Abstract
Changes in protein abundance in skeletal muscle are central to a large number of metabolic and other disorders, including, and perhaps most commonly, insulin resistance. Proteomics analysis of human muscle is an important approach for gaining insight into the biochemical basis for normal and pathophysiological conditions. However, to date, the number of proteins identified by this approach has been limited, with 107 different proteins being the maximum reported so far. Using a combination of one-dimensional gel electrophoresis and high performance liquid chromatography electrospray ionization tandem mass spectrometry, we identified 954 different proteins in human vastus lateralis muscle obtained from three healthy, nonobese subjects. In addition to a large number of isoforms of contractile proteins, we detected all proteins involved in the major pathways of glucose and lipid metabolism in skeletal muscle. Mitochondrial proteins accounted for 22% of all proteins identified, including 55 subunits of the respiratory complexes I–V. Moreover, a number of enzymes involved in endocrine and metabolic signaling pathways as well as calcium homeostasis were identified. These results provide the most comprehensive characterization of the human skeletal muscle proteome to date. These data hold promise for future global assessment of quantitative changes in the muscle proteome of patients affected by disorders involving skeletal muscle.
The percutaneous muscle biopsy technique is an important tool in the diagnosis and management of human muscle disorders and has been used widely to investigate many aspects of cell structure and metabolism in normal and abnormal human muscle (1, 2). There is a lack of comprehensive knowledge of the molecular basis of changes in protein expression that are part of normal physiologic adaptations, such as those that occur during exercise training or age-related sarcopenia, or pathophysiological changes associated with disease processes, including atrophy (caused by disuse, spinal cord injury, or weightlessness), ischemic damage, muscular dystrophies, and insulin resistance. Insulin resistance in skeletal muscle is a condition that is common to obesity, type 2 diabetes, and cardiovascular disease. These diseases affect 25–50 million individuals in the United States alone and many more worldwide. During the past decades, percutaneous needle biopsy of the vastus lateralis muscle has played an important role in defining the molecular abnormalities underlying insulin resistance (3–5). On the basis of such studies, an increasing number of genes and proteins in several signaling and metabolic pathways have been implicated in skeletal muscle insulin resistance (3–7). However, the majority of studies have focused on only a limited number of genes and proteins. Other investigators have used the needle muscle biopsy technique to study biochemical, histological, and molecular abnormalities in other common diseases, such as the muscular dystrophies (8).
There is a clear need for approaches capable of evaluating global changes in protein expression and modification. High throughput gene expression technologies such as cDNA microarrays are powerful tools for the study of physiological and pathological conditions with complex or multifactorial underlying mechanisms, proving useful in studies of age-related sarcopenia (9), muscle insulin resistance (10–13), muscular dystrophy (8), denervation (14), and endurance training (15). However, expression of muscle mRNA may not accurately reflect the abundance of proteins and can give no information regarding their posttranslational modifications (16, 17), indicating the need to correlate the results from global gene expression experiments and measurements of protein abundance.
Although human vastus lateralis muscle has been used in thousands of studies within the past decade, only a few proteomic studies have been conducted, combining protein separation by two-dimensional gel electrophoresis and protein identification by mass spectrometry (18–23). Previously, the most comprehensive assessment of the human vastus lateralis proteome reported only 107 different proteins (18). The high abundance of cytoskeletal and contractile proteins combined with dynamic range issues associated with the two-dimensional gel approach may be factors that have hampered studies on this topic.
The use of two orthogonal liquid chromatography steps for peptide separation prior to tandem mass spectrometry (MS/MS)1 (24) has yielded substantial improvements in detection limits; consequently, identification of increased numbers of proteins in mammalian tissues (25, 26). Recently, two-dimensional high performance liquid chromatography electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) was used to analyze the proteome of human beta cells, resulting in the identification of 3365 unique proteins (27). An alternative to two-dimensional gels and two-dimensional HPLC uses separation by one-dimensional SDS-PAGE and subsequent protein identification by HPLC-ESI-MS/MS (28, 29). This approach has been shown to be comparable to two-dimensional HPLC-ESI-MS/MS with respect to the number of proteins identified (30). Moreover, it is simpler from a technological point of view and is suitable for clinical studies of small tissue samples obtained in vivo by biopsy.
In the present study, we used one-dimensional gel electrophoresis and HPLC-ESI-MS/MS to characterize the proteome of human skeletal muscle obtained using percutaneous needle biopsies of the vastus lateralis muscle in healthy volunteers. We identified 954 proteins, including all enzymes participating in the major pathways of glucose and lipid metabolism, a large number of proteins involved in mitochondrial oxidative phosphorylation and calcium homeostasis, and most isoforms of the proteins that constitute the myofibrillar apparatus. As such, the proteins identified in our analyses provide a representation of the major biological functions of skeletal muscle.
EXPERIMENTAL PROCEDURES
Subjects
The skeletal muscle samples used for the proteomics analyses in this study were obtained from three healthy, nonobese volunteers (I–III; age, 30–47 years; body mass index, 24–28 kg/m2; percent body fat, 23–34%) with normal glucose tolerance and no family history of type 2 diabetes. The purpose, nature, and potential risks of the study were explained to the participants, and written consent was obtained before participation. The protocol was approved by the Institutional Review Board of Arizona State University.
Muscle Preparation, Electrophoresis, and Staining
A percutaneous needle biopsy of the vastus lateralis muscle was obtained under local anesthesia, and the muscle biopsy specimen was immediately blotted free of blood, frozen, and stored in liquid nitrogen until use. For protein analysis, the muscle biopsy specimens were homogenized while still frozen in an ice-cold buffer (10 µl/mg tissue) consisting of (final concentrations): 50 mm HEPES, pH 7.6, 150 mm NaCl, 20 mm sodium pyrophosphate, 20 mm β-glycerophosphate, 10 mm NaF, 2 mm sodium orthovanadate, 2 mm EDTA, 1% Triton, 10% glycerol, 2 mm phenylmethylsulfonyl fluoride, 1 mm MgCl2, 1 mm CaCl2, 10 µg/ml leupeptin, and 10 µg/ml aprotinin. A Polytron homogenizer (Brinkman Instruments, Westbury, NY) set on maximum speed for 30 s was used for homogenization. The homogenate was cooled on ice for 20 min and then centrifuged at 10,000 × g for 20 min at 4 °C; the resulting supernatant was frozen until use. Protein concentrations were determined by the method of Lowry (31). Two experiments were performed. Experiment 1: subject I, 75 µg of muscle lysate, proteins separated on a 10% SDS-polyacrylamide gel, and HPLC-MS/MS run in duplicate (Experiments 1–1 and 1–2). Experiments 2–1 and 2–2: 60 µg of muscle lysate proteins from subjects II and III, respectively, were separated on 4–20% gradient SDS-polyacrylamide gels; proteins were visualized with Coomassie blue (Sigma Chemical Co., St. Louis, MO).
