Abstract
Therapeutic uses of DNA functionalized gold nanoparticles (DNA-AuNPs) have shown great potential and exciting opportunities for disease diagnostics and treatment. Maintaining stable conjugation between DNA oligonucleotides and gold nanoparticles under thermally stressed conditions is one of the critical aspects for any of the practical applications. We systematically studied the thermal stability of DNA-AuNPs as affected by organosulfur anchor groups and packing densities. Using a fluorescence assay to determine the kinetics of releasing DNA molecules from DNA-AuNPs, we observed an opposite trend between the temperature-induced and chemical-induced release of DNA from DNA-AuNPs when comparing the DNA-AuNPs that were constructed with different anchor groups. Specifically, the bidentate Au–S bond formed with cyclic disulfide was thermally less stable than those formed with thiol or acyclic disulfide. However, the same bidentate Au–S bond was chemically more stable under the treatment of competing thiols (mercaptohexanol or dithiothreitol). DNA packing density on AuNPs influenced the thermal stability of DNA-AuNPs at 37 °C, but this effect was minimum as temperature increased to 85 °C. With the improved understanding from these results, we were able to design a strategy to enhance the stability of DNA-AuNPs by conjugating double-stranded DNA to AuNPs through multiple thiol anchors.
Introduction
DNA functionalized gold nanoparticles (DNA-AuNPs) have shown great potential and exciting opportunities for disease diagnostic and therapeutic treatment.1−13 DNA-AuNPs have been used for gene regulation,1−3 drug delivery,4 cancer cell imaging,5−7 and photothermal therapies.8−13 Stable conjugation between DNA oligonucleotides and gold nanoparticles is critical to any of the practical applications because extended incubation at elevated temperatures (e.g., 37 °C)1−7 or photothermal treatments to release DNA from AuNPs are commonly involved.8−13 The covalent bond between gold and sulfur (Au–S, also known as the thiolated-gold bond), usually mediated through the sulfhydryl (SH) functional group, is the most widely used interaction to achieve the stable conjugation between DNA oligonucleotides and AuNPs.14 However, such a Au–S bond is known to be subject to cleavage that can be induced by other thiols or elevated temperatures.15−17
Efforts have been made to improve the stability of DNA-AuNPs formed through the Au–S bond by using different organosulfur anchors for conjugation. These anchors include alkanethiol, acyclic disulfide, cyclic disulfide, and other types of multidentate thiolated anchor groups.16−23 However, most studies have been focused on the salt-dependent colloidal stability of DNA-AuNPs as a function of the extent of the colloidal aggregation rather than directly monitoring the cleavage of the Au–S bond. Furthermore, most comparison studies were based merely on chemical stabilities, where thiols, such as dithiothreitol (DTT) or mercaptohexanol (MCH), were used to displace the thiolated DNA from AuNP surfaces.16−18 Only limited few studies have explored the thermal stability of DNA-AuNPs at room temperature or elevated temperatures without interference from other thiols.15,20 For example, with the aim of finding the most desired conditions for the storage of purified DNA-AuNPs, Liu et al. studied the long-term DNA dissociation kinetics at 4 °C and at room temperature as a function of ionic strength, pH, and organic solvents.20 Taton et al. studied the short-term dissociation kinetics of DNA-AuNPs at elevated temperatures (40 to 95 °C).15 Both studies focused only on the Au–S bond formed with alkanethiol (RSH) as an anchor. No systematic thermal stability study for DNA-AuNPs has been reported regarding Au–S bonds formed with other types of organosulfur anchor groups. To understand the influence of different anchor groups on the thermal stability of DNA-AuNPs, we have designed DNA-AuNP probes with 3 different anchor groups (Figure 1). We report here a systematic study of their thermal stability by monitoring the kinetics of DNA release from DNA-AuNPs. We also report the effect of packing densities on the thermal stability of DNA-AuNPs. DNA packing densities on AuNPs can influence the stability of DNA-AuNPs, as a result of the strong intermolecular repulsions among the largely negatively charged phosphodiester backbones of DNA molecules.24−26 Such a density effect on the thermal stability of DNA-AuNPs has not been reported previously.
Figure 1.
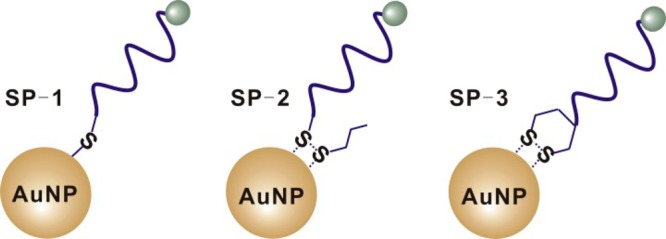
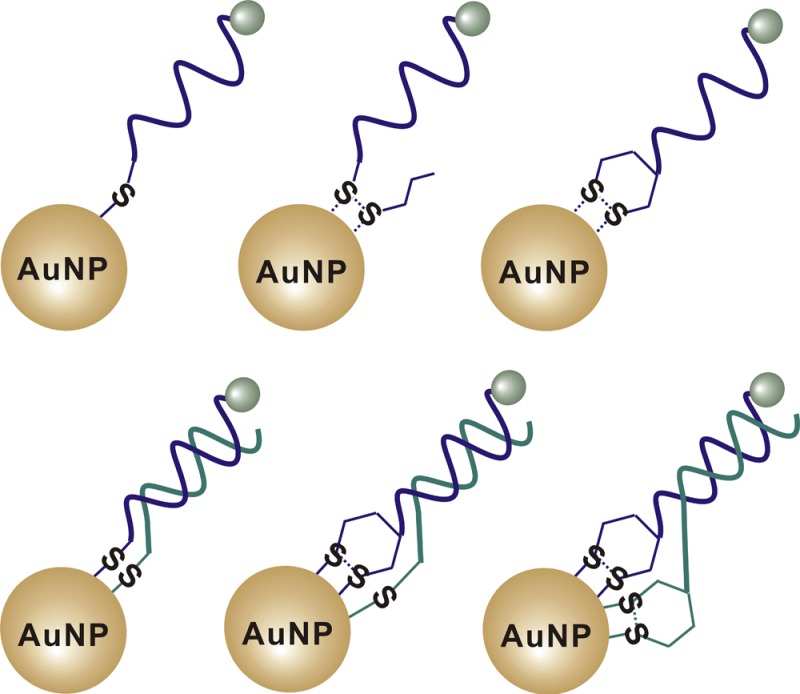
DNA-AuNP probes constructed by conjugating oligonucleotides having different organosulfur anchor groups to AuNPs. SP-1, SP-2, and SP-3 represent DNA-AuNP probes that involve conjugation through thiol, acyclic disulfide, and cyclic disulfide (DTPA), respectively.
