Abstract
Purpose
Dysregulation of the phosphoinositide 3-kinase (PI3K) and Src signaling pathways commonly occur in colorectal cancer. Mutations in the PIK3CA gene are associated with an increase in severity of disease and worse clinical outcomes. Elevated levels of Src have been identified in premalignant lesions and are suggested to play a central role in tumor progression. Because these pathways appear to enhance tumor growth and metastasis, molecularly targeted agents for both pathways are currently being evaluated in early-phase clinical trials.
Experimental Design
We used colorectal cancer cell lines and a patient-derived explant model to investigate the efficacy of saracatinib. Mutations in the PIK3CA were evaluated to examine the association between mutations in the PIK3CA gene and sensitivity to saracatinib.
Results
We have identified a subset of patients with a PIK3CA (exon 9 and 20) mutation with increased sensitivity to saracatinib. A novel 3′ untranslated region (UTR) mutation was also shown to be associated with increased sensitivity to saracatinib and have a reduced affinity for miR-520a and miR-525a. Importantly, we show that Src inhibition reduces the interaction between Src and p85, subsequently decreasing Akt-dependent signaling.
Conclusion
These results indicate that a personalized approach in targeting Src in PIK3CA-mutant patients with colorectal cancers may prove effective in a subset of patients with this genetic alteration.
Introduction
The PI3K/Akt signaling pathway plays an integral role in modulating many different cellular functions (1, 2). The serine/threonine kinase AKT is considered the focal point of this pathway whereby AKT disseminates signals to downstream targets that are essential for enhancing cellular survival and proliferation (3, 4). Activating mutations in the PIK3CA gene commonly occur in many different cancers (5) and seem to play an important role in tumorigenesis (6). In colorectal cancer, the PIK3CA gene (exon 9 or 20) has been determined to be mutated in approximately 15% to 20%of the cases (7, 8). Colon cancer cells with mutations in PIK3CA are more resistant to apoptosis (9) and have an increased capacity to metastasize as shown in an orthotopic model (10). Clinically, mutations in the PIK3CA gene have been shown to be associated with an increased risk in rectal tumor recurrence (11) and with worse survival in patients with colorectal cancers (12). Because this pathway appears to be essential for the growth and survival of many tumors, intense drug development of inhibitors of this pathway are currently being investigated.
Dysregulation of the non–receptor tyrosine kinase c-Src has been implicated to alter a wide range of tumor cell processes (13). Some of the downstream targets of Src include focal adhesion kinase (FAK), a kinase that is important for cell–cell and cell–extracellular matrix interactions and Stat-3, a kinase that enhances survival of cells and angiogenesis (14–17). The activity of Src is tightly regulated and most cells express low levels (18). Activation of Src requires the dephosphorylation of the tyrosine residue at position 527 and subsequent phosphorylation of tyrosine 416, resulting in its catalytic activation (13). Although activating mutations in Src are rare or nonexistent (19), dysregulation of Src has been shown to be important at early stages of cancer development and perhaps even more important during later stages of the disease (13, 20–22). The orally available drug saracatinib is a potent Src inhibitor and has been shown to have a wide range of antiproliferative effects in many different preclinical models (23–25). For instance, we have shown in patient-derived colorectal cancers (23) and pancreatic (25) explant models that saracatinib treatment had antitumor effects only in a subset of explants treated. Of note, biomarkers of sensitivity were identified in both studies.
Currently, early-phase clinical trials involving targeted agents for both the phosphoinositide 3-kinase (PI3K) pathway and Src pathways are being investigated. Most of the studies are unselected for biomarker(s) and consequently, either have failed (26, 27) or will not likely find statistical benefit as the majority of these compounds will certainly only affect a small subset of patients. Therefore, identifying biomarkers of sensitivity or resistance ultimately will identify patients who will most likely derive benefit from treatment. In this study, we used a patient-derived colorectal cancer explant model and have identified an association between a mutation in the PIK3CA gene, including a novel 3′ untranslated region (UTR) mutation and increased sensitivity to saracatinib (an Src inhibitor) in colorectal cancers. Furthermore, we show that Src interacts with p85 and treatment with saracatinib disrupts this interaction, subsequently decreasing the activation of AKT and downstream mediators.
Materials and Methods
Human explant xenograft model: colorectal cancer explant xenograft model
Patient-derived colorectal cancer tumor specimens were obtained from consenting patients at the University of Colorado Hospital (Aurora, CO; COMIRB # 07-0570). The tumor pieces were then implanted in mice [Institutional Animal Care and Use Committee (IACUC) # 51402007(09) 2E] and expansion was carried out as previously described (28, 29). Tumors were expanded in the left and right flanks of 5 to 6 mice (10 evaluable tumors per group). Mice were randomized into the treatment group (saracatinib) or vehicle group when tumor volumes reached approximately 200 mm3. Mice were treated daily with saracatinib 50 mg/kg and vehicle by oral gavage for 28 days. Mice were monitored daily for signs of toxicity, and tumor size was evaluated twice per week by caliper measurements with the following formula: tumor volume = (length × width2) × 0.52.
PIK3CA sequencing
We conducted sequencing to examine the mutational status of PIK3CA gene (exon 9 or 20) in colorectal cancer cell lines and explants. DNA was isolated with the Qiagen DNA Extraction Kit (Qiagen). PIK3CA exon 9 or 20 was PCR-amplified and analyzed by direct sequencing of the products as described previously (30).
SNaPshot
PCR products were analyzed for mutations with the ABI PRISM SNaPshot Multiplex Kit (Applied Biosystems), according to the protocol supplied by the manufacturer. The SNaPshot method is based on the dideoxy single-base extension of unlabeled oligonucleotide primers. For each of the mutations, a primer annealing adjacent to the potentially mutant nucleotide was developed. All primers were designed with a similar melting temperature and were checked for the absence of base pairing with other SNaPshot primers.
Evaluation of Src gene copy
An Src FISH probe was developed to evaluate Src gene copy number as previously described (30). Continuous variables for Src gene copy number were categorized and a conservative approach was used for cell lines and explants in determining a gain in Src gene copy number. An Src copy number of 3 or above was considered a gain and less than 3 was considered no gain.
Determination of sensitive and resistant molecular biomarkers
Total RNA from all colorectal cancer explants were profiled with the Affymetrix HuGene 1.0 ST Array. Sample preparation and processing procedure were done as described in the manufacturer’s manual (Affymetrix). The gene expression levels were converted to a rank-based matrix for each micro-array. Data analyses were done on this rank-based matrix. Gene set analysis was conducted as previously described (31). Gene set permutations were conducted 1,000 times for each analysis. We used the nominal P value and normalized enrichment score to sort the pathways enriched in each phenotype. We used the pathway defined by Kyoto Encyclopedia of Genes and Genomes (KEGG; ref. 32). The Gene Expression Omnibus (GEO) accession number is GSE36006.