In-gel Digestion
The gel lane resulting from each experiment was cut into 20–24 slices of approximately equal size. Each slice was cut into 1-mm cubes prior to digestion. The gel pieces were placed in a 0.6-ml polypropylene tube, washed with 400 µl of water, destained twice with 300 µl of 50% acetonitrile (ACN) in 40 mm NH4HCO3, and dehydrated with 100% ACN for 15 min. After removal of the ACN by aspiration, the gel pieces were dried in a vacuum centrifuge at 62 °C for 30 min. Trypsin (250 ng; Sigma Chemical Co.) in 30 µl of 40 mm NH4HCO3 was added and the samples were maintained at 4 °C for 15 min prior to the addition of 50 µl of 40 mm NH4HCO3. The digestion was allowed to proceed at 37 °C overnight and was terminated by the addition of 10 µl 5% formic acid (FA). After incubation at 37 °C for an additional 30 min and centrifugation for 1 min, each supernatant was transferred to a clean polypropylene tube. The extraction procedure was repeated using 80 µl of 0.5% FA, and the two extracts were combined. The sample volume was reduced to ~5 µl by vacuum centrifugation, and 20 µl of 0.05% heptafluorobutyric acid/1% FA: 2%ACN was added.
Mass Spectrometry
HPLC-ESI-MS/MS was performed on a hybrid linear ion trap (LTQ)-Fourier Transform Ion Cyclotron Resonance mass spectrometer (LTQ FT; Thermo Fisher; San Jose, CA) fitted with a PicoView nanospray source (New Objective, Woburn, MA). The mass spectrometer was calibrated weekly according to manufacturer’s instructions, achieving mass accuracy of the calibrants within 2 ppm. On-line capillary HPLC was performed using a Michrom BioResources Paradigm MS4 micro HPLC (Auburn, CA) with a PicoFrit column (New Objective; 75 µm inner diameter, packed with ProteoPep II C18 material, 300 Å). Samples were desalted using an on-line Nanotrap (Michrom BioResources) before being loaded onto the PicoFrit column. HPLC separations were accomplished with a linear gradient of 2 to 27% ACN in 0.1% FA in 70 min, a hold of 5 min at 27% ACN, followed by a step to 50% ACN, hold 5 min and then a step to 80%, hold 5 min; flow rate, 300 nl/min. A “top-10” data-dependent tandem mass spectrometry approach was utilized to identify peptides in which a full scan spectrum (survey scan) was acquired followed by collision-induced dissociation (CID) mass spectra of the 10 most abundant ions in the survey scan. The survey scan was acquired using the Fourier Transform Ion Cyclotron Resonance mass analyzer to obtain high resolution, high mass accuracy data.
Data Analysis and Bioinformatics
Tandem mass spectra were extracted from Xcalibur “RAW” files and charge states were assigned using the Extract MSN script that is a component of Xcalibur 2.0 SR2 (Thermo Fisher). Charge states and monoisotopic peak assignments were then verified using DTA-SuperCharge, part of the MS-Quant suite of software (32), before all “DTA” files from each gel lane in an experiment were combined into a single Mascot Generic format file. The fragment mass spectra were then searched against the IPI_HUMAN_v3.27 database (67,528 entries, http://www.ebi.ac.uk/IPI/) using Mascot (Matrix Science, London, United Kingdom, version 2.2). The false discovery rate was determined by selecting the option to use a “decoy” randomized search strategy that is available in Mascot, v2.2. The search parameters that were used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Probability assessment of peptide assignments and protein identifications were made through use of Scaffold (version Scaffold-01_06_17, Proteome Software Inc., Portland, OR). Only peptides with ≥95% probability were considered. Criteria for protein identification included detection of at least 2 unique identified peptides and a probability score of ≥95%. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped. Multiple isoforms of a protein were reported only if they were differentiated by at least one unique peptide with ≥ 95% probability, based on Scaffold analysis. IPI accession numbers for identified proteins were input into the UniProt database (www.pir.uniprot.org) to obtain Gene Ontology annotation.
RESULTS
Characteristics of Human Skeletal Muscle Proteome
To obtain a comprehensive proteomic characterization of the human vastus lateralis muscle, we carried out HPLC-ESI-MS/MS-based analysis of lysates of whole muscle from which proteins were first fractionated by one-dimensional gel electrophoresis. We first performed duplicate HPLC-ESI-MS/MS analyses (experiments 1–1 and 1–2) on proteins isolated from the muscle of subject I to assess the reproducibility of the approach. In these analyses, 591 and 577 proteins were identified, respectively, corresponding to a total of 668 unique proteins. There were at least two unique peptides (≥95% confidence) assigned to each identified protein, all with a confidence level ≥95% based on the Scaffold analysis. The false discovery rates as assessed by Mascot searching of a randomized database were 3.1% and 3.4%, respectively. Among these proteins, 500 were identified in both experiments 1–1 and 1–2, giving a reproducibility rate of ~75%. Data analysis showed that the three major isoforms of myosin heavy-chain (myosin 1, 2, and 7) comprised ~42% of the total spectra, indicating that the analytical space taken up by these myosin isoforms may hamper identification of other proteins that migrate similarly in SDS-polyacrylamide gels. In an attempt to minimize this, in subsequent analyses for subjects II and III, we used gradient gels (4–20%) to obtain a better separation of proteins; after separation on the gradient gels, we identified 571 and 616 proteins for subjects II and III, respectively; the proportion of spectra assigned to the 3 major myosin heavy-chain isoforms was reduced to ~32%. The false discovery rate was 5.6% and 5.2%, respectively. A total of 741 unique proteins were identified in the samples isolated from subjects II and III, with 446 of them (60%) identified in the samples from both subjects. Combining the results from all experiments for the three subjects resulted in a total of 954 proteins after redundancy reduction. To our knowledge, the proteins reported here constitute the largest catalog of the healthy human skeletal muscle proteome to date. A detailed list of all proteins identified in this study together with their IPI ID, molecular weight, sequence coverage, and number of unique peptides assigned to each protein are provided as supplemental information (supplemental Table I). For each identified peptide sequence, we have included the following in supplemental Table II: modifications, flank residues, precursor mass, charge and mass error observed, the best Mascot score, and best peptide identification probability.
Among the proteins identified in this study, numerous entries derived from the protein identification searches had multiple IPI IDs. In many cases, assignment of multiple IDs results from the potential presence of protein isoforms, which cannot be distinguished on the basis of unique peptides. In our results tables, protein with multiple IDs were assigned to a “protein group” (supplemental Table I). Proteins that were attributed a unique IPI ID are listed as a single-entry protein group. As can be seen from supplemental Table I, by performing the analysis in this manner, there were 954 protein groups. Among them, the majority (562) had one IPI ID, 230 had two associated IPI IDs and 162 had more than two associated IPI IDs. For 354 of 392 protein groups containing two or more IPI IDs, the corresponding proteins are known to be translated from the same gene. For protein groups with multiple IPI IDs, the minimum, maximum, and mean of molecular weights (MW), and the number of amino acids in each protein sequence are listed in supplemental Table I.
