Summary
The need to understand and control ester synthesis is driven by the fact that esters play a key role in the sensorial quality of fermented alcoholic beverages like beer, wine and sake. As esters are synthesized in yeast via several complex metabolic pathways, there is a need to gain a clear understanding of ester metabolism and its regulation. The individual genes involved, their functions and regulatory mechanisms have to be identified. In alcoholic beverages, there are two important groups of esters: the acetate esters and the medium‐chain fatty acid (MCFA) ethyl esters. For acetate ester synthesis, the genes involved have already been cloned and characterized. Also the biochemical pathways and the regulation of acetate ester synthesis are well defined. With respect to the molecular basis of MCFA ethyl ester synthesis, however, significant progress has only recently been made. Next to the characterization of the biochemical pathways and regulation of ester synthesis, a new and more important question arises: what is the advantage for yeast to produce these esters? Several hypotheses have been proposed in the past, but none was satisfactorily. This paper reviews the current hypotheses of ester synthesis in yeast in relation to the complex regulation of the alcohol acetyl transferases and the different factors that allow ester formation to be controlled during fermentation.
Introduction
During fermentation the yeast Saccharomyces cerevisiae produces a broad range of aroma‐active substances, which are vital for the complex flavour of fermented beverages such as beer, wine and sake. Flavour‐active substances produced by fermenting yeast cells can be divided into six main groups: organic acids, higher alcohols, carbonyl compounds, sulfur‐containing molecules, phenolic compounds and volatile esters. Although volatile esters are only trace compounds in fermented beverages, they comprise the most important set of yeast‐derived aroma‐active compounds. Volatile esters are of major industrial interest because they are responsible for the highly desired fruity, candy and perfume‐like aroma character of beer, wine and sake (Suomalainen, 1981; Nykanen and Suomalainen, 1983; Nykanen, 1986; Malcorps and Dufour, 1987; Peddie, 1990; Meilgaard, 1991; Debourg, 2000; Cristiani and Monnet, 2001; Pisarnitskii, 2001; Dufour et al., 2002; Verstrepen et al., 2003a; Aritomi et al., 2004). Esters generally have a low odour threshold in beer and wine (Tables 1 and 2). Because the concentration of most esters formed during fermentation by S. cerevisiae hovers around their respective threshold values, even small changes in the concentration of these secondary metabolites can have large effects on the sensorial quality of fermented beverages. Therefore, understanding the mechanisms of their formation in order to be able to better control their levels in the end product is of major industrial interest. As a consequence, the biochemical background of ester synthesis has been extensively studied.
Table 1.
Threshold values for esters and their concentration in lager beer (Meilgaard, 1975; Dufour and Malcorps, 1994).
| Compound | Threshold level (ppm) | Concentration range (ppm) | Flavour description |
|---|---|---|---|
| Ethyl acetate | 30 | 8–32 | Fruity, solvent‐like |
| Isoamyl acetate | 1.2 | 0.3–3.8 | Banana, pear |
| Phenyl ethyl acetate | 3.8 | 0.10–0.73 | Roses, honey |
| Ethyl hexanoate | 0.23 | 0.05–0.21 | Apple, fruity, sweetish |
| Ethyl octanoate | 0.9 | 0.04–0.53 | Apple, aniseed |
Table 2.
Threshold values for esters and their concentration in wine (Swiegers and Pretorius, 2005).
| Compound | Threshold level (ppm) | Concentration range (ppm) | Flavour description |
|---|---|---|---|
| Ethyl acetate | 7.5a | 22.5–63.5 | Nail polish, fruity |
| Isoamyl acetate | 0.03a | 0.1–3.4 | Banana, pear |
| Isobutyl acetate | 1.6b | 0.01–1.6 | Banana, fruity |
| Phenyl ethyl acetate | 0.25a | 0–18.5 | Roses, flowery |
| Hexyl acetate | 0.7c | 0–4.8 | Sweet, perfume |
| Ethyl butanoate | 0.02a | 0.01–1.8 | Floral, fruity |
| Ethyl hexanoate | 0.05a | 0.03–3.4 | Green apple |
| Ethyl octanoate | 0.02a | 0.05–3.8 | Sweet soap, apple |
| Ethyl decanoate | 0.2d | 0–2.1 | Floral, soap |
10% ethanol.
Beer.
Wine.
Synthetic wine.
There are two main categories of flavour‐active esters in fermented beverages. First, the group of the acetate esters (the acid group is acetate, the alcohol group is ethanol or a complex alcohol derived from amino acid metabolism), such as ethyl acetate (solvent‐like aroma), isoamyl acetate (banana aroma), isobutyl acetate (fruity aroma) and phenyl ethyl acetate (roses, honey) (Fig. 1). With its distinctive banana flavour, isoamyl acetate is the most influential acetate ester present in most beers, (white) wines and sake. The second group comprises the medium‐chain fatty acid (MCFA) ethyl esters (the alcohol group is ethanol, the acid group is a medium‐chain fatty acid), which includes ethyl hexanoate (aniseed, apple‐like aroma) and ethyl octanoate (sour apple aroma) (Fig. 1). Of these two groups, the acetate esters have received most attention in the past, not because they are more important, but because they are produced in much higher levels and therefore easier to measure. Also the genes involved in their synthesis have been discovered first. By contrast, until recently much less was known about ethyl ester production, despite their desirable apple‐like aromas. Recent progress has identified several genes and biochemical pathways involved in their synthesis.
Figure 1.
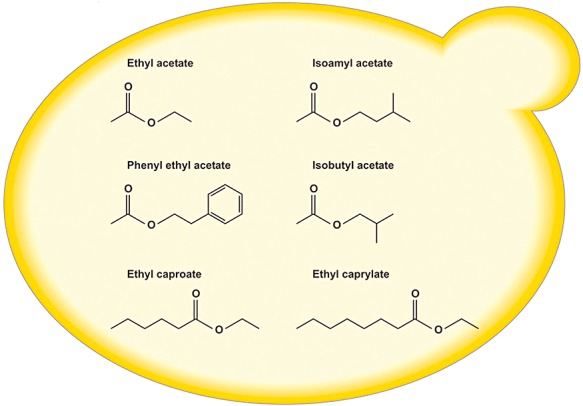
Flavour‐active esters produced by Saccharomyces yeast in wine, beer and sake. Ethyl caproate = ethyl hexanoate, ethyl caprylate = ethyl octanoate.
Aroma‐active esters are formed intracellular by fermenting yeast cells. Being lipid soluble, esters can diffuse through the plasma membrane into the fermenting medium. Unlike acetate ester diffusion, which is rapid and complete, the transfer of ethyl esters to the fermentation medium is dependent on their composition. It decreases drastically with increasing chain length, from 100% for ethyl hexanoate, to 54–68% for ethyl octanoate and 8–17% for ethyl decanoate (Nykanen and Nykanen, 1977). The rate of ester formation is dependent on three factors: the concentration of the two co‐substrates [the acyl‐coenzyme A (CoA) component and the alcohol] and the activity of the enzymes involved in their synthesis and hydrolysis. Hence, all parameters that influence substrate concentration or enzyme activity may affect ester production.
Since the discovery of the ester synthase by Nordström more than 40 years ago (Nordström, 1962), significant progress has been made in elucidating the ester flavour biochemical pathways, the genes involved in the pathways leading to ester synthesis and the factors influencing ester synthesis rates. There remain, however, several yet unresolved questions of high interest. What is the biological function of ester synthesis in yeast? Or more specifically, what is the functional role of the ester synthases in yeast? Several hypotheses have been proposed in the past. It has been suggested that esters could simply be spillover products from sugar metabolism during fermentation and that their formation may not be of any advantage to the cell (Peddie, 1990). However, the input of energy under the form of acetyl‐CoA or acyl‐CoA and especially the strict and complex regulation of the ATF1 gene strongly suggest that ester formation is not just a futile process. Instead, it is possible that the formation of volatile esters has a very specific function, maybe limited to specific life cycle stages or environmental conditions (Mason and Dufour, 2000). Furthermore, it has to be noted that ester synthases may also catalyse other reactions than the formation of volatile aroma‐active esters, such as the synthesis of more complex, non‐volatile ester compounds. It is therefore possible that volatile esters are by‐products of a more physiologically relevant process (Mason and Dufour, 2000). On the other hand, several hypotheses have been suggested for a specific role of volatile ester production which are more or less supported by different experimental results. This paper reviews the current hypotheses of ester synthesis in yeast in relation to the complex regulation of the alcohol acetyl transferases and the different factors that allow ester formation to be controlled during fermentation.