Experimental Section
Materials and Reagents
Gold nanoparticles with 20 nm diameter were purchased from Ted Pella (Redding, CA). All DNA samples were purchased from Integrated DNA Technologies (Coralville, IA) and purified by HPLC. The DNA sequences and modifications are listed in Table S1, Supporting Information. Phosphate buffered saline (PBS) 10× solution and Tween20 were purchased from Fisher Scientific. Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), 6-mercapto-1-hexanol (MCH), dl-dithiothreitol (DTT), and β-mercaptoethanol (ME) were purchased from Sigma-Aldrich (Oakville, ON, Canada). NANOpure H2O (>18.0 MÙ), purified using an Ultrapure Milli-Q water system, was used for all experiments.
Preparation of DNA-AuNPs
All organosulfur modified DNA oligonucleotides were received in disulfide or cyclic disulfide forms. For preparing the DNA-conjugated AuNP probe (SP-1) with thiol as the anchor, the disulfide modified oligonucleotide with a concentration of 50 μM was treated with 100 μM TCEP for 1 h at room temperature to reduce the disulfide bond. This solution was then added to 1 mL of 20 nm AuNP solution (concentration 1 nM), and the mixture was incubated at room temperature for 12 h. To this mixture was slowly added 20 μL of 3 M NaCl, followed by sonication for 10 s. This process was repeated 5 times at a 1 h interval. The process was repeated to maximize the oligonucleotide loading amounts.27 The final solution was stored at room temperature for 24 h. The solution was centrifuged at 13,500 rpm for 30 min to separate the AuNPs from the unreacted reagents. The DNA-AuNPs were then washed 3 times with 1/2 × PBS buffer (pH 7.4) containing 0.01% Tween20 and finally redispersed in PBS buffer. For preparing DNA-conjugated AuNP probes with the disulfide (SP-2) and DTPA (SP-3) as anchors, the same experimental procedures as those for preparing SP-1 were used, except that the initial TCEP reduction step was omitted.
Double-stranded DNA-AuNP probes (DP-1, DP-2, and DP-3) were prepared by hybridizing two complementary organosulfur modified DNA strands followed by conjugating them to AuNPs. Briefly, two complementary single-stranded DNA oligonucleotides (50 μM, 20 μL each) were mixed together with 160 μL of 1× PBS buffer. The solution was then heated up to 70 °C for 5 min and left at room temperature for 1 h. The rest of the procedure was the same as that described above for preparing SP-1.
Characterization of DNA-AuNPs
DNA-conjugated AuNPs were characterized by using UV/vis absorption spectrometry. A Lambda 35 UV/vis absorption spectrophotometer (Perkin-Elmer) was used to obtain the extinction spectra of DNA-AuNPs from 400 to 700 nm (Figure S1, Supporting Information). The average number of oligonucleotides loaded on each particle was determined by measuring the concentration of AuNPs and the concentration of fluorescent DNA in each sample according to a previously reported method.28
Monitor the Thermal Stability of DNA-AuNPs
A 200 μL solution of DNA-AuNPs was centrifuged at 13,500 rpm for 30 min to remove oligonucleotides possibly released during storage. The pellet containing DNA-AuNPs was redispersed in 200 μL of 1/2× PBS buffer and then incubated at specific temperatures. Fluorescence was monitored with a multimode microplate reader (DTX880, Beckman Coulter) at time points of 0, 1, 2, 4, 8, and 20 h. After 20 h of incubation, a 5 μL solution of β-mercaptoethanol was added to reach a final concentration of 50 mM. This solution was incubated overnight, and its fluorescence was measured with a microplate reader. The ratio of the fluorescence signal at each time point over the fluorescence signal from the β-mercaptoethanol treated solution was expressed as [released]/[total].
Monitor the Chemical-Induced Cleavage of DNA-AuNPs
A 5 μL solution of 6-mercapto-1-hexanol (MCH) was added to a 95-μL solution of AuNPs (50 μM) conjugated with FAM-labeled DNA. The mixture was then incubated at 25 °C, and its fluorescence was monitored using the multimode microplate reader. Fluorescence intensity was measured every 10 min for 400 min. Finally, a 5-μL solution of β-mercaptoethanol was added to the sample so that the concentration of β-mercaptoethanol was 50 mM. This solution was incubated overnight, and its fluorescence was measured with a microplate reader. The ratio of the fluorescence signal at each time point over the fluorescence signal from the β-mercaptoethanol treated solution was expressed as [released]/[total].
