Abstract
KIR2DL4 (CD158d) is a distinct member of the killer cell Ig-like receptor (KIR) family in human NK cells that can induce cytokine production and cytolytic activity in resting NK cells. Soluble HLA-G, normally expressed only by fetal-derived trophoblast cells, was reported to be a ligand for KIR2DL4; however, KIR2DL4 expression is not restricted to the placenta and can be found in CD56high subset of peripheral blood NK cells. We demonstrated that KIR2DL4 can interact with alternative ligand(s), expressed by cells of epithelial or fibroblast origin. A genome-wide high-throughput siRNA screen revealed that KIR2DL4 recognition of cells surface ligand(s) is directly regulated by heparan sulfate (HS) glucosamine 3-O-sulfotransferase 3B1 (HS3ST3B1). KIR2DL4 was found to directly interact with HS/heparin, and the D0-domain of KIR2DL4 was essential for this interaction. Accordingly, exogenous HS/heparin can regulate cytokine production by KIR2DL4-expressing NK cells and HEK293T cells (HEK293T-2DL4) and induces differential localization of KIR2DL4 to rab5+ and rab7+ endosomes, thus leading to down-regulation of cytokine production and degradation of the receptor. Furthermore, we showed that intimate interaction of syndecan-4 (SDC4) HS Proteo-Glycan (HSPG) and KIR2DL4 directly affects receptor endocytosis and membrane trafficking.
Introduction
Natural killer (NK) cells are innate immune cells that are capable of directly attacking tumor, virus-infected, and stressed cells (1). NK cell activation can trigger both focused target cell lysis through release of perforin and granzymes from cytolytic granules and the secretion of numerous cytokines, especially interferon (IFN)-γ and tumor necrosis factor (TNF)-α. NK cell activating receptors include activating forms of KIR [short forms (KIR2DS or KIR3DS)], 2B4, NKG2D, NKp80, and the natural cytotoxicity receptors, (NCR)-1, -2, and -3 called NKp46, NKp44, and NKp30, respectively. Selective engagement of any of these receptors can stimulate both cytotoxicity and cytokine production (2).
KIR2DL4 (2DL4; CD158d) is a structurally distinct member of the KIR family and a functionally unique NK cell receptor with expression restricted to NK cells and some T cells in higher primates (3, 4). Although early studies indicated that 2DL4 was the only KIR gene from which mRNA is expressed in every human NK cell clone tested (5, 6) and in all human subjects analyzed (7), we and others demonstrated that KIR2DL4 expression is normally restricted to a CD56high subset of NK cells (4, 8). However, the fraction expressing KIR2DL4 can be significantly increased when NK cells are cultured in the presence of IL-2 (4, 8). Due to inheritance of prevalent 2DL4 alleles that encode truncated receptors, the individuals homozygous for the “9A” allotype have consecutive series of only 9 out of 10 adenines in a portion of the exon encoding the membrane-proximal cytoplasmic domain, which encodes a truncated receptor that cannot reach the cell surface (9).
KIR2DL4 is a structurally unique receptor with distinct elements among KIR family members: 1) an extracellular domain consisting of D0 and D2 Ig-like domains, which is a feature shared only by KIR2DL5 (10, 11), 2) a cytoplasmic domain possessing a single immunoreceptor tyrosine-based inhibitory motif (ITIM) (while all inhibitory KIR have two), which can recruit only SHP-2 (12), and 3) a transmembrane domain containing a charged arginine residue, which can associate with FcεRI-γ to contribute activating function (13). Functionally, KIR2DL4 can trigger potent cytokine production (IFNγ, chemokines, and angiogenic factors), but only weak cytotoxicity in resting NK cells (14-17).
