,9,10‐14C]pyrene, 54%, and [7,10‐14C]benzo[a]pyrene, 36%, in 3 and 10 weeks respectively when inoculated into soil slurries. Eriksson and colleagues (2002) prepared enrichment cultures that originated from a polychlorinated biphenyl‐contaminated Arctic soil, Labrador, Canada, and grew biofilms on pyrene crystals. They reported that the biofilm culture degraded 20 µg ml−1 pyrene in 60 days and showed that the members of the biofilm included members of the α‐, β‐ and γ‐Proteobacteria. A pyrene‐utilizing consortium that functioned at pH 3 was documented for the first time after sampling a highly acidic soil (pH 2.4) located at a manufactured gas plant site in Belgium and was the first report of HMW PAH biodegradation at such extreme pH. DGGE analyses indicated that a dominant consortium member was most closely related to a member of the slow‐growing mycobacteria, Mycobacterium montefiorense (Uyttebroek et al., 2007).
HMW PAH degradation by consortia recovered from soils and sediments (Juhasz et al., 2000a; Yuan et al., 2000; Dries and Smets, 2002; Sartoros et al., 2005; Chang et al., 2007) and from marine and brackish saline environments (Yamada et al., 2003; Yu et al., 2005; Luan et al., 2006; Lin and Cai, 2008) have been documented in recent studies, including consortia that were psychrotolerant to at least 7°C (Eriksson et al., 2003). Arulazhagan and Vasudevan (2009) have reported pyrene and benzo[e]pyrene biodegradation by a moderately halophilic marine consortium and Cui and colleagues (2008) analysed a consortium community from the deep sea by DGGE under different conditions that was originally enriched on a PAH mixture that included pyrene.
Closing
In the last 20 years, our knowledge in regard to the bacterial biodegradation of HMW PAHs has been significantly advanced and interest in this field has continued to grow worldwide. In the last 10 years, many new HMW PAH‐degrading bacterial isolates and bacterial consortia have been recovered from different types of contaminated and uncontaminated environments and studied. During this time, advances in our understanding of in situ HMW PAH biodegradation were also achieved, and although beyond the scope of this review, HMW PAH biodegradation studies in soils (Kanaly and Bartha, 1999; Haderlein et al., 2001; 2006; Lotfabad and Gray, 2002; Fava et al., 2004; Huesemann et al., 2004; Macleod and Semple, 2006; Mohan et al., 2006; Stroud et al., 2007; Singleton et al., 2008; Mohan et al., 2009), including the use of electrokinetic methods (Shi et al., 2001; Niqui‐Arroyo and Ortega‐Calvo, 2007), in sediments (Bach et al., 2005; Boyd et al., 2005; Lei et al., 2005; Tian et al., 2008), by monitoring prokaryotic HMW PAH biodegradative activities and populations in such media through cultivation‐independent techniques, such as DGGE, real‐time PCR and stable isotope probing among others (Cheung and Kinkle, 2001; Ringelberg et al., 2001; Leys et al., 2004; 2005a,b; Viñas et al., 2005b; Ní Chadhain et al., 2006; Singleton et al., 2006; Gomes et al., 2007; Singleton et al., 2007; Cébron et al., 2008; Jones et al., 2008), by the cooperative effects of bacteria with plants and fungal species (Johnson et al., 2004; Liu et al., 2004; Jouanneau et al., 2005; Krutz et al., 2005; Parrish et al., 2005; Corgiéet al., 2006; Child et al., 2007a,b; Kim and Lee, 2007; Mueller and Shann, 2007; Byss et al., 2008; Leonardi et al., 2008; Li et al., 2008) and by the combined conditions of chemical treatment and biodegradation (Nam and Kukor, 2000; Zeng et al., 2000; Lehto et al., 2003; Piskonen and Itävaara, 2004) for example have been carried out. Indeed, combined knowledge from studies of bacterial isolates, bacterial consortia and in situ investigations will continue to shape our understanding of prokaryotic HMW PAH biodegradation.
As discussed in this review, studies involving newly isolated and previously isolated organisms have furthered our understanding of HMW PAH biodegradation pathways for four‐ and five‐ring compounds, advanced our knowledge of the enzymes involved in these transformations and have demonstrated HMW PAH biodegradation over an expanding range of temperature and pH conditions. Recently, proteomic and metabolomic approaches have been applied to the study of HMW PAH biodegradation and mark the beginning of more wide use of these techniques in the future while consortia studies and the utilization of culture‐independent techniques have begun to provide a view into the dynamics of the bacterial communities that utilize HMW PAHs. At the same time, much still needs to be investigated in regard to the complex ecological interactions of HMW PAH‐degrading communities, the roles of biofilms in HMW PAH biodegradation, the mechanisms of HMW PAH uptake by bacteria and the biodegradation of HMW PAHs in mixtures including unresolved issues of fortuitous metabolism. Research in nanotechnology is now occurring at a brisk pace and to determine the biodegradability of ‘ultimate’ HMW PAHs such as fullerenes and carbon nanotubes will present new and exciting challenges for the field in the near future.
Acknowledgments
This work was supported in part by Yokohama City University Strategic Research Grant, K2002.
References
- Achten C., Hofmann T. Native polycyclic aromatic hydrocarbons (PAHs) in coals – a hardly recognized source of environmental contamination. Sci Total Environ. 2009;407:2461–3072. doi: 10.1016/j.scitotenv.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Aitken M.D., Stringfellow W.T., Nagel R.D., Kazunga C., Chen S.‐H. Characteristics of phenanthrene‐degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can J Microbiol. 1998;44:743–752. [PubMed] [Google Scholar]
- Arenskötter M., Bröker D., Steinbüchel A. Biology of the metabolically diverse genus Gordonia. Appl Environ Microbiol. 2004;70:3195–3204. doi: 10.1128/AEM.70.6.3195-3204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulazhagan P., Vasudevan N. Role of a moderately halophilic bacterial consortium in the biodegradation of polyaromatic hydrocarbons. Mar Pollut Bull. 2009;58:256–262. doi: 10.1016/j.marpolbul.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Baboshin M., Akimov V., Baskunov B., Born T.L., Khan S.U., Golovleva L. Conversion of polycyclic aromatic hydrocarbons by Sphingomonas sp. VKM B‐2434. Biodegradation. 2008;19:567–576. doi: 10.1007/s10532-007-9162-2. [DOI] [PubMed] [Google Scholar]
- Bach Q.‐D., Kim S.‐J., Chol S.‐C., Oh Y.‐S. Enhancing the intrinsic bioremediation of PAH‐contaminated anoxic estuarine sediments with biostimulating agents. J Microbiol. 2005;43:319–324. [PubMed] [Google Scholar]
- Baek S.O., Field R.A., Goldstone M.E., Kirk P.W., Lester J.N., Perry R. A review of atmospheric polycyclic aromatic hydrocarbons: sources, fate and behavior. Water Air Soil Pollut. 1991;60:279–300. [Google Scholar]
- Balkwill D.L., Drake G.R., Reeves R.H., Frederickson J.K., White D.C., Ringelberg D.B. Taxonomic study of aromatic‐degrading bacteria from deep‐terrestrial‐subsurface sediments and description of Sphingmonas aromaticivorans sp. nov., Sphingomonas subterra sp. nov., and Sphingomonas stygia sp. nov. Int J Syst Bacteriol. 1997;47:191–201. doi: 10.1099/00207713-47-1-191. et al. [DOI] [PubMed] [Google Scholar]
- Balkwill D.L., Frederickson J.K., Romine M.F. Sphingomonas and related genera. Prokaryotes. 2006;7:605–629. [Google Scholar]
- Barkay T., Navon‐Venezia S., Ron E.Z., Rosenberg E. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl Environ Microbiol. 1999;65:2697–2702. doi: 10.1128/aem.65.6.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnsley E.A. The bacterial degradation of fluoranthene and benzo[a]pyrene. Can J Microbiol. 1975;21:1004–1008. doi: 10.1139/m75-148. [DOI] [PubMed] [Google Scholar]
- Bastiaens L., Springael D., Wattiau P., Harms H., DeWachter R., Verachtert H., Diels L. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)‐degrading bacteria using PAH‐sorbing carriers. Appl Environ Microbiol. 2000;66:1834–1843. doi: 10.1128/aem.66.5.1834-1843.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan B.W., Sullivan W.R. Physiochemical soil parameters affecting sequestration and mycobacterial biodegradation of polycyclic aromatic hydrocarbons in soil. Chemosphere. 2003;52:1717–1726. doi: 10.1016/S0045-6535(03)00455-7. [DOI] [PubMed] [Google Scholar]
- Bogan B.W., Lahner L.M., Sullivan W.R., Paterek J.R. Degradation of straight‐chain aliphatic and high‐molecular‐weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. Microbiology. 2003;94:230–239. doi: 10.1046/j.1365-2672.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- Boldrin B., Tiehm A., Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonchan S., Britz M.L., Stanley G.A. Surfactant‐enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophila. Biotechnol Bioeng. 1998;59:482–494. doi: 10.1002/(sici)1097-0290(19980820)59:4<482::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Boonchan S., Britz M.L., Stanley G.A. Degradation and mineralization of high‐molecular‐weight polycyclic aromatic hydrocarbons by defined fungal‐bacterial cocultures. Appl Environ Microbiol. 2000;66:1007–1019. doi: 10.1128/aem.66.3.1007-1019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger E.C., Kirschner P., Springer B., Zumft W. Mycobacteria degrading polycyclic aromatic hydrocarbons. Int J Syst Bacteriol. 1997;47:247. [Google Scholar]
- Bouchez M., Blanchet D., Bardin V., Haeseler F., Vandecasteele J.‐P. Efficiency of defined strains and of soil consortia in the biodegradation of polycyclic aromatic hydrocarbon (PAH) mixtures. Biodegradation. 1999;10:429–435. doi: 10.1023/a:1008382030604. [DOI] [PubMed] [Google Scholar]