Results and Discussions
Experimental Design
To systematically study the thermal stability of DNA-AuNPs, we designed a set of single-stranded DNA-AuNP probes, as shown in Figure 1. SP-1, SP-2, and SP-3 represent single-stranded DNA-AuNP probes modified with the three most widely used organosulfur anchor groups, including thiol,1−13 acyclic disulfide,16−18 and DTPA (cyclic disulfide).1,29−31 We then studied the thermal stabilities of these DNA-AuNP probes by monitoring the kinetics of release of DNA from DNA-AuNP. We also prepared DNA-AuNPs with different packing densities and compared their thermal stabilities.
We first designed a simple but sensitive fluorescence turn-on assay (Figure 2A) that was modified from the literature.20 Briefly, single-stranded DNA molecules were labeled with the fluorescent dye FAM, and these labeled DNA molecules were conjugated to AuNPs. The fluorescence of FAM-labeled DNA was quenched as a result of the close proximity between FAM and AuNPs. Once released from AuNPs, FAM-labeled DNA restored its fluorescence as a result of the increased distance between FAM and AuNPs.
Figure 2.
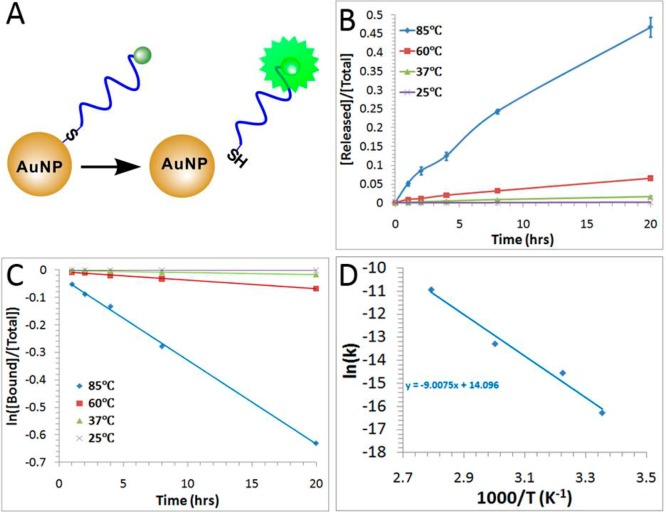
Thermal-induced release of FAM-labeled DNA from SP-1. (A) Schematic illustrating the fluorescence-based measurement of the released FAM-labeled DNA from DNA-AuNP. (B) Fraction of released DNA from SP-1 over a period of 20 h at different temperatures (25, 37, 60, and 85 °C). (C) Determination of the rate constants k based on the plots. (D) The logarithmic value of rate constants as a function of reciprocal of temperatures, providing information on the activation energy of the process. Error bars represent one standard deviation from triplicate sample analyses.
In this study, we first established the fluorescence turn-on assay using SP-1. We monitored the released DNA from SP-1 at different temperatures over a period of 20 h (Figure 2B). We then evaluated the rates of this release (Figure 2C) by determining the rate constant k from plots of the reaction rate equation: ln([bound]/[total]) = kt. Using these rate constants obtained at different temperatures (Figure 2D) and by fitting these data into the Arrhenius equation, we were able to determine the activation energy.
Effect of Anchor Groups on the Thermal Stability of DNA-AuNPs
After establishing the fluorescence turn-on method with SP-1, we studied the effect of anchor groups on the thermal stability of DNA-AuNPs by comparing the rates of releasing DNA from SP-1, SP-2, and SP-3 at different temperatures (Figure 3). After incubation at 37 °C for 20 h, the fraction of FAM-labeled DNA molecules released from SP-1, SP-2, and SP-3 were 1.6%, 1.7%, and 3.6%, respectively (Table 1 and Figure 3A). The rate constants were the same for SP-1 and SP-2 (k = 1.3 × 10–5 min–1). However, the rate constant for SP-3 (k = 2.7 × 10–5 min–1) was twice as those for SP-1 and SP-2. This observed trend in thermal stability was reproducible for at least three independently prepared DNA-AuNPs. To confirm this thermal stability trend, we repeated the experiments at 85 °C. Consistent with the results obtained at 37 °C (Figure 3A), experiments conducted at 85 °C (Figure 3B) also showed that the DNA probes in SP-1 and SP-2 were more stable than that from SP-3. Furthermore, we observed a strong colloidal aggregation for SP-3 after heating at 85 °C for 20 h (Figure 3B, inset), evidenced by changes in color of the solution from red to blue to colorless. No solution color change was observed for SP-1 or SP-2, i.e., both remained red. These observations confirmed that SP-3 was less thermally stable than SP-1 and SP-2 under the same thermal treatment. The activation energies obtained from the above kinetic data show that they were similar for SP-1 (85.3 kJ/mol) and SP-2 (82.4 kJ/mol). These were 7.6–10.5 kJ/mol higher than the activation energy for SP-3 (74.8 kJ/mol). These results were obtained under the same experimental conditions, including DNA sequences, packing densities (Table 1), buffer conditions, and incubation temperatures. Our observation of the differences in activation energy is consistent with the understanding of the Au–S bond cleavage. As has been demonstrated in previous studies of alkanethiol monolayers on Au (111),32−36 disulfide is the final product from thermal cleavage of the Au–S bonds. Below a critical surface coverage (1.8 × 1014 molecules/cm2), the Au–S bonds cleave through a thiolate radical mechanism.36 This is probably the case for the SP-1 and SP-2 probes, whose surface density of thiolated molecules is much lower (4.0 × 1012 molecules/cm2) than the critical coverage. In the case of SP-3, cyclic disulfides could be readily formed on AuNPs due to the intramolecular interactions, resulting in lower activation energy and a faster dissociation rate.
Figure 3.