Ester biosynthesis
Esters are formed via an intracellular process, catalysed by an acyl transferase or ‘ester synthase’ (Nordström, 1962). The reaction requires energy provided by the thioester linkage of the acyl‐CoA co‐substrate (Fig. 2). The most abundant acyl‐CoA is acetyl‐CoA, which can be formed either by oxidative decarboxylation of pyruvate or by direct activation of acetate with ATP. The majority of acetyl‐CoA is formed by the oxidative decarboxylation of pyruvate, while most of the other acyl‐CoAs are generated by the acylation of free CoA catalysed by acyl‐CoA synthase (fatty acid metabolism).
Figure 2.
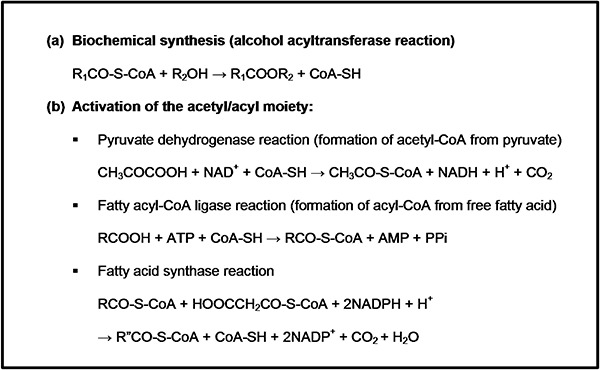
Biochemical synthesis of esters (A) and the activation of the acyl moiety (B).
Acetate esters
The alcohol acetyl transferases I and II (AATase I and II; EC 2.3.1.84), encoded by the genes ATF1 and ATF2, catalyse the synthesis of acetate esters (Yoshioka and Hashimoto, 1981; Malcorps and Dufour, 1992; Fujii et al., 1994; 1996a,b; Nagasawa et al., 1998; Yoshimoto et al., 1998). Next to ATF1, a closely related homologue Lg‐ATFI was identified in S. pastorianus strains, which is the common brewing lager strain (Yoshimoto et al., 1998). Atf1 and Atf2 catalyse the formation of acetate esters from two substrates: an alcohol and acetyl‐CoA. Although the yeast AATase was first considered to be a membrane‐bound enzyme, the results of a hydrophobicity analysis indicated that the gene products of ATF1 and ATF2 do not have a membrane‐spanning region. On the other hand, it has recently been reported that the ATF1‐encoded enzyme is localized in lipid particles (Verstrepen et al., 2004).
Several studies have been carried out to determine the role of the ATF1‐ and ATF2‐encoded alcohol acetyltransferases in acetate ester synthesis in S. cerevisiae. The role of the known S. cerevisiae alcohol acetyltransferases in volatile ester production was investigated and compared by deleting or overexpressing ATF1, Lg‐ATF1 and ATF2 in a laboratory yeast strain and a commercial brewing strain (Verstrepen et al., 2003b). Ester formation in the transformants was measured using gas chromatography, coupled with mass spectrometry. Analysis of the fermentation products confirmed that the expression levels of ATF1 and ATF2 greatly affected the production of ethyl acetate and isoamyl acetate, as shown already before also by other researchers (Fujii et al., 1994; 1996b; Nagasawa et al., 1998). Gas chromatography–mass spectrometry analysis revealed that the ATF1‐ and ATF2‐encoded enzymes are also responsible for the formation of a broad range of less volatile esters, such as propyl acetate, isobutyl acetate, pentyl acetate, hexyl acetate, heptyl acetate, octyl acetate and 2‐phenyl ethyl acetate. With respect to the esters analysed (Verstrepen et al., 2003b), the ATF2‐encoded enzyme seemed to play only a minor role compared with the ATF1‐encoded enzyme. The atf1Δ atf2Δ double deletion strain did not form any isoamyl acetate, showing that together the ATF1‐ and ATF2‐encoded alcohol acetyltransferases are responsible for the total cellular isoamyl alcohol acetyl transferase activity. However, the double deletion strain still produced considerable amounts of certain other esters, such as ethyl acetate (50% of the wild‐type strain), propyl acetate (50%), and isobutyl acetate (40%), which indicates the existence of additional, as yet unknown ester synthases in the yeast proteome. Interestingly, overexpression of alleles of ATF1 and ATF2 derived from different yeast strains led to different production rates for the individual esters, indicating that differences in the aroma profiles produced by yeast strains may be at least partially due to specific mutations in their ATF genes.
Also in wine yeasts, the ATF1‐ and ATF2‐encoded alcohol acetyltransferases play an important role in acetate ester synthesis. When ATF1 and ATF2 were overexpressed in the VIN13 wine yeast, the levels of ethyl acetate, isoamyl acetate, 2‐phenyl ethyl acetate and ethyl hexanoate were increased in wine made with this genetically modified yeast. The chemical changes had a pronounced effect on the ‘solvent/chemical’ and ‘fruity/flowery’ characters of the wine (Lilly et al., 2000; 2006). The estery/synthetic fruit flavour was overpowering in wines fermented with the yeast in which ATF1 was overexpressed, but was much more subtle in the strain overexpressing ATF2.
Ethyl esters
As an atf1Δ atf2Δ double deletion strain produces the same amount of MCFA ethyl esters as the wild‐type strain, Atf1 and Atf2 are not involved in MCFA ethyl ester synthesis (Verstrepen et al., 2003b). Mason and Dufour (2000) proposed that a fourth ester‐synthesizing enzyme, ethanol hexanoyl transferase, Eht1, is responsible for generating ethyl hexanoate from ethanol and hexanoyl‐CoA. In 2006, Saerens et al. showed that the formation of the majority of the MCFA ethyl esters in yeast is catalysed by two acyl‐CoA:ethanol O‐acyltransferases (AEATases), Eeb1 and Eht1. This means that MCFA ethyl esters are the product of an enzyme‐catalysed condensation reaction between an acyl‐CoA component and ethanol (Saerens et al., 2006). According to Saerens and colleagues (2006), the levels of ethyl butanoate, ethyl hexanoate, ethyl octanoate and ethyl decanoate produced during fermentation with an eeb1Δ strain are reduced in comparison with those produced by the wild‐type strain by respectively 36%, 88%, 45% and 40% (Table 3). Compared with the eeb1Δ strain, deletion of EHT1 does not affect the production of ethyl butanoate and ethyl decanoate, and results in only minor decreases in ethyl hexanoate formation (36%) and ethyl octanoate formation (20%). The double deletion strain eht1Δ eeb1Δ produces similar levels of ethyl butanoate, ethyl hexanoate and ethyl decanoate as the eeb1Δ single deletion strain, and a lower level of ethyl octanoate, indicating that Eht1 plays only a minor role in MCFA ethyl ester synthesis, while Eeb1 is the most important enzyme for MCFA ethyl ester synthesis. On the other hand, although the double deletion of EHT1 and EEB1 causes a pronounced drop in the production of all MCFA ethyl esters, only the production of ethyl hexanoate is virtually eliminated. Hence, yeast cells must contain one or more additional enzymes responsible for MCFA ethyl ester synthesis. In the case of ethyl octanoate and ethyl decanoate production, additional deletion of YMR210w in the eht1Δ eeb1Δ strain produced a further drop in their level. Also in the case of ethyl butanoate, one would expect the existence of one or more additional enzymes that can support its synthesis. On the other hand, chemical synthesis of ethyl butanoate might also occur.
Table 3.
Ethyl ester production in eht1Δ, eeb1Δ, ymr210wΔ (= ymrΔ) single and multiple deletion strains (Saerens et al., 2006).
| Compound | wt | eht1Δ | eeb1Δ | ymrΔ | eht1Δ eeb1Δ | eht1Δ ymrΔ | eeb1Δ ymrΔ | eht1Δ eeb1Δ ymrΔ |
|---|---|---|---|---|---|---|---|---|
| Ethyl butanoate | 1.00 | 0.97 | 0.64 | 1.06 | 0.70 | 0.91 | 0.55 | 0.54 |
| Ethyl hexanoate | 1.00 | 0.64 | 0.12 | 0.95 | 0.08 | 0.84 | 0.08 | 0.05 |
| Ethyl octanoate | 1.00 | 0.80 | 0.55 | 0.89 | 0.28 | 0.85 | 0.24 | 0.10 |
| Ethyl decanoate | 1.00 | 1.18 | 0.60 | 1.11 | 0.56 | 0.90 | 0.20 | 0.07 |
Gas chromatographic measurement of ethyl butanoate, ethyl hexanoate, ethyl octanoate and ethyl decanoate produced by the wild type (wt) and the deletion strains eht1Δ, eeb1Δ, ymr210wΔ, eht1Δ eeb1Δ, eht1Δ ymr210wΔ, eeb1Δ ymr210wΔ, and eht1Δ eeb1Δ ymr210wΔ after 96 h of fermentation. Standard deviations were typically 10% and did not exceed 20%. The level produced by the wt strain was set to 1.00 for each ester individually.
