Summary
pSILBAγ silencing vector was constructed for efficient RNA silencing triggering in the model mycorrhizal fungus Laccaria bicolor. This cloning vector carries the Agaricus bisporus gpdII promoter, two multiple cloning sites separated by a L. bicolor nitrate reductase intron and the Aspergillus nidulans trpC terminator. pSILBAγ allows an easy oriented two‐step PCR cloning of hairpin sequences to be expressed in basidiomycetes. With one further cloning step into pHg, a pCAMBIA1300‐based binary vector carrying a hygromycin resistance cassette, the pHg/pSILBAγ plasmid is used for Agrobacterium‐mediated transformation. The pHg/pSILBAγ system results in predominantly single integrations of RNA silencing triggering T‐DNAs in the fungal genome and the integration sites of the transgenes can be resolved by plasmid rescue. pSILBAγ construct and two other pSILBA plasmid variants (pSILBA and pSILBAα) were evaluated for their capacity to silence Laccaria nitrate reductase gene. While all pSILBA variants tested resulted in up to 65–76% of transformants with reduced growth on nitrate, pSILBAγ produced the highest number (65%) of strongly affected fungal strains. The strongly silenced phenotype was shown to correlate with T‐DNA integration in transcriptionally active genomic sites. pHg/pSILBAγ was shown to produce T‐DNAs with minimum CpG methylation in transgene promoter regions which assures the maximum silencing trigger production in Laccaria. Methylation of the target endogene was only slight in RNA silencing triggered with constructs carrying an intronic spacer hairpin sequence. The silencing capacity of the pHg/pSILBAγ was further tested with Laccaria inositol‐1,4,5‐triphosphate 5‐phosphatase gene. Besides its use in silencing triggering, the herein described plasmid system can also be used for transgene expression in Laccaria. pHg/pSILBAγ silencing system is optimized for L. bicolor but it should be highly useful also for other homobasidiomycetes, group of fungi currently lacking molecular tools for RNA silencing.
Introduction
RNA silencing is a sequence homology‐dependent degradation of target messenger RNAs (gene knock‐down) that exploits an ancient eukaryotic cellular mechanisms, believed to have evolved as a protection against invading viral double‐stranded RNAs (dsRNAs) (for a review see Dykxhoorn et al., 2003; Voinnet, 2005; Nakayashiki and Nguyen, 2008). The use of RNA silencing as a tool for altering gene expression has experienced a dramatic boom since the discovery of this mechanism just 10 years ago (Fire et al., 1998; Hamilton and Baulcombe, 1999). From its pioneer use in Drosophila and Caenorhabditis elegans (Kamath et al., 2003; Boutros et al., 2004) it has been successfully extended to other groups of eukaryotes including also fungi. RNA silencing is an especially attractive approach for reverse genetics studies on filamentous fungi. These eukaryotes often show extremely low homologous recombination efficiency and traditional gene knockout studies thus have not been easy to accomplish.
RNA silencing in fungi was first demonstrated in the ascomycetes Neurospora crassa, Schizosaccharomyces pombe and Magnaporthe oryzae (Cogoni et al., 1996; Kadotani et al., 2003; Raponi and Arndt 2003). Further studies on RNA silencing and related cellular processes such as heterochromatin control and assembly have turned these fungal species into important models among both fungi and other eukaryotes (Kadotani et al., 2004; Chicas et al., 2005; Martienssen et al., 2005; Nakayashiki et al., 2005; Chosed and Dent, 2007; Fulci and Macino, 2007). The early descriptive reports on RNA silencing mechanisms conducted mainly on ascomycetes and later on saprotrophic basidiomycetes are now reaching the point where this methodology is been used for large‐scale genetic studies together with information coming from fungal genome sequencing projects (Nguyen et al., 2008). Recently several filamentous basidiomycetes, these including both mycorrhizal, plant pathogenic and economically important edible species, have been reported to posses, or show indications to have RNA silencing machinery (Eastwood et al., 2008; Laurie et al., 2008; Matityahu et al., 2008; Suizu et al., 2008; Kemppainen et al., 2009).
Double‐stranded hairpin RNA (hpRNA) expression from stable integrated transgenes or from auto‐replicative elements has been shown to be a widely efficient trigger in inducing RNA silencing both on introduced marker and on endogenes in fungi. However, an efficient use of RNA silencing requires an easy to use cloning/transformation vector system compatible with the species under study and with the transformation methodology employed. For years plasmid vectors such as pHANNIBAL and pHELLSGATE (Wesley et al., 2001) or pSTARLING and pOpOff (CSIRO) have been available for producing stable RNA silencing triggers in plants. Also several vectors for viral‐induced gene silencing (VIGS) are available for transient gene silencing studies in plants (Liu et al., 2002a; Dong et al., 2007; Peretz et al., 2007). Similarly, several different silencing vector systems for mammalian cells have been reported in last years (Wadhwa et al., 2004; Wu et al., 2005; Cheng and Chang, 2007; Gou et al., 2007).
An equivalent RNA silencing vector system adapted to fungi did not exist until Nakayashiki and colleagues (2005) published pSilent‐1. This cloning vector was developed for an easy PCR‐based cloning of intronic spacer hairpin RNAs (ihpRNAs) to be expressed in ascomycetes. pSilent‐1 was also designed to function for direct plasmid transformation and carries a hygromycin resistance cassette for antibiotic selection of the transformants. This cloning/transformation vector has made RNA silencing studies in ascomycetes easy to perform and has been successfully used, with or without modifications, in M. oryzae, Colletotrichum lagenarium, Bipolaris oryzae and Acremonium chrysosgenum (syn. Cephalosporium acremonium) (Nakayashiki et al., 2005; Janus et al., 2007; Moriwaki et al., 2007). Now the second generation of silencing vectors for fungi is being designed. Further modifications of the pSilent‐1 vector, pSilent‐Dual1 (Nguyen et al., 2008) and pTroya (Shafran et al., 2008), are adapted for high‐throughput functional genomics studies in ascomycetes, the latter incorporating for the first time the GATEWAY technology in silencing vectors for fungi. Also another hpRNA‐cloning vector, pFIRD1, designed for Aspergillus niger and utilizing recombination in vitro has been recently released (Oliveira et al., 2008).
While several silencing/transformation vector systems are now available for ascomycetes, comparable molecular tools functional in basidiomycetes are still missing. Laccaria bicolor is the first ectomycorrhizal basidiomycete with the full genome sequence available (http://genome.jgi‐psf.org/Lacbi1/Lacbi1.home.html; Martin and Selosse, 2008; Martin et al., 2008). This fungus can efficiently be genetically modified by Agrobacterium‐mediated transformation (AMT) and the transgene integration occurs predominantly as a single copy and with a bias towards transcriptionally active genomic regions (Kemppainen et al., 2005; 2008).
Laccaria like many other filamentous fungi is recalcitrant to gene disruption by homologous recombination. In addition, the symbiotic phase of the life cycle in ectomycorrhizal basidiomycetes in nature is the dikaryon and therefore studies on mycorrhizal relevant genes by targeted knockouts are rather unlikely to achieve.
We have recently demonstrated that the RNA silencing pathway is active in L. bicolor and expression of an hpRNA trigger can result in dikaryotic fungal strains with altered transcript level of the target gene strongly enough to cause functional mutants (Kemppainen et al., 2009). However, a large‐scale and efficient use of RNA silencing in Laccaria requires an access to an easy to use hpRNA cloning/expression vector adapted to this organism. This cloning vector should also be combinable with AMT. Due to the lack of this molecular tool we decided to design a silencing‐AMT vector system for Laccaria.
We previously showed that the information on genomic T‐DNA integrations sites is easily and efficiently obtained by plasmid rescue (Kemppainen et al., 2008), thus this feature was also incorporated in the silencing/transformation vector system design. We constructed pHg/pSILBAγ dual‐vector system, where the pSILBAγ carries the ihpRNA expression cassette for PCR‐based oriented cloning of the hairpin arms and the Agrobacterium binary vector pHg harbours the selection marker, a hygromycin resistance cassette. The ligation of these two vectors joins the two cassettes in the T‐DNA for AMT. The pHg/pSILBAγ system also carries an ampicillin resistance marker and a replication origin for resolving the T‐DNA integration site in Laccaria genome by plasmid rescue in Escherichia coli.
We tested and optimized the system for its use in Laccaria. However, it should be applicable to any homobasidiomycete with functional RNA silencing machinery, sensitivity to hygromycin and capacity to recognize the Agaricus bisporus gpdII promoter. The latter requirement has been documented to be fulfilled at least by Suillus bovinus, Hebeloma cylindrosporum,Coprinus cinereus, Hypholoma sublateritium and Moniliophthora perniciosa (Hanif et al., 2002; Combier et al., 2003; Godio et al., 2004; Burns et al., 2005; Fagundes Lopes et al., 2008) indicating that this promoter is widely active in homobasidiomycetes. pSILBAγ also allows an easy construction of expression cassettes extending its use to transgene expression. Herein reported pHg/pSILBAγ system is designed for AMT but can certainly be used for direct plasmid introduction in species that are not susceptible to Agrobacterium transformation.
Results
Vector construction
We constructed the silencing vector pSILBA (SIL, silencing; BA, basidiomycetes) (Fig. 1) for PCR‐based cloning of ihpRNAs expressed in basidiomycetes. This vector was made by modifying pSilent‐1, a silencing/transformation vector designed for RNA silencing in ascomycetes (Nakayashiki et al., 2005). Because L. bicolor (Kemppainen et al., 2005) and several other basidiomycetes such as A. bisporus, S. bovinus, H. cylindrosporum, Paxillus involutus, Heterobasidium annosum, C. cinereus, Phanerochaete chrysosporum and Pseudozyma antarctica (Chen et al., 2000; Pardo et al., 2002; Combier et al., 2003; Samils et al., 2006; Sharma et al., 2006; Marchand et al., 2007) have shown to be susceptible to AMT, we adapted pSILBA to be used with an Agrobacterium binary vector. The silencing cassette cloning vector and transformation binary vector were kept separately as is the case of the pHANNIBAL silencing vector system designed for plants (Wesley et al., 2001). This was done in order to keep hairpin cloning steps easy to manipulate in a smaller size vector. Also maximum number of unique cloning sites was maintained available for the hairpin cloning in pSILBA.
Figure 1.
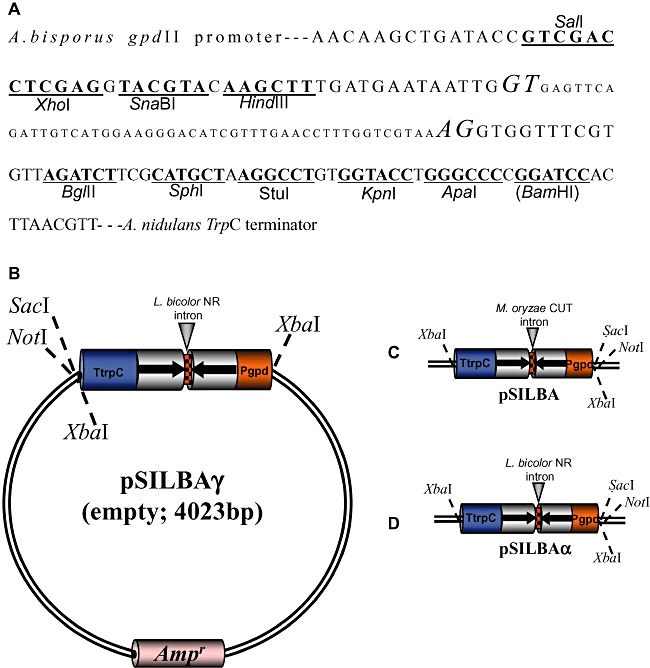
A. Sequence and unique restriction sites of pSILBAγ for inverted repeated sequence cloning. The acceptor and donor bases of L. bicolor nitrate reductase intron are in italics. The BamHI site in brackets is not unique. The same cloning sites are unique also in pSILBA and pSILBAα plasmid variants. B. pSILBAγ cloning vector. SacI and NotI linearize the plasmid; XbaI liberates the silencing triggering cassette. C. pSILBA. D. pSILBAα. Pgpd: glyceraldehide‐3‐phosphate dehydrogenase promoter of Agaricus bisporus. NR: nitrate reductase. CUT: cutinase. TtrpC: Aspergillus nidulans tryptophan synthetase terminador.
The plasmid pSILBA was constructed by removing pSilent‐1 hph cassette after which the Aspergillus nidulans trpC promoter in the silencing triggering cassette was replaced by the A. bisporus gpdII promoter. This promoter is widely recognized by other basidiomycetes such as L. bicolor while the A. nidulans trpC promoter does not produce hygromycin‐resistant strains of L. bicolor when used for AMT (Kemppainen et al., 2005). This indicates that the given ascomycete promoter is not active in Laccaria or that it is not strong enough for conferring antibiotic resistance in single integration transformants predominantly produced by AMT. However, it is worth mentioning that the A. nidulans gpd promoter has been reported functional in another homobasidiomycete, Heterobasidium annosum (Samils et al., 2006), showing that the acceptance of different ascomycete promoters in basidiomycetes can be species specific.
The A. nidulans terminator and M. oryzae CUT intron were maintained thus completing pSILBA silencing triggering cassette. The unique cloning sites available for hairpin cloning in pSILBA are presented in Fig. 1.