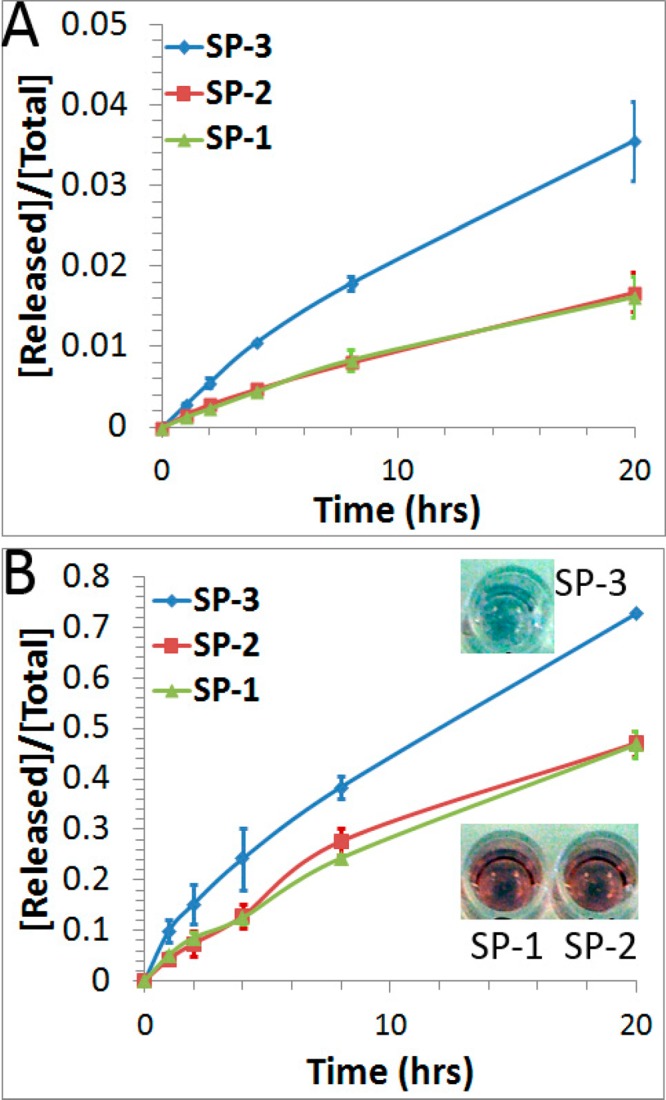
Fraction of FAM-labeled DNA released from the three DNA-AuNPs probes (SP-1, SP-2, and SP-3) over a period of 20 h at 37 °C (A) and at 85 °C (B). Error bars represent one standard deviation from triplicate sample analyses. Inset photographs in B were taken after 20 h of incubation at 85 °C, showing colors of different sample solutions at this time point.
Table 1. Summary of Three DNA-AuNP Probes (SP-1, SP-2, and SP-3), Their Packing Densities, the Rate Constant (k), and the Fraction of DNA Released after Chemical or Thermal Treatment.
| DNA
dissociation induced by MCH |
DNA
dissociation at 37 °C |
||||
|---|---|---|---|---|---|
| probe | packing densitya | ki (min–1)b | fMCHc | k (min–1)d | f37c |
| SP-1 | 207 ± 1 | 1.1 × 10–2 | 53.8% | 1.3 × 10–5 | 1.6% |
| SP-2 | 210 ± 9 | 1.3 × 10–2 | 51.4% | 1.3 × 10–5 | 1.7% |
| SP-3 | 223 ± 3 | 4.0 × 10–3 | 39.6% | 2.7 × 10–5 | 3.6% |
Packing density: defined as the number of DNA oligonucleotides per AuNP.
For DNA release induced by 50 μM mercaptohexanol (MCH), the initial rate constant ki was determined from the following equation: ln(1 – [released]/[total]) = kit, where t = 10–30 min.
Final released fraction f: fMCH = [released]/[total] at 400 min; f37 = [released]/[total] at 1200 min.
Rate constant k was determined from the first order rate equation: ln(1 – [released]/[total]) = kt, where t = 60–1200 min.
We also conducted a chemical stability study by treating DNA-AuNPs with mercaptohexanol (MCH) and monitoring the released DNA using a revised fluorescence turn-on method (Figure 4A). As shown in Figure 4B, upon the addition of 50 μM MCH, fluorescence signals for SP-1 and SP-2 quickly plateaued within 30 min, and the signal for SP-3 increased more slowly and gradually plateaued over a period of 400 min. The observed initial dissociation rates for SP-1 (ki = 1.1 × 10–2 min–1) and SP-2 (ki = 1.3 × 10–2 min–1) were two times faster than that for SP-3 (ki = 4.0 × 10–3 min–1) (Table 1). These results are consistent with previous observations that cyclic disulfide was more stable under the treatments with competing thiols, e.g., MCH or DTT.16 Thus, we conclude that the thermal stability and chemical stability of the DNA-AuNPs with these three different anchor groups follow an opposite trend.
Figure 4.