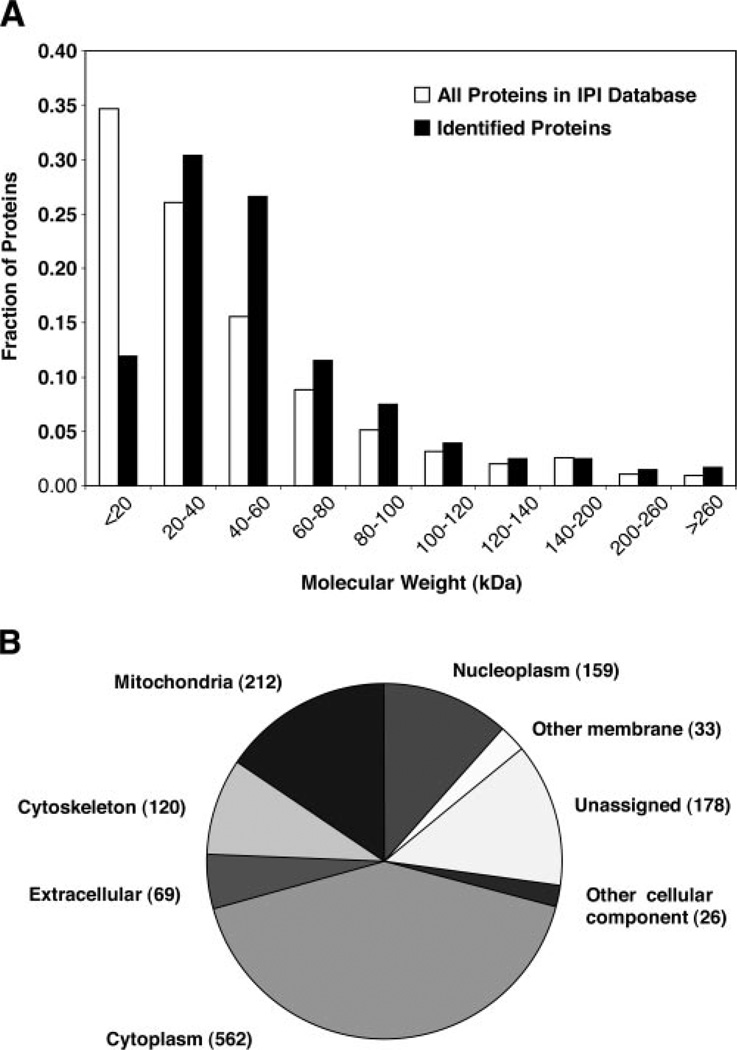
The MW distribution of all 954 identified proteins is shown in Fig. 1A. Nearly one-third of the identified proteins have a MW within the range of 20–40 kDa, and more than two-thirds are below 60 kDa. Proteins ranged from 6.1 kDa (cardiac phospholamban) to 3823 kDa (titin). We found 11 proteins with a MW less than 10 kDa and 114 greater than 100 kDa. Thus, 13% of the identified proteins were outside of the typical separation limits of two-dimensional gel electrophoresis, which has optimum resolution in the 10–100 kDa range.
Fig. 1. Molecular weight distribution and subcellular location of human skeletal muscle proteins.
A, distribution of the 954 proteins identified in human vastus lateralis muscle (closed bars) compared with all proteins in the IPI human database (open bars) relative to their predicted molecular weight. B, distribution of the 954 identified proteins according to Gene Ontology (GO) annotation. In cases where an identified protein group contained more than one IPI ID, the average molecular weight was used.
The subcellular location of 776 proteins could be assigned based on the Gene Ontology annotation information obtained from the UniProt database, while 178 remained unassigned (Fig. 1B). Fifty-nine percent of the identified proteins could be assigned to cytoplasm, representing the predominant subcellular location of all identified proteins in this study. It has been estimated that mitochondrial proteins comprise 4.8% of the total human proteome (33). However, in skeletal muscle, we found that 212 of the 954 proteins detected (22%) could be attributed to the mitochondrion. Nuclear proteins represented the third largest group, with 17% of all identified proteins.
Pathway Analysis
From the large number of proteins identified in our analyses, we decided to evaluate the potential applicability of this HPLC-ESI-MS/MS-based proteomic approach for quantitative assessment of skeletal muscle proteins in future studies aimed at elucidating the mechanism underlying changes in muscle structure and metabolism under different physiological and pathophysiological conditions. To do this, we examined the representation of the major metabolic pathways (glucose and lipid metabolism, electron transport, and oxidative phosphorylation), calcium homeostasis, myofibrillar apparatus, and components of IGF and insulin signaling among the proteins we identified.
Glucose and Lipid Metabolism
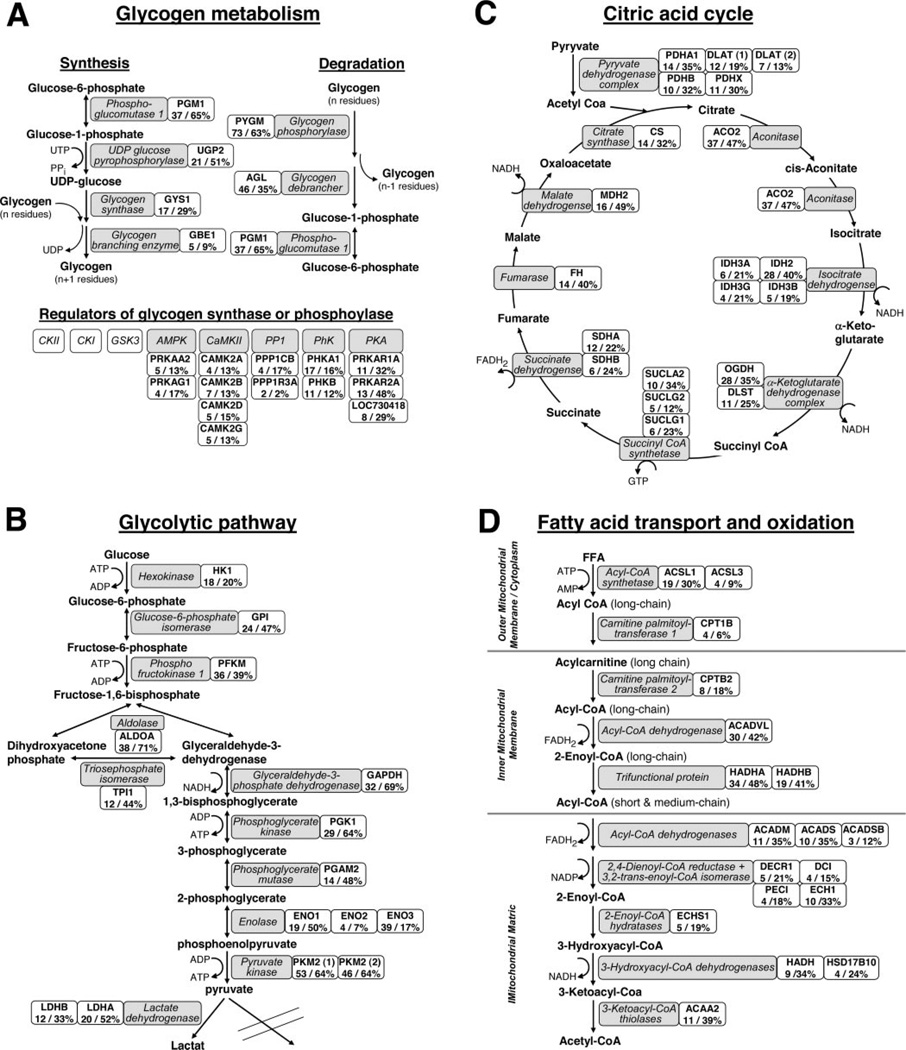
All enzymes involved in the glycolytic pathway, glycogen metabolism, and the citric acid cycle were identified in these experiments (Figs. 2A–C). Moreover, we identified several subunits of four kinases and one major phosphatase (PP1, serine/threonine-protein phosphatase) known to be involved in regulation of the phosphorylation of enzymes such as glycogen synthase or glycogen phosphorylase. In addition, the majority of proteins that are crucial for the activation and transport of fatty acids into the mitochondrion and subsequent degradation in the β-oxidation pathway were identified (Fig. 2D). These include enzymes involved in the oxidation of long-chain, medium-chain and short-chain activated fatty acids as well as enzymes required for the oxidation of unsaturated fatty acids.