Several studies have reported that KIR2DL4 recognizes a soluble form of the non-classical MHC-I molecule, HLA-G, which can trigger secretion of pro-angiogenic cytokines (6, 18). HLA-G is normally expressed only by fetal-derived trophoblast cells that invade the maternal decidua in pregnant women and create a barrier for maternal NK cell attack of the fetus (19). However, HLA-G expression has also been observed on some tumors (20, 21). Placental NK cells can express KIR2DL4 on their surface (18) and therefore KIR2DL4 may play normal physiological roles during pregnancy. Nonetheless, some have disputed the recognition of HLA-G by KIR2DL4 and its physiological significance, since women homozygous for 9A allotype appear to have normal pregnancies (22-25).
We generated a recombinant KIR2DL4 fusion protein (KIR2DL4-Ig) and observed that it can bind to the surface of several cell lines of epithelial and fibroblast origin, which lack expression of HLA-G, suggesting that these cells endogenously express alternative KIR2DL4 ligand(s). Therefore, we used a a whole genomic siRNA library screen to identify the alternative non-HLA-G ligand(s). Our studies discovered that KIR2DL4 can interact with heparan sulfate/heparin glycosaminoglycans, and these interactions can affect receptor function.
Materials and Methods
Cell culture
Human cell lines were the EBV-transformed 721.221 B cell line (CRL-1855); 721.221 transfected with HLA-G1 cDNA (721-HLA-G1; from Dr. M. Colonna, Washington University, St. Louis, MO); PC3 prostate adenocarcinoma (CRL-1435); HeLa cervical adenocarcinoma (CCL-2); Human Normal Breast Epithelium (NBE), Human Normal Kidney Epithelium (NKE) and Human Normal Fibroblasts (HNF) (obtained from the Biosample Repository Facility, Fox Chase Cancer Center, Philadelphia PA, USA); NK-92 (CRL2407) and KHYG-1 (JCRB0156; from HSRRB, Japan Health Sciences Foundation, Osaka, Japan); NK-like cell lines and those transduced with FLAG-KIR2DL4*00201 cDNA (NK92-2DL4 and KHYG-2DL4, respectively; HEK293T embryonic kidney cells (CRL-11268) and those transfected with KIR2DL4 cDNA in pCDNA3.1+ (Invitrogen; HEK293T-2DL4); CHO-K1 Chinese hamster ovarian line (CCL-61) and HS-negative CHO-677 (pgsD-677) (26). The cell line SKOV-3 human ovarian carcinoma (HTB-77), SKOV-3 transfected with mock backbone (SKOV3-V), or SKOV-3 transfected with cDNA coding for heparan sulfate 6-O-endo-sulfatase 1 (SULF-1; SKOV3-S) were cultured as recommended by ATCC for SKOV-3 cell line. Cells were cultured as recommended by ATCC or according to previously established protocols (27).
Western Blotting
Cells were lysed in standard RIPA buffer containing protease inhibitor cocktail (Pierce). Proteins were electrophoresed and transferred to PVDF membrane, blocked with 1% BSA + 0.05% Tween-20 in Ca2+- and Mg2+-free DPBS, and immunoblotted in 0.2% BSA + 0.05% Tween-20 in Ca2+- and Mg2+-free DPBS with 4H84 anti-pan-HLA-G mAb (abcam), anti-β-actin mAb (abcam) and goat anti-mouse-IgG-HRP secondary (Jackson ImmunoResearch). Membranes were developed with SuperSignal West Pico Substrate (Thermo Scientific) and imaged on an XRS+ imaging system (Bio-Rad).
Fusion proteins
Extracellular domain cDNAs were cloned into pCDNA3.1+ as fusions with the Fc domain of human IgG, as previously described (28). KIR2DL4-Ig corresponds to UniProtKB Q99706 sequence (KIR2DL4*00101). To clone independent KIR2DL4 Ig-like domains, an extra sequence and overlap at the hinge were added to improve stability, as the residues at the hinge in KIRs are making direct contacts to both domains. Hence, His24 to L122 were deleted to make D2, and V116 to Pro221 were removed to make D0. KIR2DL4-Ig, NKp46D2-Ig, LIR1-Ig or KIR2DL1-Ig accordingly fusion proteins or hFc-Ig alone were produced as previously described (28).