In addition to deletion analysis, Saerens and colleagues (2006) also evaluated Eht1 and Eeb1 for intrinsic AEATase and esterase activity in vitro. Their results obtained with purified GST‐Eht1 and GST‐Eeb1 fusion proteins clearly indicate that these proteins display enzymatic activity for both the synthesis and the hydrolysis of MCFA ethyl esters. The combined presence of MCFA ethyl ester synthase and esterase activity in the Eht1 and Eeb1 proteins raises questions as to the precise regulation of the balance between MCFA ethyl ester synthesis and hydrolysis in vivo by Eht1 and Eeb1.
To further evaluate the role of Eht1 and Eeb1 in the synthesis of MCFA ethyl esters, overexpression strains were constructed and tested in batch culture fermentations for MCFA ethyl ester production (Saerens et al., 2006). Unexpectedly, overexpression of EHT1 and EEB1 did not result in a significant increase in MCFA ethyl ester formation. Also overexpression of the EHT1 or EEB1 allele of an industrial ale strain did not increase the production of MCFA ethyl esters (Saerens et al., 2006). Even when additional substrate was added to the fermenting medium, there was no difference in MCFA ethyl ester concentration between a wild‐type yeast strain and the EHT1 or EEB1 overexpression strains of both a laboratory and an ale yeast strain. The extra esterase activity of Eht1 and Eeb1 may explain why overexpression of their genes does not enhance MCFA ethyl ester formation.
On the other hand, overexpression of EHT1 derived from wine yeast showed a slight increase in MCFA ethyl ester production. Lilly and colleagues (2006) used the EHT1 allele of a wine yeast strain (VIN13) and overexpression of this EHT1 allele in the VIN13 yeast slightly increased the concentration of all the esters, with the highest increases in the concentrations of ethyl hexanoate, ethyl octanoate and ethyl decanoate (Lilly et al., 2006). The effect of increased concentrations of ethyl hexanoate, ethyl octanoate and ethyl decanoate was evident in the sensory analysis, where EHT1 overexpression resulted in a significant enhancement of the apple aroma of the wine.
Ester hydrolysis
In contrast to ester synthases, research on the effect of ester‐hydrolysing enzymes on ethyl ester hydrolysis has been limited. The balance between ester‐synthesizing enzymes and esterases, which hydrolyse esters, was shown to be important for the net rate of ester accumulation (Fukuda et al., 1998). Esterases represent a diverse group of hydrolases that catalyse the cleavage of esters, and in some cases, the formation of ester bonds. In ester breakdown, esterases catalyse the reaction RCOOR + H2O → ROH + RCOOH (Peddie, 1990). Recently, the effect of the IAH1‐encoded ester‐degrading enzyme on the flavour profile of wine has been investigated (Lilly et al., 2006). Overexpression of IAH1 resulted in a significant decrease in ethyl acetate, isoamyl acetate, hexyl acetate and 2‐phenyl ethyl acetate. The wines produced with yeast overexpressing IAH1 showed a significant decrease in ester concentrations when compared with the control wines. The concentration of isoamyl acetate decreased 11.4–15.6‐fold and hexyl acetate was undetectable. Ethyl acetate and 2‐phenyl ethyl acetate concentrations decreased by 1.6–1.8‐fold and 3.4–3.9‐fold respectively.
The three‐dimensional structure of a wide range of esterases shows the characteristic α/β‐hydrolase fold (Ollis et al., 1992) – a definite order of α‐helices and β‐sheets. The catalytic triad is composed of Ser‐Asp‐His (Glu instead of Asp for some lipases) and usually also a consensus sequence (Gly‐x‐Ser‐x‐Gly) is found around the active site serine. This catalytic triad is also found in other enzyme types, including thioester hydrolases, proteases, haloperoxidases, haloalkane dehalogenases, epoxide hydrolases and C–C bond breaking enzymes (Holmquist, 2000). It has been shown that the dipeptide seryl‐histidine (Ser‐His) and related oligopeptides can themselves cleave DNA, protein, and the ester p‐nitrophenyl acetate over wide ranges of pH and temperature (Li et al., 2000). So, it is possible that next to esterases, also other enzyme types could be responsible for the hydrolysis of esters, e.g. serine proteases.
Physiological role of ester synthesis: hypotheses
To understand the physiological role of ester synthesis, it is important to know how both acetate and ethyl ester synthesis are regulated. Therefore, we summarize first the most important regulatory factors for acetate and ethyl ester synthesis.
Regulation of acetate ester synthesis
Basically, three factors are important for the rate of acetate ester formation: the concentration of the two substrates, acetyl‐CoA and a fusel alcohol, and the total activity of the enzymes involved in the formation and breakdown of the respective ester. While substrate concentrations certainly influence acetate ester production, in general the expression levels of the known alcohol acetyltransferases ATF1 and ATF2 are the most important factor determining acetate ester levels during fermentation (Verstrepen et al., 2003b). Indeed, as yeast strains, overexpressing the ATF1 gene, produce up to more than 30 times more ethyl acetate and up to more than 180 times more isoamyl acetate than wild‐type cells, it is clear that the availability of substrates is not a major limiting factor (Malcorps et al., 1991; Lilly et al., 2000; Verstrepen et al., 2003b). Moreover, recent work showed that the maximum expression levels of ATF1 and ATF2 during fermentation clearly correlate with the final concentration of acetate esters (Saerens et al., 2008a). This means that a higher production of acetate esters can be explained by a higher ATF1 and ATF2 expression. Much attention has therefore been paid to the genetic regulation of the ATF1 gene.
Already in 1997, it was shown by Fujii et al. that ATF1 gene expression is directly repressed by unsaturated fatty acids (UFAs) and oxygen. It was demonstrated that UFAs and oxygen repress ATF1 transcription by a different regulatory pathway and that ATF1 transcription is co‐regulated by the same mechanism as the OLE1 gene in response to UFAs (Fujiwara et al., 1998). The repression of ATF1 expression by oxygen is mainly mediated by the Rox1–Tup1–Ssn6 hypoxic repressor complex (Fujiwara et al., 1999), while the repression by UFAs is regulated through a so‐called low‐oxygen response element (Vasconcelles et al., 2001). This promoter element is activated under hypoxic conditions and selectively repressed by UFAs (Chellappa et al., 2001; Jiang et al., 2001).
In addition, other studies have demonstrated that ATF1 activity is also regulated through the protein kinases Sch9 and protein kinase A (PKA) (Fujiwara et al., 1999; Verstrepen et al., 2003c). These kinases play a central role in the transcriptional regulation of genes in response to changes in carbon, nitrogen and phosphate levels. The main targets of Sch9 are genes involved in cell growth, stress response and glycogen and trehalose metabolism (Thevelein, 1994; Crauwels et al., 1997; Rolland et al., 2002). Verstrepen and colleagues (2003c) showed that ATF1 transcription is induced by addition of glucose, regulated by the Ras/cAMP/PKA nutrient signalling pathway, and by the addition of maltose and nitrogen compounds through the ‘Fermentable Growth Medium‐induced’ pathway. ATF1 expression is also influenced by ethanol and heat stress.
In the brewing and wine fermentation industry it is already known for a very long time that fermentation variables affect ester production by yeast. The most important ones are the yeast strain used, the composition of the fermentation medium and the fermentation conditions. The influence of different substrate concentrations on acetate ester production can be explained by the regulatory mechanisms described above. It is known that the total sugar content of the medium, which is reflected in the specific gravity for brewing fermentations, together with the optimal amount of nitrogen available in the medium has an important positive influence on ester production (Younis and Stewart, 1998; 1999; Miller et al., 2007; Vilanova et al., 2007; Carrau et al., 2008; Saerens et al., 2008a,b). Other important parameters in the fermentation medium are the lipid content and the dissolved oxygen. It is generally accepted that the concentration of UFAs and the amount of dissolved oxygen in the fermentation medium are the best‐known negative regulators for acetate ester production (Anderson and Kirsop, 1974; 1975; Fujii et al., 1997).