Fig. 2. Proteomics coverage of major enzymes involved in glucose and lipid metabolism in human skeletal muscle.
A, isoforms and/or subunits of enzymes involved in glycogen metabolism. Glycolysis (B), citric acid cycle (C), and fatty acid transport (D) into and oxidation in mitochondria. Proteins identified are shown in gray boxes, and the associated gene names, maximum observed number of unique peptides, and sequence coverage are presented in adjacent white boxes.
Electron Transport and Phosphorylation
The major role of electron transport complexes I–IV is transfer of electrons coupled with proton pumping, with the ultimate goal of enabling oxidative phosphorylation (OXPHOS), the final step in energy production. Many of the subunits that make up complexes I–V are embedded in the inner membrane of mitochondria and are, therefore, often more difficult to extract from their membrane environment. In our experiments, we identified 55 (Table I) of the 88 known subunits of these complexes (34, 35). Many of the subunits in complexes I–V are of low molecular weight, which may explain why a smaller number of these subunits was identified in our experiments using 10% gels [29 in experiment 1–1 and 20 in experiment 1–2 (subject I)] compared with the experiments using the gradient (4–20%) gels [41 in experiment 2–1 (subject II) and 51 in experiment 2–2 (subject III)]. Several mitochondrial carrier and transfer proteins were also identified in our analyses.
Table I. Proteins involved in oxidative phosphorylation (OXPHOS) identified in human skeletal muscle.
Proteins were separated by one-dimensional gel electrophoresis and identified by HPLC-ESI-MS/MS in four experiments using skeletal muscle from three nonobese, healthy subjects.
| Protein | Gene name |
MW (kDa) |
Maximum unique peptidesa |
Maximum sequence coverage (%)a |
|---|---|---|---|---|
| Complex I | ||||
| NADH dehydrogenase 1 subunit C2 | NDUFC2 | 14 | 2 | 17 |
| NADH dehydrogenase 1 α subcomplex subunit 2 | NDUFA2 | 11 | 3 | 20 |
| NADH dehydrogenase 1 α subcomplex subunit 4 | NDUFA4 | 9 | 2 | 22 |
| NADH dehydrogenase 1 α subcomplex subunit 5 | NDUFA5 | 14 | 3 | 31 |
| NADH dehydrogenase 1 α subcomplex subunit 6 | NDUFA6 | 18 | 2 | 12 |
| NADH dehydrogenase 1 α subcomplex subunit 7 | NDUFA7 | 13 | 3 | 35 |
| NADH dehydrogenase 1 α subcomplex subunit 8 | NDUFA8 | 20 | 3 | 30 |
| NADH dehydrogenase 1 α subcomplex subunit 9 | NDUFA9 | 43 | 7 | 26 |
| NADH dehydrogenase 1 α subcomplex subunit 10 | NDUFA10 | 41 | 6 | 16 |
| NADH dehydrogenase 1 α subcomplex subunit 12 | NDUFA12 | 17 | 2 | 20 |
| NADH dehydrogenase 1 α subcomplex subunit 13 | NDUFA13 | 26 | 4 | 22 |
| NADH dehydrogenase 1 β subcomplex subunit 3 | NDUFB3 | 11 | 2 | 21 |
| NADH dehydrogenase 1 β subcomplex subunit 4 | NDUFB4 | 15 | 2 | 17 |
| NADH dehydrogenase 1 β subcomplex subunit 5 | NDUFB5 | 20 | 3 | 12 |
| NADH dehydrogenase 1 β subcomplex subunit 7 | NDUFB7 | 16 | 2 | 12 |
| NADH dehydrogenase 1 β subcomplex subunit 8 | NDUFB8 | 20 | 2 | 22 |
| NADH dehydrogenase 1 β subcomplex subunit 9 | NDUFB9 | 21 | 3 | 27 |
| NADH dehydrogenase 1 β subcomplex subunit 10 | NDUFB10 | 21 | 4 | 31 |
| NADH dehydrogenase 1 β subcomplex subunit 11 isoform 2 | NDUFB11 | 18 | 2 | 21 |
| NADH-ubiquinone oxidoreductase 9.6 kDa subunit | NDUFAB1 | 17 | 3 | 15 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit | NDUFS1 | 79 | 21 | 36 |
| NADH dehydrogenase iron-sulfur protein 2 | NDUFS2 | 53 | 15 | 45 |
| NADH dehydrogenase iron-sulfur protein 3 | NDUFS3 | 30 | 5 | 20 |
| NADH dehydrogenase iron-sulfur protein 4 | NDUFS4 | 20 | 2 | 15 |
| NADH dehydrogenase iron-sulfur protein 5 | NDUFS5 | 12 | 3 | 25 |
| NADH dehydrogenase iron-sulfur protein 7 | NDUFS7 | 28 | 3 | 15 |
| NADH dehydrogenase iron-sulfur protein 8 | NDUFS8 | 24 | 3 | 15 |
| NADH dehydrogenase flavoprotein 1 isoform 1 | NDUFV1 | 50 | 12 | 24 |
| NADH dehydrogenase flavoprotein 2 | NDUFV2 | 26 | 2 | 11 |
| NADH dehydrogenase flavoprotein 3 | NDUFV3 | 51 | 2 | 7 |
| Complex II | ||||
| Succinate dehydrogenase flavoprotein subunit | SDHA | 73 | 12 | 22 |
| Succinate dehydrogenase iron-sulfur protein | SDHB | 32 | 6 | 24 |
| Complex III | ||||
| Ubiquinol-cytochrome c reductase iron-sulfur subunit | UQCRFS1 | 30 | 5 | 22 |
| Cytochrome c1, heme protein | CYC1 | 35 | 5 | 16 |
| Ubiquinol-cytochrome-c reductase complex core protein I | UQCRC1 | 53 | 11 | 28 |
| Ubiquinol-cytochrome-c reductase complex core protein 2 | UQCRC2 | 48 | 13 | 41 |
| Ubiquinol-cytochrome c reductase complex 14 kDa protein | UQCRB | 13 | 3 | 32 |
| Ubiquinol-cytochrome c reductase complex 7.2 kDa protein | UCRC | 7 | 2 | 38 |
| Complex IV | ||||
| Cytochrome c oxidase subunit 2 | COX2 | 26 | 6 | 39 |
| Cytochrome c oxidase subunit 4 isoform 1 | COX4I1 | 20 | 8 | 46 |
| Cytochrome c oxidase subunit 5A | COX5A | 17 | 3 | 25 |
| Cytochrome c oxidase subunit 5B | COX5B | 14 | 2 | 9 |
| Cytochrome c oxidase polypeptide Vic | COX6C | 9 | 5 | 37 |
| Cytochrome c oxidase polypeptide VIIa-Heart | COX7A1 | 11 | 2 | 29 |
| Complex V | ||||
| ATP synthase B chain | ATP5F1 | 26 | 9 | 36 |
| ATP synthase D chain isoform 1 | ATP5H | 18 | 9 | 52 |
| ATP synthase E chain | ATP5I | 8 | 3 | 50 |
| ATP synthase F chain isoform 2 | ATP5J2 | 8 | 2 | 44 |
| ATP synthase subunit G | ATP5L | 11 | 7 | 54 |
| ATP synthase subunit α | ATP5A1 | 60 | 39 | 62 |
| ATP synthase subunit β | ATP5B | 57 | 40 | 75 |
| ATP synthase δ chain | ATP5D | 17 | 3 | 23 |
| ATP synthase γ chain isoform heart | ATP5C1 | 33 | 5 | 18 |
| ATP synthase coupling factor 6 | ATP5J | 13 | 5 | 45 |
| ATP synthase O subunit (OSCP) | ATP5O | 23 | 6 | 35 |
| Carrier/transfer proteins | ||||
| Cytochrome c | CYCS | 12 | 6 | 50 |
| Phosphate carrier protein isoform A | SLC25A3 | 35 | 4 | 12 |
| ADP/ATP translocase 1 | SLC25A4 | 33 | 10 | 35 |
| Mitochondrial 2-oxoglutarate/malate carrier protein | SLC25A11 | 34 | 7 | 26 |
| Calcium-binding mitochondrial carrier protein Aralar1 | SLC25A12 | 75 | 28 | 52 |
| Electron transfer flavoprotein subunit β | ETFA | 35 | 6 | 27 |
| Electron transfer flavoprotein subunit β | ETFB | 33 | 9 | 33 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | ETFDH | 68 | 9 | 20 |
| NAD(P) transhydrogenase | NNT | 114 | 11 | 14 |
MW, molecular weight; the mean molecular weight is shown for proteins with more than one IPI ID.