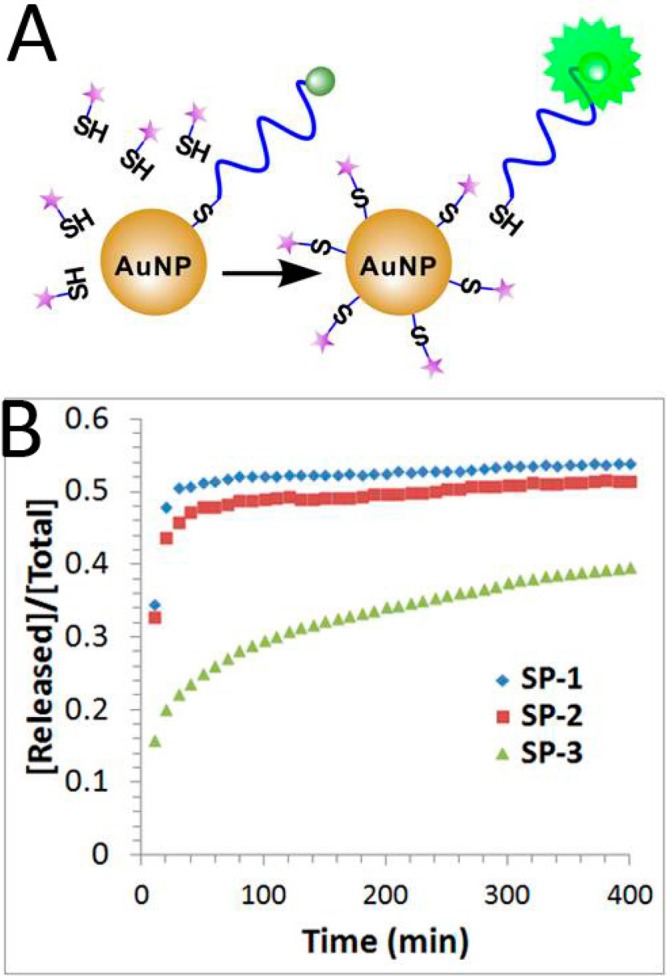
Release of FAM-labeled DNA from the DNA-AuNPs when the DNA-AuNPs were treated with 50 μM mercaptohexanol (MCH) at 25 °C. (A) Schematic illustrating the fluorescence-based measurement of the chemical release of FAM-labeled DNA from the DNA-AuNPs. (B) Fraction of FAM-labeled DNA released from DNA-AuNPs having three different anchor groups (SP-1, thiol; SP-2, acyclic disulfide; and SP-3, DTPA).
Effect of Packing Densities on the Thermal Stability of DNA-AuNPs
Packing density is another important factor that can influence the stability of DNA-AuNPs at elevated temperatures. To study this effect, we prepared a series of thiol modified DNA-AuNPs (SP-1) that have surface densities varying from 90 to 190 DNA molecules per AuNP. This DNA density range was achieved by controlling the ratio of DNA oligonucleotides to AuNPs in the conjugation reaction mixture and by adjusting the NaCl concentration during the salt aging step.
We first examined the thermal stability of DNA-AuNPs as a function of packing densities, using the established fluorescence turn-on assay (Figure 2A). As shown in Figure 5A, DNA-AuNPs with lower packing densities released DNA more slowly when incubated at 37 °C. The rate of DNA release from DNA-AuNPs with a packing density of 90 DNA molecules per AuNP was 3.4-fold slower than that of 190 DNA molecules per AuNP (Table 2). This kinetic difference is understandable because with the higher packing densities, the repulsion among DNA oligonucleotides on the surfaces of DNA-AuNPs increases.26 In addition, the increased local concentration of DNA molecules on AuNPs and the consequently decreased interaction between the surface of AuNPs and DNA molecules at a higher packing density could also contribute to the observed effect.37,38 As temperature increases, the process is dominated by the cleavage of the Au–S bond. Thus, the effect of packing density is less pronounced at 85 °C, as shown in Figure 5B and Table 2.
Figure 5.
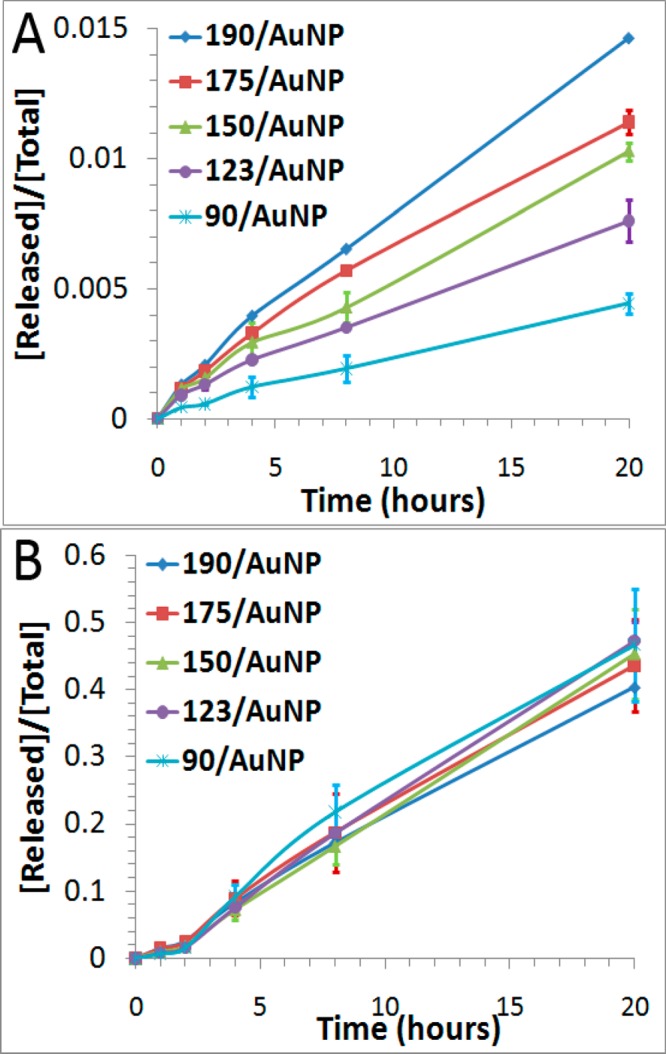
Effect of packing density on the thermal stability of DNA-AuNPs at 37 °C (A) and at 85 °C (B). The average density ranged from 90 oligonucleotide molecules per AuNP to 190 oligonucleotide molecules per AuNP. Error bars represent one standard deviation from triplicate sample analyses.