Regulation of ethyl ester synthesis
The rate of MCFA ethyl ester formation is dependent on the concentration of the two substrates (the acyl‐CoA component and ethanol) and the total activity of the enzymes involved in the synthesis and hydrolysis of the MCFA ethyl esters. However, as enhancing enzyme activity by overexpression of the ester synthesis genes only slightly affects ethyl ester production, the enzyme activity appears not to be the limiting factor for ethyl ester production, in contrast to acetate ester production. Recently, the possible correlation between ethyl ester production and gene expression levels of EEB1 and EHT1 was investigated (Saerens et al., 2008a). The results showed that the maximum expression level of EEB1 is correlated with the final concentration of ethyl hexanoate (r = 60), but that there is no correlation with the final concentration of ethyl octanoate and decanoate. For EHT1 expression levels, there seems to be a strong negative correlation with the end concentrations of ethyl octanoate and decanoate. As Eht1 has both synthesis and hydrolysis activity towards ethyl esters (Saerens et al., 2006), this appears to indicate that in vivo the esterase activity is the main factor determining ethyl octanoate and decanoate production. Moreover, the observation that addition of MCFA precursors to the fermentation medium resulted in higher ethyl ester production (Saerens et al., 2006), as opposed to overexpression of the ethyl ester synthesis genes, further confirms the important role of precursor concentration as limiting factor for ethyl ester synthesis.
To investigate if the precursor levels themselves influence the expression of the genes EEB1 and EHT1, Saerens and colleagues (2008b) investigated the effect of MCFA addition on the expression of EEB1 and EHT1 in an ale brewing strain. Only octanoic acid was able to induce expression of EEB1 and EHT1, while hexanoic, decanoic and dodecanoic acid did not affect EEB1 or EHT1 expression. If the octanoic acid precursor does not specifically inhibit the esterase activity of the Eht1 and Eeb1 proteins (compared with their ester synthesis activity), these results suggest that the cellular MCFA concentration is rate limiting for ethyl ester synthesis.
To understand how fermentation parameters influence the formation of MCFA ethyl esters, it is necessary to consider the role of MCFAs as ester precursors. During the early stages of alcoholic fermentation, S. cerevisiae releases MCFAs, and particularly octanoic and hexanoic acids (Taylor and Kirsop, 1977), which are produced by the fatty acid synthase (FAS) complex during the synthesis process of long‐chain fatty acids and not by their degradation (Marchesini and Poirier, 2003). The key enzyme in the regulation of fatty acid biosynthesis is the acetyl‐CoA carboxylase (Sumper, 1974; Wakil et al., 1983). According to the model of Dufour and colleagues (2003), the post‐transcriptional activation of this acetyl‐CoA carboxylase determines the release of MCFAs from the FAS complex.
Under brewing fermentation conditions (limiting amount of oxygen) long‐chain saturated fatty acids accumulate and inhibit the acetyl‐CoA carboxylase (Fig. 3) (Dufour et al., 2003). Acyl‐CoAs under synthesis are subsequently released from the FAS complex and as a result medium‐chain fatty acyl‐CoAs accumulate, which results in increased MCFA ethyl ester synthesis (Äyräpää and Lindström, 1977). In the presence of oxygen, UFAs are synthesized, the inhibition of acetyl‐CoA carboxylase is released, the elongation reaction proceeds to form complete long‐chain fatty acids and, as a result, the intracellular pool of medium‐chain fatty acyl‐CoAs is reduced. The effect of wort lipids (stimulation of growth with a concomitant reduction in the pool of acyl‐CoAs), the lack of oxygen or the application of a top pressure of carbon dioxide (restricted growth and accumulation of acyl‐CoA residues) can be explained with the same model.
Figure 3.
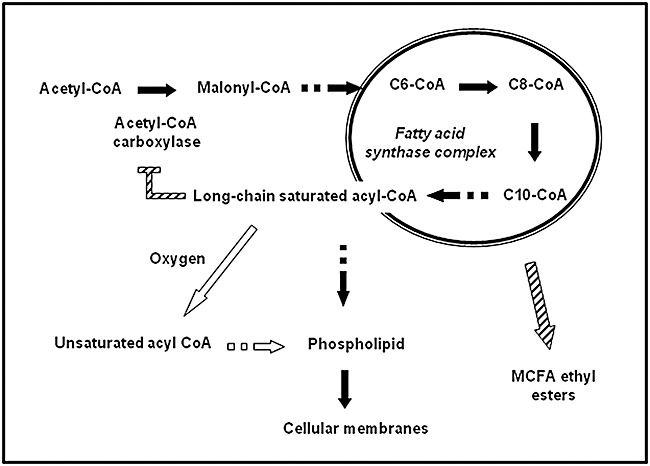
Biosynthesis of fatty acids and its relationship with medium‐chain fatty acid ester formation as proposed by Dufour and colleagues (2003). Acetyl‐CoA carboxylase initiates fatty acid synthesis and is inhibited by long‐chain saturated acyl‐CoAs. As a result, medium‐chain fatty acid CoAs are released from the fatty acid synthase complex, which can then be converted to the corresponding esters. In the presence of oxygen, long‐chain saturated acyl‐CoAs are converted to unsaturated acyl‐CoAs, which do not inhibit acetyl‐CoA carboxylase, and thus no longer cause release of medium‐chain fatty acid CoAs from the fatty acid synthase complex. Saturated and unsaturated fatty acids are used for the synthesis of phospholipids which are then incorporated into cellular membranes.
Furukawa and colleagues (2003) suggested that it is not the reduced elongation of fatty acids that promotes the release of medium‐chain fatty acyl‐CoAs, but rather the enhancement of fatty acid biosynthesis. They reported that inositol limitation increased the MCFA concentration by as much as it increased the concentration of cellular fatty acids in S. cerevisiae used for sake production. The conclusion was that the enhancement of fatty acid formation by inositol limitation in S. cerevisiae is mainly caused by the transcriptional enhancement of genes involved in fatty acid synthesis, and not by the post‐transcriptional activation of acetyl‐CoA carboxylase. By overexpression analysis, it was shown that FAS1 and FAS2 are the fatty acid synthetic genes that predominantly contribute to the enhancement of MCFA formation.
Hypothesis 1: ester formation regenerates free CoA
Under anaerobic conditions cells cannot synthesize UFAs. Hence, in the absence of UFAs in the medium, yeast is unable to accumulate UFAs under anaerobic conditions. As a result, saturated fatty acids will accumulate, causing an inhibition of acetyl‐CoA carboxylase and thus an arrest of fatty acid synthesis (Sumper, 1974; Wakil et al., 1983). In addition, the TCA‐cycle cannot be (fully) used under anaerobic conditions, so that all regular acetyl‐CoA‐consuming pathways are largely inactive. This may lead to an accumulation of acetyl‐CoA and a depletion of free CoA. Ester synthesis could therefore be a useful way for cells to regenerate free CoA without releasing high (toxic) concentrations of free acetic and medium‐chain fatty acids (Thurston et al., 1982; Malcorps and Dufour, 1992). The observation that ester synthesis is inhibited under aerobic conditions, or when UFAs are added to the medium, is consistent with this hypothesis (Peddie, 1990).
Attempts have been made in Escherichia coli to develop a CoA/acetyl‐CoA manipulation system in order to increase the productivity of ester formation. Overexpression of pantothenate kinase (panK), the key regulatory enzyme in the CoA biosynthetic pathway, with simultaneous supplementation of the CoA precursor pantothenic acid, led to an increase in the intracellular level of CoA and acetyl‐CoA (10‐fold and fivefold respectively) (Vadali et al., 2004a). In another study, a genetically engineered E. coli strain expressing both the yeast ATF2 gene and the panK gene produced sixfold more isoamyl acetate than the control strain expressing only the ATF2 gene (Vadali et al., 2004b). These results demonstrate that increasing the intracellular CoA and acetyl‐CoA concentrations leads to an increase in isoamyl acetate production. It is known that carnitine acetyltransferases (CrATs) in yeast function in a compartmental buffering system by maintaining the appropriate levels of acetyl‐CoA and CoA in cellular compartments. The most important protein, Cat2, is found in the peroxisomes and mitochondria and contributes > 95% of the total CrAT activity in galactose‐grown cells (Kispal et al., 1993). As CrATs are responsible for the modulation of the acetyl‐CoA/CoA ratio, Cordente and colleagues (2007) hypothesized that overexpression of the CAT2 gene in yeast could modify the acetyl‐CoA concentration and as a result, ester production during fermentation. The CrATs catalyse the reversible reaction between carnitine and acetyl‐CoA to form acetylcarnitine and CoA. Overexpression of the wild‐type CAT2‐encoded mitochondrial CrAT or a modified version localized in the cytosol, resulted in a decrease in the concentration of acetate esters produced during fermentation. Following the previous hypothesis, this was explained by the fact that overproduction of Cat2 favours the formation of acetylcarnitine and CoA and therefore limits the amount of precursor available for ester production (Cordente et al., 2007).