The Agrobacterium binary vector adapted to be used with pSILBA is pHg (Fig. 2). It is a modified pCAMBIA1300‐based pBGgHg vector (Chen et al., 2000) and carries a hygromycin resistance cassette for transformants selection. The original egfp cassette was removed and replaced by a fragment generating unique SacI, NotI and XbaI sites between the hph cassette and the T‐DNA RB. For AMT the two plasmids, pSILBA and pHg, are ligated into the transformation/silencing vector pHg/pSILBA by using SacI or NotI which linearize both plasmids. Care should be taken that the inverted repeated sequence used must be free of at least one of these sites. As a result the T‐DNA contains the selection marker cassette on the LB side and the silencing triggering cassette on the RB site. Once integrated in the fungal genome the pHg/pSILBA T‐DNA offers also the plasmid rescue alternative in E. coli via ampicillin resistance and replication from pSILBA backbone (Fig. S1). The information of the RB–gDNA junctions are easy to obtain by sequencing the rescued plasmids with universal sequencing primers M13/pUC reverse (−26) or T3 promoter. These primer sites are 348 and 311 bp inwards from the RB nick site making possible also sequencing of T‐DNA integrations truncated in their RB. Multiple PCR reactions and further fragment cloning for sequencing are not necessary as usually is the case when TAIL‐PCR for integration site resolving is used. Also no large‐size genomic fragments are generated thus no need for a high performance polymerase as may be the case in the inverted PCR approach. Alternatively, the silencing cassette can be removed from the pSILBA by XbaI digestion and ligated without the plasmid backbone into the unique XbaI site in pHg (Fig. 2). This could be an interesting option in order to keep the plasmid size smaller if the binary vector is being used for direct protoplast plasmid transformation instead of AMT.
Figure 2.
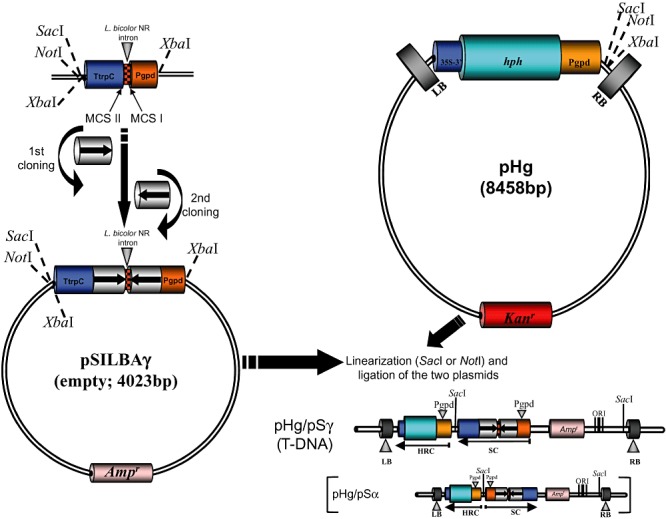
Two‐step cloning of the hairpin trigger in pSILBAγ and joining the plasmid with the binary vector pHg. The T‐DNA structures generated with pSILBAγ and pSILBAα are marked as pHg/pSγ and pHg/pSα respectively. SC: silencing cassette. HRC: hygromycin resistance cassette. MCS: multiple cloning site. LB: T‐DNA left border repeat of pCAMBIA1300. RB: T‐DNA right border repeat of pCAMBIA1300. hph: hph gene of E. coli coding for an aminocyclitol phosphotransferase that confers resistance to hygromycin B and structurally related antibiotics. Ampr: bla (ApR) gene of E. coli coding for a β‐lactamase that confers resistance to ampicillin. Kanr: aadA gene of E. coli coding for an aminoglycoside phosphotransferase that confers resistance to kanamycin. 35S‐3′: cauliflower mosaic virus 35S terminator.
It is known that for stabilizing inverted repeated structures during cloning steps in E. coli a spacer sequence must be placed between the repeats. Moreover, the incorporation of an intronic sequence as hpRNA spacer has been shown to increase silencing efficiency in plant and animals (Smith et al., 2000; Wesley et al., 2001; Lee and Carthew, 2003). This phenomenon is not well understood but it has been proposed that the passage of the ihpRNA through the splicing machinery would enhance nuclear export of the hpRNAs and thus result in higher cytosolic concentration of the silencing trigger. If the effect of an intronic spacer in fungal silencing triggers is the same as in other eukaryotes is no yet clear. Comparisons of exactly the same‐size intronic and non‐intronic sequences in the dsRNA constructs are sparse but at least in N. crassa an intronic sequence has clearly been shown to improve silencing efficiency with respect to a non‐intronic sequence (Goldoni et al., 2004). This indicates that similar mechanisms responsible for ihpRNA effect in plants and animals function at least in this ascomycete. In a RNA silencing study conducted in the basidiomycte C. cinereus no difference in silencing efficiency was observed between a non‐intronic and an intronic spacer. Unfortunately the constructs used in this study carried a transcribed intron outside the inverted repeated structure and therefore these results did not rule out the improved nuclear export of the silencing trigger by intron splicing in basidiomycetes (Wälti et al., 2006).
In Laccaria genome introns are abundant and usually relative short, of about 50–60 bp. This is clearly shorter than the 147 bp M. oryzae CUT intronic spacer used in pSilent‐1. This size difference could theoretically lead to less efficient recognition and splicing of the introduced heterologous intron in Laccaria. A similar phenomenon was proposed by pSilent‐1 designers when an untypical long M. oryzae chitin‐binding protein intron (850 bp) was used as hpRNA spacer. The silencing efficiency obtained with this construct was equivalent to one of 542 bp non‐intronic spacer. Probably this long intron had not been spliced with the same efficiency than the shorter intron (Nakayashiki et al., 2005). If the M. oryzae intron would not be recognized efficiently in Laccaria its length could be a destabilizing factor for dsRNA formation, especially if short armed hpRNAs are used for silencing triggering. Moreover, the effect of the length of non‐intronic sequence has been studied in the dimorphic pathogenic ascomycete Histoplasma capsulatum where the reduction of the spacer length clearly improved the silencing efficiency. No difference was observed between the shortest 25 and 87 bp spacers separating 709 bp inverted repeated sequences but a 165 bp spacer reduced silencing triggering efficiency (Rappleye et al., 2004). With this information available from other fungal species we considered that replacing the heterologous ascomycete intron by a Laccaria intron was justified for optimizing pSILBA vector structure for its use in this basidiomycete. We replaced the M. oryzae intron by the Laccaria nitrate reductase (NR) gene sixth intron of 52 bp together with 13 bp of surrounding exonic flanks. The construct was named pSILBAα. The same unique cloning sites for hairpin construction as in pSILBA were conserved (Fig. 1).
As the pSILBA full plasmid insertion in the T‐DNA of pHg was noticed to result always in a particular orientation accepted by E. coli the produced T‐DNA structure could be predicted (Fig. 2). The hph‐resistance cassette of pHg is driven by the same A. bisporus gpdII promoter as the silencing trigger cassettes in pSILBA and pSILBAα. The introduction of these plasmids into the T‐DNA results in an inverted repeated promoter sequence structure in pSILBA and pSILBAα (Fig. 2). This structure was known to become CpG methylated in RNA‐silenced Laccaria strains (Kemppainen et al., 2009). Whether this methylation was due to promoter sequence repetition per se by nuclear mechanisms that epigenetically modify repetitive sequences or it represented some spread methylation originated from the actively transcribed inverted repeat sequence as observed in Coprinopsis cinerea (syn. C. cinereus) (Wälti et al., 2006) was not known. It is possible that this promoter methylation has an interfering effect on the silencing trigger production and therefore on silencing efficiency. For instance, a single‐site CpG methylation within the p53 promoter has been reported to suppress its activity by 85% in mammalian cells (Pogribny et al., 2000). In order to study the possible effect of the inverted repeated promoter structure a third variant of pSILBA, pSILBAγ, was constructed by inverting the silencing triggering cassette in the plasmid backbone (Fig. 1). When pSILBAγ incorporates in pHg T‐DNA no inverted repeated promoter structure is formed. The two promoters in the T‐DNA are now in the same orientation and separated by silencing triggering and terminator sequences (Fig. 2).
Silencing of Laccaria NR by three pSILBA variants
The tree plasmid variants pSILBA (pS), pSILBAα (pSα) and pSILBAγ (pSγ) were compared by silencing Laccaria NR gene (protein ID 254 066) that allows an easy screening for silenced strains by affected growth on nitrate. Laccaria NR function is encoded by a unique gene in the genome hence is an excellent reporter for testing different RNA silencing constructs in this fungus. A 417 bp exonic sequence was used for triggering RNA silencing of the target gene (Fig. S2).
All pHg/pSILBA variants were assayed by using a dikaryotic L. bicolor strain as recipient for Agrobacterium transformation. All three constructs produced similar number of hygromycin‐resistant fungal strains indicating that the differences in the T‐DNA structures between the constructs did not affect the transformation efficiency (between 105% and 134% indicated by the number of growing points originated from the fungal colonies subjected to co‐cultivation). A preliminary evaluation of 60 strains from each construct for their growth on ammonium and nitrate solid media indicated that all constructs produced fungal strains with unaffected growth on ammonium and variable level reduced growth on nitrate. The reduced growth on nitrate was expressed as both reduced aerial and medium penetrating hyphae (data not shown).
Due to the low growth rate of L. bicolor several weeks of cultivation are needed for generating colonies big enough for visual evaluation on solid medium. The long cultivation periods can lead to variations on growth conditions between Petri dishes, especially when several strains with different growth capacities are inoculated on the same plate. To avoid these variations on solid medium a further analysis of 37 randomly selected fungal strains from each construct was carried out in liquid nitrate medium in microtitre plates. The growth was evaluated against the wild‐type strain after 2 and 4 weeks and scored as non‐affected (N), affected (A) and strongly affected (S) (Fig. 3A). After 2 weeks the growth in the controls started to saturate the wells and at 1 month all were fully grown. More variation was observed in the category affected (A) while strongly affected (S) was scored only for strains that did not generate significant hyphal growth from the initial fungal inoculum. No variation was observed between sample duplicates with any strain.
Figure 3.
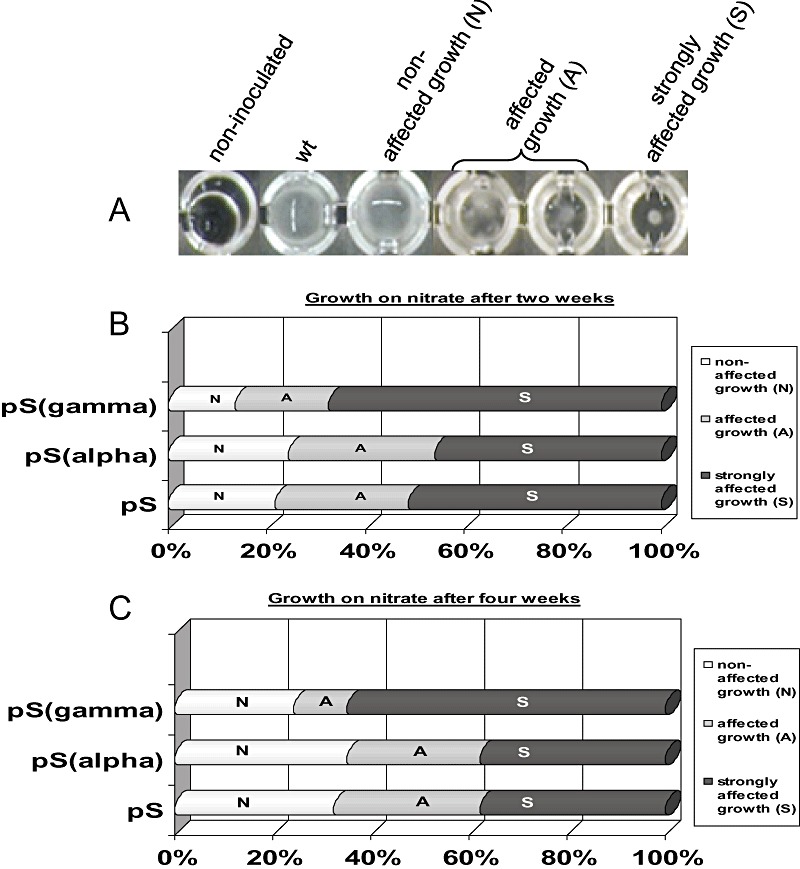
A. Growth of wild‐type dikaryon and transformed Laccaria strains after 1 month in nitrate medium in microtitre plates. Growth category group A is presented as duplicate due to wider variation within this group. B and C. Comparison of silencing efficiency between 37 randomly selected of each pHg/pSILBA/NITRLoop (pS), pHg/pSILBAα/NITRLoop (pSalpha) and pHg/pSILBAγ/NITRLoop (pSgamma) transformed strains. The growth on nitrate was compared with wild type after 2 weeks (B) and 1 month (C), and classified in three growth categories: non‐affected (N), affected (A) and strongly affected (S).
The two time point evaluations gave broadly similar results (Fig. 3B and C). The interesting observation was that the total number of pSγ strongly affected strains maintained during the complete observation period while with constructs pS and pSα some of the strongly affected (S) strains changed to the affected (A) category. Besides, the non‐affected (N) category increased. The same partial shift of A strains into N ones was also observed with the pSγ construct. After 1 month of cultivation the A and S strains represented altogether 67.6%, 64.9% and 75.7% of strains tested with constructs pS, pSα and pSγ respectively (Fig. S3A). These numbers can be considered the absolute silencing efficiency with the given constructs. The number of N strains seemed to be independent of the silencing construct used. While pSγ seemed to perform slightly better, pS and pSα produced similarly high silencing efficiency. Because the only difference between these constructs was the intron used, we conclude that the incorporation of a homologous intron did not improve the silencing efficiency of pSILBA in Laccaria. This could indicate that M. oryzae intron is recognized by Laccaria. Alternatively, the intron might not be spliced and its processing would not be important for improving the silencing efficiency in Laccaria. From our data we cannot rule out these options.
When the three constructs were compared for their capacity to generate strongly silenced strains, indicated by strongly affected growth on nitrate liquid medium, the superiority of pSγ became evident (Fig. S3A). While pS and pSα performed equally generating both 37.8% of S strains, with pSγ this category represented 64.9% of all the tested strains thus almost doubling what was observed with the two other constructs. The S strains generated by pSγ also keep up in this category during the whole culture period while the ones from the other constructs had tendency to shift to the A category.