- Boyd D.R., Sharma N.D., Agarwal R., Resnick S.M., Schocken M.J., Gibson D.T. Bacterial dioxygenase‐catalyzed dihydroxylation and chemical resolution routes to enantiopure cis‐dihydrodiols of chrysene. J Chem Soc Perkins Trans I. 1997;1:1715–1723. et al. [Google Scholar]
- Boyd D.R., Sharma N.D., Hempenstall F., Kennedy M.A., Malone J.F., Allen C.C.R. bis‐cis‐Dihydrodiols: a new class of metabolites resulting from biphenyl dioxygenase‐catalyzed sequential asymmetric cis‐dihydroxylation of polycyclic arenes and heteroarenes. J Org Chem. 1999;64:4005–4011. et al. [Google Scholar]
- Boyd T.J., Montgomery M.T., Steele J.K., Pohlman J.W., Reatherford S.R., Spargo B.J., Smith D.C. Dissolved oxygen saturation controls PAH biodegradation in freshwater estuary sediments. Microb Ecol. 2005;49:226–235. doi: 10.1007/s00248-004-0279-0. [DOI] [PubMed] [Google Scholar]
- Boyd D.R., Sharma N.D., Belhocine T., Malone J.F., McGregor S., Allen C.C.R. Dioxygenase‐catalysed dihydroxylation of arene cis‐dihydrodiols and acetonide derivatives: a new approach to the synthesis of enantiopure tetraoxygenated bioproducts from arenes. Chem Commun. 2006:4934–4936. doi: 10.1039/b612191h. [DOI] [PubMed] [Google Scholar]
- Brezna B., Khan A.A., Cerniglia C.E. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon‐degrading Mycobacterium spp. FEMS Microbiol Lett. 2003;223:177–183. doi: 10.1016/S0378-1097(03)00328-8. [DOI] [PubMed] [Google Scholar]
- Brezna B., Kweon O., Stingley R.L., Freeman J.P., Khan A.A., Polek B. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaalenii PYR‐1. Appl Microbiol Biotechnol. 2006;71:522–532. doi: 10.1007/s00253-005-0190-8. et al. [DOI] [PubMed] [Google Scholar]
- Burchiel S.W., Luster M.I. Signaling by environmental polycyclic aromatic hydrocarbons in human lymphocytes. Clin Immunol. 2001;98:2–10. doi: 10.1006/clim.2000.4934. [DOI] [PubMed] [Google Scholar]
- Byss M., Elhottová D., Tríska J., Baldrian P. Fungal bioremediation of the creosote‐contaminated soil: influence of Pleurotus ostreatus and Irpex lacteus on polycyclic aromatic hydrocarbons removal and soil microbial community composition in the laboratory‐scale study. Chemosphere. 2008;73:1518–1523. doi: 10.1016/j.chemosphere.2008.07.030. [DOI] [PubMed] [Google Scholar]
- Caldini G., Cenci G., Manenti R., Morozzi G. The ability of an environmental isolate of Pseudomonas fluorescens to utilize chrysene and other four‐ring polynuclear aromatic hydrocarbons. Appl Microbiol Biotechnol. 1995;44:225–229. [Google Scholar]
- Carmichael A.B., Wong L.‐L. Protein engineering of Bacillus megaterium CYP102. Eur J Biochem. 2001;268:3117–3125. doi: 10.1046/j.1432-1327.2001.02212.x. [DOI] [PubMed] [Google Scholar]
- Cébron A., Norini M.P., Beguiristain T., Leyval C. Real‐time PCR quantification of PAH‐ring hydroxylation dioxygenase (PAH‐RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J Microbiol Methods. 2008;73:148–159. doi: 10.1016/j.mimet.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Cenci G., Caldini G. Catechol dioxygenase expression in a Pseudomonas fluorescens strain exposed to different aromatic compounds. Appl Microbiol Biotechnol. 1997;47:306–308. [Google Scholar]
- Cerniglia C. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- Chang Y.‐T., Lee J.‐F., Hung C.‐H. PAH biodegradation in surfactant‐water systems based on the theory of cohesive energy density (CED) J Chem Technol Biotechnol. 2007;82:442–452. [Google Scholar]
- Chen S.‐H., Aitken M.D. Salicylate stimulates the degradation of high‐molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ Sci Technol. 1999;33:435–439. [Google Scholar]
- Cheung P.Y., Kinkle B.K. Mycobacterium diversity and pyrene mineralization in petroleum‐contaminated soils. Appl Environ Microbiol. 2001;67:2222–2229. doi: 10.1128/AEM.67.5.2222-2229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child R., Miller C.D., Liang Y., Narasimham G., Chatterton J., Harrison P. Polycyclic aromatic hydrocarbon‐degrading Mycobacterium isolates; their association with plant roots. Appl Microbiol Biotechnol. 2007a;75:655–663. doi: 10.1007/s00253-007-0840-0. et al. [DOI] [PubMed] [Google Scholar]
- Child R., Miller C.D., Liang Y., Sims R.C., Anderson A.J. Pyrene mineralization by Mycobacterium sp. strain KMS in a barley rhizosphere. J Environ Qual. 2007b;36:1260–1265. doi: 10.2134/jeq2007.0008. [DOI] [PubMed] [Google Scholar]
- Churchill S.A., Harper J.P., Churchill P.F. Isolation and characterization of a Mycobacterium species capable of degrading three‐ and four‐ring aromatic and aliphatic hydrocarbons. Appl Environ Microbiol. 1999;65:549–552. doi: 10.1128/aem.65.2.549-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corgié S.C., Beguiristain T., Leyval C. Differential composition of bacterial communities as influenced by phenanthrene and dibenzo[a,h]anthracene in the rhizosphere of ryegrass (Lolium perenne L. Biodegradation. 2006;17:511–521. doi: 10.1007/s10532-005-9022-x. [DOI] [PubMed] [Google Scholar]
- Cornelissen G., Rigterink H., Ferdinandy M.M.A., Van Noort P.C.M. Rapidly desorbing fractions of PAHs in contaminated sediments as a predictor of the extent of bioremediation. Environ Sci Technol. 1998;32:966–970. [Google Scholar]
- Cottin N., Merlin G. Removal of PAHs from laboratory columns simulating the humus upper layer of vertical flow constructed wetlands. Chemosphere. 2008;73:711–716. doi: 10.1016/j.chemosphere.2008.06.060. [DOI] [PubMed] [Google Scholar]
- Cui Z., Lai Q., Dong C., Shao Z. Biodiversity of polycyclic aromatic hydrocarbon‐degrading bacteria from deep sea sediments of the Middle Atlantic ridge. Environ Microbiol. 2008;10:2138–2149. doi: 10.1111/j.1462-2920.2008.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe M., Kertesz M.A. Autecological properties of soil sphingomonads involved in the degradation of polycyclic aromatic hydrocarbons. Appl Environ Biotechnol. 2006a;72:1083–1089. doi: 10.1007/s00253-006-0374-x. [DOI] [PubMed] [Google Scholar]
- Cunliffe M., Kertesz M.A. Effect of Sphingobium yanoikuyae B1 inoculation on bacterial community dynamics and polycyclic aromatic hydrocarbon degradation in aged and freshly PAH‐contaminated soils. Environ Pollut. 2006b;144:228–237. doi: 10.1016/j.envpol.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Daane L.L., Harjono I., Zylstra G.J., Häggblom M.M. Isolation and characterization of polycyclic aromatic hydrocarbon‐degrading bacteria associated with the rhizosphere of salt marsh plants. Appl Environ Microbiol. 2001;67:2683–2691. doi: 10.1128/AEM.67.6.2683-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher F., Déziel E., Lirette P., Paquette G., Bisaillon J.‐G., Villemur R. Comparative study of five polycyclic aromatic hydrocarbon degrading bacterial strains isolated from contaminated soils. Can J Microbiol. 1997;43:368–377. doi: 10.1139/m97-051. [DOI] [PubMed] [Google Scholar]
- Dandie C.E., Thomas S.M., Bentham R.H., McClure N.C. Physiological characterization of Mycobacterium sp. strain 1B isolated from a bacterial culture able to degrade high‐molecular‐weight polycyclic aromatic hydrocarbons. J Appl Microbiol. 2004;97:246–255. doi: 10.1111/j.1365-2672.2004.02087.x. [DOI] [PubMed] [Google Scholar]
- Das K., Mukherjee A.K. Differential utilization of pyrene as the sole source of carbon by Bacillus subtilis and Pseudomonas aeruginosa strains: role of biosurfactants in enhancing bioavailability. J Appl Microbiol. 2006;102:195–203. doi: 10.1111/j.1365-2672.2006.03070.x. [DOI] [PubMed] [Google Scholar]
- Daugulis A.J., McCracken C.M. Microbial degradation of high and low molecular weight polyaromatic hydrocarbons in a two‐phase partitioning bioreactor by two strains of Sphingomonas sp. Biotechnol Lett. 2003;25:1441–1444. doi: 10.1023/a:1025007729355. [DOI] [PubMed] [Google Scholar]
- Dean‐Ross D., Cerniglia C.E. Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol. 1996;46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- Dean‐Ross D., Moody J.D., Freeman J.P., Doerge D.R., Cerniglia C.E. Metabolism of anthracene by Rhodococcus species. FEMS Microbiol Lett. 2001;204:205–211. doi: 10.1111/j.1574-6968.2001.tb10886.x. [DOI] [PubMed] [Google Scholar]
- Dean‐Ross D., Moody J., Cerniglia C.E. Utilization of mixtures of polycyclic aromatic hydrocarbons by bacteria isolated from contaminated sediment. FEMS Microbiol Ecol. 2002;41:1–7. doi: 10.1111/j.1574-6941.2002.tb00960.x. [DOI] [PubMed] [Google Scholar]
- Demanèche S., Meyer C., Micoud J., Louwagie M., Willison J.C., Jouanneau Y. Identification and functional analysis of two aromatic‐ring‐hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2004;70:6714–6725. doi: 10.1128/AEM.70.11.6714-6725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derz K., Klinner U., Schuphan I., Stackebrandt E., Kroppenstedt R.M. Mycobacterium pyrenivorans sp. nov., a novel polycyclic‐aromatic‐hydrocarbon‐degrading species. Int J Syst Evol Microbiol. 2004;54:2313–2317. doi: 10.1099/ijs.0.03003-0. [DOI] [PubMed] [Google Scholar]
- Derz K., Schmidt B., Schwiening S., Schuphan I. Comparison of microbial pyrene and benzo[a]pyrene mineralization in liquid medium, soil slurry, and soil. J Environ Sci Health B. 2006;41:471–484. doi: 10.1080/03601230600701619. [DOI] [PubMed] [Google Scholar]
- Dimitriou‐Christidis P., Autenrieth R.L. Kinetics of biodegradation of binary and ternary mixtures of PAHs. Biotechnol Bioeng. 2007;97:788–800. doi: 10.1002/bit.21269. [DOI] [PubMed] [Google Scholar]
- Dimitriou‐Christidis P., Autenrieth R.L., McDonald T.J., Desai A.M. Measurement of biodegradability parameters for single unsubstituted and methylated polycyclic aromatic hydrocarbons in liquid bacterial suspensions. Biotechnol Bioeng. 2007;97:922–932. doi: 10.1002/bit.21268. [DOI] [PubMed] [Google Scholar]