Hypothesis 2: esters as UFA analogues
Yeast membranes are a lipid bilayer consisting of phospholipids and sphingolipids, whose apolar phase consists of fatty acids. The fatty acids of S. cerevisiae are mainly made up of the saturated fatty acids palmitic acid (C16) and stearic acid (C18) and the UFAs oleic acid (C18:1) and palmitoleic acid (C16:1). Changes in the membrane lipid composition can significantly disturb the membrane's function and alter the activity of membrane‐associated enzymes and transporters. Consequently, many organisms have developed mechanisms to maintain the appropriate fluidity of membrane lipids. The fluidity of the membrane is defined as the extent of molecular disorder and molecular motion within a lipid bilayer. Membranes of yeast cells with an increased unsaturation index show higher membrane fluidity. The ability of cells to alter the degree of unsaturation in their membranes is an important factor in cellular acclimatization to changing environmental temperature (Rodriguez‐Vargas et al., 2007).
During semi‐anaerobic fermentation, like wine and beer fermentations, UFA synthesis stops when oxygen is exhausted. Consequently, the membrane fluidity will be reduced. Mason and Dufour (2000) suggested that certain esters of long‐chain hydroxy fatty acids could serve as UFA analogues. Upon incorporation of the esterified hydroxy fatty acids into the membrane, the small side‐chain on the long alkyl core of these esters could disturb the membrane structure in a similar way as the double bond of UFAs. There are several arguments to support this hypothesis: (i) acetate ester synthesis is repressed by UFAs, (ii) there is a correlation between the repressive effect of UFAs and their melting temperature, (iii) Atf1 is localized in lipid particles, which are important organelles for the metabolism and storage of neutral lipids, (iv) ATF1 and the Δ9‐fatty acid desaturase‐encoding gene OLE1, are co‐regulated, (v) the long‐chain hydroxy fatty acid, 12‐hydroxy stearate can be esterified by purified AATase in vitro (Malcorps and Dufour, 1992), (vi) addition of 12‐hydroxy stearate can restore growth in anaerobic cultures without UFA supplementation (Light et al., 1962), (vii) ATF1 is repressed by heat stress (Verstrepen et al., 2003c) and (viii) the expression of ATF1 is maximal in rich medium, so that growth is not restricted by carbon or nitrogen availability, but by the inability to maintain a sufficient degree of membrane fluidity. It can be expected that, under these circumstances, yeast cells may have certain alternative mechanisms that would allow them to grow or at least survive. It is possible that esterification of certain membrane compounds can be one of these strategies.
Hypothesis 3: ester formation as a detoxification mechanism
A role for Eht1 and Eeb1 in MCFA detoxification? MCFAs can be prematurely released from the cytosolic FAS complex, when there is inhibition of fatty acid synthesis. During anaerobiosis, saturated fatty acids cause the arrest of fatty acid synthesis, which causes release of a pool of intermediate products, namely MCFAs (Sumper, 1974; Taylor and Kirsop, 1977). Like acetate and lactate, MCFAs are toxic to yeast cells, most probably because they disturb intracellular pH homeostasis by increasing plasma membrane proton permeability. The lower intracellular pH results in disturbance of pH‐dependent cellular reactions, and also causes enhanced activity of the plasma membrane H+ ATPase, which pumps protons out of the cell. The high ATP consumption by the proton pump causes a severe drop in ATP levels, causing growth arrest and possibly even cell death. Furthermore, it has been suggested that due to competitive inhibition, high concentrations of medium‐chain fatty acyl‐CoAs could interfere with cellular processes requiring long‐chain acyl‐CoA molecules (Hunkova and Fencl, 1977; Bardi et al., 1998; 1999; Pampulha and Loureiro‐Dias, 2000; Narendranath et al., 2001). As MCFA ethyl esters are much less toxic than the related acids, their synthesis has been proposed as a protection mechanism for the yeast cells against accumulation of the toxic acids. After esterification, the MCFA ethyl esters can also diffuse more easily through the plasma membrane and leak into the medium, further reducing the risk of toxic acid accumulation by shifting the equilibrium towards ester synthesis (Nordström, 1964).
The role of Atf2 in cellular detoxification of steroid‐like compounds. Atf2 has been shown to acetylate pregnenolone and other structurally similar 3 β‐hydroxysteroids, prior to excretion via the ABC transporters Pdr5 and Snq2 (Cauet et al., 1999). Elucidation of this detoxification mechanism has resulted in the alternative nomenclature of APAT (acetylCoA:pregnenolone acetyltransferase) for ATF2. No acetylation of pregnenolone could be detected in the atf2Δ mutant, even in conditions favouring the expression of ATF1, suggesting that Atf1 has no detoxifying activity on pregnenolone. Atf2 shows a remarkable specificity towards steroids. The enzyme esterifies with the same efficiency Δ5‐ or Δ4‐3β‐hydroxysteroids such as pregnenolone. The activity is dramatically reduced with pregnane structures and with estrogenic compounds such as oestradiol and estrone. On the other hand, the enzyme accepts short‐chain acyl‐CoA esters but not long‐chain acyl‐CoA esters such as oleoyl‐CoA. Based on findings from the study of Cauet and colleagues (1999), it can be suggested that Atf2 and the efflux pumps Pdr5 and Snq2 may function cooperatively to rid the yeast cell of growth‐inhibitory 3β‐hydroxysteroids generated as products of defective ergosterol biosynthesis (which could exist under limiting amount of oxygen, for example during brewery and wine fermentation). Such a scenario might provide a rational explanation for preferential expression of Atf2 under anaerobic conditions where yeast cells are unable to biosynthesize ergosterol. The high affinity of Atf2 for pregnenolone (Km = 0.5 µM) is a strong indication that the enzyme has evolved to interact with a steroidal substrate; the Km of Atf2 for isoamyl alcohol is 50 000‐fold higher (25 mM).
Recently, it was reported that an acetylation/deacetylation cycle controls the export of sterols and steroids from yeast, where sterol acetylation required the acetyltransferase Atf2 and sterol deacetylation required the steryl deacetylase Say1 (Tiwari et al., 2007). In yeast, sterols are synthesized by enzymes located in the endoplasmic reticulum membrane and are enriched in the plasma membrane where they increase the permeability barrier of the membrane and thus are important to maintain the membrane potential (Haines, 2001). Sterols are generally regarded as being stable and long‐lived lipids. However, sterol‐like molecules can also have adverse effects and act cytotoxic. Tiwari et al. state that the sterol acetylation/deacetylation cycle operates on endogenously synthesized ergosterol precursors as well as on exogenously supplied steroids, and could serve to detoxify the cells of steroid‐like compounds and hydrophobic phytochemicals such as flavonoids that are present in plants and fruits, one of the natural environments of yeast, as indicated by the sensitivity of say1Δ and atf2Δ mutant cells against eugenol. Interestingly, isoflavonoids are common phytoalexins (antimicrobial compounds) synthesized in response to bacterial or fungal infection to limit the spread of the invading pathogen, and possibly also prevent secondary infection.
The fact that Pdr5 and Snq2 can transport pregnenolone acetate means that both transporters are involved in the export of cytotoxic compounds. Only very recently, this physiological role was suggested for the multi‐drug transporter genes in yeast (Sa‐Correia et al., 2009). Yeast transporters of the major facilitator superfamily (MSF) which confer multi‐drug resistance (MDR) are able to export a significantly wide spectrum of structurally unrelated compounds. However, the prevalence and apparent redundancy of so many MSF–MDR transporters, which protect the yeast cells against toxic compounds that are unlikely to be present in its natural environment, has led to speculation concerning their natural physiological function. Based on several findings, Sa‐Correia et al. suggest a role for these transporters in the expulsion of endogenous metabolites, in addition to their function in drug extrusion. It is possible that yeast first needs a chemical conversion (acetylation) of certain endogenous metabolites, catalysed by the acetyltransferases, before these cytotoxic compounds can be exported by the MSF–MDR transporters.