Immunoprecipitation
Equal amounts of protein and volume in each group were used for the immunoprecipitation procedures. Five microliters of anti-Src or p85 antibody (Cell Signaling Technologies) was added to each sample followed by incubation (rotator) at room temperature for 1 hour. After 1 hour, 30 µL of protein G magnetic beads (Invitrogen-Dynal) was added to each sample and incubated for an additional 30 minutes. The samples were boiled for 5 minutes and loaded onto a 4% to 12% SDS-PAGE. p85 and p110 rabbit polyclonal antibodies (Cell Signaling Technologies) were used for detection at a dilution of 1:1,000.
Immunoblotting
Forty micrograms of sample was electrophoresed on Bis–Tris precast gels and transferred to nitrocellulose membrane. After blocking, Src, AKT, PDK, FAK, Stat-3, ribosomal S6, p27kip1, SGK3, actin (Cell Signaling Technology), or p-FAK Tyr861 (BioSource International), primary antibody was added at 1:1,000 in TBS with Tween (TBST) containing 5% bovine serum albumin. After washing 3 times with TBST, the membrane was incubated with anti-mouse IgG horseradish peroxidase–conjugated antibody at a final dilution of 1:50,000 in TBST. After washing 3 times with TBST, bound antibodies were detected by enhanced chemiluminescence.
miR-520a and miR-525
A dual-luciferase UTR-binding assay was conducted according to the manufacturer’s instructions (Promega). The 3′ UTR of PIK3CA containing the predicted miR-520a and miR-525 target sequence (base pairs 3,323– 3,487) was amplified from genomic DNA and cloned into the psiCHECK-2 plasmid. Perfect match primers were used in an initial round of PCR, followed by PCR amplification of the product with primers containing XhoI and NotI restriction sites for cloning into the psiCHECK-2 plasmid. PCR products were initially cloned into T-Vector (Invitrogen), sequenced and selected for wild-type and mutant alleles. Both wild-type and mutant sequences were present in the cDNA sample and thus creation of a mutation in the binding site was not necessary; rather both alleles were cloned from the same reaction and confirmed again by direct sequencing of the clones with the psiCHECK-2 reverse primer. The psiCHECK-2 vectors containing the region of PIK3CA, as described, are designated psi-PIK3CA-WT and psi-PIK3CA-MUT. Inhibition of expression of the luciferase reporter gene by miR-520a or miR-525 was assayed in HEK293 cells cotransfected with either a vector control (psiCHECK-2) or a luciferase construct psi-CHECK2-PIK3CA-UTR and a negative control mimic and mimics of miR-520a and miR-525 (Dharmacon).
Statistical analysis
An unpaired Student t test was used to determine whether the means between control and saracatinib were significant at end of treatment (~28 days). The differences were considered significant when the P value was <0.05. All error bars are represented as the SE. A tumor growth index (TGI) was calculated (mean tumor/mean control × 100) and a cutoff point of 50% was used to dichotomize treatment effects into sensitive (TGI < 50%) and resistant (TGI ≥ 50%). A 2 × 2 contingency table was used to compare the association between a mutation in the PIK3CA gene (exon 9, 20, and 3′ UTR) and sensitivity to saracatinib. Fisher’s exact test was used to calculate statistical significance. All analyses were carried out by GraphPad Prism version 5.0c for Windows (GraphPad Software).
Results
Association between PIK3CA mutations/PTEN-null and increased sensitivity to saracatinib in colorectal cancer
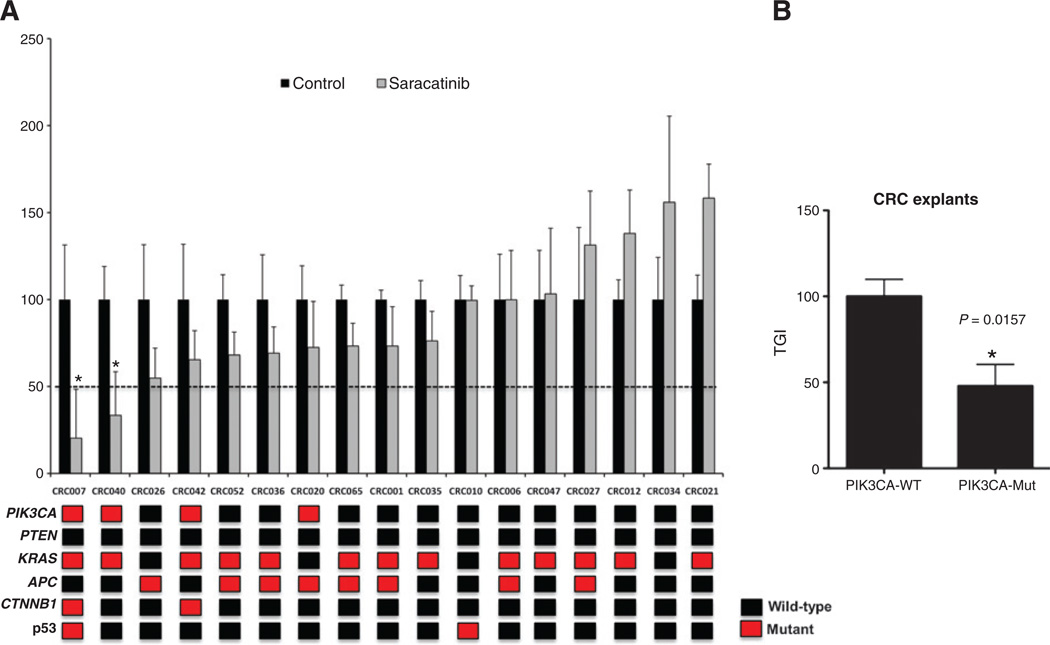
A total of 23 colorectal cancer cell lines were previously treated with saracatinib and the antiproliferative effects were examined by sulforhodamine B (SRB) assay (23). As shown in Supplementary Fig. S1, the 3 colorectal cancer cell lines LS180, LS174T, and H508 had the greatest sensitivity (IC50 < 0.5 µmol/L) to saracatinib. Examination of the PIK3CA gene revealed that all 3 cell lines harbor a mutation within exon 9 or 20 of the gene (Supplementary Table S1). Fisher’s exact test was conducted to compare whether a mutation in PIK3CA gene was associated with increased sensitivity to saracatinib. As displayed in Table 1, a significant association (P < 0.0474) between PIK3CA mutation and increased sensitivity was evident in these cell lines. We next examined the predictive power of PIK3CA mutation in the colorectal cancer cell lines in 17 colorectal cancer patient-derived explants. Supplementary Table S2 shows the type of tumor, stage of disease, previous treatment, and PIK3CA mutational status of the explants of patients with colorectal cancers. As shown in Fig. 1A, only 2 colorectal cancer explants showed sensitivity (static effects, but no regression) to saracatinib (TGI ≤ 50%). Sequencing the PIK3CA gene (exon 9 and 20) in all colorectal cancer explants revealed that both sensitive tumors CRC007 and 040 harbor a mutation. In addition, all colorectal cancer explants were evaluated by SNaPshot that tests more than 269 common mutations in cancer (Fig. 1). The relationship between PIK3CA and sensitivity to saracatinib in colorectal cancer explants was evaluated by Fisher’s exact test. As shown in Table 1, a significant association (P = 0.0441) between PIK3CA mutation and sensitivity was identified. Combining the colorectal cancer cell lines and explants resulted in a greater association (P = 0.0020) between PIK3CA mutation and increased sensitivity to saracatinib (Table 1). The overall prevalence of PIK3CA mutations in this study in the combined 40 colorectal cancer cell lines and explants was 32.5%. Finally, we compared all PIK3CA wild-type versus PIK3CA-mutant colorectal cancer explants with respect to percentage of TGI. A significant decrease (P = 0.0157) in the percentage of TGI was seen in tumors with a PIK3CA mutation that were treated with saracatinib when compared with wild-type (Fig. 1B).