Comparison of S strains produced by using pSγ and pSα constructs in microtitre plates showed that even though no remarkable mycelial growth was observed in the media fungal inocula from pSα often generated more mycelium than the ones from pSγ (Fig. S3B). Similar kind of observations was evident also during the preliminary screening on solid nitrate medium (data not shown). Considering the fact that the only difference between the pSγ and pSα construct was the orientation of the silencing triggering cassette in the T‐DNA, the variations in silencing efficiency observed and the growth pattern in liquid medium should be connected to this structural difference. The production of the silencing trigger in strongly affected strains generated with pSα construct did not seem to be as efficient as with pSγ.
One possible factor affecting the silencing efficiency could be a compromised transcription from the silencing triggering cassette due to the inverted repeated promoter structure in pSα transformants that is not present in pSγ (Fig. 4A and B). In order to reveal the epigenetic status of gpdII promoter sequences the CpG methylation was studied from two strongly affected pSα and pSγ strains. The genomic DNA was cut with methylation‐sensitive (HpaII) and ‐insensitive (MspI) restriction enzymes which both recognize the CCGG sequence. Southern blot data from the wild type and two transformants are presented in Fig. 4C. The experiment showed clearly that while the methylation of the gpdII promoter in pSγ transformed strains was minimal the pSα construct was strongly CpG methylated. This methylation may affect the transcription of the silencing trigger and would explain the lower performance of pSα compared with pSγ. The liquid medium growth experiment was carried out without hygromycin selection pressure while the mycelium for the methylation study was harvested from an antibiotic containing medium. It is possible that the lack of antibiotic in the growth medium could permit even stronger gpdII promoter methylation. With antibiotic pressure at least the hygromycin resistance cassette is forced to stay active and this could also affect the epigenetic state and transcription of the silencing triggering cassette. This possibility should be studied in future in order to asses the optimum conditions for screening of silenced Laccaria strains.
Figure 4.
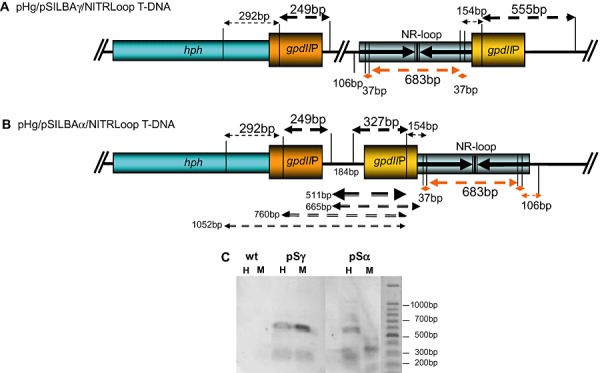
A and B. HpaII/MspI restriction map of gpdII promoters in pHg/pSILBAγ/NITRLoop (A) and pHg/pSILBAα/NITRLoop (B) T‐DNAs. CCGG restriction sites of HpaII (CpG methylation sensitive) and MspI (CpG methylation insensitive) within and around the promoter sequences are marked with vertical lines. Fragments from complete DNA digestion hybridizing to gpdIIP probe are indicated by arrows above the T‐DNAs. Detected fragments are marked with thicker arrows. Fragments from incomplete digestion of pHg/pSILBAα/NITRLoop with HpaII are indicated by black arrows below the T‐DNA in (B). Red arrows correspond to fragments from complete digestion of the nitrate reductase silencing trigger loop (not revealed in the Southern). The terminator elements of the T‐DNAs are not presented. C. Southern blot hybridized to gpdIIP probe. Results of one strongly silenced pSα, one strongly silenced pSγ and wild‐type strains are shown. If no CpG methylation is present in the target DNA both HpaII and MspI restrictions should result in identical hybridization signals. H: HpaII restriction. M: MspI restriction.
As a conclusion our evaluation on the three pSILBA constructs for their capacity to silence NR gene clearly indicates that for further silencing experiments in Laccaria and other homobasidiomycetes the vector of choice is pSILBAγ. This vector was further tested by targeting another Laccaria gene (see below).
Transgene integration in actively transcribed sites results in strong silencing phenotypes in Laccaria
Six pHg/pSγ/NITRLoop‐transformed Laccaria strains were used for further analysis including biomass production, genomic Southern blot, semi‐quantitative reverse transcription polymerase chain reaction (RT‐PCR), integration site by plasmid rescue and target endogene methylation. We selected one strain from N (strain 62), one strain from A (strain 32) and four strains from S (strain 1, 21, 26 and 30) categories. The growth on solid ammonium and nitrate media indicated that all the strains grew properly and equally on ammonium and the phenotypes on nitrate were the same to the observed in liquid medium (Fig. 5). The biomass production in liquid nitrate medium resulted in almost equivalent growth (72%) between the wild type and the strain 62 (N). Even though the affected strain 32 (A) produced visually more mycelium than the strongly affected strains on both solid and liquid medium (Fig. 5A and B), the mycelial dry weight was not distinguishable from the strongly affected strains which produced only 5.6–16.7% of the wild‐type dry weight (Fig. 5C).
Figure 5.
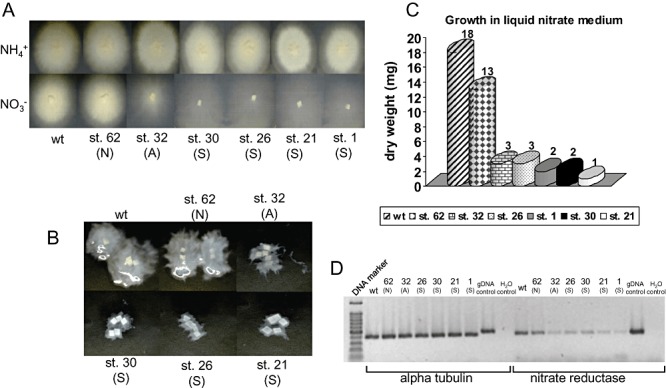
A. Laccaria wild‐type dikaryon and six pHg/pSILBAγ/NITRLoop‐transformed strains grown for 23 days on solid medium with ammonium or nitrate as N sources. Growth categories: N, non‐affected; A, affected; S, strongly affected. B. Growth of wild type and transformants in liquid nitrate medium after 22 days. C. Dry weight of mycelia produced by wild type and six pHg/pSILBAγ/NITRLoop‐transformed strains in liquid nitrate medium after 22 days. D. Reverse transcription polymerase chain reaction (RT‐PCR) expression analysis of L. bicolor nitrate reductase‐encoding gene (protein ID 254066). Total RNA from mycelia of Laccaria S238N wild type and six pHg/pSILBAγ/NITRLoop‐transformed strains was isolated and used for first‐strand cDNA synthesis. A PCR was performed with first‐strand cDNA as template and between 25 and 30 cycles of amplification with alpha tubulin (control gene, protein ID 192524) and nitrate reductase primers. The picture shows fragments amplified after 30 cycles of PCR. For details see Experimental procedures.
Semi‐quantitative RT‐PCR showed that the NR transcript levels correlated with the dry weight produced by the fungal strains (Fig. 5D). The strain 62 (N) had almost the wild‐type level of target gene transcript and the other studied strains showed a clear reduction of NR mRNA (approximately 10% of wild‐type level) connecting the observed affected growth phenotype to reduced target gene transcript level.
Southern blot analysis was carried out with five of these strains in order to establish a possible correlation between the T‐DNA copy number and the silencing strength. Laccaria genomic DNA was hybridized with an amp probe that detects the pSILBA component in the T‐DNA. The T‐DNA was present in all studied strains as full size (minimum size expected 3.14 kb) and mostly as a single‐copy integration (Fig. 6). No correlation was observed between the silencing trigger copy number and the silenced phenotype as most of the strongly silenced strains carried a clear single integration. Only the strain 26 (S) presented a putative second full‐size T‐DNA integration.
Figure 6.
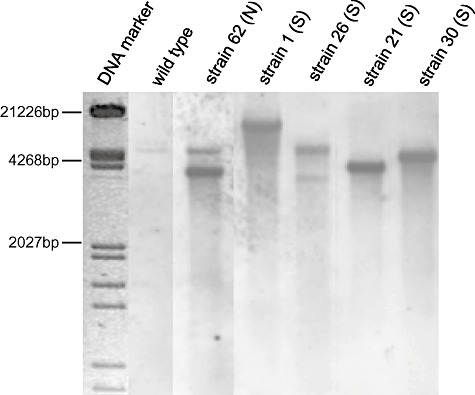
Southern blot of wild type and five pHg/pSILBAγ/NITRLoop transformants. Genomic DNA was cut with BamHI, blotted and probed with amp probe. N: non‐affected growth; and S: strongly affected growth on nitrate medium. DNA marker: λ/EcoRV/HindIII.
In order to resolve the genomic integration sites of the silencing triggering constructs in the six chosen fungal transgenic strains the T‐DNA RB–gDNA junctions were plasmid rescued and sequenced. All the strains produced plasmids replicable in E. coli under ampicillin selection. The strain 26 (S) produced plasmids of two different sizes which were in concordance with the Southern blot data suggesting that this strain harbour two independent T‐DNA integrations. The growth of this double integration strain on nitrate was however not noticed to be more affected than other single‐integration transgenic strains indicating that the transgene copy number is not a factor affecting Laccaria RNA silencing strength. The rescued plasmids were sequenced with M13‐reverse primer and genomic sequences of the T‐DNA integration sites were recovered for five fungal strains belonging to N, A and S growth categories. The genomic sites were evaluated for the following features: the presence or absence of a putative protein coding sequence, EST evidence for the gene, the exact integration site in the gene (exon, intron, UTR integration, putative promoter or terminator integration) and in the case of gene integration the silencing cassette orientation with respect to the interrupted gene promoter (Table S1). The genomic sequences revealed that all the S category fungal strains carried at least one T‐DNA integration in a protein‐coding gene with EST support. The only strains with intergenic integrations were the N and A category strains 62 and 32. The gene integrations were in putatively transcribed regions of the genes, but the majority (three‐fourths) in opposite orientation to the transcription of the interrupted gene. This suggests that the strongly silenced phenotype was not a result of the higher silencing triggering transcription from endogenous Laccaria promoters and instead depended on other characteristics of the integration sites. From these data we conclude that the transcriptional activity of the integration site, and not the integration copy number, of the silencing triggering construct is fundamental for the strong silencing phenotype in Laccaria. This may indicate that an actively transcribed genomic site has an open chromatin structure which creates the optimal transcription conditions also for the production of the silencing trigger. Observation of the intergenic genomic integration sites of the N and A category strains did not reveal correlation between the two categories. Actually the N strain 62 had an integration in a gene‐richer zone than the A category strain 32. The former carried its T‐DNA inserted 414 bp downstream of a gene with EST support (protein ID18940) and 341 bp upstream of a putative ORF (protein ID 315853) while the latter had the silencing triggering cassette inserted 1902 bp upstream (protein ID 334135) and 5179 downstream (protein ID 396132) of genes with EST support.
IhpRNA‐triggered silencing and target endogene methylation status in Laccaria
We previously detected target endogene CpG methylation in NR‐silenced dikaryotic Laccaria strains. This methylation was restricted to the gene sequence present in the hairpin silencing trigger and did not affect the entire genomic DNA (Kemppainen et al., 2009). The RNA silencing in that study was produced with a trigger that did not carry a spliceable intron. We were interested to see if the same pronounced target gene methylation was present in fungal strains where RNA silencing was launched with an ihpRNA. The study was performed with HpaII and MspI enzymes that recognize the same sequence but show different methylation sensitivity using two strongly silenced strains obtained with pSα and pSγ. A map corresponding to the NR gene with the HpaII restriction sites and three different genomic NR probes used together with the fragment corresponding to hpRNA trigger are presented in Fig. 7A. The hybridization signals rising from a complete DNA digestion (solid arrows) and possible signals from methylated NR gene digested with HpaII (dashed arrows) are indicated in the figure. The NR probes 1 and 2 hybridize also with the silencing triggering construct (683 bp) (Fig. 4) while the probe 3 detects only the target endogene (1448 bp) when unmethylated. In the case of endogene methylation, within the sequence used for constructing the silencing trigger, we would expect signals corresponding to fragments bigger than 1448 bp in HpaII digestions (1485 bp, not distinguishable from 1448 bp; and 1832 bp). If the methylation was present also outside the NR sequence used in the transgene construct a bigger fragment (such as 2271bp) should also be detected in HpaII digestions. The hybridization results of wild type, one pSα and one pSγ transformants are presented in Fig. 7B–D. None of such fragments was present (not shown). However, a very low level of CpG methylation in the target endogene cannot be excluded and in fact it may occur as differences in lower‐size fragment signals between the wild type and transgenic strains are observed. On the other hand, Fig. 7B and C shows that methylation of the transgene hairpin occurred.
Figure 7.
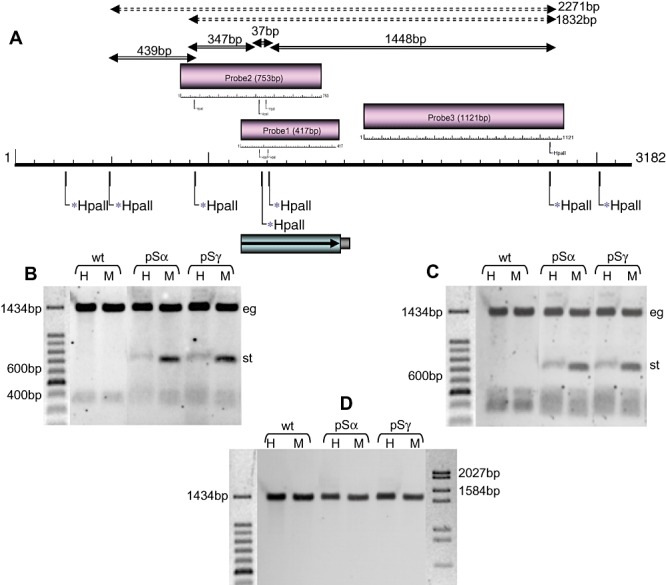
A. HpaII (CpG methylation sensitive)/MspI (CpG methylation insensitive) restriction map of the Laccaria nitrate reductase gene. The probes used for hybridization (1–3) and expected fragments from complete digestion of the DNA are represented above the gene with solid two‐headed arrows (37 bp under the detection level). The two smallest possible fragments originating from CpG‐methylated DNA digested with HpaII are represented by dashed two‐headed arrows. The 417 bp sequence used for triggering RNA silencing is presented below the gene. B–D. CpG methylation analysis of the Laccaria nitrate reductase gene in wild type and in silenced strains. (B) NR probe 1. (C) NR probe 2. (D) NR probe 3. pSα refers to a strain transformed with pHg/pSILBAα/NITRLoop construct. pSγ refers to strain transformed with pHg/pSILBAγ/NITRLoop construct. H: HpaII restriction. M: MspI restriction. eg; main endogene signal. st: main silencing trigger signal.