Hypothesis 4: ester formation as a mean for the dissipation of yeast in nature
Field studies of Drosophila species have documented that these insects feed on yeast growing on rotting fruit or plant parts (Fogleman et al., 1981). Directly onto this food, eggs are deposited by female flies, who choose optimal sites for oviposition based on the quality of the available food. Recently, it was shown that larvae of Drosophila melanogaster are strongly attracted by the ester ethyl butyrate (Asahina et al., 2009). As larvae hatch directly on their food source, it is essential that they can tolerate high odour concentrations and remain attracted to them without being distracted by low‐odour stimuli (Asahina et al., 2008). In the case of ethyl butyrate, it was shown that wild‐type larvae were strongly attracted to this odour across a 500‐fold range of concentration.
As S. cerevisiae is found on fermenting fruit, the production of esters could be a mechanism to attract insects like Drosophila species to ensure yeast dissipation in nature. As Drosophila species feed on S. cerevisiae, several cells could stick onto the flies after a visit of the fly to the fruit. It is possible that not only the larvae, but also the fly itself is attracted by potent odorants, released by fermenting fruit. Yeast cells producing more fruity esters would then have a better chance to stick to the fly and in this way to be dissipated in nature.
Conclusions and perspectives
Volatile aroma‐active esters constitute the largest and arguably the most important class of flavour compounds produced by fermenting yeast cells. Their aromas are essential for the fruity character of high‐quality alcoholic beverages, like beer, wine and sake. Significant progress has been made in elucidating the biochemical pathways responsible for the synthesis of these flavour compounds and in identifying the genes that encode the different enzymes of these pathways. However, our knowledge in this respect is not complete yet. The biochemical components and regulation of acetate ester synthesis have been defined in greatest detail, although some components remain to be identified. On the other hand, the genes and biochemical pathways of MCFA ethyl ester synthesis have been revealed only recently and here too, additional components remain to be discovered.
As acetate ester production is largely determined by the activity of the synthesizing enzymes, Atf1 and Atf2, any cellular factor influencing the expression of the ATF1 and ATF2 genes is a potential target to modulate acetate ester synthesis. This opens up perspectives to construct novel genetically modified brewer's or wine strains that produce a desired concentration of acetate esters. In the case of ethyl ester synthesis, substrate concentration is a major limiting factor and we need to search for parameters that can change the substrate concentration (e.g. the modification of expression of fatty acid synthesis genes) if we want to modulate ethyl ester production during fermentation. However, not only the substrate concentration is important for ethyl ester production, but also the balance between synthesis and hydrolysis activity is a crucial factor that can be modified when we want to optimize the ethyl ester concentration in the final fermentation product. Almost all work carried out has been focused on ester synthases and little information regarding ester hydrolysing enzymes is available. Future work concerning the optimization of the ester profile of yeast should include identification and characterization of esterases involved in the breakdown of specific ester compounds.
Next to straight‐chain fatty acid ethyl esters, also branched‐chain fatty acid ethyl esters and fatty acid acetate esters are found in fermented beverages like beer and wine. The biosynthesis pathways of these components are unknown. A search for yeast strains with different profiles of such esters and investigation of their formation in deletion strains of the known ester synthases, as well as other candidate enzymes, could be a first step to identify the enzymes responsible for their synthesis. This would allow subsequent exploration of their physiological role.
Finally, the true physiological role of ester synthesis is still an unanswered question. Several hypotheses have been proposed in the past, but recent results point to additional possibilities. The involvement of Eeb1 and Eht1 in esterification of MCFAs and the role of Atf2 in acetylation of sterols suggests a possible role of ester synthesis in cellular detoxification metabolism. As also some MSF–MDR transporters are believed to export endogenous toxic compounds, this hypothesis suggests that some compounds need first an acetylation, catalysed by the acetyltransferases, before the cytotoxic compounds can be exported. Another possibility is that some esters can serve as UFA analogues in the cell membrane to ensure optimal membrane fluidity during fermentation. However, as most esters (and certainly the shorter ones) are volatile compounds, this hypothesis can only be valid for the longer esters like the ethyl esters. Because of the volatility of the esters, an ecological function like the attraction of insects to fermenting fruits to ensure the spreading of yeast in nature also seems a plausible hypothesis. Finally, it cannot be excluded that esters actually play several different roles.
In conclusion, there still remain many unanswered questions in the field of aroma ester synthesis, especially with regard to the biological function. Continued research in this field is required to further improve our understanding of the pathways of ester biosynthesis, their regulation and physiological role. This knowledge will ultimately lead to more insight into the intriguing question why yeast produces aroma compounds like esters.
Acknowledgments
This work was supported by a grant from the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT Flanders) (SBO project IWT050148) to J.M. Thevelein and F.R. Delvaux.
References
- Anderson R.G., Kirsop B.H. Oxygen as a regulator of ester accumulation during fermentation of wort of high specific gravity. J Inst Brew. 1975;81:111–115. [Google Scholar]
- Anderson R.J., Kirsop B.H. The control of volatile ester synthesis during the fermentation of wort of high specific gravity. J Inst Brew. 1974;80:48–55. [Google Scholar]
- Aritomi K., Hirosawa I., Hoshida H., Shiigi M., Nishizawa Y., Kashiwagi S., Akada R. Self‐cloning yeast strains containing novel FAS2 mutations produce a higher amount of ethyl caproate in Japanese sake. Biosci Biotechnol Biochem. 2004;68:206–214. doi: 10.1271/bbb.68.206. [DOI] [PubMed] [Google Scholar]
- Asahina K., Pavlenkovich V., Vosshall L.B. The survival advantage of olfaction in a competitive environment. Curr Biol. 2008;18:1153–1155. doi: 10.1016/j.cub.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina K., Louis M., Piccinotti S., Vosshall L.B. A circuit supporting concentration‐invariant odor perception in Drosophila. J Biol. 2009;8:9. doi: 10.1186/jbiol108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Äyräpää T., Lindström I. Aspects of the influence of exogenous fatty acids on the fatty acid metabolism of yeast. Proc Eur Brew Conv. 1977;16:507–517. [Google Scholar]
- Bardi L., Crivelli C., Marzona M. Esterase activity and release of ethyl esters of medium‐chain fatty acids by Saccharomyces cerevisiae during anaerobic growth. Can J Microbiol. 1998;44:1171–1176. [PubMed] [Google Scholar]
- Bardi L., Cocito C., Marzona M. Saccharomyces cerevisiae cell fatty acid composition and release during fermentation without aeration and in absence of exogenous lipids. Int J Food Microbiol. 1999;47:133–140. doi: 10.1016/s0168-1605(98)00203-7. [DOI] [PubMed] [Google Scholar]
- Carrau F.M., Medina K., Farina L., Boido E., Henschke P.A., Dellacassa E. Production of fermentation aroma compounds by Saccharomyces cerevisiae wine yeasts: effects of yeast assimilable nitrogen on two model strains. FEMS Yeast Res. 2008;8:1196–1207. doi: 10.1111/j.1567-1364.2008.00412.x. [DOI] [PubMed] [Google Scholar]