Table 2. Effects of Packing Density on the Thermal Dissociation Kinetics of DNA-AuNPs.
| DNA
dissociation at 37 °C |
DNA
dissociation at 85 °C |
|||
|---|---|---|---|---|
| packing density | k37a (min–1) | f37b | k85a (min–1) | f85b |
| 190/AuNP | 1.2 × 10–5 | 1.5% | 4.5 × 10–4 | 40.4% |
| 175/AuNP | 9.0 × 10–6 | 1.1% | 5.0 × 10–4 | 43.6% |
| 150/AuNP | 8.0 × 10–6 | 1.0% | 5.3 × 10–4 | 45.3% |
| 123/AuNP | 5.8 × 10–6 | 0.8% | 5.7 × 10–4 | 47.2% |
| 90/AuNP | 3.5 × 10–6 | 0.4% | 5.6 × 10–4 | 46.7% |
Rate constant k was determined from the first order reaction rate equation: ln(1 – [released]/[total]) = kt, where t = 60–1200 min.
Final released fraction f: f = [released]/[total] at 1200 min.
Enhancing the Stability of DNA-AuNPs Using Double-Stranded DNA and Multiple Thiol Anchors
It has been demonstrated that DNA-AuNPs that were formed with double-stranded DNA improved the rate of target hybridization and DNA-mediated AuNP assembly.31,39,40 Here, we explored the possibility of using double-stranded DNA to enhance the stability of DNA-AuNPs. As shown in Figure 6A, we designed three double-stranded DNA to construct a set of double-stranded DNA-AuNP probes (DP-1, DP-2, and DP-3). These double-stranded DNA were modified with different combinations of organosulfur anchor groups. We reasoned that modifying both complementary strands with anchor groups could allow each double-stranded DNA to carry multiple thiol anchors and thus could potentially be used to enhance the stability of DNA-AuNPs.
Figure 6.
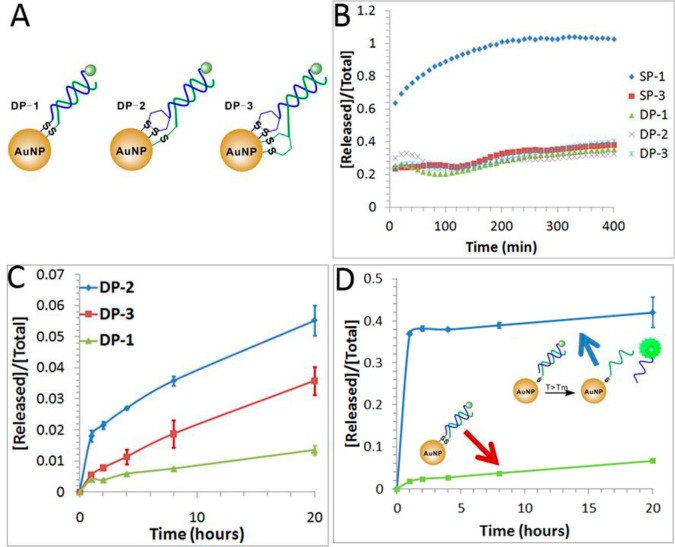
Design of double-stranded DNA-AuNP probes to enhance the stability of the short internal complementary DNA (sicDNA). (A) Schematic showing the double-stranded DNA-AuNP probes constructed by conjugating both of the complementary strands with different combinations of organosulfur anchors. DP-1 represents DNA-AuNP constructed by conjugating both DNA strands with thiol anchors; DP-2 represents DNA-AuNP by conjugating one DNA strand with the thiol anchor and the other strand with the DTPA anchor; DP-3 represents DNA-AuNP by conjugating DNA strands with DTPA anchors. (B) Release of DNA from DNA-AuNPs induced by 2 mM DTT at room temperature. (C) Release of DNA from double-stranded DNA-AuNP probes at a temperature of 37 °C, which is lower than the melting temperature (48.6 °C) of the double-stranded DNA (dsDNA). (D) Comparison of the thermal stability of DP-1 (two thiol anchors) with its control (a single thiol anchor) at a temperature of 60 °C, which is higher than the melting temperature (48.6 °C) of the dsDNA. Error bars represent one standard deviation from triplicate sample analyses.
It is necessary to maintain a stable DNA duplex during the conjugation process. To achieve this, we designed double-stranded DNA probes to have a melting temperature (Tm = 48.6 °C at [NaCl] = 50 mM) much higher than the temperature (T = 25 °C) used for the conjugation procedures. To further examine the formation of double-stranded DNA-AuNP probes, we studied their chemical stabilities by treating DNA-AuNPs with 2 mM DTT. As shown in Figure 6B, all three double-stranded DNA-AuNPs have improved chemical stability as compared to the single-stranded DNA-AuNP (SP-1). These results indicate that the generation of multiple thiol anchors by using double-stranded DNA can enhance the chemical stability of DNA-AuNPs. Additionally, the increasing number of thiol anchors from 2 (DP-1) to 4 (DP-3) did not result in kinetic differences.
We then studied the thermal stability of this set of DNA-AuNP probes by incubating them at 37 °C for a period of 20 h. As shown in Figure 6C, DP-1 has the best thermal stability, which is consistent with our previous results (Figure 3), confirming that the thiol anchor is more thermally stable than the cyclic disulfide anchor on DNA-AuNPs. We further examined the thermal stability of DP-1 at 60 °C, a temperature higher than the melting temperature of the double-stranded DNA (Tm = 48.6 °C). As can be seen from Figure 6D, DP-1 remained stable at this elevated temperature. Only a small fraction (less than 10%) of FAM-labeled DNA was released from DP-1 after 20 h of incubation at 60 °C. In contrast, more than 40% of the DNA was released within 1 h from the DNA-AuNPs that was formed with double-stranded DNA having a single thiol anchor at one strand. These results verify the conjugation of both strands of double-stranded DNA to AuNPs in DP-1, DP-2, and DP-3. The conjugation of both strands to AuNPs can substantially increase the local effective concentrations of complementary strands, which favors the hybridization of these complementary strands and increases the melting temperature of these double-stranded DNA.41,42 This approach may be applied to maintain DNA hybridization under thermally stressed conditions.