In a previous work (Kemppainen et al., 2009) we observed a more marked endogene methylation by using a non‐intronic hairpin construct for RNA silencing. Thus our data would indicate that nuclear epigenetic modifications occur when the silencing is launched with a hairpin that does not carry an intronic spacer. This effect could be related to a more prolonged presence and higher concentration of the silencing triggering RNA within the nucleus when it is not exported to the cytoplasm as efficiently as an intron containing silencing trigger.
Silencing of Laccaria synaptojanin‐like 5‐Pase gene
We wanted to test the pHg/pSILBAγ plasmid system with another Laccaria endogene. A random T‐DNA integration study carried out in our laboratory on monokaryotic Laccaria strain H82 produced a fungal strain with a remarkable phenotype. No smooth aerial hyphae were formed and the branching pattern was clearly altered in this T‐DNA‐tagged strain. A young colony shows straight poorly branched hyphae in the border. More remarkably the whole colony of the mutant strain folds in a spiral like pattern (Fig. 8A). This fungal strain carries a single T‐DNA integration in a gene homologous to an inositol‐1,4,5‐triphosphate 5‐phosphatase (5‐Pase) which is a synaptojanin‐like protein (protein ID 306121). A RT‐PCR study showed that this gene is strongly expressed in the wild type when growing on ammonium while in the strain with the gene interruption the messenger is absent (data not shown).
Figure 8.
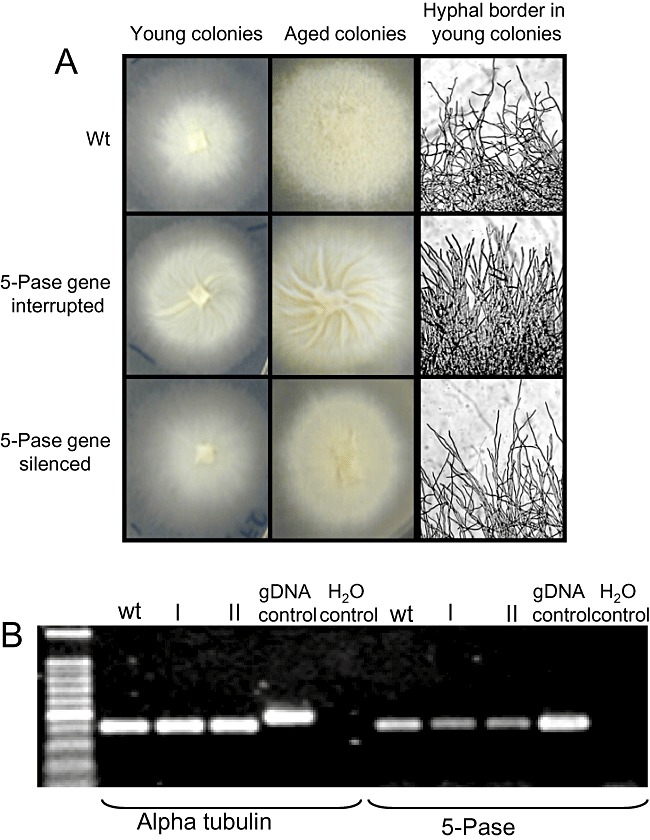
A. Laccaria wild‐type monokaryon (wt), mutant with 5‐Pase gene interruption and 5‐Pase‐silenced strains. B. Reverse transcription polymerase chain reaction (RT‐PCR) expression analysis of L. bicolor synaptojanin‐like 5‐Pase‐encoding gene (protein ID 306121). Total RNA from mycelia of Laccaria H82 wild type and two pHg/pSILBAγ/INOLoop transformants (I, II) was isolated and used for first‐strand cDNA synthesis. A PCR was performed with first‐strand cDNA as template and between 25 and 30 cycles of amplification with alpha tubulin (control gene, protein ID 192524) and 5‐Pase gene primers. The picture shows fragments amplified after 27 cycles. For details see Experimental procedures.
The synaptojanin proteins are phosphatases that participate in turnover and control of cellular distribution of phosphatidylinositols (PIs). These compounds control several cellular processes including cell signalling and growth, vesicular trafficking, transcription and actin cytoskeletal arrangements (Fruman et al., 1998; Martin, 2001; Simonsen et al., 2001; Stefan et al., 2002). Especially phosphatidylinositol (4,5)‐biphosphate (PIP2) has been shown to control several cellular processes such as actin organization and membrane trafficking via PIP2‐binding proteins (Janmey, 1994; Martin, 2001). These signalling effects of PIP2 are initiated by specific kinases and ended by synanptojanin PI 5‐phosphatases in mammalian cells. These are proteins that carry an N‐terminal SacI‐like PPIPase domain and a C‐terminal 5‐Pase domain. The ascomycete Saccharomyces cerevisiae has three structurally mammalian‐like synaptojanins (Inp51p, Inp52p and Inp53) and the fourth PI 5‐Pase, Inp54, that only has the 5‐Pase activity domain (Guo et al., 1999; Wiradjaja et al., 2001) (Fig. S4). While single deletions of these genes are viable the simultaneous deletion of all three synaptojanin‐like genes is lethal in yeast indicating an essential cellular function of these proteins (Srinivasan et al., 1997; Stolz et al., 1998).
Laccaria genome encodes one structurally similar to yeast and mammalian synaptojanin‐like gene (protein ID 183121). In other filamentous fungi, both ascomycetes and basidiomycetes, such as N. crassa, Trichoderma reesei and P. chrysosporum, only one yeast‐like synaptojanin with both SacI‐like and 5‐Pase domains can be found (protein ID NCU03298.3, 122419 and 129093 respectively). However, three putative synaptojanin‐like N‐terminal (protein ID 305813, a SAC1‐like protein; ID 243126, putative Laccaria SAC1 homology; ID 184407, putative Laccaria FIG4 homology) and three Pase‐domain‐encoding genes (protein ID 297312, 306121 and 295589) are present in Laccaria genome. Of these Pases two are putative synaptojanin‐like 5‐Pases (protein ID 297312 and 306121) (Fig. S5). In our Laccaria mutant strain one of these synaptojanin‐like 5‐Pase‐encoding genes (protein ID 306121), which is proposed to participate in PI turnover and control, is interrupted. The detected phenotype with an altered hyphal growth pattern is therefore most probably related to an altered cytoskeletal assembly or vesicle trafficking. Because the 5‐Pase gene interruption had produced an easily detectable growth phenotype this sequence was used for RNA silencing with pHg/pSILBAγ vector system in Laccaria H82 monokaryotic strain. A 387 bp armed dsRNA trigger cassette was constructed in pSILBAγ. Forty hygromycin‐resistant fungal strains were evaluated for an expected phenotype in presence and absence of antibiotic pressure. Ten strains (20%) with slight reduction on aerial hyphae production were detected. These strains showed also some reduction on hyphal branching compared with wild type (Fig. 8A). No fungal strains with exactly the same phenotype as the gene‐interrupted Laccaria strain were detected. Two transformants with an altered phenotype were subjected to semi‐quantitative RT‐PCR analysis and the results showed the silencing of the target gene launched by the pHg/pSILBAγ construct (Fig. 8B).
Discussion
We have recently demonstrated RNA silencing in a mycorrhizal fungus for the first time (Kemppainen et al., 2009). However, in order to exploit RNA silencing to its full extent for further gene function studies in Laccaria and other basidiomycetes silencing vectors functional in this group of fungi should be developed. Recent advances in AMT of fungi have made possible the genetic manipulation of several members of filamentous basidiomycetes, these including Laccaria (Kemppainen et al., 2005). Therefore, RNA silencing tools adapted to function in concert with AMT were especially needed.
Double‐stranded RNAs are highly efficient triggers of RNA silencing in plants, animals and fungi (Smith et al., 2000; Tavernarakis et al., 2000; Kalidas and Smith, 2002; Paddison et al., 2002; Kadotani et al., 2003; Goldoni et al., 2004). In this article we have described the pHg/pSILBAγ‐AMT plasmid system which offers an efficient RNA silencing triggering by ihpRNA expression in Laccaria. It is most likely applicable to other hygromycin‐sensible homobasidiomycetes with functional RNA silencing machinery. Besides AMT the vector system described here can also be used for direct transformation of protoplast if that methodology is routinely used with the fungal species under study.
The hairpin arms in pSILBAγ are created by PCR and carry appropriated restriction sites for cloning. This can be accomplished with a minimum of three primers, one of these being a blunt‐end site primer, and the hairpin can be completed with two oriented ligation reactions. The length of the hairpin arms used for efficient triggering of RNA silencing varies between fungal species and the target gene. Most studies have found that dsRNA arms of 300–700 bp are highly efficient even though shorter dsRNAs have been used for launching RNA silencing in fungi (Goldoni et al., 2004; Rappleye et al., 2004; Heneghan et al., 2007; Yamada et al., 2007). The silencing efficiencies obtained with these shorter triggers have however been significantly lower than with longer dsRNAs. This could indicate that fungal Dicers have lower activity on shorter dsRNAs or that the population of siRNAs generated from longer dsRNAs is higher in number and thus can act more efficiently on the target mRNA. Human Dicer has been shown to have higher activity on 500 bp than on 200 bp dsRNAs (Bernstein et al., 2001). However, a very recent report on C. cinerea shows that as short as 19 bp dsRNA trigger can initiate a specific RNA silencing response (Costa et al., 2008). This would indicate that fungal cells posses a Dicer‐like activity that can process as short as 19 bp hpRNAs into siRNAs. Therefore, the limiting step with shorter dsRNAs as silencing triggers may be more related to their expression level and thus to the cytosolic concentration of siRNAs targeting the homologous mRNA and not to the incapacity of Dicer to act on short dsRNAs. We have cloned variable length‐inverted repeated loops in pSILBAγ. This plasmid tolerates well repetitions of 800 bp while loop arms of 1000 bp seem to generate some structural vector instability and thus are not recommended.
The third ligation of the complete pSILBAγ‐hairpin plasmid into the Agrobacterium binary vector pHg joins the hairpin expression cassette and the hygromycin resistance cassette in the binary vector's T‐DNA. This ligation is also oriented by the compatibility of the two vectors and the selection of the correct ligation products in E. coli is enhanced by the use of two different antibiotic resistances, kanamycin and ampicillin, from both vector's backbones. It is worth mentioning that despite the relative big size of the produced pHg/pSILBAγ transformation vectors (minimum size 12.5 kb) they purify with standard plasmid miniprep protocols as efficiently as the smaller‐size pSILBAγ making their manipulation easy. The Agrobacterium strain used in this study, AGL1, is resistant to ampicillin and the transformation vectors were maintained just under kanamycin selection. The plasmids are stable under single antibiotic selection and can be purified efficiently also from Agrobacterium with standard plasmid miniprep protocols used for E. coli. Once integrated into the fungal genome part of the T‐DNA of pHg/pSILBAγ can be recovered together with the gDNA–RB junction by plasmid rescue and sequenced with universal primers.
We analysed the pHg/pSILBA system by evaluating three different pSILBA variants (pSILBA, pSILBAα and pSILBAγ) for their capacity to silence NR (protein ID 254066). These three constructs varied in the intron spacer used and the orientation of the silencing cassette with respect to the hygromycin resistance cassette after ligation with pHg. The pSILBAγ performed clearly better than the two other plasmid variants. This was traced back to nearly CpG methylation‐free promoter of the silencing cassette in genome integrated T‐DNAs of pHg/pSILBAγ. This most likely resulted in a maximum dsRNA trigger production from pSILBAγ. On the other hand, both endogenic and heterologous intron spacers performed equally well and only a minimum CpG endogene methylation, corresponding to the sequence used for constructing the silencing trigger, was detected in strongly silenced fungal strains. Besides, no promoter sequences were used in silencing triggering thus most likely the detected reduction on target gene mRNA level in this study would be due to a post‐transcriptional gene silencing mechanism (PTGS) rather than to transcriptional gene silencing (TGS). This differs from our previously reported more pronounced target gene methylation in Laccaria, triggered with hpRNA separated by a non‐intronic spacer (Kemppainen et al., 2009). In that case a possible participation of TGS mechanisms cannot be excluded at least for the transformed nucleus in the dikaryon.