- Dries J., Smets B.F. Transformation and mineralization of benzo[a]pyrene by microbial cultures enriched on mixtures of three‐ and four‐ring polycyclic aromatic hydrocarbons. J Ind Microbiol Biotechnol. 2002;28:70–73. doi: 10.1038/sj/jim/7000211. [DOI] [PubMed] [Google Scholar]
- Ehrenfreund P., Charnley S.B. Organic molecules in the interstellar medium, comets, and meteorites: a voyage from dark clouds to the early earth. Annu Rev Astron Astrophys. 2000;38:427–483. [Google Scholar]
- Ehrenfreund P., Sephton M.A. Carbon molecules in space: from astrochemistry to astrobiology. Faraday Discuss. 2006;133:277–288. doi: 10.1039/b517676j. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Dalhammar G., Mohn W.W. Bacterial growth and biofilm production on pyrene. FEMS Microbiol Ecol. 2002;40:21–27. doi: 10.1111/j.1574-6941.2002.tb00932.x. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Sodersten E., Yu Z., Dalhammar G., Mohn W.W. Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate‐reducing conditions in enrichment cultures from Northern soils. Environ Microbiol. 2003;69:275–284. doi: 10.1128/AEM.69.1.275-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava F., Berselli S., Conte P., Piccolo A., Marchetti L. Effects of humic substances and soya lecithin on the aerobic bioremediation of a soil historically contaminated by polycyclic aromatic hydrocarbons (PAHs) Biotechnol Bioeng. 2004;88:214–223. doi: 10.1002/bit.20225. [DOI] [PubMed] [Google Scholar]
- Feitkenhauer H., Müller R., Märkl H. Degradation of polycyclic aromatic hydrocarbons and long chain alkanes at 60–70°C by Thermus and Bacillus spp. Biodegradation. 2003;14:367–372. doi: 10.1023/a:1027357615649. [DOI] [PubMed] [Google Scholar]
- Ferraro D.J., Brown E.N., Yu C.‐L., Parales R.E., Gibson D.T., Ramaswamy S. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3‐dioxygenase from Sphingobium yanoikuyae B1. BMC Struct Biol. 2007;7:10. doi: 10.1186/1472-6807-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson‐Pitts B., Pitts J.N. Tropospheric air pollution: ozone, airborne toxics, polycyclic aromatic hydrocarbons, and particles. Science. 1997;276:1045–1052. doi: 10.1126/science.276.5315.1045. [DOI] [PubMed] [Google Scholar]
- Fismes J., Perrin‐Ganier C., Empereur‐Bissonnet P., Morel J.L. Soil‐to‐root transfer and translocation of polycyclic aromatic hydrocarbons by vegetables grown on industrial contaminated soils. J Environ Qual. 2002;31:1649–1656. doi: 10.2134/jeq2002.1649. [DOI] [PubMed] [Google Scholar]
- Frederickson J.K., Balkwill D.L., Drake G.R., Romine M.F., Ringelberg D.B., White D.C. Aromatic‐degrading Sphingomonas isolates from the deep subsurface. Appl Environ Microbiol. 1995;61:1917–1922. doi: 10.1128/aem.61.5.1917-1922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin S., Bentham R. Comparison of enrichment methods of the isolation of pyrene‐degrading bacteria. Int Biodeterior Biodeg. 2005;56:80–85. [Google Scholar]
- Gauthier E., Déziel E., Villemur R., Juteau P., Lépine F., Beaudet R. Initial characterization of new bacteria degrading high‐molecular weight polycyclic aromatic hydrocarbons isolated from a 2‐year enrichment in a two‐liquid‐phase culture system. J Appl Microbiol. 2003;94:301–311. doi: 10.1046/j.1365-2672.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Geiselbrecht A.D., Hedlund B.P., Tichi M.A., Staley J.T. Isolation of marine polycyclic aromatic hydrocarbon (PAH)‐degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl Environ Microbiol. 1998;64:4703–4710. doi: 10.1128/aem.64.12.4703-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.T. Beijerinckia sp strain B1: a strain by any other name. . . . . J Ind Microbiol Biotechnol. 1999;23:284–293. doi: 10.1038/sj.jim.2900715. [DOI] [PubMed] [Google Scholar]
- Gibson D.T., Roberts R.L., Wells M.C., Kobal V.M. Oxidation of biphenyl by a Beijerinckia species. Biochem Biophys Res Commun. 1973;50:211–219. doi: 10.1016/0006-291x(73)90828-0. [DOI] [PubMed] [Google Scholar]
- Gibson D.T., Mahadevan V., Jerina D.M., Yagi H., Yeh H.J.C. Oxidation of the carcinogens benzo[a]pyrene and benzo[a]anthracene to dihydrodiols by a bacterium. Science. 1975;189:295–297. doi: 10.1126/science.1145203. [DOI] [PubMed] [Google Scholar]
- Gomes N.C.M., Borges L.R., Paranhos R., Pinto F.N., Krögerrecklenfort E., Mendonça‐Hagler L.C.S., Smalla K. Diversity of ndo genes in mangrove sediments exposed to different sources of polycyclic aromatic hydrocarbon pollution. Appl Environ Microbiol. 2007;73:7392–7399. doi: 10.1128/AEM.01099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L., Dobson A.D.W. Fluoranthene degradation in Pseudomonas alcaligenes PA‐10. Biodegradation. 2001;12:393–400. doi: 10.1023/a:1015029519142. [DOI] [PubMed] [Google Scholar]
- Guthrie E.A., Bortiatynski J.M., Van Heemst J.D.H., Richman J.E., Hardy K.S., Kovach E.M., Hatcher P.G. Determination of [13C]pyrene sequestration in sediment microcosms using flash pyrolysis‐GC‐MS and 13C NMR. Environ Sci Technol. 1999;33:119–125. [Google Scholar]
- Guthrie‐Nichols E., Grasham A., Kazunga C., Sangaiah R., Gold A., Bortiatynski J. The effect of aging on pyrene transformation in sediments. Environ Toxicol Chem. 2003;22:40–49. et al. [PubMed] [Google Scholar]
- Habe H., Kanemitsu M., Nomura M., Takemura T., Iwata K., Nojiri H. Isolation and characterization of an alkaliphilic bacterium utilizing pyrene as a carbon source. J Biosci Bioeng. 2004;98:306–308. doi: 10.1016/S1389-1723(04)00287-7. et al. [DOI] [PubMed] [Google Scholar]
- Habe H., Chung J.‐S., Ishida A., Kasuga K., Ide K., Takemura T. The fluorene catabolic linear plasmid in Terrabacter sp. strain DBF63 carries the β‐ketoadipate pathway genes, pcaRHGBDCFIJ, also found in proteobacteria. Microbiology. 2005;151:3713–3722. doi: 10.1099/mic.0.28215-0. et al. [DOI] [PubMed] [Google Scholar]
- Haderlein A., Legros R., Ramsay B. Enhancing pyrene mineralization in contaminated soil by the addition of humic acids or composted contaminated soil. Appl Microbiol Biotechnol. 2001;56:555–559. doi: 10.1007/s002530000520. [DOI] [PubMed] [Google Scholar]
- Haderlein A., Legros R., Ramsay B.A. Pyrene mineralization capacity increases with compost maturity. Biodegradation. 2006;17:293–302. doi: 10.1007/s10532-005-4217-8. [DOI] [PubMed] [Google Scholar]
- Han X., Naeher L.P. A review of traffic‐related air pollution exposure assessment studies in the developing world. Environ Int. 2006;32:106–120. doi: 10.1016/j.envint.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Harford‐Cross C.F., Carmichael A.B., Allan F.K., England P.A., Rouch D.A., Wong L.‐L. Protein engineering of cytochrome P450cam (CYP101) for the oxidation of polycyclic aromatic hydrocarbons. Protein Eng. 2000;13:121–128. doi: 10.1093/protein/13.2.121. [DOI] [PubMed] [Google Scholar]
- Heitkamp M.A., Cerniglia C.E. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl Environ Microbiol. 1988;54:1612–1614. doi: 10.1128/aem.54.6.1612-1614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitkamp M.A., Franklin W., Cerniglia C.E. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene‐degrading bacterium. Appl Environ Microbiol. 1988a;54:2549–2555. doi: 10.1128/aem.54.10.2549-2555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitkamp M.A., Freeman J.P., Miller D.W., Cerniglia C.E. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988b;54:2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herwijnen R., Springael D., Slot P., Govers H.A.J., Parsons J.R. Degradation of anthracene by Mycobacterium sp. strain LB501T proceeds via a novel pathway, through o‐phthalic acid. Appl Environ Microbiol. 2003a;69:186–190. doi: 10.1128/AEM.69.1.186-190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herwijnen R., Wattiau P., Bastiaens L., Daal L., Jonker L., Springael D. Elucidation of the metabolic pathways of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res Microbiol. 2003b;154:199–206. doi: 10.1016/S0923-2508(03)00039-1. et al. [DOI] [PubMed] [Google Scholar]
- Van Herwijnen R., Joffe B., Ryngaert A., Hausner M., Springael D., Govers H.A.J. Effect of bioaugmentation and supplementary carbon sources on degradation of polycyclic aromatic hydrocarbons by a soil‐derived culture. FEMS Microbiol Ecol. 2006;55:122–135. doi: 10.1111/j.1574-6941.2005.00001.x. et al. [DOI] [PubMed] [Google Scholar]
- Hickey A.M., Gordon L., Dobson A.D.W., Kelly C.T., Doyle E.M. Effect of surfactants on fluoranthene degradation by Pseudomonas alcaligenes PA‐10. Appl Microbiol Biotechnol. 2007;74:851–856. doi: 10.1007/s00253-006-0719-5. [DOI] [PubMed] [Google Scholar]
- Hilyard E.J., Jones‐Meehan J.M., Spargo B.J., Hill R.T. Enrichment, isolation, and phylogenetic identification of polycyclic aromatic hydrocarbon‐degrading bacteria from Elizabeth River Sediments. Appl Environ Microbiol. 2008;74:1176–1182. doi: 10.1128/AEM.01518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Jackson M., Yang Y., Mueller J.G., Pritchard P.H. Characterization of fluoranthene‐ and pyrene‐degrading bacteria isolated from PAH‐contaminated soils and sediments. J Ind Microbiol Biotechnol. 2000;24:100–112. [Google Scholar]
- Holman H.‐Y.N., Nieman K., Sorensen D.L., Miller C.D., Martin M.C., Borch T. Catalysis of PAH biodegradation by humic acid shown in synchrotron infrared studies. Environ Sci Technol. 2002;36:1276–1280. doi: 10.1021/es0157200. et al. [DOI] [PubMed] [Google Scholar]
- Howsam M., Jones K.C., Ineson P. PAHs associated with the leaves of three deciduous tree species. I – Concentrations and profiles. Environ Pollut. 2000;108:413–424. doi: 10.1016/s0269-7491(99)00195-5. [DOI] [PubMed] [Google Scholar]
- Hu Y.‐T., Zhou P.‐J., Zhou Y.‐G., Liu Z.‐H., Liu S.‐J. Saccharothrix xinjiangensis sp. nov., a pyrene‐degrading actinomycete isolated from Tianchi Lake, Xinjiang, China. Int J Syst Evol Microbiol. 2004;54:2091–2094. doi: 10.1099/ijs.0.63143-0. [DOI] [PubMed] [Google Scholar]