- Cauet G., Degryse E., Ledoux C., Spagnoli R., Achstetter T. Pregnenolone esterification in Saccharomyces cerevisiae– a potential detoxification mechanism. Eur J Biochem. 1999;261:317–324. doi: 10.1046/j.1432-1327.1999.00282.x. [DOI] [PubMed] [Google Scholar]
- Chellappa R., Kandasamy P., Oh C.S., Jiang Y., Vemula M., Martin C.E. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression – fatty acid‐mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem. 2001;276:43548–43556. doi: 10.1074/jbc.M107845200. [DOI] [PubMed] [Google Scholar]
- Cordente A.G., Swiegers J.H., Hegardt F.G., Pretorius I.S. Modulating aroma compounds during wine fermentation by manipulating carnitine acetyltransferases in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2007;267:159–166. doi: 10.1111/j.1574-6968.2006.00548.x. [DOI] [PubMed] [Google Scholar]
- Crauwels M., Donaton M.C.V., Pernambuco M.B., Winderickx J., DeWinde J.H., Thevelein J.M. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable‐growth‐medium‐induced (FGM) pathway. Microbiol-UK. 1997;143:2627–2637. doi: 10.1099/00221287-143-8-2627. [DOI] [PubMed] [Google Scholar]
- Cristiani G., Monnet V. Food micro‐organisms and aromatic ester synthesis. Sci Aliments. 2001;21:211–230. [Google Scholar]
- Debourg A. Yeast flavour metabolites. Eur Brew Conv Monogr. 2000;28:60–73. [Google Scholar]
- Dufour J.‐P., Malcorps P. Ester synthesis during fermentation: enzymes characterization and modulation mechanism. In: Campbell I., Priest F. G., editors. The Institute of Brewing; 1994. pp. 137–151. , and . In Proceedings of the 4th Aviemore Conference on Malting, Brewing and Distilling, and (eds). London, UK: , pp. [Google Scholar]
- Dufour J.‐P., Verstrepen K.J., Derdelinckx G. Brewing yeasts. In: Boekhout T., Robert V., editors. Behr's Verlag Gmbh & Co; 2002. pp. 347–388. , and . In Yeasts in Food, and (eds). Hamburg, Germany: , pp. [Google Scholar]
- Dufour J.‐P., Malcorps P., Silcock P. Control of ester synthesis during brewery fermentation. In: Smart K., editor. Blackwell Publishing; 2003. pp. 213–233. , and . In Brewing Yeast Fermentation Performance (ed.). Oxford, UK: , pp. [Google Scholar]
- Fogleman J.C., Starmer W.T., Heed W.B. Larval selectivity for yeast species by Drosophila mojavensis in natural substrates. Proc Natl Acad Sci. 1981;78:4435–4439. doi: 10.1073/pnas.78.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Nagasawa N., Iwamatsu A., Bogaki T., Tamai W., Hamachi M. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl Environ Microbiol. 1994;60:2786–2792. doi: 10.1128/aem.60.8.2786-2792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Yoshimoto H., Nagasawa N., Bogaki T., Tamai Y., Hamachi M. Nucleotide sequences of alcohol acetyltransferase genes from lager brewing yeast, Saccharomyces carlsbergensis. Yeast. 1996a;12:593–598. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C593::AID-YEA593%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fujii T., Yoshimoto H., Tamai Y. Acetate ester production by Saccharomyces cerevisiae lacking the ATF1 gene encoding the alcohol acetyltransferase. J Ferment Bioeng. 1996b;81:538–542. [Google Scholar]
- Fujii T., Kobayashi O., Yoshimoto H., Furukawa S., Tamai Y. Effect of aeration and unsaturated fatty acids on expression of the Saccharomyces cerevisiae alcohol acetyltransferase gene. Appl Environ Microbiol. 1997;63:910–915. doi: 10.1128/aem.63.3.910-915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara D., Yoshimoto H., Sone H., Harashima S., Tamai Y. Transcriptional co‐regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1 and Delta‐9 fatty acid desaturase gene, OLE1 by unsaturated fatty acids. Yeast. 1998;14:711–721. doi: 10.1002/(SICI)1097-0061(19980615)14:8<711::AID-YEA263>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Fujiwara D., Kobayashi O., Yoshimoto H., Harashima S., Tamai Y. Molecular mechanism of the multiple regulation of the Saccharomyces cerevisiae ATF1 gene encoding alcohol acetyltransferase. Yeast. 1999;15:1183–1197. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1183::AID-YEA444>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Fukuda K., Yamamoto N., Kiyokawa Y., Yanagiuchi T., Wakai Y., Kitamoto K. Balance of activities of alcohol acetyltransferase and esterase in Saccharomyces cerevisiae is important for production of isoamyl acetate. Appl Environ Microbiol. 1998;64:4076–4078. doi: 10.1128/aem.64.10.4076-4078.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Yamada T., Mizoguchi H., Hana S. Increased ethyl caproate production by inositol limitation in Saccharomyces cerevisiae. J Biosci Bioeng. 2003;95:448–454. doi: 10.1016/s1389-1723(03)80043-9. [DOI] [PubMed] [Google Scholar]
- Haines T.H. Do sterols reduce proton and sodium leaks through lipid bilayers? Prog Lipid Res. 2001;40:299–324. doi: 10.1016/s0163-7827(01)00009-1. [DOI] [PubMed] [Google Scholar]
- Holmquist M. Alpha/Beta‐hydrolase fold enzymes: structures, functions and mechanisms. Curr Prot Pep Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Hunkova Z., Fencl Z. Toxic effects of fatty acids on yeast cells – dependence of inhibitory effects on fatty acid concentration. Biotechnol Bioeng. 1977;19:1623–1641. doi: 10.1002/bit.260191103. [DOI] [PubMed] [Google Scholar]
- Jiang Y.D., Vasconcelles M.J., Wretzel S., Light A., Martin C.E., Goldberg M.A. MGA2 is involved in the low‐oxygen response element‐dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:6161–6169. doi: 10.1128/MCB.21.18.6161-6169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Sumegi B., Dietmeier K., Bock I., Gajdos G., Tomcsanyi T., Sandor A. Cloning and sequencing of a cDNA encoding Saccharomyces cerevisiae carnitine acetyltransferase – use of the cDNA in gene disruption studies. J Biol Chem. 1993;268:1824–1829. [PubMed] [Google Scholar]
- Li Y.S., Zhao Y.F., Hatfield S., Wan R., Zhu Q., Li X.H. Dipeptide seryl‐histidine and related oligopeptides cleave DNA, protein, and a carboxyl ester. Bioorgan Med Chem. 2000;8:2675–2680. doi: 10.1016/s0968-0896(00)00208-x. et al. [DOI] [PubMed] [Google Scholar]
- Light R.J., Lennarz W.J., Bloch K. The metabolism of hydroxy stearic acids in yeast. J Biol Chem. 1962;237:1793–1800. [PubMed] [Google Scholar]
- Lilly M., Lambrechts M.G., Pretorius I.S. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol. 2000;66:744–753. doi: 10.1128/aem.66.2.744-753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M., Bauer F.F., Lambrechts M.G., Swiegers J.H., Cozzolino D., Pretorius I.S. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast. 2006;23:641–659. doi: 10.1002/yea.1382. [DOI] [PubMed] [Google Scholar]
- Malcorps P., Dufour J.P. Ester synthesis by Saccharomyces cerevisiae– localization of the acetyl‐CoA isoamyl alcohol acetyltransferase. J Inst Brew. 1987;93:160. [Google Scholar]
- Malcorps P., Dufour J.P. Short‐chain and medium‐chain aliphatic ester synthesis in Saccharomyces cerevisiae. Eur J Biochem. 1992;210:1015–1022. doi: 10.1111/j.1432-1033.1992.tb17507.x. [DOI] [PubMed] [Google Scholar]
- Malcorps P., Cheval J.M., Jamil S., Dufour J.P. A new model for the regulation of ester synthesis by alcohol acetyltransferase in Saccharomyces cerevisiae. J Am Soc Brew Chem. 1991;49:47–53. [Google Scholar]
- Marchesini S., Poirier Y. Futile cycling of intermediates of fatty acid biosynthesis toward peroxisomal beta‐oxidation in Saccharomyces cerevisiae. J Biol Chem. 2003;278:32596–32601. doi: 10.1074/jbc.M305574200. [DOI] [PubMed] [Google Scholar]
- Mason A.B., Dufour J.P. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast. 2000;16:1287–1298. doi: 10.1002/1097-0061(200010)16:14<1287::AID-YEA613>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Meilgaard M.C. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. MBAA Techn Quart. 1975;12:151–168. [Google Scholar]
- Meilgaard M.C. The flavor of beer. MBAA Techn Quart. 1991;28:132–141. [Google Scholar]