Whole Genome siRNA screen
The siGENOME siRNA library (from Dharmacon RNAi Technologies, Fisher Scientific) consists of individual siRNA pools targeting 21,300 human genes, arranged in 267 96-well plates. Transfections utilized DharmaFECT 2 (Fisher Scientific) according to manufacturer specifications. The following controls were used on each of the 267 96-well plates: siGENOME β2m targeting siRNA was used as a positive control of transfection efficiency, while siGENOME luciferase-targeting siRNA siGL2 was a negative control. Cycloheximide (CHX) was used as a positive control to down-modulate KIR2DL4-Ig surface staining (100μM for 24h).
The whole-genome siRNA screen was performed in duplicate run at the High Throughput Screening Facility at Fox Chase Cancer Center (FCCC) using a “reverse transfection” protocol that was optimized for use with the PC3 cell line. Automated pipetting was carried out with a CyBi Well Vario (CyBio) unit. DharmaFECT 2 transfection reagent (1:100), and RPMI-1640 medium (Gibco) were first dispensed into separate 96-well plates, then siRNAs solution (50nM) was added, mixed and incubated for 15 min. PC3 cells (5000 per well) were added in complete RPMI-1640 medium supplemented with 10% FBS (Hyclone), 20U penicillin, 20mg streptomycin, 0.1mM non-essential amino acids and 1mM sodium pyruvate, 2mM L-Glutamine, and 10 mM HEPES, pH 7.4 (Gibco). Each well was transfected with a separate SMARTPool containing four distinct siRNAs targeting a single mRNA. Plates were incubated at 37 °C in 5% CO2 for 48 hours.
Fluorescent microscopy screening and siRNA screen data analysis
Based on the kinetics of HLA-A,B,C knockdown after transfection with β2m -targeting siRNA, PC3 cells were scored for KIR2DL4 ligand expression 48 hours after siRNA transfection. PC3 cells were labeled with Cell Tracker Green - CMFDA (Invitrogen; per manufacturer specifications), washed with ice cold Ca2+- and Mg2+-free DPBS, fixed with 4% PFA for 10′ at 4°C, washed again and stained with 20μg/ml of KIR2DL4-Ig, followed by staining with APC-anti-human-IgG-Fc Ab (Jackson ImmunoResearch). β2m mAb (Biolegend) staining was followed by APC-anti-mouse-IgG-Fc Ab (Jackson ImmunoResearch). Plates were covered with mounting solution of 0.5% N-Propyl Gallate and 80% Glycerol in Ca2+- and Mg2+-free DPBS, pH7.4. Cell-associated fluorescence was acquired on six separate image fields from each individual well with an automated HTS-Fluorescent microscope (ImageXpress Micro, Molecular Devices) using MetaXpress software, and data were analyzed in AcuityXpress software (Molecular Devices). The capture of six sites per well allowed analysis of up to 1000 cells in a total 6 biological replica images per siRNA pool screened. Automated cell counts and recognition were based upon both nuclear staining and cytoplasmic area as both determined with Cell Tracker Green staining. KIR2DL4-Ig staining score was based on the parameters of Pit Count, Pit Integrated Intensity, and Pit Total Area, as determined by the AcuityXpress software (Molecular Devices) and normalized to the total number of cells in each biological replicate. Data analysis was done using the cellHTS2 package in Bioconductor, and the median of siGL2 as used as a normalization control. Hits were identified based on: Hit /siGL2 ratio <70% (0.7) of CHX/siGL2 ratio for all 3 parameters for primary screen and <80% (0.8) for secondary screen, and Hit/siGL2 < Nase/siGL2 should apply for all 3 parameters. To assess the efficiency and reproducibility of the screening protocol, a Z’-factor value of 0.64 was calculated using cellHTS2, which established validity of the assay.