- Huesemann M.H., Hausmann T.S., Fortman T.J. Does bioavailability limit biodegradation? A comparison of hydrocarbon biodegradation and desorption rates in aged soils. Biodegradation. 2004;15:261–274. doi: 10.1023/b:biod.0000042996.03551.f4. [DOI] [PubMed] [Google Scholar]
- Husain S. Effect of surfactants on pyrene degradation by Pseudomonas fluorescens 29L. World J Microbiol Biotechnol. 2008;24:2411–2419. doi: 10.1007/s00284-008-9198-5. [DOI] [PubMed] [Google Scholar]
- Hylland K. Polycyclic aromatic hydrocarbon (PAH) ecotoxicology in marine ecosystems. J Toxicol Environ Health. 2006;69:109–123. doi: 10.1080/15287390500259327. [DOI] [PubMed] [Google Scholar]
- Iwabuchi T., Harayama S. Biochemical and genetic characterization of trans‐2′carboxybenzalpyruvate hydratase‐aldolase from a phenanthrene‐degrading Nocardioides strain. J Bacteriol. 1998a;180:945–949. doi: 10.1128/jb.180.4.945-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi T., Harayama S. Biochemical and molecular characterization of 1‐hydroxy‐2‐naphthoate dioxygenase from Nocardioides sp. KP7. J Biol Chem. 1998b;273:8332–8336. doi: 10.1074/jbc.273.14.8332. [DOI] [PubMed] [Google Scholar]
- Iwabuchi T., Venkateswaran K., Harayama S., Tanaka H. Low growth yield of a marine Pseudomonas grown on phenanthrene: a general phenomenon in bacteria grown on polycyclic aromatic hydrocarbons. J Mar Biotechnol. 1995;2:11–14. [Google Scholar]
- Jakoncic J., Jouanneau Y., Meyer C., Stojanoff V. The catalytic pocket of the ring‐hydroxylating dioxygenase from Sphingomonas CHY‐1. Biochem Biophys Res Commun. 2007a;352:861–866. doi: 10.1016/j.bbrc.2006.11.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoncic J., Jouanneau Y., Meyer C., Stojanoff V. The crystal structure of the ring‐hydroxylating dioxygenase from Sphingomonas CHY‐1. FEBS J. 2007b;274:2470–2481. doi: 10.1111/j.1742-4658.2007.05783.x. [DOI] [PubMed] [Google Scholar]
- Jerina D.M., Van Bladeren P.J., Yagi H., Gibson D.T., Mahadevan V., Neese A.S. Synthesis and absolute configuration of the bacterial cis‐1,2‐, cis‐8,9‐, and cis‐10,11‐dihydrodiol metabolites of benz[a]anthracene formed by a strain of Beijerinckia. J Org Chem. 1984;49:3621–3628. et al. [Google Scholar]
- Johnsen A., Karlson U. Diffuse PAH contamination of surface soils: environmental occurrence, bioavailability and microbial degradation. Appl Microbiol Biotechnol. 2007;76:533–543. doi: 10.1007/s00253-007-1045-2. [DOI] [PubMed] [Google Scholar]
- Johnsen A.R., Karlson U. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons. Appl Microbiol Biotechnol. 2004;63:452–459. doi: 10.1007/s00253-003-1265-z. [DOI] [PubMed] [Google Scholar]
- Johnsen A.R., Schmidt S., Hybholt T.K., Henriksen S., Jacobsen C.S., Andersen O. Strong impact of the polycyclic aromatic hydrocarbon (PAH)‐degrading community of a PAH‐polluted soil but marginal effect on PAH degradation when priming with bioremediated soil dominated by Mycobacteria. Appl Environ Microbiol. 2007;73:1474–1480. doi: 10.1128/AEM.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.L., Maguire K.L., Anderson D.R., McGrath S.P. Enhanced dissipation of chrysene in planted soil: the impact of a rhizobial inoculum. Soil Biol Biochem. 2004;36:33–38. [Google Scholar]
- Jones M.D., Singleton D.R., Carstensen D.P., Powell S.N., Swanson J.S., Pfaender F.K., Aitken M.D. Effect of incubation conditions on the enrichement of pyrene‐degrading bacteria identified by stable‐isotope probing in an aged, PAH‐contaminated soil. Microb Ecol. 2008;56:341–349. doi: 10.1007/s00248-007-9352-9. [DOI] [PubMed] [Google Scholar]
- Jouanneau Y., Meyer C. Purification and characterization of an arene cis‐dihydrodiol dehydrogenase endowed with broad substrate specificity toward polycyclic aromatic hydrocarbon dihydrodiols. Appl Environ Microbiol. 2006;72:4726–4734. doi: 10.1128/AEM.00395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau Y., Willison J.C., Meyer C., Krivobok S., Chevron N., Besombes J.‐L., Blake G. Stimulation of pyrene mineralization in freshwater sediments by bacterial and plant bioaugmentation. Environ Sci Technol. 2005;39:5729–5735. doi: 10.1021/es050412d. [DOI] [PubMed] [Google Scholar]
- Jouanneau Y., Meyer C., Jakoncic J., Stojanoff V., Gaillard J. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry. 2006;45:12380–12391. doi: 10.1021/bi0611311. [DOI] [PubMed] [Google Scholar]
- Jouanneau Y., Micoud J., Meyer C. Purification and characterization of a three‐component salicylate 1‐hydroxylase from Sphingomonas sp. strain CHY‐1. Appl Environ Microbiol. 2007;73:7515–7521. doi: 10.1128/AEM.01519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz A.L., Naidu R. Bioremediation of high molecular weight polycyclic aromatic hydrocarbons; a review of the microbial degradation of benzo[a]pyrene. Int Biodeterior Biodegradation. 2000;45:57–88. [Google Scholar]
- Juhasz A.L., Britz M.L., Stanley G.A. Degradation of High molecular weight polycyclic aromatic hydrocarbons by Pseudomonas cepacia. Biotechnol Lett. 1996;8:577–582. [Google Scholar]
- Juhasz A.L., Britz M.L., Stanley G.A. Degradation of fluoranthene, pyrene, benz[a]anthracene and dibenz[a,h]anthracene by Burkholderia cepacia. J Appl Microbiol. 1997;83:189–198. [Google Scholar]
- Juhasz A.L., Britz M.L., Stanley G.A. Evaluation of a creosote‐based medium for the growth and preparation of a PAH‐degrading bacterial community for bioaugmentation. J Ind Microbiol Biotechnol. 2000a;24:277–284. [Google Scholar]
- Juhasz A.L., Stanley G.A., Britz M.L. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10,003. Lett Appl Microbiol. 2000b;30:396–401. doi: 10.1046/j.1472-765x.2000.00733.x. [DOI] [PubMed] [Google Scholar]
- Juhasz A.L., Stanley G.A., Britz M.L. Metabolite repression inhibits degradation of benzo[a]pyrene and dibenz[a,h]anthracene by Stenotrophomonas maltophilia VUN 10,003. J Ind Microbiol Biotechnol. 2002;28:88–96. doi: 10.1038/sj/jim/7000216. [DOI] [PubMed] [Google Scholar]
- Kanaly R.A., Bartha R. Cometabolic mineralization of benzo[a]pyrene caused by hydrocarbon additions to soil. Environ Toxicol Chem. 1999;18:2186–2190. doi: 10.1002/etc.5620181010. [DOI] [PubMed] [Google Scholar]
- Kanaly R.A., Harayama S. Biodegradation of high‐molecular‐weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol. 2000;182:2059–2067. doi: 10.1128/jb.182.8.2059-2067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R.A., Watanabe K. Multiple mechanisms contribute to the biodegradation of benzo[a]pyrene by petroleum‐derived multi‐component NAPLs. Environ Toxicol Chem. 2004;23:850–856. doi: 10.1897/03-191. [DOI] [PubMed] [Google Scholar]
- Kanaly R.A., Bartha R., Fogel S., Findlay M. Biodegradation of [14C]benzo[a]pyrene added in crude oil to uncontaminated soil. Appl Environ Microbiol. 1997;63:4511–4515. doi: 10.1128/aem.63.11.4511-4515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R.A., Bartha R., Watanabe K., Harayama S. Rapid mineralization of benzo[a]pyrene by a microbial consortium growing on diesel fuel. Appl Environ Microbiol. 2000;66:4205–4211. doi: 10.1128/aem.66.10.4205-4211.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R.A., Bartha R., Watanabe K., Harayama S. Enhanced mineralization of benzo[a]pyrene in the presence of nonaqueous phase liquids. Environ Toxicol Chem. 2001;20:498–501. [PubMed] [Google Scholar]
- Kanaly R.A., Harayama S., Watanabe K. Rhodanobacter sp. strain BPC1 in a benzo[a]pyrene‐mineralizing bacterial consortium. Appl Environ Microbiol. 2002;68:5826–5833. doi: 10.1128/AEM.68.12.5826-5833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R.A., Watanabe K., Matsui S. Bioavailability of benzo[a]pyrene during NAPL‐enhanced biodegradation in soil and in liquid culture. Water Sci Technol. 2006;53:17–25. doi: 10.2166/wst.2006.333. [DOI] [PubMed] [Google Scholar]
- Kästner M., Breuer‐Jammali M., Mahro B. Enumeration and characterization of the soil microflora from hydrocarbon‐contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH) Appl Microbiol Biotechnol. 1994;41:267–273. [Google Scholar]
- Kästner M., Breuer‐Jammali M., Mahro B. Impact of inoculation protocols, salinity, and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH‐degrading bacteria introduced into soil. Appl Environ Microbiol. 1998;64:359–362. doi: 10.1128/aem.64.1.359-362.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K., Suzuki I. Ice core record of polycyclic aromatic hydrocarbons over the past 400 years. Naturwissenchaften. 1994;81:502–505. [Google Scholar]
- Kazunga C., Aitken M.D. Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon‐degrading bacteria. Appl Environ Microbiol. 2000;66:1917–1922. doi: 10.1128/aem.66.5.1917-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazunga C., Aitken M.D., Gold A., Sangaiah R. Fluoranthene‐2,3‐ and ‐1,5‐diones are novel products from the bacterial transformation of fluoranthene. Environ Sci Technol. 2001;35:917–922. doi: 10.1021/es001605y. [DOI] [PubMed] [Google Scholar]
- Kelley I., Freeman J.P., Evans F.E., Cerniglia C.E. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacterium sp. Appl Environ Microbiol. 1991;57:636–641. doi: 10.1128/aem.57.3.636-641.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley I., Freeman J.P., Evans F.E., Cerniglia C.E. Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR‐1. Appl Environ Microbiol. 1993;59:800–806. doi: 10.1128/aem.59.3.800-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.A., Wang R.‐F., Cao W.‐W., Doerge D.R., Wennerstrom D., Cerniglia C.E. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR‐1. Appl Environ Microbiol. 2001;67:3577–3585. doi: 10.1128/AEM.67.8.3577-3585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.A., Wang R.‐F., Cao W.‐W., Franklin W., Cerniglia C.E. Reclassification of a polycyclic aromatic hydrocarbon‐metabolizing bacterium, Beijerinckia sp. strain B1, as Sphingomonas yanoikuyae by fatty acid analysis, protein pattern analysis, DNA‐DNA hybridization, and 16S ribosomal DNA sequencing. Int J Syst Evol Microbiol. 1996;46:466–469. doi: 10.1099/00207713-46-2-466. [DOI] [PubMed] [Google Scholar]