- Miller A.C., Wolff S.R., Bisson L.F., Ebeler S.E. Yeast strain and nitrogen supplementation: dynamics of volatile ester production in Chardonnay juice fermentations. Am J Enol Vitic. 2007;58:470–483. [Google Scholar]
- Nagasawa N., Bogaki T., Iwamatsu A., Hamachi M., Kumagai C. Cloning and nucleotide sequence of the alcohol acetyltransferase II gene (ATF2) from Saccharomyces cerevisiae Kyokai No. 7. Biosci Biotechnol Biochem. 1998;62:1852–1857. doi: 10.1271/bbb.62.1852. [DOI] [PubMed] [Google Scholar]
- Narendranath N.V., Thomas K.C., Ingledew W.M. Acetic acid and lactic acid inhibition of growth of Saccharomyces cerevisiae by different mechanisms. J Am Soc Brew Chem. 2001;59:187–194. [Google Scholar]
- Nordström K. Formation of ethyl acetate in fermentation with brewer's yeast III. Participation of coenzyme A. J Inst Brew. 1962;68:398–407. [Google Scholar]
- Nordström K. Formation of esters from acids by brewer's yeast. IV. Effect of higher fatty acids and toxicity of lower fatty acids. J Inst Brew. 1964;70:233–238. [Google Scholar]
- Nykanen I., Suomalainen H. Formation of aroma compounds by yeast. In: Nykanen I., Suomalainen H., editors. Reidel Publishing Company; 1983. pp. 3–16. , and . In Aroma of Beer, Wine and Distilled Beverages, and (eds). Dordrecht, The Netherlands: , pp. [Google Scholar]
- Nykanen L. Formation and occurence of flavor compounds in wine and distilled beverages. Am J Enol Vitic. 1986;37:84–96. [Google Scholar]
- Nykanen L., Nykanen I. Production of esters by different yeast strains in sugar fermentations. J Inst Brew. 1977;83:30–31. [Google Scholar]
- Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M. The alpha/beta‐hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. et al. [DOI] [PubMed] [Google Scholar]
- Pampulha M.E., Loureiro‐Dias M.C. Energetics of the effect of acetic acid on growth of Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;184:69–72. doi: 10.1111/j.1574-6968.2000.tb08992.x. [DOI] [PubMed] [Google Scholar]
- Peddie H.A.B. Ester formation in brewery fermentations. J Inst Brew. 1990;96:327–331. [Google Scholar]
- Pisarnitskii A.F. Formation of wine aroma: tones and imperfections caused by minor components (review) Appl Biochem Microbiol. 2001;37:552–560. [PubMed] [Google Scholar]
- Rodriguez‐Vargas S., Sanchez‐Garcia A., Martinez‐Rivas J.M., Prieto J.A., Randez‐Gil F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol. 2007;73:110–116. doi: 10.1128/AEM.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Winderickx J., Thevelein J.M. Glucose‐sensing and ‐signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Sa‐Correia I., Dos Santos S.C., Teixeira M.C., Cabrito T.R., Mira N.P. Drug:H(+) antiporters in chemical stress response in yeast. Trends Microbiol. 2009;17:22–31. doi: 10.1016/j.tim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Saerens S.M.G., Verstrepen K.J., Laere V., Voet S.D.M., Van Dijck A.R.D., Delvaux P. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium‐chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem. 2006;281:4446–4456. doi: 10.1074/jbc.M512028200. et al. [DOI] [PubMed] [Google Scholar]
- Saerens S.M., Verbelen P.J., Vanbeneden N., Thevelein J.M., Delvaux F.R. Monitoring the influence of high‐gravity brewing and fermentation temperature on flavour formation by analysis of gene expression levels in brewing yeast. Appl Microbiol Biotechnol. 2008a;80:1039–1051. doi: 10.1007/s00253-008-1645-5. [DOI] [PubMed] [Google Scholar]
- Saerens S.M., Delvaux F., Verstrepen K.J., Van Dijck P., Thevelein J.M., Delvaux F.R. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl Environ Microbiol. 2008b;74:454–461. doi: 10.1128/AEM.01616-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M. Control of fatty acid biosynthesis by long‐chain acyl‐CoAs and by lipid membranes. Eur J Biochem. 1974;49:469–475. doi: 10.1111/j.1432-1033.1974.tb03851.x. [DOI] [PubMed] [Google Scholar]
- Suomalainen H. Yeast esterases and aroma esters in alcoholic beverages. J Inst Brew. 1981;87:296–300. [Google Scholar]
- Swiegers J.H., Pretorius I.S. Yeast modulation of wine flavor. Adv Appl Microbiol. 2005;57:131–175. doi: 10.1016/S0065-2164(05)57005-9. [DOI] [PubMed] [Google Scholar]
- Taylor G.T., Kirsop B.H. The origin of medium chain length fatty acids present in beer. J Inst Brew. 1977;83:241–243. [Google Scholar]
- Thevelein J.M. Signal‐transduction in yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- Thurston P.A., Quain D.E., Tubb R.S. Lipid‐metabolism and the regulation of volatile ester synthesis in Saccharomyces cerevisiae. J Inst Brew. 1982;88:90–94. [Google Scholar]
- Tiwari R., Koffel R., Schneiter R. An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 2007;26:5109–5119. doi: 10.1038/sj.emboj.7601924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadali R.V., Bennett G.N., San K.Y. Cofactor engineering of intracellular CoA/acetyl‐CoA and its effect on metabolic flux redistribution in Escherichia coli. Metab Eng. 2004a;6:133–139. doi: 10.1016/j.ymben.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Vadali R.V., Bennett G.N., San K.Y. Applicability of CoA/acetyl‐CoA manipulation system to enhance isoamyl acetate production in Escherichia coli. Metab Eng. 2004b;6:294–299. doi: 10.1016/j.ymben.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Vasconcelles M.J., Jiang Y., McDaid K., Gilooly L., Wretzel S., Porter D.L. Identification and characterization of a low oxygen response element involved in the hypoxic induction of a family of Saccharomyces cerevisiae genes. Implications for the conservation of oxygen sensing in eukaryotes. J Biol Chem. 2001;276:14374–14384. doi: 10.1074/jbc.M009546200. et al. [DOI] [PubMed] [Google Scholar]
- Verstrepen K.J., Derdelinckx G., Dufour J.P., Winderickx J., Thevelein J.M., Pretorius I.S., Delvaux F.R. Flavor‐active esters: adding fruitiness to beer. J Biosci Bioeng. 2003a;96:110–118. [PubMed] [Google Scholar]
- Verstrepen K.J., Van Laere S.D.M., Vanderhaegen B.M.P., Derdelinckx G., Dufour J.P., Pretorius I.S. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg‐ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol. 2003b;69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen K.J., Derdelinckx G., Dufour J.P., Winderickx J., Pretorius I.S., Thevelein J.M., Delvaux F.R. The Saccharomyces cerevisiae alcohol acetyl transferase gene ATF1 is a target of the cAMP/PKA and FGM nutrient‐signalling pathways. FEMS Yeast Res. 2003c;4:285–296. doi: 10.1016/S1567-1356(03)00166-1. [DOI] [PubMed] [Google Scholar]
- Verstrepen K.J., Van Laere S.D.M., Vercammen J., Derdelinckx G., Dufour J.P., Pretorius I.S. The Saccharomyces cerevisiae alcohol acetyl transferase Atf1p is localized in lipid particles. Yeast. 2004;21:367–376. doi: 10.1002/yea.1100. et al. [DOI] [PubMed] [Google Scholar]
- Vilanova M., Ugliano M., Varela C., Siebert T., Pretorius I.S., Henschke P.A. Assimilable nitrogen utilisation and production of volatile and non‐volatile compounds in chemically defined medium by Saccharomyces cerevisiae wine yeasts. Appl Microbiol Biotechnol. 2007;77:145–157. doi: 10.1007/s00253-007-1145-z. [DOI] [PubMed] [Google Scholar]
- Wakil S.J., Stoops J.K., Joshi V. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Fujiwara D., Momma T., Ito C., Sone H., Kaneko Y., Tamai Y. Characterization of the ATF1 and Lg‐ATF1 genes encoding alcohol acetyltransferases in the bottom fermenting yeast Saccharomyces pastorianus. J Ferment Bioeng. 1998;86:15–20. [Google Scholar]
- Yoshioka K., Hashimoto N. Ester formation by alcohol acetyltransferase from brewers yeast. Agr Biol Chem. 1981;45:2183–2190. [Google Scholar]
- Younis O.S., Stewart G.G. Sugar uptake and subsequent ester and higher alcohol production by Saccharomyces cerevisiae. J Inst Brew. 1998;104:255–264. [Google Scholar]
- Younis O.S., Stewart G.G. Effect of malt wort, very‐high‐gravity malt wort, and very‐high‐gravity adjunct wort on volatile production in Saccharomyces cerevisiae. J. Am Soc Brew Chem. 1999;57:39–45. [Google Scholar]