Table 1.
Association between PIK3CA mutation and sensitivity to saracatinib in colorectal cancer cell lines, colorectal cancer explants, and colorectal cancer cell lines and explants combined.
| Saracatinib- resistant |
Saracatinib- sensitive |
|
|---|---|---|
| Colorectal cancer cell lines | ||
| PIK3CA wild-type | 14 | 0 |
| PIK3CA mutant | 6 | 3 |
| P = 0.0474 | ||
| Colorectal cancer explants | ||
| PIK3CA wild-type | 13 | 0 |
| PIK3CA mutant | 2 | 2 |
| P = 0.0441 | ||
| Combined (colorectal cancer cell lines and explants) | ||
| PIK3CA wild-type | 27 | 0 |
| PIK3CA mutant | 8 | 5 |
| P = 0.0020 | ||
NOTE: P < 0.05 was considered significant.
Figure 1.
Saracatinib effects on tumor growth in colorectal cancer explants. A, seventeen colorectal cancer explants were treated with saracatinib 50 mg/kg/d by oral gavage for 28 days. Tumor size was evaluated twice per week by caliper measurements by the formula: tumor volume = (length × width2) × 0.52. TGI was calculated by relative tumor growth of treated mice divided by relative tumor growth of control mice × 100. Cases with a TGI < 50% were considered sensitive, TGI > 50% were considered resistant to saracatinib. Two xenografts (CRC007 and CRC040) were sensitive to saracatinib (TGI ≤ 50%) and 15 xenografts were resistant to saracatinib (TGI > 50%). Columns, mean (n = 8–10 tumors per group); bars, SE; and *, significance (P < 0.05) compared with vehicle-treated tumors. Mutational status of PIK3CA, PTEN, KRAS, APC, CTNNB1, and p53 are shown for each explant. Mutations in the PIK3CA gene were identified in CRC007 [3′ UTR; (c.*19T>C)], CRC040 (E542K), CRC042 [3′ UTR; (c.*19T>C)], and CRC020 (E542G). B, the relationship between PIK3CA mutation and saracatinib effects (TGI) in colorectal cancer explants. A significant association (P = 0.0157) between PIK3CA mutants and sensitivity was identified. CRC, colorectal cancer; WT, wild-type.
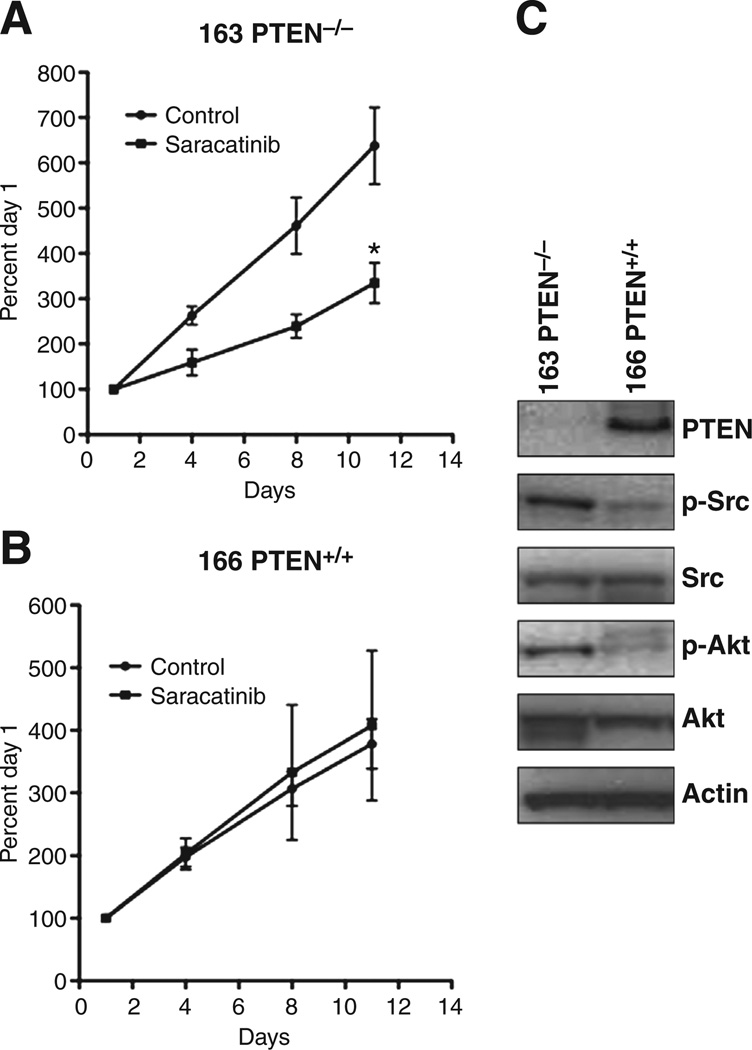
We next were interested to see that whether a loss of PTEN also resulted in enhanced sensitivity to saracatinib. The PTEN isogenic (PTEN null and PTEN wild-type) HCT116 cell line was provided to us from Dr. Volgestein’s laboratory at Johns Hopkins (Baltimore, MD). Both the PTEN-null and PTEN wild-type cell lines were evaluated in a xenograft model. As displayed in Fig. 2, treatment with saracatinib (50 mg/kg/d) resulted in a significant decrease in tumor growth in the PTEN-null cell line (Fig. 2A). In contrast, no effect with treatment was seen in the PTEN wild-type (Fig. 2B) cell line. As expected, an increase in the activation of AKT was seen in the PTEN-null tumor when compared with the PTEN wild-type tumor (Fig. 2C). Interestingly, Src activation was upregulated in the PTEN-null cell line when compared with the PTEN wild-type cell line. In addition to the PTEN isogenic cell line, we analyzed the explants for PTEN mutations and response to saracatinib. However, all colorectal cancer explants were PTEN wild-type (Fig. 1A) and protein levels of PTEN by Western blotting showed protein visible on the immunoblot (data not shown). Taken together, these results indicate that mutations in PIK3CA or PTEN have enhanced sensitivity to saracatinib in colorectal cancer preclinical models.
Figure 2.