Maximum values for one analysis.
The phosphocreatine/creatine (PCr/Cr) pool is the major source of high energy phosphate bonds for ATP replenishment in skeletal muscle in response to energy-dependent activities such as muscle contraction. Regulation of this reservoir involves a number of enzymes, all of which were identified in this study (36). These enzymes included the catalytic α-2 and the regulatory γ-1 subunits of AMPK kinase, two cytosolic isoforms of creatine kinase (M and B), the mitochondrial creatine kinase isoform, two adenylate kinase isoforms, ADP/ATP translocase, and three isoforms of porin (supplemental Table I). Together these enzymes have been suggested to comprise a PCr/Cr shuttle, which ensures an efficient mechanism by which PCr can be resynthesized by mitochondrial ATP synthesis (36).
Calcium Homeostasis
Calcium homeostasis and calcium-dependent signaling in skeletal muscle play major roles in a number of cellular processes such as contraction, apoptosis, the adiponectin-AMPK signaling pathway, insulin-mediated glucose uptake, and mitochondrial biogenesis and function (37–41). In the present study, we identified three members of the voltage-dependent calcium channel complex, the ryanodine receptor 1, calmodulin, and several important calcium/calmodulin binding or regulated proteins (Table II). These proteins include two subunits of the phosphatase, calcineurin A, all four subunits of calcium/calmodulin dependent kinase II (CaMKII), cardiac phospholamban, and the two major calcium ATPases (SERCA1 and 2). We also identified a calcium-regulated receptor, T-cadherin, which has been suggested to be a potential adiponectin receptor (42), and the calcium binding protein, MO25, which is a member of the LKB1-STRADA-MO25 complex regulating phosphorylation of AMPK.
Table II. Proteins involved in calcium homeostasis identified in human skeletal muscle.
Proteins were separated by one-dimensional gel electrophoresis and identified by HPLC-ESI-MS/MS in four experiments using skeletal muscle from three nonobese, healthy subjects.
| Protein | Gene name |
MW (kDa) |
Maximum unique peptidesa |
Maximum sequence coverage (%)a |
|---|---|---|---|---|
| Calcium Channels/Receptors | ||||
| Dihydropyridine receptor α 2 | CACNA2D1 | 125 | 10 | 13 |
| Voltage-dependent l-type calcium channel α -S1 | CACNA1S | 212 | 2 | 2 |
| Voltage-dependent l-type calcium channel β -1 | CACNB1 | 64 | 9 | 23 |
| Ryanodine receptor 1 | RYR1 | 565 | 8 | 3 |
| Calcium-Adiponectin-AMPK | ||||
| Cadherin 13 (T-cadherin) | CDH13 | 78 | 4 | 8 |
| 5′-AMP-activated protein kinase catalytic α -2 | PRKAA2 | 62 | 5 | 13 |
| 5′-AMP-activated protein kinase regulatory γ-1 | PRKAG1 | 38 | 4 | 17 |
| Calcium-binding protein 39 (Protein Mo25) | CAB39 | 40 | 6 | 18 |
| Calcium regulated enzymes | ||||
| Calmodulin | CALM1 | 17 | 4 | 42 |
| Calmodulin-dependent calcineurin A α | PPP3CA | 58 | 3 | 8 |
| Calmodulin-dependent calcineurin A β | PPP3CB | 58 | 4 | 10 |
| Calcium/calmodulin-dependent protein kinase II α | CAMK2A | 54 | 4 | 13 |
| Calcium/calmodulin-dependent protein kinase II β | CAMK2B | 73 | 7 | 13 |
| Calcium/calmodulin-dependent protein kinase II δ | CAMK2D | 56 | 5 | 15 |
| Calcium/calmodulin-dependent protein kinase II γ | CAMK2G | 59 | 5 | 13 |
| Cardiac phospholamban | PLN | 6 | 3 | 23 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | ATP2A1 | 103 | 41 | 36 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | ATP2A2 | 110 | 42 | 33 |
| GTP-binding protein RAD | RRAD | 33 | 2 | 10 |
| Calpain-1 catalytic subunit | CAPN1 | 82 | 12 | 22 |
| Calpain-2 catalytic subunit | CAPN2 | 80 | 3 | 5 |
| Calpain-3 | CAPN3 | 94 | 7 | 13 |
| Calpain small subunit 1 | CAPNS1 | 31 | 4 | 18 |
| Calcium binding proteins | ||||
| Calreticulin | CALR | 42 | 2 | 9 |
| Calnexin | CANX | 68 | 5 | 11 |
| Sarcalumenin | SRL | 97 | 20 | 26 |
| Sarcoplasmic reticulum histidine-rich calcium-binding protein | HRC | 81 | 4 | 12 |
| Calsequestrin-1 | CASQ1 | 45 | 15 | 28 |
| Calsequestrin-2 | CASQ2 | 46 | 10 | 29 |
| Annexin A1 | ANXA1 | 39 | 7 | 28 |
| Annexin A2 | ANXA2 | 40 | 15 | 48 |
| Annexin A4 | ANXA4 | 35 | 2 | 8 |
| Annexin A5 | ANXA5 | 36 | 14 | 42 |
| Annexin A6 | ANXA6 | 76 | 32 | 53 |
| Annexin A7 | ANXA7 | 52 | 4 | 11 |
| Annexin A11 | ANXA11 | 54 | 8 | 22 |
| S-100 calcium-binding protein A1 | S-100A1 | 11 | 2 | 38 |
| S-100 calcium-binding protein A6 | S-100A6 | 10 | 2 | 17 |
| Copine-3 | CPNE3 | 60 | 3 | 7 |
| Gelsolin | GSN | 83 | 12 | 25 |
MW, molecular weight; the mean molecular weight is shown for proteins with more than one IPI ID.
Maximum values for one analysis.