Validation siRNA screen
Subsequent validation testing of top Hits was performed on PC3 cells 48 hours after transfection with individual siRNAs (Dharmacon) from the corresponding SmartPool as described above. The validation siRNA screen was performed in four independent duplicate runs. Detection of cell surface staining with KIR2DL4-Ig was quantified on an HTS-equipped FACSCanto II (BD Biosciences) analyzer in a 96-well format.
FACS staining
Cells were incubated with various Ig fusion proteins (20 μg/ml; human IgG-Fc alone, KIR2DL4, NKp46D2, LIR1 or KIR2DL1 human IgG-Fc fusion proteins - hFc-Ig, KIR2DL4-Ig, NKp46D2-Ig, LIR1-Ig or KIR2DL1-Ig accordingly). Cells were washed and stained with APC-conjugated anti-human-IgG-Fc Ab (5 μg/ml; Jackson Immuno Research). All steps were performed on ice or in a chilled centrifuge (4°C). In all blocking experiments, following Ig fusion proteins were pre-incubated for 30′ on ice with indicated concentrations of HS, CS or antibody before incubation with Propidium iodide (PI) was added to all staining samples for exclusion of dead cells. The following antibodies were used to stain surface markers: unconjugated 53.1 anti-KIR2DL4 mAb (hybridoma generously provided by Dr. Marco Colonna, Washington University, St. Louis, MO); unconjugated and APC-conjugated #33 anti-KIR2DL4 mAb (Biolegend); PE-conjugated HCD56 anti-CD56a mAb (Biolegend); HP-MA4 anti-KIR2DL1 mAb (Biolegend); anti human syndecan-1 mAb (eBioscience); goat Anti human syndecan-4 (R&D); rat anti human syndecan-2 (R&D). Following secondary antibodies were used for cell surface markers: APC-conjugated anti-mouse-IgG-Fc (Jackson). Following secondary antibodies were used for syndecan expression profile: FITC-conjugated anti-goat, anti-rat or anti-mouse-IgG-Fc Ab (Jackson). In all blocking experiments 20 μg/ml of hIg-Fc fusion proteins were pre-incubated with 5μg/ml of either HS (Heparin - low molecular weight) or CS (Chondroitin Sulfate A) (Sigma) for 30′ at 4°C. Then the mixture was used to stain cells. Flow cytometry was performed on FACSCanto II (BD Biosciences) and data was analyzed with FlowJo software (Tree Star).
Enzymatic treatment
1 × 106 cells were treated with 2U/mL Heparinase I or III (Sigma-Aldrich) in DPBS supplemented with 0.5% BSA or Proteinase K 1U/mL (Sigma-Aldrich) in plain DPBS. Each treatment was carried out for 60 min at 37°C, followed by two washes with ice-cold DPBS supplemented with 0.5% BSA.
Confocal imaging and co-localization assay
Cells were cultured on 8-chamber coverglass μ-slides (iBidi) as described above in 300μl of complete culture medium alone or medium supplemented with 1μg/ml of either HS (Heparin - low molecular weight) or CS (Chondroitin Sulfate A) (Sigma) and incubated at 37°C in 5% CO2 for 2h. All images were acquired at these standard cell culture conditions on a FluoVew FV1000 confocal system (Olympus) using a 63× UPLSAPO objective (Olympus). The co-localization analysis was performed using the ImageJ software package supplemented with the JACoP plugin. Manders’ overlap coefficients M1 and M2 were used to evaluate the degree of co-localization between two fluorescent labels (29-31). The threshold value was calculated from the background intensity of the noisiest images (32). This value of threshold was applied in all subsequent M1 and M2 calculations, setting all below threshold pixels to zero. M1 was defined as the proportion of pixel with non-zero intensities from the green image (GFP alone; Rab5, Rab7, SDC4 and CD55 GFP fusion proteins), for which the intensity in the red channel is also above zero. Conversely, M2 was defined as the proportion of pixel with non-zero intensities from the red image (KIR2DL4-mCherry fusion protein), for which the intensity in the green channel is also above zero. Thereby M2 coefficient corresponds to the proportion of KIR2DL4-mCherry co-localized with the appropriate GFP-labeled marker.