The RNA silencing machinery has been demonstrated to be needed for heterochromatin formation in plants, animals and fungi (for review see Lippman and Martienssen, 2004). While the role of the RNA silencing pathway in recognition and in epigenetic modification of genomic and transgene repetitions has been widely studied in the fission yeast S. pombe and in the filamentous fungus N. crassa (Tamuru and Selker, 2001; Selker et al., 2002; Volpe et al., 2002; Schramke and Allshire, 2003; Freitag et al., 2004; Chicas et al., 2005; Bühler and Moazed, 2007), data from other filamentous fungi are sparse. The silencing locus and target gene sequence methylation as a response to RNA silencing seem to vary between species. In the filamentous ascomycete M. oryzae RNA silencing of eGFP triggered by dsRNA expression does not induce de novo cytosine methylation in CCGG sequences of the target transgene or in the silencing locus (Kadotani et al., 2003). However, in the homobasidomycete C. cinerea constitutive dsRNA‐triggered silencing of both eGFP transgene and target endogene correlates with increased cytosine methylation of the silencing trigger and target gene sequence (Wälti et al., 2006). Transgene‐triggered cytosine methylation of the silencing locus and the target gene has been reported also in Schizophyllum commune (Schuurs et al., 1997). We have observed a variation on the target endogene methylation status related to the presence or absence of intronic sequences in the hpRNA trigger in Laccaria (Kemppainen et al., 2009; this paper). An intron‐free trigger seems to launch more marked nuclear epigenetic modifications, detected as CpG methylation of the target gene genomic sequence, than an ihpRNA. Similar kind of response has been reported in the fission yeast S. pombe in which the presence or absence of an intronic sequence in a gene‐coding sequence dsRNA has been connected to possible epigenetic effects. While an intron containing silencing trigger clearly initiated post‐transcriptional silencing (Sigova et al., 2004) a construct with a coding sequence repeat separated by a non‐intronic spacer was reported to result in transcriptional silencing of the target gene (Schramke and Allshire, 2003). Moreover, the presence of an intronic sequence has been shown to improve the silencing efficiency in plants and animals (Smith et al., 2000; Kalidas and Smith, 2002). This phenomenon has been proposed to be related to a possible higher nuclear accumulation of an intron‐free trigger while the intron containing trigger would be more efficiently exported to the cytoplasm via the nuclear splicing pathway. In filamentous fungi and yeasts efficient RNA silencing has been triggered with dsRNAs carrying both non‐intronic (Liu et al., 2002b; Kadotani et al., 2003; Schramke and Allshire, 2003; Fitzgerald et al., 2004; Goldoni et al., 2004; Mouyna et al., 2004; Rappleye et al., 2004; Hammond and Keller, 2005; McDonald et al., 2005; Nakayashiki et al., 2005; Namekawa et al., 2005; Tanguay et al., 2006; Bohse and Woods, 2007; Eastwood et al., 2008; Kemppainen et al., 2009) and intronic spacers (Goldoni et al., 2004; Sigova et al., 2004; Nakayashiki et al., 2005; Cardoza et al., 2006; Gong et al., 2007; Moriwaki et al., 2007; Matityahu et al., 2008). Also constructs carrying recognizable introns outside the dsRNA spacer have been used for efficient RNA silencing in S. commune, C. cinereus and Ophiostoma piceae (de Jong et al., 2006; Tanguay et al., 2006; Wälti et al., 2006). Even though strong silencing phenotypes have been obtained in fungi with both intronic and non‐intonic spacers of hpRNAs, it is not conclusive whether the cellular processes responsible for the observed phenotypes with different triggers are the same. PTGS is characterized by siRNA‐dependent destruction of the target transcripts and TGS by reduction of the gene transcription due to RNA silencing pathway‐mediated epigenetic modifications of the target gene genomic sequence. These are usually triggered by dsRNAs carrying sequences homologous to gene promoter sequences. The fungal silencing studies published thus far have used hpRNA trigger homologous to target gene‐coding regions. However, transcriptional activity studies on the target genes in silenced fungi have not been performed and siRNAs are rarely demonstrated. Moreover, when siRNAS are detected their abundance is not compared with the strength of the silencing phenotype. Therefore, it is possible that simultaneous involvement of both PTGS and TGS mechanisms in RNA silencing of fungi is more common than originally thought. An example of this comes from the oomycete Phytophtora infestans where RNA silencing triggered with ihpRNA has recently been shown to result in partially silenced phenotypes with PTGS characteristics and strongly silenced phenotypes produced by TGS. Also the amount of siRNAs did not correlate with the strength of the partial silencing phenotypes and a mix of PTGS and TGS mechanisms was proposed (Audrey et al., 2008). If a similar simultaneous PTGS/TGS mechanism is active in silenced fungi should be further studied.
Besides the characteristics of the silencing triggering construct itself and its effect on silencing efficiency, we focused on another well‐known but less studied aspect of RNA silencing: the silencing strength variation (SSV) observed among different strains carrying the same silencing trigger. All RNA silencing studies, in fungi or in other eukaryotes, where gene knock‐down is launched from stable silencing trigger expression, show variation on the level of silencing between independent transformants. The reason for this phenomenon is not well understood and different factors have been proposed to be responsible for SSV. These include the nuclear transgene copy number, different epigenetic effects related to transgene integration site (euchromatin versus heterochromatin) and post‐transformational modifications and genomic rearrangements that might affect the transgene integrity. Silencing strength variation has been reported in yeast and filamentous fungi and both with integrative and auto‐replicative silencing vectors (Liu et al., 2002a;Kadotani et al., 2003; Rappleye et al., 2004; Tanguay et al., 2006; Bohse and Woods, 2007). It is also possible that foreign DNA, independently of its localization in cells, is destined to be detected and modified, for example by methylation. These modifications could occur to a different extent in different strains resulting in variable silencing trigger production and SSV. A variable transgene copy number could theoretically generate variable cellular silencing trigger production and affect silencing strength. However, non‐silenced and silenced fungal strains are often reported to carry full‐size silencing vectors and the copy number of silencing triggering constructs do not necessarily correlate with higher degree of silencing (Nicolás et al., 2003; Fitzgerald et al., 2004; Nakayashiki et al., 2005; Tanguay et al., 2006; Wälti et al., 2006; Henry et al., 2007; Moriwaki et al., 2007; Yamada et al., 2007; Oliveira et al., 2008). It is thus unlikely that the silencing trigger copy number is a fundamental factor causing SSV in fungi. Our previous observations indicated that a single T‐DNA transgene integration could produce a silencing phenotype as strong as double transgene integration in Laccaria (Kemppainen et al., 2009). An analysis of genomic integration sites of three strongly silenced Laccaria strains revealed that all of them carried the silencing triggering cassette in a putative gene (Kemppainen et al., 2009). These findings were of special interest because they suggested that the silencing cassette integration in active genomic sites could be one of the factors causing SSV in this fungus. Further confirmation of the transgene site effect could come however from resolving more genomic integration sites in silenced Laccaria strains. This type of study has not been yet conducted on fungi, and to our knowledge, neither on other silenced eukaryotes. Methodologies that utilize direct introduction of transgenes such as electroporation, PEG‐assisted protoplast transformation and particle bombardment generally result in several copies, and frequently in tandem repeats of the transgenes. These transformation methods are traditionally used in genetic manipulation of fungi, especially on filamentous model ascomycetes. However, resolving transgene integration sites in strains that carry multiple transgene copies is not easy. Moreover, the obtained information would be rather difficult to relate to SSV. On the other hand, AMT usually results in single T‐DNA integrations in L. bicolor and these can be easily resolved by plasmid rescue from strains transformed with the pHg/pSILBA system. In this work we resolved genomic integrations of non‐silenced, intermediately silenced and strongly NR‐silenced fungal strains. All studied strongly silenced strains had their T‐DNA integration within a gene with EST support while intermediately silenced and non‐silenced strains had intergenic integrations. Together with our previous results (Kemppainen et al., 2009) it becomes clear that for the maximum silencing effect the transgene ought to integrate in an actively transcribed genomic region, preferably within a transcriptional unit in Laccaria genome.
Taking into account the importance of the active transcriptional status of genomic integration sites, transformation methods that could direct transgenes to transcriptionally active sites would have higher probability in creating strongly silenced fungal strains. The T‐DNA integration bias to active genomic regions is well known from studies in plants. An analysis of T‐DNA insertion site sequences in Arabidopsis indicates that the density of T‐DNA insertion events correlates with gene density along Arabidopsis chromosomes (Alonso et al., 2003). Part of this bias was later on explained by the selection pressure generated by the use of an antibiotic resistance marker which can hinder the recovery of non‐resistant plant lines that carry epigenetically silenced T‐DNA integrations in transcriptionally inactive genomic regions (Francis and Spiker, 2005). We have previously studied the T‐DNA integration pattern in Laccaria and detected predominantly single integrations that were also biased towards putatively active genomic sites like in plants (Kemppainen et al., 2008). A similar bias was reported also for another fungus, Leptosphaeria maculans, where the T‐DNA preferentially integrated in gene‐rich regions of the fungal genome (Blaise et al., 2007). Considering the information on the T‐DNA integration bias in Laccaria and the data on the effect of the integration site on silencing strength, the use of AMT seems an optimal strategy for obtaining strongly silenced single integration carrying fungal strains.
We demonstrated the functionality of pHg/pSILBAγ with two Laccaria endogenes, NR (protein ID 254066) and a synaptojanin‐like 5‐Pase (protein ID 306121). The silencing of NR in a dikaryotic background resulted in a high percentage of strongly silenced fungal strains (65%) while the silencing of the 5‐Pase gene in a monokaryotic strain produced intermediately silenced strains (20%) and no strongly silenced strains were detected. The difference observed with the silencing efficiency between the two genes tested was surprising as both silencing constructs carried a hairpin of the same size range (NR: 417 bp, 5‐Pase: 387 bp). Also with both genes the hairpins were designed to target the central part of the transcripts and the silencing trigger for 5‐Pase carried a higher percentage of its complete target transcript sequence (30%) than the NR trigger (16%). Similar variation of silencing efficiency in targeting different genes has been reported also in other fungi (Mouyna et al., 2004). Most intriguing is the fact that some genes seem to be resistant to RNA silencing (Fraser et al., 2000). Later on we have also efficiently silenced another Laccaria gene with the pHg/pSILBAγ system obtaining strongly silenced transgenic strains (M.J. Kemppainen and A.G. Pardo, unpubl. data).
Because the silencing cassette in pSILBAγ contains a multiple cloning site between a basidiomycete promoter and a fungal terminator, it can be used also as a gene expression vector. We have successfully expressed a heterologous gene in Laccaria by using the pHg/pSILBA vector system (M.J. Kemppainen and A.G. Pardo, unpubl. data). Also the presence of an intron in the pSILBA could be useful for gene expression experiments. It is known that an intronic sequence is often needed for an efficient transgene expression in homobasidiomycetes (Lugones et al., 1999; Ma et al., 2001; Scholtmeijer et al., 2001; Burns et al., 2005; Yamazaki et al., 2006).
RNA silencing makes possible to study fungal genes, whose knockouts would be lethal, in combination with inducible silencing trigger promoters (Mouyna et al., 2004). Moreover, in many filamentous fungi in which gene knockout studies are difficult to perform due to low rate of homologous recombination, RNA silencing offers a fast approach for studying gene function, also of genes with limited sequence information. Moreover, simultaneous silencing of gene families sharing sequence homology is possible (Wälti et al., 2006). While this is a benefit it is also the biggest drawback of the RNA silencing technology. Unwanted off‐site effects originated from even short sequence homologies are possible. These off‐site effects were recently studied by Nguyen and colleagues in RNA‐silenced M. oryzae. Genes sharing sequence similarity with an E‐value of only about e−3 or e−4 were sometimes silenced at similar levels and only 13 nt perfect homology between genes was enough for simultaneous silencing of an unwanted target (Nguyen et al., 2008). Also in HeLa cells only a 7 nt sequence complementarity between the unwanted target and a siRNA or a short‐hairpin RNA trigger has been reported to launch off‐site effects (Jackson et al., 2006). Special attention should thus be pay in future for designing RNA silencing triggers. It seems that RNA silencing is not as sequence specific as it was previously believed and probable off‐site effects cannot be completely avoided.
We were able to detect only intermediate silencing of Laccaria 5‐Pase (synaptojanin‐like) gene. One reason for the absence of strongly silenced fungal strains could come from possible off‐target effects launched by the silencing trigger. In yeast knockout of all synaptojanin‐like genes is lethal. If our target gene silencing could cause simultaneous silencing of other Laccaria synaptojanin‐related 5‐phosphatases this would most probably lead to reduced viability. Strong silencing of the target gene maintained by a constitutive expression of the silencing trigger would also generate more pronounced off‐site effect. This could explain the lack of strong silenced strains. Comparison of nucleotide sequences of three Laccaria synaptojanin‐like 5‐Pase revealed that even though no long exact match exists between our gene of interest (protein ID 306121) and two other synaptojanin‐like 5‐Pase domain genes (protein ID 183121 and 297312), 7 nt perfect matches surrounded by partially matching nucleotides were found between the target gene and both of the homologous genes. Also longer near‐perfect matches were detected. These could theoretically create conditions for a cascade of off‐site effects on all the Laccaria synaptojanin‐like genes. We were not expecting this type of response and the preliminary screening of the strains was performed based on the altered but strongly growing phenotype similar to target gene disruption. Strongly silenced and weakly growing strains could have been masked by the less silenced and stronger growing strains during transformants selection.
Another important aspect considering possible silencing trigger off‐site effect in fungi is transitive silencing. The secondary cascade of siRNAs, produced by RdRP activity from mRNA sequences outside the target sequence, has been demonstrated in plants, C. elegans and in the zygomycete Mucor circinelloides (Sijen et al., 2001; Braunstein et al., 2002; Han and Grierson, 2002; Klahre et al., 2002; Sanders et al., 2002; Vaistij et al., 2002; Alder et al., 2003; Nicolás et al., 2003; Van Houdt et al., 2003). These secondary siRNAs can act on other mRNAs sharing sequence homology and result in simultaneous silencing of several genes independently of the primary silencing trigger sequence used. In plants transitive silencing acts mostly on transgenes (Vaistij et al., 2002; Himber et al., 2003; Kościańska et al., 2005; Miki et al., 2005; Petersen and Albrechtsen, 2005) while in C. elegans and in Mucor both transgene and endogene are targets of the secondary siRNAs (Sijen et al., 2001; Alder et al., 2003; Nicolás et al., 2003). The existence of transitive silencing in different fungal species has not yet been studied. However, filamentous basidiomycetes seem to have a higher number of RdRPs, proteins shown to be responsible for transitive silencing in plants and C. elegans, compared with ascomycetes (Nakayashiki and Kadotani, 2006). Also in Laccaria genome the abundance of silencing machinery proteins is higher encoding for six putative RdRPs‐like proteins (Kemppainen et al., 2009). This variability on the RNA silencing protein repertoire could indicate that also the capacity for transitive silencing varies within the fungal kingdom. If the set of numerous RdRPs in basidiomycetes is involved in production of secondary siRNAs from endogene mRNAs like in the zycomycete, Mucor should be investigated in order to evaluate the true RNA silencing trigger sequence/target gene specificity in Laccaria. Nevertheless, in organisms where targeted gene disruptions are difficult to produce RNA silencing offers a way for gene function studies which were not possible before.