PTEN-null cells exhibit sensitivity to saracatinib in a xenograft model. The (A) PTEN-null (−/−) and (B) wild-type (+/+) cell lines were injected in axenograft model to examine the antitumor effects of saracatinib in vivo. When tumor volumes reached approximately 200 mm3, mice were randomized and treated with saracatinib 50 mg/kg/d by oral gavage for 12 days. Tumor size was evaluated twice per week by caliper measurements by the formula: tumor volume = (length × width2) × 0.52. Treatment with saracatinib resulted in a significant decrease in growth in the PTEN-nul cell line (*, P < 0.05). C, evaluation of the activation of Src and Akt in PTEN wild-type and -null tumors. Baseline levels of p-Src and p-Akt were increased in the PTEN-null tumor when compared with wild-type.
Effects of saracatinib on the Src and PI3K/AKT pathways
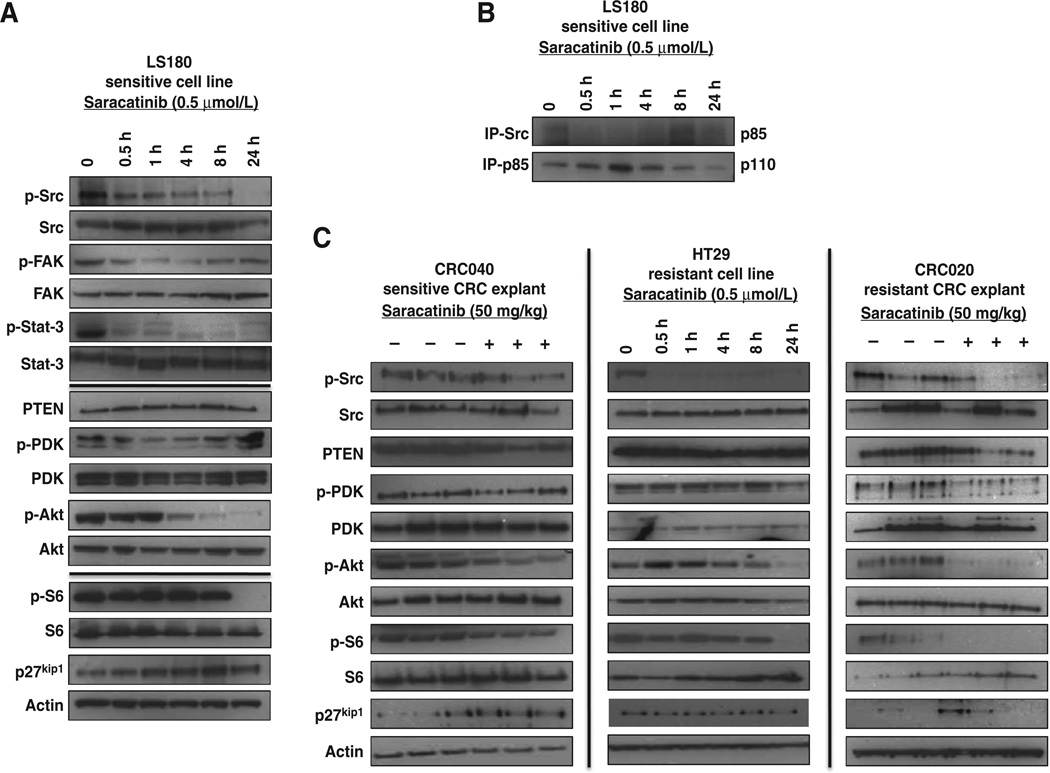
To gain further insight into the mechanism whereby treatment with saracatinib reduces the activation of the PI3K pathway, we assessed the effects of Src inhibition on the Akt and Src pathways by immunoblotting in the LS180 sensitive cell line. As expected, saracatinib treatment reduced the activity of Src, FAK, and STAT-3 at all time points (0.5, 1, 4, 8, and 24 hours) examined in the sensitive LS180 cell line (Fig. 3A). In addition, treatment altered the phosphorylation of PDK-1 (1 hour maximal decrease) and significantly decreased the activation of Akt at 4, 8, and 24 hours after treatment (Fig. 3A). There were no differences identified with respect to saracatinib treatment and modulation of phosphorylated/total PTEN levels and with the activation of the AKT-independent PDK-1 target SGK-3 (data not shown). In addition, treatment increased the levels of p27KIP1, an Akt target protein involved in cell-cycle regulation and the phosphorylation of ribosomal S6 at 24 hours following treatment (Fig. 3A).
Figure 3.
Effect of saracatinib on the Src and PI3K signaling pathways. A, treatment with saracatinib (0.5 µmol/L) at 30 minutes, 1, 4, 8, and 24 hours resulted in a decrease in the phosphorylation of Src, FAK, and Stat-3 in the LS180 sensitive cell line (N = 3). Src inhibition also decreased the activation of PDK-1, AKT, and S6 and increased protein levels of p27kip1. B, co-immunoprecipitation (IP) of Src showed a decrease in binding to p85 at 0.5, 1, and 4 hours after treatment with saracatinib. Conversely, immunoprecipitation of p85 resulted in an increase in binding the p110-α subunit (N = 3). C, the activation of the PI3K pathway (Akt and S6) was decreased with saracatinib treatment in CRC040 (sensitive), CRC020 (resistant), and HT29 (resistant). CRC, colorectal cancer.
We next examined the interaction between Src and PI3K (p85 and p110) in our LS180 sensitive cell line by co-immunoprecipitation studies. As displayed in Fig. 3B, treatment with saracatinib resulted in a decrease in the interaction between Src and p85 at 30 minutes, 1, and 4 hours after treatment. Conversely, saracatinib treatment increased the interaction between p85 and p110 at 30 minutes, 1, and 4 hours after treatment, suggesting that Src inhibition decreases the binding of Src to p85, resulting in an increase in the interaction between p85 and p110.
We also investigated the AKT pathway in our CRC040 sensitive explant, HT29 resistant cell line, and CRC020 resistant explant (Fig. 3C). Of note, HT29, CRC020, and CRC040 have mutations in PIK3CA exon 9 or 20. Treatment with saracatinib (50 mg/kg/d) decreased the activation of Src, Akt, and ribosomal S6 in both explants when evaluated 28 days after treatment. Similar results were identified with respect to the activation of Src, Akt, and ribosomal S6 in the resistant HT29 cell line. Overall, these results indicate that saracatinib treatment not only inhibits the Src pathway but also downregulates the activation of PI3K/AKT pathway.
Association between increased Src gene copy number and resistance to saracatinib
Previous work from our group showed a trend toward significance with SRC gene copy number and resistance to saracatinib (23). In this study, we evaluated SRC gene copy number in additional 7 colorectal cancer explants by FISH. Together with our previous work, a significant association (P < 0.05) is now evident with respect to an increased Src gene copy number and resistance to saracatinib (Supplementary Table S3). Of note, a gain in Src gene copy was identified in conjunction with an increase in chromosome 20, showing that the Src gene was not amplified.