Cell stimulation and ELISA assays
For IFN-γ and IL-8 experiments, cells were stimulated in 96-well U-bottom plates (NUNC) pre-coated with the following antibodies at 5 μg/ml concentration for 18h at 4°C: anti-KIR2DL4 mAb (clone 181703, 53.1, or 33), anti-2B4 mAb (clone c1.7, Biolegend), anti-KIR2DL1 mAb (clone HP-MA4) or HCD56 anti-CD56a mAb. Antibodies were diluted in Na2HPO4 buffer, pH 9 for plate coating. For IL-8 experiments HEK293T cells were instantly mixed and co-incubated with either HS or CS as above. For IFN-γ assays NK cells were preincubated in complete culture medium (supplemented with 20 U/mL rhIL-2) with HS or with CS for 2h at 37°C before plating. For all experiments 5×105 of either NK cells or HEK293T cells were added to each well in 150μl of complete culture medium and incubated for 18 h (37°C, 5% CO2). These cells were stimulated with antibodies, HS, or PC3 target cells at the indicated concentration or E:T ratio. In all cases the cytokine concentration in 100μl of supernatant was assayed by standard ELISA assay according to manufacturer specifications (ELISA MAX, Biolegend).
Primary NK cell purification
A human negative NK cell isolation kit (Miltenyi Biotec) was used to purify NK cells from peripheral blood of healthy volunteer donors, who were recruited by informed consent as approved by the FCCC and BGU Institutional Review Boards. NK cell purity was >90% (CD3-CD56+). Purified NK cells were cultured in CellGro stem cell serum-free growth medium (CellGenix) supplemented with 10% heat-inactivated human plasma from healthy donors, 50U penicillin, 50mg streptomycin, 0.1mM non-essential amino acids, 1mM sodium pyruvate, 2mM L-Glutamine, 10 mM HEPES, pH 7.4, 0.1mM 2-mercaptoethanol (Gibco) and 300 IU/ml human IL-2 (Biological Industries).
BIAcore
A BIACORE 3000 device fitted with CM5 sensor chips (BIACORE AB, Uppsala, Sweden) was used for studying the interactions between heparin and recombinant KIR2DL4 in conjunction with BIAevaluation software (v4.1). Running buffer was Ca2+- and Mg2+-free DPBS pH 7.4, supplemented with 0.005% Tween 20, at a flow rate of 5 μl/min at 25°C. To activate the chip, the EDC/NHS amine coupling procedure was used according to the manufacturer’s protocol (BIAcore), followed by addition of 10 μg/ml neutravidin (Pierce), which was immobilized in the different flow cells, followed by blocking the free active groups with 1 M ethanolamine. Following neutravidin binding, heparin-biotin (Sigma) (10 μg/ml) was injected to achieve up to 20RU in flow cell 2, while flow cell 1 served as a control (neutravidin was bound but not heparin) and was subtracted from the responses obtained from the flow cells with bound heparin. Kinetic measurements were performed at a flow rate of 30 μl/min. Different analyte concentrations were injected, each followed by regeneration of the surface using 10 mM NaOH. Data were analyzed using a 1:1 Langmuir binding model. The χ2 values were less than 1.
3D modeling
The extracellular domain of KIR2DL4 was modeled by sequence homology modeling to the 3D crystal structures of KIR3DL1 (PDB code 3VH8, 58% identity), KIR2DL3 (PDB code 1B6U, 58%identity), and NKp46 (PDB code 1P6F, 32% identity) using MolIDE and SCWRL software (33, 34). The model based on KIR2DL3 was chosen as it had a better E-value and longer alignment length. After first generating a PQR file of the KIR2DL4 model with assigned atomic charges using the CHARMM force field, the Coulombic charge surface was calculated in Chimera (35), as shown in Figure 3A. Inspection of the surface revealed a very basic patch (blue residues) lying within the D0 domain that we postulated as containing the binding site for HS. The same procedure with identical parameters was used to calculate a Coulombic surface for the known structures of 2DL1 (PDB code 1IM9) and LIR1 (PDB code 1UGN), and shown in Figure 3 B & C.