Experimental procedures
Fungal and bacterial strains, and growth conditions
Laccaria bicolor (Maire) Orton dikaryotic strain S238N and monokaryotic strain S238N‐H82 (Crater Lake National Park, Oregon) were maintained on solid modified P5 (MP5) medium at 22°C and transformed with Agrobacterium tumefaciens strain AGL‐1 according to Kemppainen and colleagues (2005). Both dikaryotic and monokaryotic transformed strains were passed through one selection round on 600 µg ml−1 cefotaxime and 300 µg ml−1 hygromycin B (Invitrogen) and two rounds on 150 µg ml−1 hygromycin before further phenotypic and molecular evaluation. Escherichia coli strain TOP10 (Invitrogen) was used for molecular cloning. Plasmids were introduced into E. coli and A. tumefaciens by a standard electroporation protocol (Sambrook et al., 1989).
PCR conditions
PCR reactions in this work were performed with a T‐personal thermocycler (Biometra®) by using PCR chemicals from Fermentas. The conditions for 20–30 µl reaction volume were: 1× reaction buffer with (NH4)2SO4; dNTPs, 200 µM each; MgCl2, 1.5 mM; primers, 1 µM each; and 1 U Taq DNA polymerase. The thermocycling programmes consisted of an initial cycle of 94°C for 5 min, followed by 25–30 cycles of 94°C for 30 s, annealing temperature adequate for each set of primers, 30 s (temperatures are mentioned elsewhere), elongation at 72°C for 30 s per 500 bp amplicon and a final extension at 72°C for 5 min. PCR reactions were separated in 1% agarose gels, stained with ethidium bromide and observed under UV light. When needed DNA was purified from agarose gels with the QIAEX®II Gel extraction kit (Qiagen).
Plasmid constructs
The ligation reactions were performed with T4 DNA ligase and reagents from Promega. The pSILBA (SIL, silencing; BA, basidiomycetes) cloning vector was constructed by modifying pSilent‐1, a pBluescript SK+ II (Stratagene) based RNA silencing trigger cloning vector designed for ascomycetes (Nakayashiki et al., 2005). First the pSilent‐1 hygromycin resistance cassette was removed with NotI followed by replacing the A. nidulans trpC promoter by the A. bisporus gpdII promoter. Agaricus promoter was amplified from plasmid pBGgHg (Chen et al., 2000) with primers gpdFH/gpdRC (5′‐GAAGAAGCTTTAAGAGGTCCGC‐3′/5′‐CAATCGATGGCGATAAGCTTGTTGTG‐3′; HindIII restriction sequence underlined) at 60°C of annealing temperature and cut with HindIII. The A. nidulans promoter in pSilent‐1 was removed with SpeI and ClaI and the Agaricus promoter was blunt end‐ligated in the vector creating pSILBA (Fig. 1C). For hairpin cloning the sequence between the promoter and M. oryzae CUT intron includes unique SalI, XhoI, SnaBI and HindIII sites while the 3′ multiple cloning site between the spacer intron sequence and the trpC terminator has unique BglII, SphI, StuI, KpnI and ApaI sites (Fig. 1A). The restriction sites SacI and NotI are unique in the vector and can be used for linearization of pSILBA, and XbaI can be used for liberating the silencing triggering cassette from the vector (Fig. 1B). When designing the inverted repeated loop structure attention should be pay as the sequences used must be free of SacI or NotI restriction sites in order to introduce pSILBA into pHg.
The pSILBAα vector was made by modifying pSILBA further by replacing the M. oryzae CUT intron by L. bicolor NR sixth intron. This Laccaria intron consists of 52 bp and was introduced in pSILBA together with 13 bp of neighbouring exonic sequences on both sides of the intron. The original pSilent‐1 intron was excised from pSILBA with HindIII and BglII. Laccaria NR intron was amplified from genomic DNA with IN‐HindFor/IN‐BglRev primers (5′‐TGTGAAGCTTTGATGAATAATTGGTGAGTTCAG‐3′/5′‐GAAGAGATCTAACACGAAACCACCTTTACG‐3′; restriction sites are underlined, donor and acceptor bases of the intron are in italics) which create the same restriction sites at 55°C of annealing temperature and ligated into pSILBA to create pSILBAα (Fig. 1D).
The pSILBAγ was constructed by modifying pSILBAα further. In pSILBAγ the silencing triggering cassette was inverted with respect to pSILBAα which results in different T‐DNA structure in the further cloning step in the binary vector pHg (Fig. 2). The inversion of the cassette was performed by digesting pSILBAα with XbaI. This enzyme cuts upstream of the A. bisporus gpdII promoter and downstream of the A. nidulans trpC terminator liberating the silencing cassette intact. The plasmid backbone and the silencing cassette were re‐ligated and a plasmid with an inverted cassette orientation, pSILBAγ, was selected (Fig. 1B).
The Agrobacterium binary vector pHg was made by modifying pBGgHg (Chen et al., 2000), a pCAMBIA1300‐based plasmid. The original egfp expression cassette of pBGgHg was removed with SacI and XbaI and replaced by a SacI/XbaI fragment of pBluescript KS+ (Stratagene) multiple cloning site. The plasmid pHg carries in its T‐DNA a hygromycin resistance cassette under the A. bisporus gpdII promoter and unique SacI, NotI and XbaI cloning sites (Fig. 2). The SacI and NotI sites can be used for introducing a complete pSILBA plasmid together with the hairpin construct into the T‐DNA of pHg while XbaI can be used for introducing only the silencing triggering cassette (Fig. 2).
The pSILBA/NITRLoop, pSILBAα/NITRLoop and pSILBAγ/NITRLoop are plasmids that carry a hairpin of Laccaria NR gene sequence constructed in different variants of the pSILBA cloning vector (Fig. S2). In all three constructs a 417 bp exonic sequence of Laccaria NR gene (protein ID 254066) was cloned into SnaBI/HindIII sites. The genomic PCR fragment was amplified with nitrEcoRV/nitrHind primers which carry EcoRV/HindIII restriction sequences (5′‐TGTGGATATCATATATCATTCGCGAGTTGAATC‐3′/5′‐GAAGAAGCTTATTCATCATTCCAGTCGCATTC‐3′; restriction sites are underlined, genomic sequences are in italics) at 60°C of annealing temperature. The second hairpin arm repeat on the 3′ side of the intronic sequence in pSILBA variants was cloned in SphI/StuI sites. This NR PCR fragment was amplified with nitEcoRv/nitSph primers (see above/5′‐GAAGCATGCATTCATCATTCCAGTCGCATTC‐3′; restriction site is underlined) at 60°C of annealing temperature.
The pSILBAγ/INOLoop carries an inverted repeated hairpin of 387 bp targeting the Laccaria 5‐Pase (protein ID 306121). The PCR fragments were cloned in pSILBAγ SnaBI/HindIII and SphI/StuI sites. Primers used were INOSnaBFor/INOHindRev (5′‐GAAGTACGTAATTCGTCAACTGTCATCTCG‐3′/5′‐TGTGAAGCTTCTATACTTGTCAGTTTCGCC‐3′; restriction sites are underlined) and INOSnaBFor/INOSphRev (see above/5′‐TGTGGCATGCCTATACTTGTCAGTTTCGCC‐3′; restriction sites are underlined). These primer sets amplify the third exon of the target gene and were used on a genomic DNA template at 60°C of annealing temperature.
The pHg/pSILBA(α,γ)/Loop transformation vectors were constructed by ligating pSILBA plasmids carrying Laccaria NR or 5‐Pase gene hairpin sequences respectively to pHg (Fig. 2). The plasmids were linearized with SacI, electrophoresed and purified from a 1% agarose gel with the QIAEX®II Gel extraction kit (Qiagen). Plasmid pHg was dephosphorylated (Shrimp alkaline phosphatase, Fermentas) before ligation. The resulting plasmids were electroporated into E. coli by a standard protocol (Sambrook et al., 1989) and bacterial colonies were selected by both kanamycin and ampicillin resistance (100 µg ml−1 each). The double‐resistance conferring plasmids were purified and checked by digestion with BamHI. Plasmid pHg is not cut with this enzyme while pSILBA and its variants are (Fig. S2). The only viable ligation orientation of the two plasmids consists of pSILBA ampicillin resistance cassette ligated towards the T‐DNA right border and the silencing cassette towards the hph‐resistance cassette in pHg (Fig. 2). Digestion of the pHg/pSILBA vector with BamHI should always, independently of the sequence cloned in the silencing cassette, result in the release of a DNA fragment clearly bigger than linear pHg.
Phenotypic evaluation of transformants
Sixty fungal transformants produced with each of the three pHg/pSILBANITRLoop variants (Fig. S2) were evaluated for their unaffected growth on ammonium using solid MP5 medium with 150 µg ml−1 hygromycin. The same preliminary evaluation for the growth on nitrate was performed on modified P5 medium with 4 mM KNO3 (MP5NO3) as N source without hygromycin after 1 month of growth. The fungal colonies of the transformants were evaluated compared with the wild type for their capacity to produce aerial and medium‐penetrating hyphae. Thirty‐seven randomly selected strains of each construct showing normal growth on ammonium were further evaluated for their growth on nitrate liquid medium. Small fragments of mycelia were used for inoculating 96‐well microtitre plates with 150 µl of liquid MP5NO3 medium. Each strain was gown in duplicates and Laccaria wild‐type dikaryon control in seven replicas per microtitre plate. The plates were incubated covered from light at 24°C and fungal growth was evaluated at 2 and 4 weeks.
The biomass production of six pHg/pSILBAγ/NITRLoop transformants and the wild type was evaluated further in liquid medium. Erlenmeyer flasks (50 ml) with 25 ml of MP5NO3 medium were inoculated with five small agar plugs taken from the margin of 7‐day‐old cultures on solid MP5 medium and the mycelia were let to grow as static cultures in the dark at 24°C for 22 days. The mycelia were photographed, harvested, dried at 70°C for 24 h and weighted. Data were evaluated by anova (P < 0.05).
Laccaria H82‐derived transgenic strains with pHg/pSILBAγ/INOLoop were evaluated for their growth on solid MP5 medium with and without hygromycin for detecting consistent phenotypes independent of antibiotic pressure. The capacity of 40 strains to produce aerial hyphae was compared with the wild type and the branching pattern of hyphae was studied under light microscope (Nikon Eclipse E600 light microscope with a Nikon Coolpix950 Digital Camera incorporated).
Southern blot
Genomic DNA was extracted from mycelia grown on MP5 medium (in the case of transformants supplemented with 150 µg ml−1 hygromycin) with the DNeasy Plant mini kit (Qiagen) according to manufacturer's instructions. Approximately 5 µg of genomic DNA was digested with BamHI and separated in 1% agarose gel, stained with ethidium bromide and depurinated before alkaline transfer to a Hydrobond‐N+ nylon membrane (Amersham Biosciences). The Southern blot was probed with an alkaline phosphatase‐labelled DNA fragment hybridizing to the ampicillin resistance gene of pSILBA. The DNA probe was produced by PCR from plasmid pSILBA with AMP‐For/AMP‐Rev primers (5′‐CCCAAGCTTTGCAAGCAGCAGATTACGCG‐3′/5′‐CGCGGATCCGCTCATGAGACAATAACCC‐3′). These primers amplify a 1131 bp fragment corresponding to the ampicillin resistance cassette. The hybridization was carried out at 55°C and the chemiluminescent signal was revealed with the CDP‐star detection reagent of the Gene Images AlkPhos Direct Labelling and Detection system (Ammersham Biosciences) and exposure of the membrane to a film (Agfa new medical X‐ray film).
Plasmid rescue
Fungal genomic DNA (0.5–1 µg) was cut with BamHI and used for plasmid rescue according to Kemppainen and colleagues (2008). Five rescued plasmid clones rising from each fungal transgenic strain were purified and checked by linearization with BamHI before sequencing (Macorgen sequencing service, Seoul) with primer M13Rev‐pUC (Fig. S1).
Semi‐quantitative RT‐PCR analysis
The biomass for RNA extraction from pHg/pSILBAγ/NITRLoop‐transformed fungal strains and wild‐type dikaryon control was produced by a N source shift. Modified P5 medium plates with 150 µg ml−1 hygromycin for transformed fungal strains and medium free of antibiotic for the wild type were covered with cellophane disks and inoculated with mycelial fragments. After 7 days of growth on ammonium the fungal colonies were moved with the cellophane support to MP5NO3 medium with 4 mM KNO3 as the sole N source supplemented with 150 µg ml−1 hygromycin in the case of transformed strains. The colonies were let to grow further for 9 days on this NR‐inducing medium before mycelia were harvested and freezed in liquid nitrogen.
Mycelia for RT‐PCR analysis of Laccaria monokaryon H82 wild type and pHg/pSILBAγ/INOLoop transformants were grown on solid MP5 medium. The medium was supplemented with 150 µg ml−1 hygromycin in the case of transformed strains. The produced biomass was harvested after 18 days of growth and processed for RNA extraction in the same way as dikaryotic strains.