- Khan A.A., Kim S.‐J., Paine D.D., Cerniglia C.E. Classification of a polycyclic aromatic hydrocarbon‐metabolizing bacterium, Mycobacterium sp. strain PYR‐1, as Mycobacterium vanbaalenii sp. nov. Int J Syst Evol Microbiol. 2002;52:1997–2002. doi: 10.1099/00207713-52-6-1997. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Weber W.J. Preferential surfactant utilization by a PAH‐degrading strain: effects on micellar solubilization phenomena. Environ Sci Technol. 2003;15:3574–3580. doi: 10.1021/es0210493. [DOI] [PubMed] [Google Scholar]
- Kim J.‐D., Lee C.‐G. Microbial degradation of polycyclic aromatic hydrocarbons in soil by bacterium‐fungus co‐cultures. Biotechnol Bioprocess Eng. 2007;12:410–416. [Google Scholar]
- Kim S.‐J., Jones R.C., Cha C.‐J., Kweon O., Edmondson R.D., Cerniglia C.E. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR‐1 using two‐dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics. 2004a;4:3899–3909. doi: 10.1002/pmic.200400872. [DOI] [PubMed] [Google Scholar]
- Kim S.‐J., Kweon O., Freeman J.P., Jones R.C., Adjei M.D., Jhoo J.‐W. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low‐ and high‐molecular weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR‐1. Appl Environ Microbiol. 2006;72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.‐J., Kweon O., Jones R.C., Freeman J.P., Edmondson R.D., Cerniglia C.E. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR‐1 based on systems biology. J Bacteriol. 2007;189:464–472. doi: 10.1128/JB.01310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.J., Lee E.Y., Kim Y.J., Cho K.S., Ryu H.W. Degradation of polyaromatic hydrocarbons by Burkholderia cepacia 2A‐12. World J Microbiol Biotechnol. 2003a;19:411–417. [Google Scholar]
- Kim Y.‐H., Engesser K.‐H., Cerniglia C.E. Two polycyclic aromatic hydrocarbon o‐quinone reductases from a pyrene‐degrading Mycobacterium. Arch Biochem Biophys. 2003b;416:209–217. doi: 10.1016/s0003-9861(03)00297-2. [DOI] [PubMed] [Google Scholar]
- Kim Y.‐H., Moody J.D., Freeman J.P., Brenzna B., Engesser K.‐H., Cerniglia C.E. Evidence for the existence of PAH‐quinone reductase and catechol‐O‐methyltransferase in Mycobacterium vanbaalenii PYR‐1. J Ind Microbiol Biotechnol. 2004b;31:507–516. doi: 10.1007/s10295-004-0178-x. [DOI] [PubMed] [Google Scholar]
- Kim Y.‐H., Engesser K.‐H., Cerniglia C.E. Numerical and genetic analysis of polycyclic aromaic hydrocarbon‐degrading mycobacteria. Microb Ecol. 2005a;50:110–119. doi: 10.1007/s00248-004-0126-3. [DOI] [PubMed] [Google Scholar]
- Kim Y.‐H., Freeman J.P., Moody J.D., Engesser K.H., Cerniglia C.E. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR‐1. Appl Microbiol Biotechnol. 2005b;67:275–285. doi: 10.1007/s00253-004-1796-y. [DOI] [PubMed] [Google Scholar]
- Kleespies M., Kroppenstedt R.M., Rainey F.A., Webb L.E., Stackebrandt E. Mycobacterium holderi sp. nov., a new member of the fast‐growing mycobacteria capable of degrading polycyclic aromatic hydrocarbons. Int J Syst Evol Microbiol. 1996;46:683–687. doi: 10.1099/00207713-46-3-683. [DOI] [PubMed] [Google Scholar]
- Krivobok S., Kuony S., Meyer C., Louwagie M., Willison J.C., Jouanneau Y. Identification of pyrene‐induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring‐hydroxylation dioxygenases. J Bacteriol. 2003;185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutz L.J., Beyrouty C.A., Gentry T.J., Wolf D.C., Reynolds C.M. Selective enrichment of a pyrene degrader population and enhanced pyrene degradation in Bermuda grass rhizosphere. Biol Fertil Soils. 2005;41:359–364. [Google Scholar]
- Kweon O., Kim S.‐J., Jones R.C., Freeman J.P., Adjei M.D., Edmondson R.D., Cerniglia C.E. A polyomic approach to elucidate the fluoranthene‐degradative pathway in Mycobacterium vanbaalenii PYR‐1. J Bacteriol. 2007;189:4635–4647. doi: 10.1128/JB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.‐K., Lee H.‐S., Jung H.‐B., Kang J.‐H., Kim S.‐J. Yeosuana aromativorans gen. nov., sp. nov., a mesophilic marine bacterium belonging to the family Flavobacteriaceae, isolated from estuarine sediment of the South Sea, Korea. Int J Syst Evol Microbiol. 2006;56:727–732. doi: 10.1099/ijs.0.64073-0. [DOI] [PubMed] [Google Scholar]
- Lafortune I., Juteau P., Déziel E., Lépine F., Beaudet R., Villemur R. Bacterial diversity of a consortium degrading high‐molecular‐weight polycyclic aromatic hydrocarbons in a two‐liquid phase biosystem. Microb Ecol. 2009;57:455–468. doi: 10.1007/s00248-008-9417-4. [DOI] [PubMed] [Google Scholar]
- Lang C., Tao S., Liu W., Zhang Y., Simonich S. Atmospheric transport and outflow of polycyclic aromatic hydrocarbons from China. Environ Sci Technol. 2008;42:5196–5201. doi: 10.1021/es800453n. [DOI] [PubMed] [Google Scholar]
- Launen L.A., Dutta J., Turpeinen R., Eastep M.E., Dorn R., Buggs V.H. Characterization of the indigenous PAH‐degrading bacteria of Spartina dominated salt marshes in the New York/New Jersey Harbor. Biodegradation. 2008;19:347–363. doi: 10.1007/s10532-007-9141-7. et al. [DOI] [PubMed] [Google Scholar]
- Lee S.‐E., Seo J.‐S., Kuem Y.‐S., Lee K.‐J., Li Q.X. Fluoranthene metabolism and associated proteins in Mycobacterium sp. JS14. Proteomics. 2007;7:2059–2069. doi: 10.1002/pmic.200600489. [DOI] [PubMed] [Google Scholar]
- Lehto K.‐M., Puhakka J.A., Lemmetyinen H. Biodegradation of selected UV‐irradiated and non‐irradiated polycyclic aromatic hydrocarbons (PAHs) Biodegradation. 2003;14:249–263. doi: 10.1023/a:1024704505832. [DOI] [PubMed] [Google Scholar]
- Lei L., Khodadoust A.P., Suidan M.T., Tabak H.H. Biodegradation of sediment‐bound PAHs in field‐contaminated sediment. Water Res. 2005;39:349–361. doi: 10.1016/j.watres.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Leonardi V., Giubilei M.A., Federici E., Spaccapelo R., Sasek V., Novotny C. Mobilizing agents enhance fungal degradation of polycyclic aromatic hydrocarbons and affect diversity of indigenous bacteria in soil. Biotechnol Bioeng. 2008;101:273–285. doi: 10.1002/bit.21909. et al. [DOI] [PubMed] [Google Scholar]
- Leys N.M.E.J., Ryngaert A., Bastiaens L., Verstraete W., Top E.M., Springael D. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2004;70:1944–1955. doi: 10.1128/AEM.70.4.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys N.M., Bastiaens L., Verstraete W., Springael D. Influence of the carbon/nitrogen/phosphorus ratio on polycyclic aromatic hydrocarbon degradation by Mycobacterium and Sphingomonas in soil. Appl Microbiol Biotechnol. 2005a;66:726–736. doi: 10.1007/s00253-004-1766-4. [DOI] [PubMed] [Google Scholar]
- Leys N.M., Ryngaert A., Bastiaens L., Top E.M., Verstraete W., Springael D. Culture independent detection of Sphingomonas sp. EPA 505 related strains in soils contaminated with polycyclic aromatic hydrocarbons (PAHs) Microbial Ecol. 2005b;49:443–450. doi: 10.1007/s00248-004-0011-0. [DOI] [PubMed] [Google Scholar]
- Li A., Tanabe S., Jiang G., Giesy J.P., Lam P.K.S. Elsevier; 2007. , and Persistent Organic Pollutants in Asia: Sources, Distributions, Transport and Fate. Amsterdam, the Netherlands: [Google Scholar]
- Li X., Li P., Lin X., Zhang C., Li Q., Gong Z. Biodegradation of aged polycyclic aromatic hydrocarbons (PAHs) by microbial consortia in soil and slurry phases. J Haz Mater. 2008;150:21–26. doi: 10.1016/j.jhazmat.2007.04.040. [DOI] [PubMed] [Google Scholar]
- Li X.‐F., Le X.‐C., Simpson C.D., Cullen W.R., Reimer K.J. Bacterial transformation of pyrene in a marine environment. Environ Sci Technol. 1996;30:1115–1119. [Google Scholar]
- Liang Y., Gardner D.R., Miller C.D., Chen D., Anderson A.J., Weimer B.C., Sims R.C. Study of biochemical pathways and enzymes involved in pyrene degradation by Mycobacterium sp. strain KMS. Appl Environ Microbiol. 2006;72:7821–7828. doi: 10.1128/AEM.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Britt D.W., McLean J.E., Sorensen D.L., Sims R.C. Humic acid effect on pyrene degradation: finding an optimal range for pyrene solubility and mineralization enhancement. Appl Microbiol Biotechnol. 2007;74:1368–1375. doi: 10.1007/s00253-006-0769-8. [DOI] [PubMed] [Google Scholar]
- Lin Y., Cai L.‐X. PAH‐degrading microbial consortium and its pyrene‐degrading plasmids from mangrove sediment samples in Huian, China. Mar Pollut Bull. 2008;57:703–706. doi: 10.1016/j.marpolbul.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Liu S.L., Luo Y.M., Cao Z.H., Wu L.H., Ding K.Q., Christie P. Degradation of benzo[a]pyrene in soil with arbuscular mycorrhizal alfalfa. Environ Geochem Health. 2004;26:285–293. doi: 10.1023/b:egah.0000039592.80489.e5. [DOI] [PubMed] [Google Scholar]
- López Z., Vila J., Grigoll M. Metabolism of fluoranthene by mycobacterial strains isolated by their ability to grow in fluoranthene or pyrene. J Ind Microbiol Biotechnol. 2005;32:455–464. doi: 10.1007/s10295-005-0022-y. [DOI] [PubMed] [Google Scholar]
- López Z., Vila J., Minguillón C., Grifoll M. Metabolism of fluoranthene by Mycobacterium sp. strain AP1. Appl Microbiol Biotechnol. 2006;70:747–756. doi: 10.1007/s00253-005-0120-9. [DOI] [PubMed] [Google Scholar]
- Lotfabad S.K., Gray M.R. Kinetics of biodegradation of mixtures of polycyclic aromatic hydrocarbons. Appl Microbiol Biotechnol. 2002;60:361–365. doi: 10.1007/s00253-002-1104-7. [DOI] [PubMed] [Google Scholar]