Insulin and IGF-I Signaling
Insulin and IGF-I have significant metabolic and growth promoting roles in skeletal muscle, and their respective signaling pathways are involved in skeletal muscle insulin resistance. Among the proteins currently believed to mediate the effect of insulin and IGF-I on glucose transport, glycogen synthesis, and protein synthesis in skeletal muscle (3–7, 43), we identified the glycogen-targeting subunit of PP1 (PP1g), several Rab proteins involved in GLUT4 translocation, and a number of eukaryotic translation initiation and elongation factors and ribosomal protein S6 kinase involved in protein synthesis (supplemental Table I). Moreover, a number of kinases, phosphatases and other proteins known to modulate insulin and IGF-I signaling (3) were found. In addition to those already mentioned, three subunits of another phosphatase, PP2A, MAP kinase, MAP kinase kinases, and serine/threonine kinases, were identified. A number of cytosolic and mitochondrial chaperones were also found. Of these, HSP90 and its co-chaperone cdc37, in particular, have been suggested to interact with PDK1 and Akt (44); both are elements of insulin and IGF-I signaling. Moreover, four isoforms of the phosphoserine/phosphothreonine binding 14–3-3 proteins were identified. They have been reported to bind to insulin receptor substrate (IRS)-1, IRS-2, and PDK1 and to modulate the activities of IRS-1 associated PI3-kinase and PDK1 (3).
Extracellular Matrix (ECM) and Contractile Proteins
Muscle adaptation in response to a number of physiological and pathophysiological conditions involves changes in ECM and contractile proteins (13, 14, 45). Among ECM proteins, we found three α-subunits of type VI collagen, fibronectin, decorin, lumican, and prolargin. Multiple isoforms of proteins constituting the myofibrillar apparatus were identified, including four actin isoforms, three α-actinin isoforms, nine myosin heavy chain isoforms, eight myosin light chains isoforms, two myomesin isoforms, and titin. Moreover, both slow and fast skeletal muscle isoforms of troponin I, C, and T in the troponin complex were identified together with several tropomyosin and tropomodulin isoforms.
Phosphorylation Sites
Although no attempt was made to enrich phosphopeptides, 35 phosphorylation sites in 24 proteins were detected with confidence (95% using Scaffold analysis, Table III). According to available databases (www.phosphosite.org, www.phospho.elm.eu.org, www.mitocheck.org, www.phosida.com and www.expasy.ch) and existing literature, 16 of these phosphorylation sites have been reported before, whereas 19 phosphorylation sites in 13 proteins appear to be novel. These new phosphorylation sites were identified on proteins critical for muscle metabolism such as aldolase A, alpha/beta-enolase, glyceraldehyde-3-phosphate dehydrogenase, creatine kinase M, and glycogen phosphorylase as well as on muscle-specific proteins such as myoglobin, myomesin 1, myosin binding protein C, and LIM domain-binding protein 3.
Table III. Phosphorylation sites in human skeletal muscle proteins identified by HPLC-ESI-MS/MS.
Proteins were separated by one-dimensional gel electrophoresis and phosphorylation sites were identified by HPLC-ESI-MS/MS in four experiments using skeletal muscle from three nonobese, healthy subjects. No phosphopeptide enrichment strategies were used.
| Protein | Gene name | Sequencea | Mono- phosphorylation site(s) |
|---|---|---|---|
| Aldolase A | ALDOA | 29GILAADESTGSIAK42 | S36, T37,b S39 |
| Alpha crystallin family protein | CRYAB | 29RASAPLPGLSAPGR42 | S31 |
| Alpha/beta-enolase | ENO1/3 | 33AAVPSGASTGIYEALELR50 | S40,b T41,b Y44 |
| cAMP-dependent protein kinase regulatory α −1 subunit | PRKAR2A | 75TDSREDEISPPPPNPVVK92 | S83 |
| Carbonic anhydrase 3 | CA3 | 40HDPSLQPWSVSYDGGSAK57 | S48b |
| Cardiac phospholamban | PLN | 14RASTIEMPQQAR25 | S16 |
| Cardiac phospholamban | PLN | 14ASTIEMPQQAR25 | T17 |
| Creatine kinase M | CKM | 157LSVEALNSLTGEFK170 | S164b |
| Creatine kinase M | CKM | 321GTGGVDTAAVGSVFDVSNADR341 | T322,b T327,b S332b |
| Gamma filamin variant | FLNC | 2211LGSFGSITR2219 | S2213 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 163VIHDNFGIVEGLMTTVHAITATQK186 | T182, T184b |
| Glycogen phosphorylase, muscle form | PYGM | 467TIFKDFYELEPHK479 | Y473b |
| Glycogen synthase, muscle | GYS1 | 708RNSVDTATSSSLSTPSEPLSPTSSLGEERN737 | S710 |
| Heat-shock protein beta-1 | HSPB1 | 80QLSSGVSEIR89 | S82 |
| Phosphorylase B kinase regulatory α subunit | PHKA1 | 972SVRPTDSNVSPAISIHEIGAVGATK996 | S972 |
| LIM domain-binding protein 3 | LDB3 | 189DLAVDSASPVYQAVIK204 | Y199b |
| MYC box-dependent-interacting protein 1 | BIN1 | 296SPSPPDGSPAATPEIR311 | S298 |
| Myoglobin | MB | 65HGATVLTALGGILK78 | T68,b T71b |
| Myomesin 1 | MYOM1 | 63RASASSSQQQASQHALSSEVSR84 | S65b |
| Myosin binding protein C, slow type | MYBPC1 | 57KDSDWTLVETPPGEEQAK74 | S59b |
| Myosin binding protein C, fast type | MYBPC2 | 37EAPPEDQSPTAEEPTGVFLK56 | S44b |
| Pyruvate dehydrogenase, E1 component α subunit | PDHA1 | 289YHGHSMSDPGVSYR302 | Y289, T293 |
| RAS GTPase-activating protein-binding protein 1 | G3BP1 | 230SSSPAPADIAQTVQEDLR247 | S232 |
| Sarcoplasmic reticulum His-rich calcium-binding protein | HRC | 113VGDEGVSGEEVFAEHGGQAR132 | S119b |
| Sarcoplasmic reticulum His-rich calcium-binding protein | HRC | 157SHSHQDEDEDEVVSSEHHHHILR179 | S170b |
| Smoothelin-like 2 | SMTNL2 | 342SQSFGVASASSIK354 | S344b |
The sequence location of each peptide is indicated in superscript.
Sites that appear to be novel.
DISCUSSION
A number of previous proteomic studies of human skeletal muscle have been accomplished using protein separation by two-dimensional gel electrophoresis with subsequent identification using MALDI-TOF/MS and/or HPLC-ESI-MS/MS analyses (18–23). Most are comparative studies reporting only proteins that are differentially regulated in conditions such as type 2 diabetes (23), obesity (22), Tibetans at high altitude (20), and aging (21), or in comparisons between different muscle fiber types (19). The proteins reported in those studies represent mainly highly abundant structural or metabolic proteins. Of the 62 different proteins reported in these five studies (19–23), 59 were also identified in the present investigation. In comparative experiments that employ two-dimensional gel electrophoresis, differences in spot staining intensity are used as a measure of changes in protein expression. However, since a large number of proteins are present in more than one differently migrating protein spot (due to the presence of different isoforms or post-translational modifications), it is difficult to get a reliable estimate of the total content of a specific protein by the two-dimensional gel approach (19, 23). Furthermore, those studies focus primarily on differentially regulated proteins; thus, many other proteins that are not altered in abundance or are present at too low a level to be easily quantified by spot analysis often are not identified.