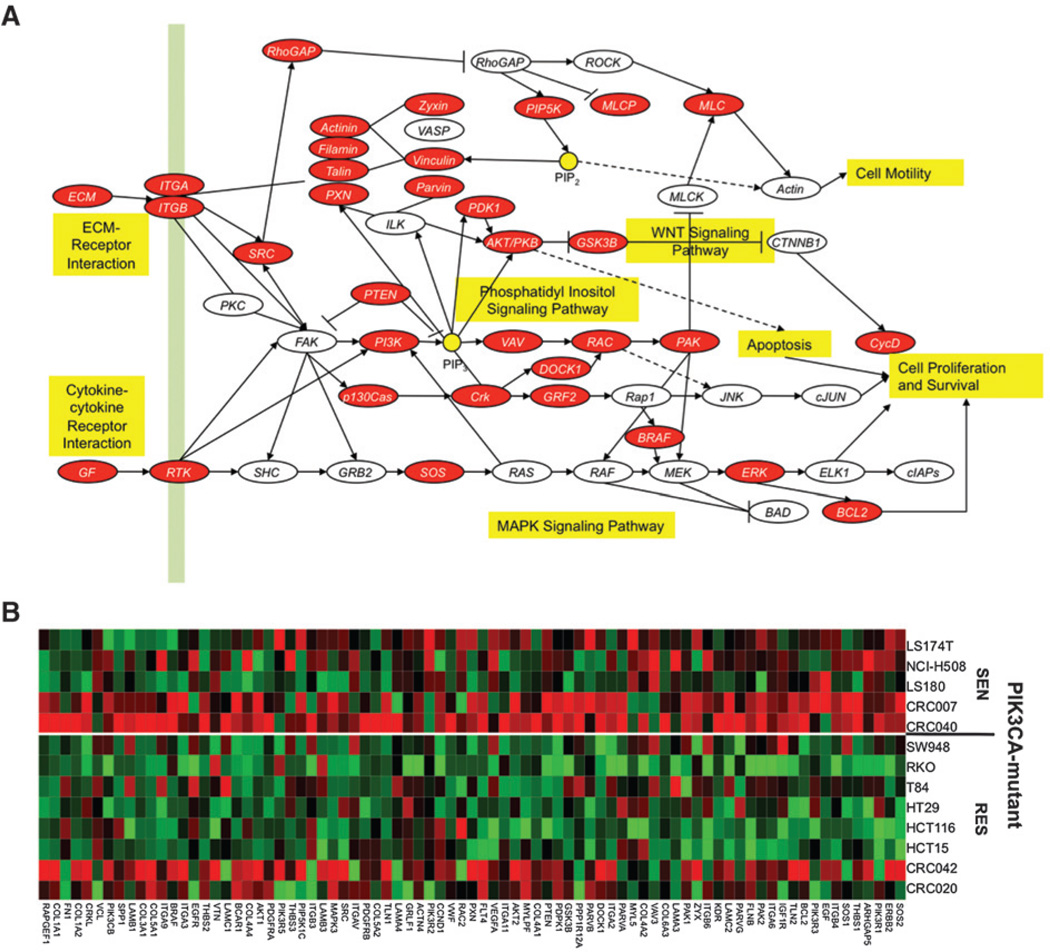
Src and PI3K pathways are upregulated in PIK3CA mutant sensitive
To investigate the differences between PIK3CA-mutant–sensitive versus PIK3CA-mutant–resistant cell lines and explants, we conducted gene array and pathway analysis. As shown in Fig. 4, many components of the Src and PI3K pathways were upregulated (highlighted in red; A, pathway and B, heatmap) in the PIK3CA-mutant–sensitive cell lines and explants when compared with those PIK3CA-mutant and -resistant. Interestingly, b-Raf and extracellular signal— regulated kinase (Erk)1/2 also had increased gene expression in PIK3CA-mutant–sensitive (data not shown). In contrast, expression of genes involved in cell-cycle control were increased in PIK3CA-mutant–resistant when compared with PIK3CA-mutant–sensitive (data not shown).
Figure 4.
Src pathway is enriched in sensitive cell lines and explants. A, pathways enriched in the sensitive (SEN) cell lines and explants, and focal adhesion is one of the pathways with false discovery rate less than 1%. B, heatmap of the core genes of the focal adhesion pathway. Red and green represent over- and underexpressed genes, respectively. RES, resistant.
A novel PIK3CA 3′ UTR (c.*19T>C) identified in colorectal cancer explants
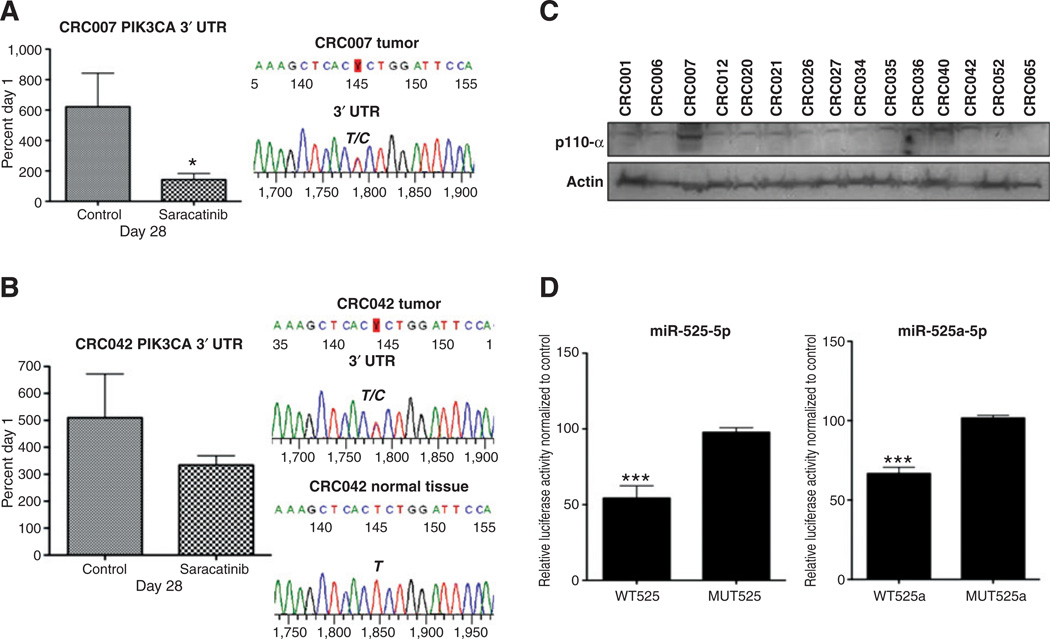
Analysis of the PIK3CA sequence in our colorectal cancer explants revealed a novel mutation in the 3′ UTR of the PIK3CA gene in 2 of our explants (CRC007 and CRC042). Whereas CRC042 showed some sensitivity to saracatinib (TGI = 65%), CRC007 had the greatest sensitivity (TGI = 23%) among all of the colorectal cancer explants tested (Fig. 5A). Comparison of normal versus tumor tissue in CRC042 revealed that this mutation was the result of a somatic mutation, as the mutation was not identified in the germ line (Fig. 5B). Unfortunately, no normal tissue was available for analysis on CRC007. Evaluation of p110-α protein levels in the colorectal cancer explants showed that CRC007 had the highest levels of p110 protein (Fig. 5C) when compared with other explants. Because we identified this mutation in the 3′ UTR (c* 19T>C) of the PIK3CA gene, an online search (www.targetscan.org) was conducted to identify miRNAs that have the potential to bind this region where the mutation is located. miR-520a-5p and miR-525-5p were predicted to be targets of this region in the 3′ UTR (c.*19T>C). We next set out to determine whether this mutation altered the binding of miR-520a and miR-525. We cloned the 164-bp wild-type sequence and the 164-bp mutant sequence separately into a psiCHECK-2 vector and conducted a dual luciferase assay 24 hours after the addition of the miR-520 and miR-525. As shown in Fig. 5D, the addition of miR-520a and miR-525 significantly reduced the luciferase signal in the wild-type sequence, whereas no differences from control were observed in the mutant miR-520a and miR-525. These results suggest that a mutation in the 3′ UTR (c* 19T>C) reduces the binding of miR-520a and miR-525, resulting in an increase in protein levels of the p110-α subunit of PI3K.