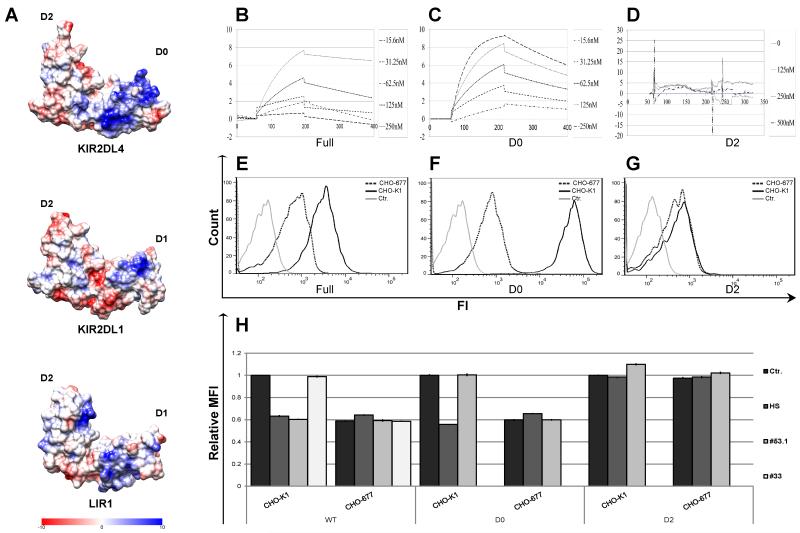
Figure 3. Mapping of the HS/heparin binding site on the KIR2DL4 extracellular domain.
(A) Coulombic charge map of a 3D model of the KIR2DL4 extracellular domain structure. B-D. BIAcore binding-kinetic plots are shown for the interactions of the full structure of KIR2DL4-Ig (B) and individual D0 (C) or D2 (D) Ig-like domains with HS bound on a CM5 sensor chip. Individual lines represent analyses at 2DL4-Ig input concentrations in nM. E-G. Direct binding of KIR2DL4-Ig WT (E), D0 (F) or D2 (G) proteins to parental CHO-K1 cell line (bold line) or the HS-deficient CHO-677 mutant cell line (dotted line) was assayed by FACS. Data represent one of n=4 independent experiments. H. Blocking of FACS-based binding of KIR2DL4-Ig to CHO-K1 or CHO-677 cell lines by HS and anti-KIR2DL4 mAbs. 20μg of KIR2DL4-Ig fusion protein was pre-incubated with either 5μg of HS, CS or 30μg of anti-KIR2DL4 mAb (clones 53.1 or 33) or control mAb (2B4; Ctr.) prior to addition of the mixture to cells. Data represent normalized to Ctr. mean ± s.d. of n=3 independent experiments for (H).
Results
To test for alternative KIR2DL4 ligand(s) we produced a soluble form of KIR2DL4 extracellular domain fused to the CH2 + CH3 domains of human IgG1 (KIR2DL4-Ig) in HEK293T cells, as previously described (28). We then screened a number of HLA-G deficient cell lines and primary cells for specific staining with KIR2DL4-Ig fusion protein (Supplemental Fig. 1). PC3 and HeLa cell lines manifested the highest staining with KIR2DL4-Ig (Supplemental Fig. 1; overlaid in Supplemental Fig. 2A) and were confirmed to lack detectable HLA-G expression in Western Blot (Supplemental Figure 2B). Furthermore, co-culture with PC3 target cells resulted in a considerably higher secretion of IFN-γ by KHYG-1 cells transduced to overexpress surface KIR2DL4 (KHYG1-2DL4) as compared to the parental KHYG-1 NK cell line (Supplemental Fig. 2C). Therefore, the PC3 cell line was chosen for further study of novel KIR2DL4 ligand(s) for the following characteristics: 1) high and uniform cell surface staining with KIR2DL4-Ig, 2) no detectable expression of HLA-G protein, 3) capacity to stimulate cytokine secretion in KIR2DL4 overexpressing NK cell line, and 4) PC3 is an adherent cell line that can be efficiently cultured and transfected.