- Luan T.G., Yu K.S.H., Zhong Y., Zhou H.W., Lan C.Y., Tam N.F.Y. Study of metabolites from the degradation of polycyclic aromatic hydrocarbons (PAHs) by bacterial consortium enriched from mangrove sediments. Chemosphere. 2006;65:2289–2296. doi: 10.1016/j.chemosphere.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Lundstedt S., White P.A., Lemieux C.L., Lynes K.D., Lambert I.B., Öberg L., Heglund P. Sources, fate, and toxic hazards of oxygenated polycyclic aromatic hydrocarbons (PAHs) at PAH‐contaminated sites. Ambio. 2007;36:475–485. doi: 10.1579/0044-7447(2007)36[475:sfatho]2.0.co;2. et al. [DOI] [PubMed] [Google Scholar]
- Luning Prak D.J., Pritchard P.H. Degradation of polycyclic aromatic hydrocarbons dissolved in Tween 80 surfactant solutions by Sphingomonas paucimobilis EPA 505. Can J Microbiol. 2002;48:151–158. doi: 10.1139/w02-004. [DOI] [PubMed] [Google Scholar]
- McLellan S.L., Warshawsky D., Shann J.R. The effect of polycyclic aromatic hydrocarbons on the degradation of benzo[a]pyrene by Mycobacterium sp. strain RJGII‐135. Environ Toxicol Chem. 2002;21:253–259. [PubMed] [Google Scholar]
- Macleod C.T., Daugulis A.J. Biodegradation of polycyclic aromatic hydrocarbons in a two‐phase partitioning bioreactor in the presence of a bioavailable solvent. Appl Microbiol Biotechnol. 2003;62:291–296. doi: 10.1007/s00253-003-1297-4. [DOI] [PubMed] [Google Scholar]
- Macleod C.T., Daugulis A.J. Interfacial effects in a two‐phase partitioning bioreactor: degradation of polycyclic aromatic hydrocarbons (PAHs) by a hydrophobic Mycobacterium. Process Biochem. 2005;40:1799–1805. [Google Scholar]
- Macleod C.J.A., Semple K.T. The influence of single and multiple applications of pyrene on the evolution of pyrene catabolism in soil. Environ Pollut. 2006;139:455–460. doi: 10.1016/j.envpol.2005.06.014. [DOI] [PubMed] [Google Scholar]
- McNally D.L., Mihelcic J.R., Lueking D.R. Degradation of 3‐ and 4‐ring polycyclic aromatic hydrocarbons under denitrification conditions. Environ Sci Technol. 1998;32:2633–2639. [Google Scholar]
- McNally D.L., Mihelcic J.R., Lueking D.R. Biodegradation of mixtures of polycyclic aromatic hydrocarbons under aerobic and nitrate‐reducing conditions. Chemosphere. 1999;38:1313–1321. [Google Scholar]
- Mahaffey W.R., Gibson D.T., Cerniglia C.E. Bacterial oxidation of chemical carcinogens: formation of polycyclic aromatic acids from benz[a]anthracene. Appl Environ Microbiol. 1988;54:2415–2423. doi: 10.1128/aem.54.10.2415-2423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux J., Déziel E., Villemur R., Lépine F., Bisaillon J.‐G., Beaudet R. Optimization of high‐molecular‐weight polycyclic aromatic hydrocarbons' degradation in a two‐liquid‐phase bioreactor. J Appl Microbiol. 2000;88:655–662. doi: 10.1046/j.1365-2672.2000.01011.x. [DOI] [PubMed] [Google Scholar]
- Mastral A.M., Callén M. A review on polycyclic aromatic hydrocarbon (PAH) emissions from energy generation. Environ Sci Technol. 2000;34:3051–3057. [Google Scholar]
- Meador J.P. Bioaccumulation of pahs in marine invertebrates. In: Douben P.E.T., editor. John Wiley and Sons; 2003. pp. 147–171. . In PAHs: An Ecotoxicological Perspective (ed.). London, UK: , pp. [Google Scholar]
- Miller C.D., Hall K., Liang Y.N., Nieman K., Sorensen D., Issa B. Isolation and characterization of polycyclic aromatic hydrocarbon‐degrading Mycobacterium isolates from soil. Microbial Ecol. 2004;48:230–238. doi: 10.1007/s00248-003-1044-5. et al. [DOI] [PubMed] [Google Scholar]
- Miller C.D., Child R., Hughes J.E., Benscai M., Der J.P., Sims R.C., Anderson A.J. Diversity of soil mycobacterium isolates from three sites that degrade polycyclic aromatic hydrocarbons. J Appl Microbiol. 2007;102:1612–1624. doi: 10.1111/j.1365-2672.2006.03202.x. [DOI] [PubMed] [Google Scholar]
- Mohan S.V., Kisa T., Ohkuma T., Kanaly R.A., Shimizu Y. Bioremediation technologies for treatment of PAH‐contaminated soil and strategies to enhance process efficiency. Rev Environ Sci Biotechnol. 2006;5:347–374. [Google Scholar]
- Mohan S.V., Reddy B.P., Sarma P.N. Ex situ slurry phase bioremediation of chrysene contaminated soil with the function of metabolic function: process evaluation by data enveloping analysis (DEA) and Taguchi design of experimental methodology (DOE) Bioresour Technol. 2009;100:164–172. doi: 10.1016/j.biortech.2008.06.020. [DOI] [PubMed] [Google Scholar]
- Molina M., Araujo R., Hodson R.E. Cross‐induction of pyrene and phenanthrene in a Mycobacterium sp. isolated from polycyclic aromatic hydrocarbon contaminated river sediments. Can J Microbiol. 1999;45:520–529. [PubMed] [Google Scholar]
- Moody J.D., Freeman J.P., Doerge D.R., Cerniglia C.E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR‐1. Appl Environ Microbiol. 2001;67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody J.D., Fu P.P., Freeman J.P., Cerniglia C.E. Regio‐ and stereoselective metabolism of 7,12‐dimethylbenz[a]anthracene by Mycobacterium vanbaalenii PYR‐1. Appl Environ Microbiol. 2003;69:3924–3931. doi: 10.1128/AEM.69.7.3924-3931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody J.D., Freeman J.P., Fu P.P., Cerniglia C.E. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR‐1. Appl Environ Microbiol. 2004;70:340–345. doi: 10.1128/AEM.70.1.340-345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody J.D., Freeman J.P., Cerniglia C.E. Degradation of benz[a]anthracene by Mycobacterium vanbaalenii strain PYR‐1. Biodegradation. 2005;16:513–526. doi: 10.1007/s10532-004-7217-1. [DOI] [PubMed] [Google Scholar]
- Mueller J.G., Chapman P.J., Pritchard P.H. Action of a fluoranthene‐utilizing bacterial community on polycyclic aromatic hydrocarbon components of creosote. Appl Environ Microbiol. 1989;55:3085–3090. doi: 10.1128/aem.55.12.3085-3090.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J.G., Chapman P.J., Blattmann B.O., Pritchard P.H. Isolation and characterization of a fluoranthene‐utilizing strain of Pseudomonas paucimobilis. Appl Environ Microbiol. 1990;56:1079–1086. doi: 10.1128/aem.56.4.1079-1086.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J.G., Devereux R., Santavy D.L., Lantz S.E., Willis S.G., Pritchard P.H. Phylogenetic and physiological comparisons of PAH‐degrading bacteria from geographically diverse soils. Antonie van Leeuwenhoek. 1997;71:329–343. doi: 10.1023/a:1000277008064. [DOI] [PubMed] [Google Scholar]
- Mueller K.E., Shann J.R. Effects of tree root‐derived substrates and inorganic nutrients on pyrene mineralization in rhizosphere and bulk soil. J Environ Qual. 2007;36:120–126. doi: 10.2134/jeq2006.0130. [DOI] [PubMed] [Google Scholar]
- Mutnuri S., Vasudevan N., Kaestner M. Degradation of anthracene and pyrene supplied by microcrystals and non‐aqueous‐phase liquids. Appl Microbiol Biotechnol. 2005;67:569–576. doi: 10.1007/s00253-005-1905-6. [DOI] [PubMed] [Google Scholar]
- Nam J.J., Sweetman A.J., Jones K.C. Polynuclear aromatic hydrocarbons (PAHs) in global background soils. J Environ Monit. 2009;11:45–48. doi: 10.1039/b813841a. [DOI] [PubMed] [Google Scholar]
- Nam K., Kukor J.J. Combined ozonation and biodegradation fro remediation of mixtures of polycyclic aromatic hydrocarbons in soil. Biodegradation. 2000;11:1–9. doi: 10.1023/a:1026592324693. [DOI] [PubMed] [Google Scholar]
- Navarro R.R., Ichikawa H., Iimura Y., Tatsumi K. Removal of polycyclic aromatic hydrocarbons from contaminated soil by aqueous DNA solution. Environ Sci Technol. 2007;41:4240–4245. doi: 10.1021/es0624523. [DOI] [PubMed] [Google Scholar]
- Navarro R.R., Iimura Y., Ichikawa H., Tatsumi K. Treatment of PAHs in contaminated soil by extraction with aqueous DNA followed by biodegradation with a pure culture of Sphingomonas sp. Chemosphere. 2008;78:1414–1419. doi: 10.1016/j.chemosphere.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Neff J.M. Elsevier Press; 2002. Bioaccumulation in Marine Organisms: Effect of Contaminants From Oil Well Produced Water. Amsterdam, the Netherlands: [Google Scholar]
- Ní Chadhain S.M.N., Norman R.S., Pesce K.V., Kukor J.J., Zylstra G.J. Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation. Appl Environ Microbiol. 2006;72:4078–4087. doi: 10.1128/AEM.02969-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ní Chadhain S.M., Moritz E.M., Kim E., Zylstra G.J. Identification, cloning and characterization of a multicomponent biphenyl dioxygenase from Sphingobium yanoikuyae B1. J Ind Microbiol Biotechnol. 2007;34:605–613. doi: 10.1007/s10295-007-0235-3. [DOI] [PubMed] [Google Scholar]
- Nieman J.K.C., Holz R.C., Sims R.C. 13C NMR analysis of biologically produced pyrene residues by Mycobacterium sp. KMS in the presence of humic acid. Environ Sci Technol. 2007;41:242–249. doi: 10.1021/es0614464. [DOI] [PubMed] [Google Scholar]
- Niqui‐Arroyo J.‐L., Ortega‐Calvo J.‐J. Integrating biodegradation and electroosmosis for the enhanced removal of polycyclic aromatic hydrocarbons from creosote‐polluted soils. J Environ Qual. 2007;36:1444–1451. doi: 10.2134/jeq2006.0516. [DOI] [PubMed] [Google Scholar]
- Obayori O.S., Ilori M.O., Adebusoye S.A., Oyetibo G.O., Amund O.O. Pyrene‐degradation potentials of Pseudomonas species isolated from polluted tropical soils. World J Microbiol Biotechnol. 2008;24:2639–2646. [Google Scholar]
- Ohkouchi N., Kawamura K., Kawahata H. Distributions of three‐ to seven‐ring polynuclear aromatic hydrocarbons on the deep sea floor in the central pacific. Environ Sci Technol. 1999;33:3086–3090. [Google Scholar]
- Osterreicher‐Ravid D., Ron E.Z., Rosenberg E. Horizontal transfer of an exopolymer complex from one bacterial species to another. Environ Microbiol. 2000;2:366–372. doi: 10.1046/j.1462-2920.2000.00110.x. [DOI] [PubMed] [Google Scholar]