In the past, there are only a limited number of reports in the literature on characterization of the human skeletal muscle proteome (18, 46). To date, the most comprehensive list of proteins of human vastus lateralis muscle has been reported by Gelfi et al. (18) who used two-dimensional gel electrophoresis, MALDI-TOF/MS, and HPLC-ESI-MS/MS. Approximately 500 protein spots were visualized by silver staining, and 150 spots were excised, resulting in identification of 107 different proteins. Of those 107 proteins, 101 were identified in the present study. In a more recent report using two-dimensional difference gel electrophoresis to characterize the effects of aging on the human skeletal muscle, ~2700 protein spots were visualized, 52 of which were differentially expressed, and 39 proteins were identified (21). However, in addition to difficulties in identifying proteins at the extremes of the MW and pI ranges, a major weakness in the two-dimensional gel approach is in detection and subsequent identification of low-abundance proteins. Moreover, identification of proteins in all spots detected in a two-dimensional gel, such as the 2700 reported by Gelfi et al. (21), would be extremely labor-intensive, even with robotic processing.
Nuclear extracts from human muscle specimens of unspecified origin obtained from surgery or autopsies have been analyzed by two-dimensional HPLC-ESI-MS/MS, resulting in the identification of 192 different muscle proteins (46). In our analysis of whole cell lysates of human vastus lateralis muscle, we identified 116 of these nuclear proteins. Among those we did not identify, half were either hypothetical proteins or were assigned to the mouse database.
For our analysis of the human muscle protein, we elected to apply a relatively simple approach based on one-dimensional gel electrophoresis and HPLC-ESI-MS/MS analysis, a strategy that has been successful in studies of human cell lines (28, 29) and can be used with very small amounts of tissue in clinical studies. Using this approach, we identified 954 different proteins, which to our knowledge, represents the most comprehensive identification of proteins in human vastus lateralis muscle. This list includes a large number of membraneassociated and low molecular weight proteins, indicating less issues in this regard compared with two-dimensional gel electrophoresis. In Fig. 1B is a comparison of the MW range of proteins identified in our study with those listed in the IPI human database. If the MW distribution of proteins in human muscle is the same as the in the human proteome catalogued in the IPI human database, we identified a lower than expected proportion of proteins under 20 kDa. This suggests that either there is a lack of detection of these proteins by our approach or else the molecular weights of proteins in human muscle do not parallel those of the entire proteome.
Mechanisms underlying changes in muscle structure and metabolism during different physiological and pathophysiological conditions have traditionally been studied by focusing on small numbers of genes or proteins. Transcriptional profiling has been applied to define the molecular signature of denervation, immobilization, exercise-training, age-related sarcopenia, insulin resistance and muscular dystrophy (8–15, 45). However, global profiling of temporal changes in metabolic enzymes and structural proteins and their posttranslational modifications in skeletal muscle in vivo has been limited by the lack of appropriate proteomic technology. The present study shows that it is feasible to obtain a comprehensive identification of human skeletal muscle proteins with full representation of enzymes involved in glucose and lipid metabolism, a high number of mitochondrial proteins, including 55 OXPHOS subunits and carrier/transport proteins involved in ATP synthesis, and multiple proteins involved in calcium homeostasis including calmodulin, CaMKII, phospholamban and calcineurin A, which have been shown to play a role in insulin-mediated glucose transport, mitochondrial biogenesis and transcriptional regulation of lipid oxidation genes (37–41). As expected, a diversity of isoforms of myofibrillar and cytoskeletal proteins was found, reflecting the mixed fiber type composition of the human vastus lateralis muscle. Of potential importance to future studies regarding insulin resistance, we were able to identify several kinases, phosphatases, and enzymes known to regulate glycogen synthase, glucose transport, and protein synthesis and to modulate proximal insulin and IGF-I signaling (3–7, 43). Moreover, 16 known and 19 previously unreported phosphorylation sites were detected without phosphopeptide enrichment.
An analysis of the activity and subunit composition of mitochondria plays a central role in the diagnosis of mitochondrial myopathies, the pathophysiology of type 2 diabetes and age-related sarcopenia, and physiological responses to exercise. In addition to the citric acid cycle, β-oxidation and OXPHOS, mitochondrial proteins are involved in a variety of cellular processes, including intracellular calcium homeostasis, programmed cell death (apoptosis), and ion homeostasis. The 212 mitochondrial proteins identified in the present study represent, to our knowledge, the most comprehensive proteomic identification of mitochondrial proteins of human skeletal muscle reported to date. In a proteomic study of rat mitochondria purified from skeletal muscle, heart, and liver, 689 proteins were identified, with only small differences found between skeletal muscle and heart (47). The most extensive evaluation of human mitochondrial proteins identified 680 proteins in purified heart mitochondria (48), utilizing 40 mg of purified mitochondria as starting material and 701 HPLC-ESI-MS/MS analyses. Based on those results, it is likely that we would improve our coverage of the estimated 1500 human mitochondrial proteins (33) if muscle mitochondria were isolated prior to one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. The finding that mitochondrial proteins accounted for 22% of all skeletal muscle proteins identified in the present study, compared with 4.8% in the total human proteome (33), may reflect the critical role that mitochondria play for energy metabolism in skeletal muscle and the enormous demand for ATP elicited by muscle contraction.
In conclusion, this study represents the first application of one-dimensional gel electrophoresis and HPLC-ESI-MS/MS for analysis of the human skeletal muscle proteome. Using only 60–70 µg of total protein from 3 lean subjects in <90 HPLC-ESI-MS/MS runs, we provide the most comprehensive proteome coverage of human skeletal muscle to date. This includes the largest catalog of mitochondrial proteins in human skeletal muscle. It is important to emphasize that the quantity of protein from each subject that was used in these experiments represents a small fraction of the muscle obtained by percutaneous needle biopsy. These data demonstrate the utility of this relatively simple proteomic approach for analysis of small human tissue samples, making it a potentially valuable tool in elucidating changes in the proteome associated with human disease.
Supplementary Material
Acknowledgment
We thank Michael Sweet, an undergraduate student in our group, for initial data analysis.
Footnotes
The abbreviations used are: FA, formic acid; ACN, acetonitrile; CID, collision-induced dissociation, OXPHOS, oxidative phosphorylation; MW, molecular weight; MS/MS, tandem mass spectrometry; HPLC, high performance liquid chromatography; ESI electrospray ionization.
This work was supported by National Institutes of Health Grants R01DK47936 and R01DK66483 (to L. J. M.) and a Mentor-Based Postdoctoral Fellowship Award from the American Diabetes Association (to L. J. M.).
The on-line version of this article (available at http:www.mcponline.org) contains supplemental material.
Funded by grants from the Danish Diabetes Association and the Danish Medical Research Council.