Conclusions
We have systematically studied the thermal stability of DNA functionalized gold nanoparticles (DNA-AuNPs) as a function of anchor types and packing densities. By monitoring the kinetics of releasing DNA from DNA-AuNPs that have different designs, we demonstrated that thermal stability of DNA-AuNPs varied with both anchor types and packing densities. Further analyses on the effects of anchor types revealed an opposite trend between the thermal stability and the chemical stability among the DNA-AuNPs that have different anchor types. These results can potentially be used to guide the design of DNA-AuNPs for different therapeutic applications. Finally, we designed a series of double-stranded DNA-AuNP probes by conjugating both DNA strands with organosulfur anchors and demonstrated their abilities to enhance both the chemical stability and thermal stability of DNA-AuNPs.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research, the Canada Research Chairs Program, Alberta Health, and Alberta Innovates.
Supporting Information Available
DNA sequences used for constructing different DNA-AuNPs; UV/vis extinction spectra of bare gold nanoparticles (AuNPs) and DNA functionalized AuNPs (DNA-AuNPs); and evaluation of the inner filter effect from DNA-AuNPs. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K.; Han M. S.; Mirkin C. A. (2006) Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027–1030. [DOI] [PubMed] [Google Scholar]
- Giljohann D. A.; Seferos D. S.; Prigodich A. E.; Patel P. C.; Mirkin C. A. (2009) Gene regulation with polyvalent siRNA-nanoparticle conjugates. J. Am. Chem. Soc. 131, 2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. C.; Giljohann D. A.; Daniel W. L.; Zheng D.; Prigodich A. E.; Mirkin C. A. (2010) Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem. 21, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S.; Daniel W. L.; Giljohann D. A.; Mirkin C. A.; Lippard S. J. (2009) Polyvalent oligonucleotide gold nanoparticle conjugates as delivery vehicles for platinum(IV) warheads. J. Am. Chem. Soc. 131, 14652–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos D. S.; Giljohann D. A.; Hill H. D.; Prigodich A. E.; Mirkin C. A. (2007) Nano-flares: probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 129, 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D.; Seferos D. S.; Giljohann D. A.; Patel P. C.; Mirkin C. A. (2009) Aptamer nano-flares for molecular detection in living cells. Nano Lett. 9, 3258–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigodich A. E.; Seferos D. S.; Massich M. D.; Giljohann D. A.; Lane B. C.; Mirkin C. A. (2009) Nano-flares for mRNA regulation and detection. ACS Nano 3, 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhoumi A.; Huschka R.; Bardhan R.; Knight M. W.; Halas N. J. (2009) Light-induced release of DNA from plasmon-resonant nanoparticles: Towards light-controlled gene therapy. Chem. Phys. Lett. 482, 171–179. [Google Scholar]
- Braun G. B.; Pallaoro A.; Wu G. H.; Missirlis D.; Zasadzinski J. A.; Tirrell M.; Reich N. O. (2009) Laser-activated gene silencing via gold nanoshell-siRNA conjugates. ACS Nano 3, 2007–2015. [DOI] [PubMed] [Google Scholar]
- Poon L.; Zandberg W.; Hsiao D.; Erno Z.; Sen D.; Gates B. D.; Branda N. R. (2010) Photothermal release of single-stranded DNA from the surface of gold nanoparticles through controlled denaturating and Au-S bond breaking. ACS Nano 4, 6395–6403. [DOI] [PubMed] [Google Scholar]
- Jain P. K.; Qian W.; El-Sayed M. A. (2006) Ultrafast cooling of photoexcited electrons in gold nanoparticle-thiolated DNA conjugates involves the dissociation of the gold-thiol bond. J. Am. Chem. Soc. 128, 2426–2433. [DOI] [PubMed] [Google Scholar]
- Huschka R.; Neumann O.; Barhoumi A.; Halas N. J. (2010) Visualizing light-triggered release of molecules inside living cells. Nano Lett. 10, 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huschka R.; Zuloaga J.; Knight M. W.; Brown L. V.; Nordlander P.; Halas N. J. (2011) Light-induced release of DNA from gold nanoparticles: nanoshells and nanorods. J. Am. Chem. Soc. 133, 12247–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisselier E.; Astruc D. (2009) Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 38, 1759–1782. [DOI] [PubMed] [Google Scholar]
- Herdt A. R.; Drawz S. M.; Kang Y. J.; Taton T. A. (2006) DNA dissociation and degradation at gold nanoparticle surfaces. Colloids Surf., B 51, 130–139. [DOI] [PubMed] [Google Scholar]
- Letsinger R. L.; Elghanian R.; Viswanadham G.; Mirkin C. A. (2000) Use of a steroid cyclic disulfide anchor in constructing gold nanoparticle-oligonucleotide conjugates. Bioconjugate Chem. 11, 289–291. [DOI] [PubMed] [Google Scholar]
- Li Z.; Jin R. C.; Mirkin C. A.; Letsinger R. L. (2002) Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 30, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan J. A.; Karlsson C.; Smith W. E.; Graham D. (2007) Enhanced oligonucleotide-nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res. 35, 3668–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela F.; Ding P.; Budow S. (2011) DNA gold nanoparticle conjugates incorporating thiooxonucleosides: 7-deaza-6-thio-2′-deoxyguanosine as gold surface anchor. Bioconjugate Chem. 22, 794–807. [DOI] [PubMed] [Google Scholar]