- Pagnout C., Rast C., Veber A.‐M., Poupin P., Férard J.‐F. Ecotoxicological assessment of PAHs and their dead‐end metabolites after degradation by Mycobacterium sp. Strain SNP11. Ecotoxicol Environ Safety. 2006;65:151–158. doi: 10.1016/j.ecoenv.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Pagnout C., Frache G., Poupin P., Maunit B., Muller J.‐F., Férard J.‐F. Isolation in Mycobacterium sp. strain SNP11: expression in Mycobacterium smegmatis mc2155. Res Microbiol. 2007;158:175–186. doi: 10.1016/j.resmic.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Parrish Z.D., Banks M.K., Schwab A.P. Effect of root death and decay on dissipation of polycyclic aromatic hydrocarbons in the rhizosphere of yellow sweet clover and tall fescue. J Environ Qual. 2005;34:207–216. doi: 10.2134/jeq2005.0207. [DOI] [PubMed] [Google Scholar]
- Phillips D.H. Polycyclic aromatic hydrocarbons in the diet. Mut Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Piskonen R., Itävaara M. Evaluation of chemical pretreatment of contaminated soil for improved PAH bioremediation. Appl Microbiol Biotechnol. 2004;65:627–634. doi: 10.1007/s00253-004-1679-2. [DOI] [PubMed] [Google Scholar]
- Pizzul L., Del Pilar Castillo M., Stenström J. Characterization of selected actinomycetes degrading polyaromatic hydrocarbons in liquid culture and spiked soil. World J Microbiol Biotechnol. 2006;22:745–752. [Google Scholar]
- Pizzul L., Sjögren A., Del Pilar Castillo M., Stenström J. Degradation of polycyclic aromatic hydrocarbons in soil by two‐step sequential treatment. Biodegradation. 2007;18:607–616. doi: 10.1007/s10532-006-9093-3. [DOI] [PubMed] [Google Scholar]
- Quann R.J. Modeling the chemistry of complex petroleum mixtures. Environ Health Perspect. 1998;106:1441–1448. doi: 10.1289/ehp.98106s61441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann K., Noll H.P., Steinberg C.E.W., Kettrup A.A. Pyrene degradation by Mycobacterium sp. strain KR2. Chemosphere. 1998;36:2977–2992. doi: 10.1016/s0045-6535(97)10240-5. [DOI] [PubMed] [Google Scholar]
- Rehmann K., Hertkorn N., Kettrup A.A. Fluoranthene metabolism in Mycobacterium sp. strain KR20: identity of pathway intermediates during degradation and growth. Microbiology. 2001;147:2783–2794. doi: 10.1099/00221287-147-10-2783. [DOI] [PubMed] [Google Scholar]
- Rentz J.A., Alvarez P.J.J., Schnoor J.L. Benzo[a]pyrene co‐metabolism in the presence of plant root extracts and exudates: implications for phytoremediation. Environ Pollut. 2005;136:477–484. doi: 10.1016/j.envpol.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Rentz J.A., Alvarez P.J.J., Schnoor J.L. Benzo[a]pyrene degradation by Sphingomonas yanoikuyae JAR02. Environ Pollut. 2008;151:669–677. doi: 10.1016/j.envpol.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Ringelberg D.B., Talley J.W., Perkins E.J., Tucker S.G., Luthy R.G., Bouwer E.J., Fredrickson H.L. Succession of phenotypic, genotypic, and metabolic community characteristics during in vitro bioslurry treatment of polycyclic aromatic hydrocarbon‐contaminated sediments. Appl Environ Microbiol. 2001;67:1542–1550. doi: 10.1128/AEM.67.4.1542-1550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Iwabuchi T., Harayama S. A novel phenanthrene dioxygenase from Nocardiodes sp. strain KP7: expression in Escherichia coli. J Bacteriol. 2000;182:2134–2141. doi: 10.1128/jb.182.8.2134-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P.M., Bhattacharya D., Krishnan S., Lal B. Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl Environ Microbiol. 2004;70:3163–3166. doi: 10.1128/AEM.70.5.3163-3166.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartoros C., Yerushalmi L., Béron P., Guiot S.R. Effects of surfactant and temperature on biotransformation kinetics of anthracene and pyrene. Chemosphere. 2005;61:1042–1050. doi: 10.1016/j.chemosphere.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Schneider J., Grosser R., Jayasimhulu K., Xue W., Warshawsky D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII‐135, isolated from a former coal gasification site. Appl Environ Microbiol. 1996;62:13–19. doi: 10.1128/aem.62.1.13-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.‐S., Keum Y.‐S., Harada R.M., Li Q.X. Isolation and characterization of bacteria capable of degrading polycyclic aromatic hydrocarbons (PAHs) and organophosphorus pesticides from PAH‐contaminated soil in Hilo, Hawaii. J Agric Food Chem. 2007;55:5383–5889. doi: 10.1021/jf0637630. [DOI] [PubMed] [Google Scholar]
- Sepic E., Bricelj M., Leskovsek H. Biodegradation studies of polyaromatic hydrocarbons in aqueous media. J Appl Microbiol. 1997;83:561–568. doi: 10.1046/j.1365-2672.1997.00261.x. [DOI] [PubMed] [Google Scholar]
- Sepic E., Bricelj M., Leskovsek H. Degradation of fluoranthene by Pasteurella sp. IFA and Mycobacterium sp. PYR‐1: isolation and identification of metabolites. J Appl Microbiol. 1998;85:746–754. doi: 10.1111/j.1365-2672.1998.00587.x. [DOI] [PubMed] [Google Scholar]
- Sepic E., Bricelj M., Leskovsek H. Toxicity of fluoranthene and its biodegradation metabolites to aquatic organisms. Chemosphere. 2003;52:1125–1133. doi: 10.1016/S0045-6535(03)00321-7. [DOI] [PubMed] [Google Scholar]
- Shi T., Frederickson J.K., Balkwill D.L. Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas strains isolated from terrestrial subsurface. J Ind Microbiol Biotechnol. 2001;26:283–289. doi: 10.1038/sj.jim.7000130. [DOI] [PubMed] [Google Scholar]
- Sho M., Hamel C., Greer C.W. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol Ecol. 2004;48:209–220. doi: 10.1016/j.femsec.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Shuler L., Ní Chadhain S.M., Jouanneau Y., Meyer C., Zylstra G.J., Hols P., Agathos S.N. Characterization of a novel angular dioxygenase from fluorene‐degrading Sphingomonas sp. strain LB126. Appl Environ Microbiol. 2008;74:1050–1057. doi: 10.1128/AEM.01627-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth K.L., Sung J., Kim E., Cerniglia C.E. Physiological and genetic comparison of two aromatic hydrocarbon‐degrading Sphingomonas strains. Mol Cells. 2000;10:199–205. doi: 10.1007/s10059-000-0199-x. [DOI] [PubMed] [Google Scholar]
- Singleton D.R., Sangiah R., Gold A., Ball L.M., Aitken M.D. Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH‐contaminated soil. Environ Microbiol. 2006;10:1736–1745. doi: 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- Singleton D.R., Hunt M., Powell S.N., Frontera‐Suau R., Aitken M.D. Stable‐isotope probing with multiple growth substrates to determine substrate specificity of uncultivated bacteria. J Microbiol Methods. 2007;69:180–187. doi: 10.1016/j.mimet.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Singleton D.R., Richardson S.D., Aitken M.D. Effects of enrichment with phthalate on polycyclic aromatic hydrocarbon biodegradation in contaminated soil. Biodegradation. 2008;19:577–587. doi: 10.1007/s10532-007-9163-1. [DOI] [PubMed] [Google Scholar]
- Sisler F.D., ZoBell C.E. Microbial utilization of carcinogenic hydocarbons. Science. 1947;106:521–522. doi: 10.1126/science.106.2761.521. [DOI] [PubMed] [Google Scholar]
- Sohn J.H., Kwon K.K., Kang J.‐H., Jung H.‐B., Kim S.‐J. Novosphingobium pentaromativorans sp. nov., a high‐molecular‐mass polycyclic aromatic hydrocarbon‐degrading bacterium isolated form estuarine sediment. Int J Syst Evol Microbiol. 2004;54:1483–1487. doi: 10.1099/ijs.0.02945-0. [DOI] [PubMed] [Google Scholar]
- Somtrakoon K., Suanjit S., Pokethitiyook P., Kruatrachue M., Lee H., Upatham S. Phenanthrene stimulates the degradation of pyrene and fluoranthene by Burkholderia sp. VUN10013. World J Microbiol Biotechnol. 2008;24:523–531. [Google Scholar]
- Stach J.E.M., Burns R.G. Enrichment versus biofilm culture: a functional and phylogenetic comparison of polycyclic aromatic hydrocarbon‐degrading microbial communities. Environ Microbiol. 2002;4:169–182. doi: 10.1046/j.1462-2920.2002.00283.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Rainey F.A., Ward‐Rainey N.L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- Stingley R.L., Brezna B., Khan A.A., Cerniglia C.E. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR‐1. Microbiology. 2004a;150:3749–3761. doi: 10.1099/mic.0.27263-0. [DOI] [PubMed] [Google Scholar]
- Stingley R.L., Khan A.A., Cerngilia C.E. Molecular characterization of a phenanthrene degradation operon of Mycobacterium vanbaalenii PYR‐1. Biochem Biophys Res Commun. 2004b;322:133–146. doi: 10.1016/j.bbrc.2004.07.089. [DOI] [PubMed] [Google Scholar]
- Stolz A. Molecular characteristics of xenobiotic‐degrading sphingomonads. Appl Microbiol Biotechnol. 2009;81:793–811. doi: 10.1007/s00253-008-1752-3. [DOI] [PubMed] [Google Scholar]
- Story S.P., Parker S.H., Kline J.D., Tzeng T.‐R.J., Mueller J.G., Kline E.L. Identification of four structural genes and two putative promoters necessary for utilization of naphthalene, phenanthrene, and fluoranthene by Sphingomonas paucimobilis var. EPA505. Gene. 2000;260:155–169. doi: 10.1016/s0378-1119(00)90445-1. [DOI] [PubMed] [Google Scholar]
- Story S.P., Parker S.H., Hayasaka S.S., Riley M.B., Kline E.L. Convergent and divergent points in catabolic pathways involved in utilization of fluoranthene, naphthalene, anthracene, and phenanthrene by Sphingomonas paucimobilis var. EPA505. J Ind Microbiol Biotechnol. 2001;26:369–382. doi: 10.1038/sj.jim.7000149. [DOI] [PubMed] [Google Scholar]
- Story S.P., Kline E.L., Hughes T.A., Riley M.B., Hayasaka S.S. Degradation of aromatic hydrocarbons by Sphingomonas paucimobilis strain EPA505. Arch Environ Contam Toxicol. 2004;47:168–176. doi: 10.1007/s00244-004-3069-2. [DOI] [PubMed] [Google Scholar]