Total RNA was extracted from liquid nitrogen grounded mycelia with the Nucleospin®RNA plant kit (Macherey‐Nagel) by using RAP lysis buffer according to manufacturer's instructions. Two micrograms of RNA was treated with RQ1 DNase free of RNases (Promega). The absence of contaminating genomic DNA was checked from 200 ng of DNase‐treated RNA for lack of amplification with alphaTub1BFor/alphaTub1BRev primers (5′‐TGTCGGGAAAGAACTCATCGAC‐3′/5′‐CAGGGATCCGTCAAATCGGAG‐3′) (Kemppainen et al., 2009). These primers were designed for Laccaria alpha‐1B tubulin gene (protein ID 192524) to be used as an internal reference gene for RT‐PCR analysis. The primers produce a 471 bp amplicon from genomic DNA and a 418 bp product from cDNA template thus revealing possible genomic DNA contaminations in the analysis. These tubulin primers are specific to their target gene and they do not amplify a second Laccaria alpha tubulin gene (protein ID 192523). Total RNA (550 ng) was used for cDNA synthesis with the first‐strand cDNA Kit (Fermentas) by using oligo(dT)18 primer in 20 µl reaction volume. Synthesized cDNA (0.5–1 µl) was used as template in 20 µl of PCR reactions of 25–30 cycles at 60°C of annealing temperature. Tubulin alpha‐1B chain, NR and 5‐Pase gene expression levels in wild type and transformants were detected with primers alphaTub1BFor/alphaTub1BRev; nitrEcoRV/nitrHind and INOSnaBFor/INOHindRev respectively (see plasmids construction). Genomic DNA (20 ng) and ddH2O were used as positive and negative PCR controls respectively. Equal volumes of RT‐PCR reactions were separated in a 1% agarose gel, stained with ethidium bromide and observed under UV light. Images were analysed with the Image J software (http://rsb.info.nih.gov/ij/).
Methylation analysis of NR endogene, NR hairpin and gpdII promoter
Genomic DNA from wild type, two pHg/pSILBAα/NITRLoop and two pHg/pSILBAγ/NITRLoop‐transformed fungal strains with strongly affected growth on nitrate was digested with MspI or its CpG methylation‐sensitive isoschizomer HpaII. When the external C in the recognition sequence CCGG is methylated neither MspI nor HpaII can cleave. However, MspI can cleave the sequence when the internal C residue is methylated. Digested DNA was processed for Southern blot without depurination and hybridized with different NR and gpdII promoter sequence probes. NR probe1: sequence used in NR‐loop construction and produced with primers nitrEcoRV/nitrHind (see plasmid constructs) (417 bp amplicon). NR probe2: genomic NR gene region used in the silencing triggering loop together with a sequence upstream produced with NITR‐FXb/nitrHind (5′‐TGCGTCTAGAGCTATCCTGGGCGATGAATGGTC‐3′/see above) at 60 °C of annealing temperature (753 bp amplicon). NR probe3: NR gene sequence downstream of the region used for the silencing construct. This probe was made by amplifying a fragment of NR gene with nitrEcoRV/NITR‐RKp (see above/5′‐GAATGGTACCTGACGTCCTTGCTCAGTGGGTC‐3′) primers and digesting the amplicon with HindIII which cuts into the NR gene downstream of the sequence used for the silencing trigger. The DNA fragment of interest (1121 bp) was purified from a 1% agarose gel with the QIAEX®II Gel extraction kit (Qiagen) and used for probe labelling. gpdII promoter probe: was produced with primers gpdFH/gpdRC using pSILBAγ as template (see plasmid constructs).
Acknowledgments
The Laccaria genome sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/. This work was supported by Universidad Nacional de Quilmes, CONICET and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina. We would like to thank two anonymous reviewers and editorial board who contributed to improve this work.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Schematic representation showing the resolution of genomic integration sites of the transgenes from pH/pSILBAγ/Loop‐transformed fungal strains by BamHI plasmid rescue.
Inverted repeated loops for triggering RNA silencing of Laccaria nitrate reductase gene constructed in three different pSILBA variants (pSILBA, pSILBAα and pSILBAγ). Inverted repeated sequences represent a 417 bp span of Laccaria nitrate reductase gene exonic sequence. Ori: replication origin of pBluescript KS+. Priming sites for M13‐R, T3, T4 and M13‐F are indicated.
A. Differences in silencing efficiency between pSILBA constructs. B. Variation observed between growth category groups strongly affected (S) of pHg/pSα/NITRLoop and pHg/pSγ/NITRLoop transformants. The category S of pSγ was clearly more affected in their growth on nitrate than the ones produced with the pSα construct.
Schematic representation of Saccharomyces cerevisiae synaptojanin‐like proteins Sjl1p/Inp5, Sjl2p/Inp52, Sjl3p/Inp53 and Inp54p. SacI‐like PPIPase and 5‐Pase domains are indicated. In Sjl1p/Inp5 the SacI‐like PPIPase domain is not active (according to Stefan et al., 2002).
Schematic representation of Laccaria proteins with synaptojanin‐related SacI‐like PPIPase and 5‐Pase domains.
Genomic integration sites in L. bicolor pHg/pSILBAγ/NITRLoop transgenic strains resolved by plasmid rescue. Growth categories: N, non‐affected; A, affected; and S, strongly affected growth on nitrate growth medium.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alder M.N., Dames S., Gaudet J., Mango S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P. Genome‐wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. et al. [DOI] [PubMed] [Google Scholar]
- Audrey M.V., Bormann‐Chung C.A., Judelson H.S. Optimization of transgene‐mediated silencing in Phytophtora infestans and its association with small‐interfering RNAs. Fungal Genet Biol. 2008;45:1197–1205. doi: 10.1016/j.fgb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.H., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Blaise F., Remy E., Meyer M., Zhou L., Narcy J.P., Roux J. A critical assessment of Agrobacterium tumefaciens‐mediated transformation as a tool for pathogenicity gene discovery in the phytopathogenic fungus Leptosphaeria maculans. Fungal Genet Biol. 2007;44:123–138. doi: 10.1016/j.fgb.2006.07.006. et al. [DOI] [PubMed] [Google Scholar]
- Bohse M.L., Woods J.P. RNA interference‐mediated silencing of the YPS3 gene of Hisoplasma capsulatum reveals virulence defects. Infect Immun. 2007;75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Kiger A., Armknecht S., Kerr K., Hild M., Koch B. Genome‐wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. et al. [DOI] [PubMed] [Google Scholar]
- Braunstein T.H., Moury B., Johannessen M., Albrechtsen M. Specific degradation of 3′ regions of GUS mRNA in posttranscriptionally silenced tobacco lines may be related to 5′–3′ spreading of silencing. RNA. 2002;8:1034–1044. doi: 10.1017/s1355838202026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M., Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Burns C., Gregory K.E., Kirby M., Cheung M.K., Riquelme M., Elliott T.J. Efficient GFP expression in the mushroom Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol. 2005;42:191–199. doi: 10.1016/j.fgb.2004.11.005. et al. [DOI] [PubMed] [Google Scholar]
- Cardoza R.E., Vizcaino J.A., Hermosa M.R., Sousa S., Gonzalez F.J., Llobell A. Cloning and characterization of the erg1 of Trichoderma harzianum: effect of the erg1 silencing on ergosterol biosíntesis and resistance to terbinafine. Fungal Genet Biol. 2006;43:164–178. doi: 10.1016/j.fgb.2005.11.002. et al. [DOI] [PubMed] [Google Scholar]
- Chen X., Stone M., Schlagnhaufer C., Romaine C.P. A fruiting body tissue method for efficient Agrobacterium‐mediated transformation of Agaricus bisporus. Appl Environ Microbiol. 2000;66:4510–4513. doi: 10.1128/aem.66.10.4510-4513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.L., Chang W.T. Construction of simple and efficient DNA vector‐based short hairpin RNA expression system for specific gene silencing in mammalian cells. Methods Mol Biol. 2007;408:223–241. doi: 10.1007/978-1-59745-547-3_13. [DOI] [PubMed] [Google Scholar]
- Chicas A., Forrest E.C., Sepich S., Cogoni C., Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation is necessary for transgenic tandem repeat stabilization in Neurospora crassa. Mol Cell Biol. 2005;25:3793–3801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R., Dent S.Y.R. A two‐way street: LSD1 regulates chromatin boundary formation in S. pombe and Drosophila. Mol Cell. 2007;26:160–162. doi: 10.1016/j.molcel.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Irelan J.T., Schumacher M., Schmidhauser T.J., Selker E.U., Macino G. Transgene silencing of the al‐1 gene in vegetative cells of Neurospora is mediated by cytoplasmic effector and does not depend on DNA–DNA interaction or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Melayah D., Raffier C., Gay G., Marmeisse R. Agrobacterium tumefaciens‐mediated transformation as a tool for insertional matagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Micorbiol Lett. 2003;220:141–148. doi: 10.1016/S0378-1097(03)00089-2. [DOI] [PubMed] [Google Scholar]
- Costa A.M., Mills P.R., Bailey A.M., Foste G.D., Challen M.P. Oligonucleotide sequences forming short self‐complimentary hairpins can expedite the down‐regulation of Coprinopsis cinerea genes. J Microbiol Methods. 2008;75:205–208. doi: 10.1016/j.mimet.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Dong Y., Burch‐Smith T.M., Liu Y., Mamillapalli P., Dinesh‐Kumur S.P. A ligation‐independent cloning tobacco rattle virus vector for high‐throughput virus‐induced gene silencing identifies roles for NbMADS4‐1 and ‐2 in floral development. Plant Physiol. 2007;145:1161–1170. doi: 10.1104/pp.107.107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: short RNAs that silence gene expression. Nat Rev. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Eastwood D.C., Challen M.P., Zhang C., Jenkins H., Henderson J., Burton K.S. Hairpin‐mediated down‐regulation of the urea cycle enzyme argininosucchinate lyase in Agaricus bisporus. Mycol Res. 2008;112:708–716. doi: 10.1016/j.mycres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Fagundes Lopes F.J., Marisa Vieira de Queiroz M., Oliveira Lima J., Oliveira Silva V.A., Fernandes de Araújo E. Restriction enzyme improves the efficiency of genetic transformations in Moniliophthora perniciosa, the causal agent of witches broom disease in Theobroma cacao. Braz Arch Biol Technol. 2008;51:27–34. [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double‐stranded RNA in Caenorhabditidis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fitzgerald A., Van Kan J.A.L., Plummer K.M. Simultaneous silencing of multiple genes in the apple scab fungus Venturia inaequalis by expression of RNA with chimeric inverted repeats. Fungal Genet Biol. 2004;41:963–971. doi: 10.1016/j.fgb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Francis K.E., Spiker S. Identification of Arabidopsis thaliana transformants without selection reveals a high occurrence of silenced T‐DNA integrations. Plant J. 2005;41:464–477. doi: 10.1111/j.1365-313X.2004.02312.x. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez‐Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Freitag M.D., Lee W., Kothe G.O., Pratt R.J., Aramayo R., Selker E.U. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Meyers R.E., Cantley L.C. Phophoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Fulci B., Macino G. Quelling: post‐transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol. 2007;10:199–203. doi: 10.1016/j.mib.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Godio R., Fouces R., Guniña E., Martin J. Agrobacterium tumefaciens‐mediated transformation of an antitumor clavaric acid‐producing basidiomycete Hypholoma sublateritium. Curr Genet. 2004;45:287–294. doi: 10.1007/s00294-004-0533-5. [DOI] [PubMed] [Google Scholar]
- Goldoni M., Azzalin G., Macino G., Cogoni C. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet Biol. 2004;41:1016–1024. doi: 10.1016/j.fgb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Gong X., Fu Y., Jiang D., Li G., Yi X., Peng Y. l‐arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet Biol. 2007;44:1368–1379. doi: 10.1016/j.fgb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Gou D., Weng T., Wang Y., Wang Z., Zhang H., Gao L. A novel approach for the construction of multiple shRNA expression vectors. J Gene Med. 2007;9:751–763. doi: 10.1002/jgm.1080. et al. [DOI] [PubMed] [Google Scholar]
- Guo S., Stolz L.E., Lemrow S.M., York J.D. SAC1‐like domains of yeast SAC1INP52 and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J., Baulcombe D.C.A. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond T.M., Keller N.P. RNA silencing in Aspergillus nidulans is independent of RNA dependent RNA polymerases. Genet. 2005;169:607–617. doi: 10.1534/genetics.104.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Grierson D. The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol Genet Genomics. 2002;267:629–635. doi: 10.1007/s00438-002-0696-z. [DOI] [PubMed] [Google Scholar]
- Hanif M., Pardo A.G., Gorfer M., Raudaskoski M. T‐DNA transfer and integration in the ectomycorrhizal fungus Suillus bovinus using hygromycin B as a selectable marker. Curr Genet. 2002;41:183–188. doi: 10.1007/s00294-002-0297-8. [DOI] [PubMed] [Google Scholar]
- Heneghan M.N., Costa A.M., Challen M.P., Mills P.R., Mailey A., Foster G.D. A comparison of methods for successful triggering of gene silencing in Coprinus cinereus. Mol Biotechnol. 2007;35:283–296. doi: 10.1007/BF02686014. [DOI] [PubMed] [Google Scholar]
- Henry C., Mouyna I., Latgé J.P. Testing the efficacy of RNA interference constructs in Aspergillus fumigatus. Curr Genet. 2007;51:277–284. doi: 10.1007/s00294-007-0119-0. [DOI] [PubMed] [Google Scholar]
- Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O. Transitivity‐dependent and ‐independent cell‐to‐cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA ‘off‐target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P.A. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Janus D., Hoff B., Hofmann E., Kück U. An efficient fungal RNA‐silencing system using the DsRed reporter gene. Appl Environ Microbiol. 2007;73:962–970. doi: 10.1128/AEM.02127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong J.F., Deelstra H.J., Wösten H.A.B., Lugones L.G. RNA‐mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl Environ Microbiol. 2006;72:1267–1269. doi: 10.1128/AEM.72.2.1267-1269.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadotani N., Nakayashiki H., Tosa Y., Mayama S. RNA silencing in the phytophatogenic fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2003;16:769–776. doi: 10.1094/MPMI.2003.16.9.769. [DOI] [PubMed] [Google Scholar]
- Kadotani N., Nakayashiki H., Tosa Y., Mayama S. One of the two Dicer‐like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA‐triggered RNA silencing and related small interfering RNA accumulation. J Biol Chem. 2004;279:44467–44474. doi: 10.1074/jbc.M408259200. [DOI] [PubMed] [Google Scholar]