High throughput whole genome siRNA screen
To identify the novel ligand(s) mediating KIR2DL4-Ig binding to the PC3 cell line, a whole genome siRNA library screen was performed using a High Throughput Screening (HTS) epifluorescence microscope in a 96-well format (Fig. 1A). The Dharmacon siGENOME siRNA library, which consists of individual pools of four siRNAs targeting each of 21,300 human genes, was introduced using a fully automated reverse transfection protocol to knockdown expression of specific mRNAs in individual wells (see Methods). We hypothesized that siRNAs targeting the ligand or proteins important for surface expression of the ligand would significantly diminish KIR2DL4-Ig staining. Efficacy of siRNA knockdown was exemplified by a β2m-specific siRNA pool (siβ2m), that significantly down-regulated surface β2m expression on PC3 cells (Fig. 1B), while a negative control siRNA pool targeting luciferase (siGL2) did not affect β2m expression (siGL2; Fig. 1B).
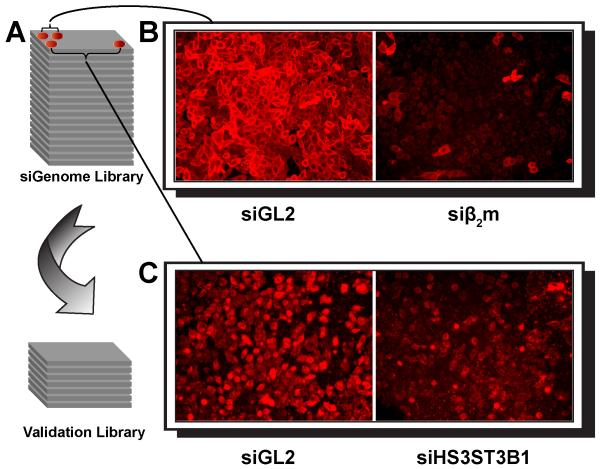
Figure 1. Genomic siRNA library screening strategy and representative images of immunofluorescence analysis.
(A) siGenome siRNA library (Dharmacon) screening testing involved transfecting PC3 cells with individual SMARTPools containing 4 distinct siRNAs targeting a single mRNA and arranged in 96-well plates format. Validation screen tested individual siRNAs (Dharmacon) from the corresponding SmartPool “hits” and arranged in 96-well plates format. (B) As an internal negative control, cells were introduced with SMARTPool siRNA targeting luciferase (siGL2). As a positive control, siRNA silencing with β2m specific siRNA SMARTPool significantly reduced β2m surface expression on PC3 cells (siβ2m) as compared to siGL2 control. Cells were stained with anti-β2m mAb and APC-conjugated secondary Ab. (C) siRNA silencing with SH3ST3B1 specific siRNA SMARTPool significantly reduced KIR2DL4 ligand(s) surface expression on PC3 cells as compared to siGL2 control. Cells were stained with KIR2DL4-Ig and APC-conjugated secondary Ab.
PC3 cells were screened for loss of immunofluorescent KIR2DL4-Ig staining after transfection with individual wells of the whole genome siRNA library, consisting of 267 96-well plates. Silencing of detectable cell surface marker expression was optimal 48 hours post transfection (as exemplified by siβ2m). Indeed, KIR2DL4-Ig staining was significantly reduced after siRNA-mediated knockdown of several genes, as exemplified for heparan sulfate glucosamine 3-O-sulfotransferase 3B1 specific siRNA pool (siHS3ST3B1) in Figure 1C (compared to siGL2) was employed to test specificity. In total a primary hit list of 134 genes significantly reduced binding of KIR2DL4-Ig.