- Bhatt N.; Huang P. J. J.; Dave N.; Liu J. (2011) Dissociation and degradation of thiol-modified DNA on gold nanoparticles in aqueous and organic solvents. Langmuir 27, 6132–6137. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Servos M. R.; Liu J. (2012) Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and sufactant-free route. J. Am. Chem. Soc. 134, 7266–7269. [DOI] [PubMed] [Google Scholar]
- Barrett L.; Dougan J. A.; Faulds K.; Graham D. (2011) Stable dye-labelled oligonucleotide-nanoparticle conjugates for nucleic acid detection. Nanoscale 3, 3221–3227. [DOI] [PubMed] [Google Scholar]
- Xu S.; Yuan H.; Xu A.; Wang J.; Wu L. (2011) Rapid synthesis of stable and functional conjugates of DNA/Gold nanoparticles mediated by Tween 80. Langmuir 27, 13629–13634. [DOI] [PubMed] [Google Scholar]
- Heinz H.; Vaia R. A.; Farmer B. L. (2008) Relation between packing density and thermal transitions of alkyl chains on layered silicate and metal surfaces. Langmuir 24, 3727–3733. [DOI] [PubMed] [Google Scholar]
- Mei B. C.; Oh E.; Susumu K.; Farrell D.; Mountziaris T. J.; Mattoussi H. (2009) Effects of ligand coordination number and surface curvature on the stability of gold nanoparticles in aqueous solutions. Langmuir 25, 10604–10611. [DOI] [PubMed] [Google Scholar]
- Cederquist K. B.; Keating C. D. (2009) Curvature effects in DNA:Au nanoparticle conjugates. ACS Nano 3, 256–260. [DOI] [PubMed] [Google Scholar]
- Hurst S. J.; Lytton-Jean A. K. R.; Mirkin C. A. (2006) Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 78, 8313–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers L. M.; Mirkin C. A.; Mucic R. C.; Reynolds R. A.; Letsinger R. L.; Elghanian R.; Viswanadham G. (2000) A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 72, 5535–5541. [DOI] [PubMed] [Google Scholar]
- Wen Y. Q.; McLaughlin C. K.; Lo P. K.; Yang H.; Sleiman H. F. (2010) Stable gold nanoparticle conjugation to internal DNA positions: facile generation of discrete gold nanoparticle-DNA assemblies. Bioconjugate Chem. 21, 1413–1416. [DOI] [PubMed] [Google Scholar]
- Lee J. S.; Lytton-Jean A. K. R.; Hurst S. J.; Mirkin C. A. (2007) Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 7, 2112–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.; Chen P.; Sun Y. W.; Xing Y. Z.; Yang Y.; Dong Y. C.; Xu L. J.; Yang Z. Q.; Liu D. S. (2011) A new strategy improves assembly efficiency of DNA mono-modified gold nanoparticles. Chem. Commun. 47, 5774–5776. [DOI] [PubMed] [Google Scholar]
- Vericat C.; Vela M. E.; Carro P.; Salvarezza R. C. (2010) Self-assembled monolayers of thiols and dithiols on gold: new challenges for a well-known system. Chem. Soc. Rev. 39, 1805–1834. [DOI] [PubMed] [Google Scholar]
- Fenter P.; Eberhardt A.; Eisenberger P. (1994) Self-assembly of N-alkyl thiols as disulfides on Au(111). Science 266, 1216–1218. [DOI] [PubMed] [Google Scholar]
- Kluth G. J.; Carraro C.; Maboudian R. (1999) Direct observation of sulfur dimers in alkanethiol self-assembled monolayers on Au(111). Phys. Rev. B 59, 10449–10452. [Google Scholar]
- Schlenoff J. B.; Li M.; Ly H. (1995) Stability and self-exchange in alkanethiol monolayers. J. Am. Chem. Soc. 117, 12528–12536. [Google Scholar]
- Kondoh H.; Kodama C.; Sumida H.; Nozoye H. (1999) Molecular processes of adsorption and desorption of alkanethiol monolayers on Au(111). J. Chem. Phys. 111, 1175–1184. [Google Scholar]
- Li F.; Li J.; Wang C.; Zhang J.; Li X.-F.; Le X. C. (2011) Competitive protection of aptamer-functionalized gold nanoparticles by controlling the DNA assembly. Anal. Chem. 83, 6464–6467. [DOI] [PubMed] [Google Scholar]
- Schreiner S. M.; Hatch A. L.; Shudy D. F.; Howard D. R.; Howell C.; Zhao J.; Koelsch P.; Zharnikov M.; Petrovykh D. Y.; Opdahe A. (2011) Impact of DNA-surface interactions on the stability of DNA hybrids. Anal. Chem. 83, 4288–4295. [DOI] [PubMed] [Google Scholar]
- Prigodich A. E.; Lee O. S.; Daniel W. L.; Seferos D. S.; Schatz G. C.; Mirkin C. A. (2010) Tailoring DNA structure to increase target hybridization kinetics on surfaces. J. Am. Chem. Soc. 132, 10638–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maye M. M.; Nykypanchuk D.; Lelie D. V.; Gang O. (2006) A simple method for kinetic control of DNA-induced nanoparticle assembly. J. Am. Chem. Soc. 128, 14020–14021. [DOI] [PubMed] [Google Scholar]
- Li F.; Zhang H.; Lai C.; Li X.-F.; Le X. C. (2012) A molecular translator that acts by binding-induced DNA strand displacemenet for a homogeneous protein assay. Angew. Chem., Int. Ed. 51, 9317–9320. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Li F.; Dever B.; Wang C.; Li X.-F.; Le X. C. (2013) Assembling DNA through affinity recognitions for protein detection. Angew. Chem., Int. Ed. 52, 10698–10705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.