- Stroud J.L., Paton G.I., Semple K.T. Importance of chemical structure on the development of hydrocarbon catabolism in soil. FEMS Microbiol Lett. 2007;272:120–126. doi: 10.1111/j.1574-6968.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Hamana K., Hiraishi A. Proposal of the genus Sphingomonas sensu stricto and three genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol. 2001;51:1405–1417. doi: 10.1099/00207713-51-4-1405. [DOI] [PubMed] [Google Scholar]
- Tian Y., Liu H.J., Zheng T.L., Kwon K.K., Kim S.J., Yan C.L. PAHs contamination and bacterial communities in mangrove surface sediments of the Jiulong River Estuary, China. Mar Pollut Bull. 2008;57:707–715. doi: 10.1016/j.marpolbul.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Toledo F.L., Calvo C., Rodelas B., González‐López J. Selection and identification of bacteria isolated from waste crude oil with polycyclic aromatic hydrocarbons removal capacities. Int J Syst Evol Microbiol. 2006;29:244–252. doi: 10.1016/j.syapm.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Uyttebroek M., Vermeir S., Wattiau P., Ryngaert A., Springael D. Characterization of cultures enriched from acidic polycyclic aromatic hydrocarbon‐contaminated soil for growth on pyrene at low pH. Appl Environ Microbiol. 2007;73:3159–3164. doi: 10.1128/AEM.02837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Meer J.R., De Vos W.M., Harayama S., Zehnder A.J.B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeer K.D., Daugulis A.J. Enhanced degradation of a mixture of polycyclic aromatic hydrocarbons by a defined microbial consortium in a two‐phase partitioning bioreactor. Biodegradation. 2007;18:211–221. doi: 10.1007/s10532-006-9056-8. [DOI] [PubMed] [Google Scholar]
- Van Hamme J.D., Singh A., Ward O.P. Recent advances in petroleum microbiology. Microbiol Mol Biol Rev. 2003;67:503–549. doi: 10.1128/MMBR.67.4.503-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Metre P.C., Mahler B.J., Furlong E.T. Urban sprawl leaves its PAH signature. Environ Sci Technol. 2000;34:4064–4070. [Google Scholar]
- Van de Wiele T., Vanhaecke L., Boeckaert C., Peru K., Headley J., Verstraete W., Siciliano S. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ Health Perspect. 2005;113:6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J., López Z., Sabaté J., Minguillón C., Solanas A.M., Grifoll M. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two‐ and three‐ring polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2001;67:5497–5505. doi: 10.1128/AEM.67.12.5497-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R., Déziel E., Benachenhou A., Marcoux J., Gauthier E., Lépine F. Two‐liquid‐phase slurry bioreactors to enhance the degradation of high‐molecular‐weight polycyclic aromatic hydrocarbons in soil. Biotechnol Prog. 2000;16:966–972. doi: 10.1021/bp000118j. et al. [DOI] [PubMed] [Google Scholar]
- Viñas M., Sabaté J., Guasp C., Lalucat J., Solanas A.M. Culture‐dependent and ‐independent approaches to establish the complexity of a PAH‐degrading microbial consortium. Can J Microbiol. 2005a;51:897–909. doi: 10.1139/w05-090. [DOI] [PubMed] [Google Scholar]
- Viñas M., Sabaté J., Espuny M.J., Solanas A.M. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote‐contaminated soil. Appl Environ Microbiol. 2005b;71:7008–7018. doi: 10.1128/AEM.71.11.7008-7018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagrowski D.M., Hites R.A. Polycyclic aromatic hydrocarbon accumulation in urban, suburban, and rural vegetation. Environ Sci Technol. 1997;31:279–282. [Google Scholar]
- Walter U., Beyer M., Klein J., Rehm H.‐J. Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol. 1991;34:671–676. [Google Scholar]
- Wang B., Lai Q., Cui Z., Tan T., Shao Z. A pyrene‐degrading consortium from deep‐sea sediment of the West Pacific and its key member Cycloclasticus sp. Environ Microbiol. 2008;10:1948–1963. doi: 10.1111/j.1462-2920.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- Wang R.‐F., Wennerstrom D., Cao W.‐W., Khan A.A., Cerniglia C.E. Cloning, expression, and characterization of the katG gene, encoding catalase‐peroxidase, from the polycyclic aromatic hydrocarbon‐degrading bacterium Mycobacterium sp. strain PYR‐1. Appl Environ Microbiol. 2000;66:4300–4304. doi: 10.1128/aem.66.10.4300-4304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenfels W.D., Beyer M., Klein J., Rehm H.J. Microbial metabolism of fluoranthene: isolation and identification of ring fission products. Appl Microbiol Biotechnol. 1991;34:528–535. [Google Scholar]
- Wick L., Colangelo T., Harms H. Kinetics of mass transfer‐limited bacterial growth on solid PAHs. Environ Sci Technol. 2001;35:354–361. doi: 10.1021/es001384w. [DOI] [PubMed] [Google Scholar]
- Wick L.Y., Wattiau P., Harms H. Influence of the growth substrate on mycolic acid profiles of mycobacteria. Environ Microbiol. 2002;4:612–616. doi: 10.1046/j.1462-2920.2002.00340.x. [DOI] [PubMed] [Google Scholar]
- Wick L.Y., Pasche N., Bernasconi S.M., Pelz O., Harms H. Characterization of multiple‐substrate utilization by anthracene‐degrading Mycobacterium frederiksbergense LB501T. Appl Environ Microbiol. 2003a;69:6133–6142. doi: 10.1128/AEM.69.10.6133-6142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick L.Y., Pelz O., Bernasconi S.M., Andersen N., Harms H. Influence of the growth substrate on ester‐linked phospho‐ and glycolipid fatty acids of Mycobacterium sp. LB501T. Environ Microbiol. 2003b;5:672–680. doi: 10.1046/j.1462-2920.2003.00455.x. [DOI] [PubMed] [Google Scholar]
- Wilcke W. Polycyclic aromatic hydrocarbons (PAHs) in soil – a review. J Plant Nutr Soil Sci. 2000;163:229–248. [Google Scholar]
- Willison J.C. Isolation and characterization of a novel sphingomonad capable of growth on chrysene as sole carbon and energy source. FEMS Microbiol Lett. 2004;241:143–150. doi: 10.1016/j.femsle.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Willumsen P., Karlson U., Stackebrandt E., Kroppenstedt R.M. Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon‐degrading Mycobacterium species. Int J Syst Evol Microbiol. 2001;51:1715–1722. doi: 10.1099/00207713-51-5-1715. [DOI] [PubMed] [Google Scholar]
- Willumsen P.A., Arvin E. Kinetics of degradation of surfactant‐solubilized fluoranthene by a Sphingomonas paucimobilis. Environ Sci Technol. 1999;33:2571–2578. [Google Scholar]
- Willumsen P.A., Karlson U. Effect of calcium on the surfactant tolerance of a fluoranthene degrading bacterium. Biodegradation. 1998;9:369–379. doi: 10.1023/a:1008357904624. [DOI] [PubMed] [Google Scholar]
- Yamada M., Takada H., Toyoda K., Yoshida A., Shibata A., Nomura H. Study on the fate of petroleum‐derived polycyclic aromatic hydrocarbons (PAHs) and the effect of chemical dispersant using an enclosed ecosystem, mescosm. Mar Pollut Bull. 2003;47:105–113. doi: 10.1016/S0025-326X(03)00102-4. et al. [DOI] [PubMed] [Google Scholar]
- Ye D., Siddiqi M.A., Maccubbin A.E., Kumar S., Sikka H.C. Degradation of polynuclear aromatic hydrocarbons by Sphingomonas paucimobilis. Environ Sci Technol. 1996;30:136–142. [Google Scholar]
- Yu C.L., Liu W., Ferraro D.J., Brown E.N., Parales J.V., Ramaswamy S. Purification, characterization, and crystallization of the components of a biphenyl dioxygenase system from Sphingobium yanoikuyae B1. J Ind Microbiol Biotechnol. 2007;34:311–324. doi: 10.1007/s10295-006-0199-8. et al. [DOI] [PubMed] [Google Scholar]
- Yu S.H., Ke L., Wong Y.S., Tam N.F.Y. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a bacterial consortium enriched from mangrove sediments. Environ Int. 2005;31:149–154. doi: 10.1016/j.envint.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Yuan S.Y., Wei S.H., Chang B.V. Biodegradation of polycyclic aromatic hydrocarbons by a mixed culture. Chemosphere. 2000;41:1463–1468. doi: 10.1016/s0045-6535(99)00522-6. [DOI] [PubMed] [Google Scholar]
- Zakaria M.P., Takada H., Tsutsumi S., Ohno K., Yamada J., Kouno E., Kumata H. Distribution of polycyclic aromatic (PAHs) in rivers and estuaries in Malaysia: a widespread input of petrogenic PAHs. Environ Sci Technol. 2002;36:1907–1918. doi: 10.1021/es011278+. [DOI] [PubMed] [Google Scholar]
- Zeinali M., Vossoughi M., Ardestani S.K. Characterization of a moderate thermophilic Nocardia species able to grow on polycyclic aromatic hydrocarbons. Lett Appl Microbiol. 2007;45:622–628. doi: 10.1111/j.1472-765X.2007.02241.x. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Hong P.K.A., Wavrek D.A. Integrated chemical‐biological treatment of benzo[a]pyrene. Environ Sci Technol. 2000;34:854–862. [Google Scholar]
- Zhang H., Kallimanis A., Koukkou A.I., Drainas C. Isolation and characterization of novel bacteria degrading polycyclic aromatic hydrocarbons form polluted Greek soils. Appl Microbiol Biotechnol. 2004;65:124–131. doi: 10.1007/s00253-004-1614-6. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Luan T., Zhou H., Lan C., Tam N.F.Y. Metabolite production in degradation of pyrene alone or in a mixture with another polycyclic aromatic hydrocarbon by Mycobacterium sp. Environ Toxicol Chem. 2006;25:2853–2859. doi: 10.1897/06-042r.1. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Luan T., Wang X., Lan C., Tam N.F.Y. Influence of growth media on cometabolic degradation of polycyclic aromatic hydrocarbons by Sphingomonas sp. strain PheB4. Appl Microbiol Biotechnol. 2007;75:175–186. doi: 10.1007/s00253-006-0789-4. [DOI] [PubMed] [Google Scholar]
- Zhou H.W., Guo C.L., Wong Y.S., Tam N.F.Y. Genetic diversity of dioxygenase genes in polycyclic aromatic hydrocarbon‐degading bacteria isolated from mangrove sediments. FEMS Microbiol Lett. 2006;262:148–157. doi: 10.1111/j.1574-6968.2006.00379.x. [DOI] [PubMed] [Google Scholar]
- Zhou H.W., Luan T.G., Zou F., Tam N.F.Y. Different bacterial groups for biodegradation of three‐ and four‐ring PAHs isolated from a Hong Kong mangrove sediment. J Hazard Mater. 2008;152:1179–1185. doi: 10.1016/j.jhazmat.2007.07.116. [DOI] [PubMed] [Google Scholar]