Figure 5.
A novel mutation in the PIK3CA 3′ UTR affects the binding of miR-520a-5p and miR-525-5p. CRC007 (A) and CRC042 (B) had increased sensitivity to saracatinib. Data are presented as an average of treated (T)/control (C) × 100 at 28 days (end of study). A novel PIK3CA mutation in the 3′ UTR was identified in both explants by sequencing the PIK3CA gene in (A) CRC007 tumor and (B)CRC042 normal and tumor. C, Western blot analysis of the p110-α protein. Protein levels of the p110-α subunit were the greatest in CRC007. D, cotransfection of miR-520a or miR-525 mimic and wild-type psi-PIK3CA-T plasmid significantly decreases the luciferase activity from HEK293 cell lysates compared with control group. While cotransfection with the mutant identified in these studies psi-PIK3CA-C was not significantly different from the empty vector control. ***P < 0.001. CRC, colorectal cancer. CRC, colorectal cancer; WT, wild-type.
Discussion
The heterodimeric (regulatory and catalytic subunits) PI3K lipid kinase phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to produce phosphatidylinositol-3,4,5-triphosphate (PIP3). This triggers a cascade of events whereby PIP3 phosphorylates PDK-1 that in turn activates Akt, resulting in the potentiation of cellular growth and proliferation. The PIK3CA gene (p110-α subunit) is located on chromosome 3q26.3 and 3 common mutations have been described that include 2 helical mutations at E542K and E545K (exon 9), which alters the inhibitory effects of p85 and one kinase domain mutation at H1047R (exon 20) that enhances the catalytic lipid kinase activity (6). All 3 mutations have been shown to be functionally relevant in the dysregulation of Akt-dependent and -independent signaling (33, 34). In this preclinical study, we showed that a PIK3CA mutation in exon 9 or 20 as well as a novel 3′ UTR (c.*19T>C) mutation was associated with enhanced sensitivity to saracatinib in both colorectal cancer cell lines and a patient-derived colorectal cancer explant model.
In our colorectal cancer preclinical model, we have identified a subset of tumors that harbor PIK3CA mutations that have increased sensitivity to saracatinib. In particular, PIK3CA mutations in exon 9 or exon 20 were identified in 3 sensitive cell lines (LS180, LS174T, and H508) and 2 sensitive colorectal cancer explants (CRC007 and CRC040). Of note, gene amplification of the PIK3CA region was identified in the resistant colorectal cancer explants CRC006 and CRC012. Evaluation of colorectal cancer explants showed a significant decrease in TGI in PIK3CA mutants when compared with wild-type tumors. These findings indicate that the Src pathway plays an important role in regulating the growth of tumors with PIK3CA mutations. More recently, the importance of PIK3CA mutations and clinical outcomes has been investigated. In particular, mutations in the PIK3CA gene have been shown to be associated with local recurrences in rectal cancer and predictive of poor outcomes in patients with colorectal cancers (11, 12). He and colleagues (11) examined PIK3CA mutations in 240 rectal tumors and showed that the patients with a mutation in PIK3CA gene (exon 9 or 20) were more likely to have disease recurrence. In addition, Kato and colleagues (12) showed PIK3CA mutations to be independent predictors of worse survival among 158 patients with colorectal cancers. These studies, as well as our findings in our preclinical study, may have important implications for treatment of patients with colorectal cancers; saracatinib treatment may only be effective in patients with colorectal cancers with PIK3CA mutations and treatment may ultimately decrease disease recurrence and improve outcomes in this subset of patients.
Both Fak and Stat-3 are well-known targets of Src and are involved at controlling many biologic processes involved in cell survival and metastasis (14, 15, 17). Previously, we have shown in the sensitive cell lines LS174T and H508 that saracatinib treatment reduced the activation of Src, Fak (Tyr861), and Stat-3 (Tyr 705; ref. 23). Interestingly, these cell lines also exhibited greater Src activation than in the resistant cell lines SW620 and SW480 (23). As expected, in this study, we found similar results in the LS180 sensitive cell line; treatment with saracatinib resulted in a decrease in the phosphorylation of Src, Fak, and Stat-3. Because we identified a significant association between PIK3CA mutants and sensitivity to saracatinib, we next explored the interaction between Src and the PI3K/Akt pathway. We showed that Src inhibition resulted in a decrease in the activation of PDK-1 and Akt. As previously investigated, oncogenic mutations in the PIK3CA gene also have Akt-independent effects, mainly through the activation of SGK-3 (33); however, we did not observe Akt-independent effects, as saracatinib did not alter the activation of SGK-3. Interestingly, saracatinib treatment also inhibited the Akt pathway in the resistant cell line HT29 and resistant CRC020 explant, suggesting that although PIK3CA is mutated in this cell line and explant, other genetic abnormalities may be present and more important at driving the growth of these tumors. Identifying these genetic differences will provide a stronger PIK3CA marker of sensitivity.
Previously, we have shown that the main effects of saracatinib on cellular proliferation are by inducing cell-cycle arrest and not apoptosis (23). Therefore, we examined the Akt downstream targets ribosomal S6 and p27KIP1. Both p27KIP1 and ribosomal S6 have been shown to be important regulators of the cell cycle and cell growth, respectively (35, 36). Saracatinib treatment decreased the phosphorylation of ribosomal S6 and increased levels of p27KIP1. Furthermore, Stat-3 and Fak have been shown to be involved in cellular proliferation and cell-cycle control (37, 38) and, as discussed above, saracatinib also reduced the activation of Stat-3 and Fak. Taken together, these results indicate that saracatinib may induce cell-cycle arrest in PIK3CA-mutant tumors through inhibition of the downstream targets of the Src and PI3K signaling pathways.