- Kalidas S., Smith D.P. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Polin G., Durbin R., Gotta M. Systemic functional analysis of the Caenorhabdilis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. et al. [DOI] [PubMed] [Google Scholar]
- Kemppainen M., Circosta A., Tagu D., Martin F., Pardo A.G. Agrobacterium‐mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N. Mycorrhiza. 2005;16:19–22. doi: 10.1007/s00572-005-0008-7. [DOI] [PubMed] [Google Scholar]
- Kemppainen M., Duplessis S., Martin F., Pardo A.G. T‐DNA insertion, plasmid rescue and integration analysis in the model mycorrhizal fungus Laccaria bicolor. Microb Biotechnol. 2008;1:258–269. doi: 10.1111/j.1751-7915.2008.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen M., Duplessis S., Martin F., Pardo A.G. RNA silencing in the model mycorrhizal fungus Laccaria bicolor. Gene knock‐down of nitrate reductase results in inhibition of symbiosis with Populus. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.01912.x. [DOI] [PubMed] [Google Scholar]
- Klahre U., Crété P., Leuenberger S.A., Iglesias V.A., Meins F., Jr High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc Natl Acad Sci USA. 2002;99:11561–11563. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kościańska E., Kalantidis K., Wypijewski K., Sadowski J., Tabler M. Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum. Plant Mol Biol. 2005;59:647–661. doi: 10.1007/s11103-005-0668-x. [DOI] [PubMed] [Google Scholar]
- Laurie J.D., Linning R., Bakkeren G. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr Genet. 2008;53:49–58. doi: 10.1007/s00294-007-0165-7. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Liu H., Cottrell T., Pierini L.M., Goldman W.E., Doering T. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genet. 2002a;160:463–470. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh‐Kumar S.P. Virus‐induced gene silencing in tomato. Plant J. 2002b;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- Lugones L.G., Scholtmeijer K., Klootwijk R., Wessels J.G.H. Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol. 1999;32:681–689. doi: 10.1046/j.1365-2958.1999.01373.x. [DOI] [PubMed] [Google Scholar]
- Ma B., Mayweld M.B., Gold M.H. The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium. Appl Environ Microbiol. 2001;67:948–955. doi: 10.1128/AEM.67.2.948-955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald T., Brown D., Keller N.P., Hammond T.M. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact. 2005;18:539–545. doi: 10.1094/MPMI-18-0539. [DOI] [PubMed] [Google Scholar]
- Marchand G., Fortier E., Neveu B., Bolduc S., Belzile F., Belenger R.R. Alternative methods for genetic transformation of Pseudozyma antarctica, a basidiomycetous yeast‐like fungus. J Microbiol Methods. 2007;70:519–527. doi: 10.1016/j.mimet.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A., Zaratiegui M., Goto D.B. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Martin F., Selosse M.A. The Laccaria genome: a symbiont blueprint decoded. New Phytol. 2008;180:296–310. doi: 10.1111/j.1469-8137.2008.02613.x. [DOI] [PubMed] [Google Scholar]
- Martin F., Aerts A., Ahrén D., Brun A., Danchin E.G., Duchaussoy F. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. et al. [DOI] [PubMed] [Google Scholar]
- Martin T.F. PI(4,5)P2 regulation on surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- Matityahu A., Hadar Y., Dosoretz C.G., Belinky P.A. Gene silencing by RNA interference in the white‐rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 2008;74:5359–5365. doi: 10.1128/AEM.02433-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Itoh R., Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki A., Ueno M., Arase S., Kihara J. RNA‐mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol Lett. 2007;269:85–89. doi: 10.1111/j.1574-6968.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- Mouyna I., Henry C., Doering T.L., Latgé J.‐P. Gene silencing by RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol Lett. 2004;237:317–324. doi: 10.1016/j.femsle.2004.06.048. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Kadotani N.S.M. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Nguyen Q.B. RNA interference: roles in fungal biology. Curr Opin Microbiol. 2008;11:494–502. doi: 10.1016/j.mib.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Hanada S., Nguyen B.Q., Kadotani N., Tosa Y., Mayama S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol. 2005;42:275–283. doi: 10.1016/j.fgb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Namekawa S.H., Iwabata K., Sugawara H., Hamada F.K., Koshiyama A., Chiku H. Knockdown of LIM/15DMC1 in the mushroom Coprinus cinereus by double‐stranded RNA‐mediated gene silencing. Microbiology. 2005;151:3669–3678. doi: 10.1099/mic.0.28209-0. et al. [DOI] [PubMed] [Google Scholar]
- Nguyen Q.B., Kadotani N., Kasahara Y.T., Mayama S., Nakayashiki H. Systematic functional analysis of calcium‐signalling proteins in the genome of the rice‐blast fungus, Magnaporthe oryzae, using a high‐throughput RNA‐silencing system. J Microbiol. 2008;68:1348–1365. doi: 10.1111/j.1365-2958.2008.06242.x. [DOI] [PubMed] [Google Scholar]
- Nicolás F.E., Torres‐Martínez S., Ruiz‐Vázquez R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non‐integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J.M., Van Der Veen D., De Graaff L.H., Qui L. Efficient cloning system for construction of gene silencing vectors in Aspergillus niger. Appl Microbiol Biotechnol. 2008;80:917–924. doi: 10.1007/s00253-008-1640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon J.G., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence‐specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo A.G., Hanif M., Raudaskoski M., Gorfer M. Genetic transformation of ectomycorrhizal fungi mediated by Agrobacterium tumefaciens. Mycol Res. 2002;106:132–137. [Google Scholar]
- Peretz Y., Mozes‐Koch R., Akad F., Tanne E., Czosnek H., Sela I. A universal expression/silencing vector in plants. Plant Physiol. 2007;145:1251–1263. doi: 10.1104/pp.107.108217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Albrechtsen M. Evidence implying only unprimed RdRP activity during transitive gene silencing in plants. Plant Mol Biol. 2005;58:575–583. doi: 10.1007/s11103-005-7307-4. [DOI] [PubMed] [Google Scholar]
- Pogribny I.P., Pogribna M., Christman J.K., James S.J. Single‐site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: possible in vivo relevance during tumorigenesis. Cancer Res. 2000;60:588–594. [PubMed] [Google Scholar]
- Raponi M., Arndt M.G. Double‐stranded RNA‐mediated gene silencing in fission yeast. Nucleic Acids Res. 2003;31:4481–4489. doi: 10.1093/nar/gkg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C.A., Engle J.T., Coldman W.E. RNA interference in Hitoplasma capsulatum demonstrates a role for alpha‐(1,3)‐glucan in virulence. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory; 1989. , and Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY, USA: [Google Scholar]
- Samils N., Elfstrand M., Lindner Czederpiltz D.L., Fahleson J., Olson Å., Dixelius C., Stenlid J. Development of a rapid and simple Agrobacterium tumefaciens mediated transformation system for the fungal pathogen Heterobasidium annosum. FEMS Microbiol Lett. 2006;255:82–88. doi: 10.1111/j.1574-6968.2005.00069.x. [DOI] [PubMed] [Google Scholar]
- Sanders M., Maddelein W., Depicker A., Van Montagu M., Cornelissen M., Jacobs J. An active role for endogenous beta‐1,3‐glucanase genes in transgene‐mediated co‐suppression in tobacco. EMBO J. 2002;21:5824–5832. doi: 10.1093/emboj/cdf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtmeijer K., Wösten H.A.B., Springer J., Wessels J.G.H. Effect of introns and AT‐rich sequences on expression of the bacterial hygromycin B resistance gene in the basidiomycete Schizophyllum commune. Appl Environ Microbiol. 2001;67:481–483. doi: 10.1128/AEM.67.1.481-483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V., Allshire R. Hairpin RNAs and retrotransposons LTRs effect RNAi and chromatin‐based gene silencing. Science. 2003;301:1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- Schuurs T.A., Schaeffer E.A.M., Wessels J.G.H. Homology‐dependent silencing of the SC3 gene in Schizophyllum commune. Genet. 1997;147:589–596. doi: 10.1093/genetics/147.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E.U., Freitag M., Kothe G.O., Margolin B.S., Rountree M.R., Allis C.D., Tamuru H. Induction and maintenance of nonsymmetrical DNA methylation in Neurospora. Proc Natl Acad Sci USA. 2002;99:16485–16490. doi: 10.1073/pnas.182427299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafran H., Miyara I., Eshed R., Prusky D., Sherman A. Development of new tools for studying gene function in fungi based on the Gateway system. Fungal Genet Biol. 2008;45:1147–1154. doi: 10.1016/j.fgb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Sharma K.K., Gupta S., Kuhad R.C. Agrobacterium‐mediated delivery of marker genes to Phanerochaete chrysosporium mycelial pellets: a model transformation system for white‐rot fungi. Biotechnol Appl Biochem. 2006;43:181–186. doi: 10.1042/BA20050160. [DOI] [PubMed] [Google Scholar]
- Sigova A., Rhind N., Zamore P. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359–2367. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L. On the role of RNA amplification in dsRNA‐triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. et al. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Wurmser A.E., Emr S.D., Stenmark H. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- Smith N.A., Singh S.P., Wang M.B., Stoutjesdijk P.A., Green A.G., Waterhouse P.M. Total silencing by intron‐spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Seaman M., Nemoto Y., Daniell L., Suchy S.F., Emr S. Disruption of three phosphatidylinositol‐polyphosphate 5‐phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. et al. [PubMed] [Google Scholar]
- Stefan C.J., Audhya A., Emr S.D. The yeast synaptojanin‐like proteins control the cellular distribution of phosphatidylinositol (4,5)‐bisphosphate. Mol Biol Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz L.E., Kuo W.J., Longchamps J., Sekho M.K., York J.D. INP51, a yeast inositol polyphosphate 5‐phosphatase required for phosphatidylinositol 4,5‐bisphosphate homeostasis and whose absence confers a cold‐resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Suizu T., Zhou G.L., Oowatari Y., Kawamukai M. Analysis of expressed sequence tags (ESTs) from Lentinula edodes. Appl Microbiol Biotechnol. 2008;79:461–470. doi: 10.1007/s00253-008-1441-2. [DOI] [PubMed] [Google Scholar]
- Tamuru H., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tanguay P., Bozza S., Breui C. Assessing RNAi frequency and efficiency in Ophiostoma floccosum and O. piceae. Fungal Genet Biol. 2006;43:804–812. doi: 10.1016/j.fgb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Wang S.L., Dorovkov M., Ryazanov A., Driscoll M. Heritable and inducible genetic interference by double‐stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones L., Baulcombe D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA‐dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H., Bleys A., Depicker A. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 2003;131:245–253. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kinder C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine‐9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wadhwa R., Kaul S.C., Miyagishi M., Taira K. Vectors for RNA interference. Curr Opin Mol Ther. 2004;6:367–372. [PubMed] [Google Scholar]
- Wälti M.A., Villalba C., Buser R.M., Grünler A., Aebi M., Künzler M. Targeted gene silencing in the model mushroom Coprinus cinereaCoprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5:732–744. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., Helliwell C.A., Smith N.A., Wang M.B., Rouse D.T., Liu Q. Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. et al. [DOI] [PubMed] [Google Scholar]
- Wiradjaja F., Ooms L.M., Whisstock J.C., McColl B., Helfenbaum L., Sambrook J.F. The yeast inositol polyphosphate 5‐phosphatase Inp54p localizes to the endoplasmic reticulum via a C‐terminal hydrophobic anchoring tail: regulation of secretion from the endoplasmic reticulum. J Biol Chem. 2001;276:7643–7653. doi: 10.1074/jbc.M010471200. et al. [DOI] [PubMed] [Google Scholar]
- Wu M.T., Wu R.H., Hung C.F., Cheng T.L., Tsai W.H., Chang W.T. Simple and efficient DNA vector‐based RNAi system in mammalian cells. Biochem Biophys Res Commun. 2005;330:53–59. doi: 10.1016/j.bbrc.2005.02.129. [DOI] [PubMed] [Google Scholar]
- Yamada O., Ikeda R., Ohkita Y., Hayashi R., Sakamoto K., Akita O. Gene silencing by RNA interference in the koji mold Aspergillus oryzae. Biosci Biotechnol Biochem. 2007;71:138–144. doi: 10.1271/bbb.60405. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Okajima Y., Kawashima H., Tsukamoto A., Sugiera J., Shishido K. Intron‐dependent accumulation of mRNA in Coriolus hirsutus of lignin peroxidase gene the product of which is involved in conversion/degradation of polychlorinated aromatic hydrocarbons. Biosci Biotechnol Biochem. 2006;70:1293–1299. doi: 10.1271/bbb.50471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation showing the resolution of genomic integration sites of the transgenes from pH/pSILBAγ/Loop‐transformed fungal strains by BamHI plasmid rescue.
Inverted repeated loops for triggering RNA silencing of Laccaria nitrate reductase gene constructed in three different pSILBA variants (pSILBA, pSILBAα and pSILBAγ). Inverted repeated sequences represent a 417 bp span of Laccaria nitrate reductase gene exonic sequence. Ori: replication origin of pBluescript KS+. Priming sites for M13‐R, T3, T4 and M13‐F are indicated.
A. Differences in silencing efficiency between pSILBA constructs. B. Variation observed between growth category groups strongly affected (S) of pHg/pSα/NITRLoop and pHg/pSγ/NITRLoop transformants. The category S of pSγ was clearly more affected in their growth on nitrate than the ones produced with the pSα construct.
Schematic representation of Saccharomyces cerevisiae synaptojanin‐like proteins Sjl1p/Inp5, Sjl2p/Inp52, Sjl3p/Inp53 and Inp54p. SacI‐like PPIPase and 5‐Pase domains are indicated. In Sjl1p/Inp5 the SacI‐like PPIPase domain is not active (according to Stefan et al., 2002).
Schematic representation of Laccaria proteins with synaptojanin‐related SacI‐like PPIPase and 5‐Pase domains.
Genomic integration sites in L. bicolor pHg/pSILBAγ/NITRLoop transgenic strains resolved by plasmid rescue. Growth categories: N, non‐affected; A, affected; and S, strongly affected growth on nitrate growth medium.