REFERENCES
- 1.Bergstrom J. Percutaneous needle biopsy of skeletal muscle in physiological and clinical research. Scand. J. Clin. Lab. Invest. 1975;35:609–616. [PubMed] [Google Scholar]
- 2.Edwards R, Young A, Wiles M. Needle biopsy of skeletal muscle in the diagnosis of myopathy and the clinical study of muscle function and repair. N. Engl. J. Med. 1980;302:261–271. doi: 10.1056/NEJM198001313020504. [DOI] [PubMed] [Google Scholar]
- 3.Hojlund K, Beck-Nielsen H. Impaired glycogen synthase activity and mitochondrial dysfunction in skeletal muscle: markers or mediators in type 2 diabetes. Curr. Diabetes Rev. 2006;2:375–395. doi: 10.2174/1573399810602040375. [DOI] [PubMed] [Google Scholar]
- 4.Krebs M, Roden M. Molecular mechanisms of lipid-induced insulin resistance in muscle, liver and vasculature. Diabetes Obes. Metab. 2005;7:621–632. doi: 10.1111/j.1463-1326.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- 5.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 6.Pirola L, Johnston AM, Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47:170–184. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 7.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 8.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 9.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol. Genomics. 2005;21:253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 10.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 12.Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51:1913–1920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 13.Richardson DK, Kashyap S, Bajaj M, Cusi K, Mandarino SJ, Finlayson J, DeFronzo RA, Jenkinson CP, Mandarino LJ. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J. Biol. Chem. 2005;280:10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 14.Batt J, Bain J, Goncalves J, Michalski B, Plant P, Fahnestock M, Woodgett J. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006;20:115–117. doi: 10.1096/fj.04-3640fje. [DOI] [PubMed] [Google Scholar]
- 15.Teran-Garcia M, Rankinen T, Koza RA, Rao DC, Bouchard C. Endurance training-induced changes in insulin sensitivity and gene expression. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1168–E1178. doi: 10.1152/ajpendo.00467.2004. [DOI] [PubMed] [Google Scholar]
- 16.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 17.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelfi C, De Palma S, Cerretelli P, Begum S, Wait R. Two-dimensional protein map of human vastus lateralis muscle. Electrophoresis. 2003;24:286–295. doi: 10.1002/elps.200390025. [DOI] [PubMed] [Google Scholar]
- 19.Capitanio D, Vigano A, Ricci E, Cerretelli P, Wait R, Gelfi C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics. 2005;5:2577–2586. doi: 10.1002/pmic.200401183. [DOI] [PubMed] [Google Scholar]
- 20.Gelfi C, De Palma S, Ripamonti M, Eberini I, Wait R, Bajracharya A, Marconi C, Schneider A, Hoppeler H, Cerretelli P. New aspects of altitude adaptation in Tibetans: a proteomic approach. FASEB J. 2004;18:612–614. doi: 10.1096/fj.03-1077fje. [DOI] [PubMed] [Google Scholar]
- 21.Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P. The human muscle proteome in aging. J. Proteome Res. 2006;5:1344–1353. doi: 10.1021/pr050414x. [DOI] [PubMed] [Google Scholar]
- 22.Hittel DS, Hathout Y, Hoffman EP, Houmard JA. Proteome analysis of skeletal muscle from obese and morbidly obese women. Diabetes. 2005;54:1283–1288. doi: 10.2337/diabetes.54.5.1283. [DOI] [PubMed] [Google Scholar]
- 23.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 24.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs JM, Mottaz HM, Yu LR, Anderson DJ, Moore RJ, Chen WN, Auberry KJ, Strittmatter EF, Monroe ME, Thrall BD, Camp DG, 2nd, Smith RD. Multidimensional proteome analysis of human mammary epithelial cells. J. Proteome Res. 2004;3:68–75. doi: 10.1021/pr034062a. [DOI] [PubMed] [Google Scholar]
- 26.Pan Y, Kislinger T, Gramolini AO, Zvaritch E, Kranias EG, MacLennan DH, Emili A. Identification of biochemical adaptations in hyper- or hypocontractile hearts from phospholamban mutant mice by expression proteomics. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2241–2246. doi: 10.1073/pnas.0308174101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metz TO, Jacobs JM, Gritsenko MA, Fontes G, Qian WJ, Camp DG, 2nd, Poitout V, Smith RD. Characterization of the human pancreatic islet proteome by two-dimensional LC/MS/MS. J. Proteome Res. 2006;5:3345–3354. doi: 10.1021/pr060322n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schirle M, Heurtier MA, Kuster B. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics. 2003;2:1297–1305. doi: 10.1074/mcp.M300087-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Rezaul K, Wu L, Mayya V, Hwang SI, Han D. A systematic characterization of mitochondrial proteome from human T leukemia cells. Mol. Cell. Proteomics. 2005;4:169–181. doi: 10.1074/mcp.M400115-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, Eling WM, Hall N, Waters AP, Stunnenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 31.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 32.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J. Proteome Res. 2006;5:64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- 33.Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 34.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 35.Scheffler IE. The Human OXPHOS system: Structure, Function, Physiology. Landes Bioscience and Kluwer Academic/Plenum Publishers; 2004. [Google Scholar]
- 36.Neumann D, Schlattner U, Wallimann T. A molecular approach to the concerted action of kinases involved in energy homoeostasis. Biochem. Soc. Trans. 2003;31:169–174. doi: 10.1042/bst0310169. [DOI] [PubMed] [Google Scholar]
- 37.Gunter TE, Yule DI, Gunter KK, Eliseev RA, Salter JD. Calcium and mitochondria. FEBS Lett. 2004;567:96–102. doi: 10.1016/j.febslet.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 38.Konstantopoulos N, Marcuccio S, Kyi S, Stoichevska V, Castelli LA, Ward CW, Macaulay SL. A purine analog kinase inhibitor, calcium/calmodulin-dependent protein kinase II inhibitor 59, reveals a role for calcium/calmodulin-dependent protein kinase II in insulin-stimulated glucose transport. Endocrinology. 2007;148:374–385. doi: 10.1210/en.2006-0446. [DOI] [PubMed] [Google Scholar]
- 39.Lanner JT, Katz A, Tavi P, Sandstrom ME, Zhang SJ, Wretman C, James S, Fauconnier J, Lannergren J, Bruton JD, Westerblad H. The role of Ca2+ influx for insulin-mediated glucose uptake in skeletal muscle. Diabetes. 2006;55:2077–2083. doi: 10.2337/db05-1613. [DOI] [PubMed] [Google Scholar]
- 40.Long YC, Glund S, Garcia-Roves PM, Zierath JR. Calcineurin regulates skeletal muscle metabolism via coordinated changes in gene expression. J. Biol. Chem. 2007;282:1607–1614. doi: 10.1074/jbc.M609208200. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 42.Hug C, Wang J, Ahmad NS, Bogan JS, Tsao TS, Lodish HF. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10308–10313. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon AM, Bouloux PM. Modifying muscle mass: the endocrine perspective. J. Endocrinol. 2006;191:349–360. doi: 10.1677/joe.1.06837. [DOI] [PubMed] [Google Scholar]
- 44.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 45.Urso ML, Scrimgeour AG, Chen YW, Thompson PD, Clarkson PM. Analysis of human skeletal muscle after 48 h immobilization reveals alterations in mRNA and protein for extracellular matrix components. J. Appl. Physiol. 2006;101:1136–1148. doi: 10.1152/japplphysiol.00180.2006. [DOI] [PubMed] [Google Scholar]
- 46.Cagney G, Park S, Chung C, Tong B, O’Dushlaine C, Shields DC, Emili A. Human tissue profiling with multidimensional protein identification technology. J. Proteome Res. 2005;4:1757–1767. doi: 10.1021/pr0500354. [DOI] [PubMed] [Google Scholar]
- 47.Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using two-dimensional blue-native/SDS-PAGE. J. Proteome Res. 2006;5:1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- 48.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.