We also set out to determine the upstream effects of Src inhibition on the PI3K pathway. A study by Lu and colleagues (39) showed that Src interacts with the tumor suppressor PTEN to regulate the PI3K pathway. For instance, greater activity of Src reduces the stability of PTEN, resulting in an increase in the Akt pathway (39). In addition, Zhang and colleagues (40) identified a mechanism whereby PTEN downregulates Src activity. In this study, we did not observe any effects with treatment on the phosphorylated/total PTEN levels. Therefore, we conducted co-immunoprecipitation studies with Src and p85 and showed that saracatinib reduces the interaction between Src and p85 and, as a result, enhances the binding of p85 and p110. Given that Src inhibition increases p85/p110 heterodimerization and the p85 subunit of PI3K in its inactivated form stabilizes and prevents the activation of the p110 subunit (41) indicates a possible mechanism where saracatinib decreases the PI3K/AKT pathway in tumors with PIK3CA mutations.
In this study, we identified a subset of PIK3CA-mutant cell lines and explants that were sensitive to saracatinib. To have a better understanding of the genetic differences between PIK3CA-mutant–sensitive and -resistant cell lines and explants, we conducted gene array and pathway analysis. We show that the Src and PI3K pathways were enriched in the PIK3CA-mutant saracatinib–sensitive cell lines and explants, suggesting that the growth of these tumors were more dependent on Src and PI3K pathways. These results were consistent with our previous study showing that the activation of Src was the greatest in cell lines and explant tumors that were sensitive to saracatinib. Additional genetic studies on PIK3CA-mutant–resistant tumors are needed to further define the important aberrations driving tumor growth in this subset of tumors.
In this study, we identified a novel mutation (T–C transition) in CRC007 and CRC042 in the 3′ UTR that is located 19 base pairs from the stop codon of the PIK3CA gene. Similar to the PIK3CA helical and kinase domain mutations (CRC040, LS180, H508, and LS174T), CRC007 (TGI = 23%) and CRC042 (TGI = 65%) exhibited sensitivity to saracatinib. Of note, although, saracatinib had some treatment effects on CRC042 in our study, this explant was classified as resistant. Analysis of p110 protein levels revealed an increase in protein expression in CRC007 (highest) and CRC042, when compared with other colorectal cancer explants. These findings indicate that this mutation in the 3′ UTR (c* 19T>C) may alter the binding of miRNA (s), resulting in an increase in p110 protein translation. Given this observation, we searched for potential miRNAs with a predicted sequence match for this region and identified miR-520a-5p and miR-525-5p. Herein, we determined that both miR-520a-5p and miR-525-5p decreased luciferase reporter gene activity arising from wild-type PIK3CA but did not have an effect on the activity from the mutant PIK3CA sequence. These results show that we identified the PIK3CA gene as a target for miR-520a-5p and miR-525-5p and further show that the mutation in the 3′ UTR (c.*19T>C) of the PIK3CA gene reduces the binding of both miRNAs. Whether this increase in p110 protein expression translates to enhanced PI3K activity remains to be determined. Perhaps, with increased extracellular stimuli promoting tumor growth in conjunction with enhanced levels of p110 in the PIK3CA-mutant 3′ UTR (c.*19T>C) tumors may enhance the activity of this pathway.
Our colorectal cancer explant model has been used previously to predict biomarkers of sensitivity and resistance (23, 25, 42–44). Although, we feel that this model more closely reflects tumor heterogeneity and growth characteristics seen in patients when compared with cell lines, one of the main limitations with this model is the limited dynamic range. As is the case with using IC50 arbitrary cutoff values in in vitro drug potency studies, there are also limitations using TGI cutoff values (TGI ≤ 50%, sensitive and TGI > 50%, resistant) in our colorectal cancer explant model. One of the main difficulties is how to interpret small differences in TGI. For example, it is highly probable that a TGI of 49% (we classify as sensitive) and 51% (we classify as resistant) are not very different with respect to treatment effects. Thus, as there is a limited dynamic range in this model, we generally do not use tumors with intermediate sensitivity and typically compare tumors with a TGI ≤ 50% (sensitive) to the most resistant tumors with a TGI of approximately >80% for biomarker development. Whether this is the best way of categorizing tumors based on sensitive and resistance remains to be determined.
Although the benefit of saracatinib in clinical trials has been disappointing, identification of biomarkers of sensitivity may further define a population of patients who will benefit from treatment with this compound, or point the way toward effective combination strategies. In our colorectal cancer preclinical model, saracatinib reduced tumor growth in a subset of tumors with a PIK3CA mutation. Our data show that in addition to inhibiting the Src pathway, saracatinib disrupts the activation of the Akt pathway thereby enhancing cell-cycle arrest in tumors with PIK3CA mutations. Because saracatinib was not effective on all tumors with PIK3CA mutations, additional studies are warranted to further define the genetic differences between PIK3CA-mutant–sensitive tumors and PIK3CA-mutant–resistant tumors. This will ultimately provide a more robust biomarker for patient selection. Whether these results translate to the clinical setting remains to be determined. These findings are being translated into a biomarker-driven clinical trial, which is currently under development.
Supplementary Material
Translational Relevance.
The Src and phosphoinositide 3-kinase (PI3K) pathways are frequently dysregulated in colorectal cancer and play an important role in tumorigenesis. The objective of this study was to investigate the effects of the Src inhibitor, saracatinib, in a preclinical colorectal cancer model and to identify biomarkers of sensitivity. Here, we show that a mutation in the PIK3CA gene is associated with enhanced sensitivity to saracatinib. In addition to the common helical (E542K and E545K) and kinase (H1047R) mutations, a novel 3’ untranslated region mutation in the PIK3CA gene was also shown to be associated with increased sensitivity to saracatinib. This mutation resulted in a reduced affinity for miR-520a and miR-525a as well as increased levels of p110-a when compared with wild-type. Importantly, treatment with saracatinib inhibits the Src pathway and the activation of Akt-dependent signaling by altering Src and p85 interaction. Thus, these results indicate that a subset of patients with colorectal cancer with a PIK3CA mutation may derive benefit from treatment with the Src inhibitor saracatinib.
Acknowledgments
Grant Support
This work was supported by grant 1RO1CA152303-01.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
S.G. Eckhardt has commercial research grant from AstraZeneca. J.J. Arcaroli and W.A. Messersmith are consultants/advisory board members for AstraZeneca. No potential conflicts of interests were disclosed by other authors.
Authors' Contributions
Conception and design: J.J. Arcaroli, W.A. Messersmith
Development of methodology: J.J. Arcaroli, M. Varella-Garcia, L. Bemis, A. Dasari, S.G. Eckhardt
Acquisition of data (provided animals,acquired and managed patients, provided facilities, etc.): J.J. Arcaroli, K.S. Quackenbush, R.W. Powell, T.M. Pitts, A. Spreafico, M. Varella-Garcia, B.M. Touban, A. Dasari, S.G. Eckhardt, W.A. Messersmith
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.J. Arcaroli, M. Varella-Garcia, L. Bemis, A.C. Tan, A. Dasari, S.G. Eckhardt, W. A. Messersmith
Writing, review, and/or revision of the manuscript: J.J. Arcaroli, M. Varella-Garcia, A.C. Tan, A. Dasari, S.G. Eckhardt, W.A. Messersmith
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.J. Arcaroli, T.M. Pitts, J.M. Reinemann, A. Dasari, W.A. Messersmith
Study supervision: W.A. Messersmith.
References
- 1.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Wymann MP, Zvelebil M, Laffargue M. Phosphoinositide 3-kinase signalling–which way to target? Trends Pharmacol Sci. 2003;24:366–376. doi: 10.1016/S0165-6147(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 3.Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 4.Franke TF, Kaplan DR, Cantley LC. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 5.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 6.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 7.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Jr, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–2259. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Kuropatwinski K, Hauser J, Rossi MR, Zhou Y, Conway A, et al. Colon carcinoma cells harboring PIK3CA mutations display resistance to growth factor deprivation induced apoptosis. Mol Cancer Ther. 2007;6:1143–1150. doi: 10.1158/1535-7163.MCT-06-0555. [DOI] [PubMed] [Google Scholar]
- 10.Guo XN, Rajput A, Rose R, Hauser J, Beko A, Kuropatwinski K, et al. Mutant PIK3CA-bearing colon cancer cells display increased metastasis in an orthotopic model. Cancer Res. 2007;67:5851–5858. doi: 10.1158/0008-5472.CAN-07-0049. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Van't Veer LJ, Mikolajewska-Hanclich I, van Velthuysen ML, Zeestraten EC, Nagtegaal ID, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15:6956–6962. doi: 10.1158/1078-0432.CCR-09-1165. [DOI] [PubMed] [Google Scholar]
- 12.Kato S, Iida S, Higuchi T, Ishikawa T, Takagi Y, Yasuno M, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer. 2007;121:1771–1778. doi: 10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 13.Yeatman T. A renaissance for src. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 14.Hauck C, Hsia D, Schlaepfer D. The focal adhesion kinase: a regulator of cell migration and invasion. IUBMB Life. 2002;53:115–119. doi: 10.1080/15216540211470. [DOI] [PubMed] [Google Scholar]
- 15.Niu G, Wright K, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 16.Schaller MD. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim Biophys Acta. 2001;1540:1–21. doi: 10.1016/s0167-4889(01)00123-9. [DOI] [PubMed] [Google Scholar]
- 17.Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 18.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 19.Irby R, Mao W, Coppola D, Kang J, Loubeau J, Trudeau W, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 20.Cartwright C, Meisler A, Eckhart W. Activation of the pp60c-src protein kinase is an early event in colonic carcinogenesis. Proc Natl Acad Sci U S A. 1990;87:558–562. doi: 10.1073/pnas.87.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malek RL, Irby RB, Guo QM, Lee K, Wong S, He M, et al. Identification of Src transformation fingerprint in human colon cancer. Oncogene. 2002;21:7256–7265. doi: 10.1038/sj.onc.1205900. [DOI] [PubMed] [Google Scholar]
- 22.Talamonti M, Roh M, Curley S, Gallick G. Increase in activity and level of pp60c-src in progressive stages of human colorectal cancer. J Clin Invest. 1993;91:53–60. doi: 10.1172/JCI116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arcaroli JJ, Touban BM, Tan AC, Varella-Garcia M, Powell RW, Eckhardt SG, et al. Gene array and fluorescence in situ hybridization biomarkers of activity of saracatinib (AZD0530), a Src inhibitor, in a preclinical model of colorectal cancer. Clin Cancer Res. 2010;16:4165–4177. doi: 10.1158/1078-0432.CCR-10-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green TP, Fennell M, Whittaker R, Curwen J, Jacobs V, Allen J, et al. Preclinical anticancer activity of the potent, oral Src inhibitor AZD0530. Mol Oncol. 2009;3:248–261. doi: 10.1016/j.molonc.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajeshkumar NV, Tan AC, De Oliveira E, Womack C, Wombwell H, Morgan S, et al. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin Cancer Res. 2009;15:4138–4146. doi: 10.1158/1078-0432.CCR-08-3021. [DOI] [PubMed] [Google Scholar]
- 26.Fury MG, Baxi S, Shen R, Kelly KW, Lipson BL, Carlson D, et al. Phase II study of saracatinib (AZD0530) for patients with recurrent or metastatic head and neck squamous cell carcinoma (HNSCC) Anticancer Res. 2011;31:249–253. [PMC free article] [PubMed] [Google Scholar]
- 27.Mackay HJ, Au HJ, McWhirter E, Alcindor T, Jarvi A, Macalpine K, et al. A phase II trial of the Src kinase inhibitor saracatinib (AZD0530) in patients with metastatic or locally advanced gastric or gastro esoph-ageal junction (GEJ) adenocarcinoma: a trial of the PMH phase II consortium. Invest New Drugs. 2011 Mar 12; doi: 10.1007/s10637-011-9650-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Dona-hue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 29.Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, et al. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 30.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetux-imab. Cancer Res. 2008;68:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd RV, Erickson LA, Jin L, Kulig E, Qian X, Cheville JC, et al. p27kip1: a multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am J Pathol. 1999;154:313–323. doi: 10.1016/S0002-9440(10)65277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson RT, Schreiber SL. Translation control: connecting mitogens and the ribosome. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- 37.Fukada T, Ohtani T, Yoshida Y, Shirogane T, Nishida K, Nakajima K, et al. STAT3 orchestrates contradictory signals in cytokine-induced G1 to S cell-cycle transition. EMBO J. 1998;17:6670–6677. doi: 10.1093/emboj/17.22.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Yu Q, Liu JH, Zhang J, Wang H, Koul D, et al. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J Biol Chem. 2003;278:40057–40066. doi: 10.1074/jbc.M303621200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuevas BD, Lu Y, Mao M, Zhang J, LaPushin R, Siminovitch K, et al. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J Biol Chem. 2001;276:27455–27461. doi: 10.1074/jbc.M100556200. [DOI] [PubMed] [Google Scholar]
- 42.Tentler JJ, Nallapareddy S, Tan AC, Spreafico A, Pitts TM, Morelli MP, et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in K-ras-mutated colorectal cancer. Mol Cancer Ther. 2010;9:3351–3362. doi: 10.1158/1535-7163.MCT-10-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messersmith WA, Rajeshkumar NV, Tan AC, Wang XF, Diesl V, Choe SE, et al. Efficacy and pharmacodynamic effects of bosutinib (SKI-606), a Src/Abl inhibitor, in freshly generated human pancreas cancer xenografts. Mol Cancer Ther. 2009;8:1484–1493. doi: 10.1158/1535-7163.MCT-09-0075. [DOI] [PubMed] [Google Scholar]
- 44.Pitts TM, Tan AC, Kulikowski GN, Tentler JJ, Brown AM, Flanigan SA, et al. Development of an integrated genomic classifier for a novel agent in colorectal cancer: approach to individualized therapy in early development. Clin Cancer Res. 2010;16:3193–3204. doi: 10.1158/1078-0432.CCR-09-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.