From this primary hit list, we prioritized the 24 most physiologically relevant genes to follow up in a secondary validation screen (Table 1). Since the primary screen was performed by transfecting individual wells with pools of 4 siRNAs targeting a specific gene, we employed the “single siRNA” validation method to test individual siRNAs from each of the prioritized siRNA pools. The prioritized hits were scored by the number of “positive” individual siRNAs targeting each gene that effectively reduced KIR2DL4-Ig binding in the validation screen. The validation screen was performed on an HTS-equipped FACSCanto II (BD Biosciences) analyzer in a 96-well format (see Methods). This secondary analysis eliminated any primary screen data collection artifacts or “off-target” effects due to sequence similarities with non-target mRNA, enhanced miRNA-mediated suppression in the absence of a shared siRNA-targeted mRNA, or due to a cytotoxic effect that results in a broad loss of protein expression. Any “off-target” effect of individual siRNAs within the pools should yield a low score with only one or two out of the four single siRNAs downregulating KIR2DL4-Ig binding. This is exemplified by our low scores for individual siRNAs targeting HLA-A and HLA-E (score 0/4 in validation testing) or HLA-G (score 1/4). PC3 cells were found to be HLA-A positive, but lack detectable HLA-E and HLA-G protein, and only 1 of 12 siRNAs targeting these genes was found to suppress KIR2DL4 binding, which we conclude to be a single off-target effect. In the validation testing stage, the best scoring hit was HS3ST3B1 (NM_006041; 3/4 validation score), which encodes heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (H3-OST-3B protein, Swiss-Prot) (Table 1 and Figure 1C). HS3ST3B1 is widely expressed, unlike HS3ST2 and HS3ST4, which are neuronal lineage-specific (36, 37). Importantly, no other sulfotransferase (e.g. for chondroitin sulfate (CS) GAG) was identified as a high-score hit in the primary screen.
Table 1. siGENOME and Validation screen: prioritized Hit list.
Each gene is characterized by the average primary score (Whole Genome SMARTPool siRNA screen) and average score for each individual siRNA from the corresponding SmartPool. Validation index is indicating the number of “positive” out of 4 individual siRNAs, targeting each gene, which effectively reduced KIR2DL4-Ig binding in the validation screen.
| Gene Symbol | Accession No. |
Av. Primary Score |
Av. Score siRNA I |
Av. Score siRNA II |
Av. Score siRNA III |
Av. Score siRNA IV |
Validation Index |
|---|---|---|---|---|---|---|---|
| BSG | NM_001728 | 0.61 | 0.70 | 1.14 | 0.98 | 0.89 | 1/4 |
| CD63 | NM_001780 | 0.69 | 0.75 | 1.06 | 1.01 | 0.91 | 1/4 |
| CDH11 | NM_001797 | 0.63 | 1.31 | 1.09 | 1.08 | 1.60 | 0/4 |
| CDH3 | NM_001793 | 0.67 | 1.15 | 1.29 | 1.40 | 2.02 | 0/4 |
| CEACAM4 | NM_001817 | 0.67 | 1.22 | 1.45 | 0.75 | 1.82 | 1/4 |
| CEECAM1 | NM_016174 | 0.61 | 1.18 | 1.14 | 0.64 | 0.89 | 1/4 |
| CNTN5 | NM_014361 | 0.59 | 1.02 | 0.89 | 1.00 | 1.12 | 0/4 |
| DKFZP761O0113 | NM_018409 | 0.42 | 1.13 | 1.08 | 0.98 | 0.90 | 0/4 |
| FSTL3 | NM_005860 | 0.52 | 1.03 | 1.21 | 0.74 | 1.19 | 1/4 |
| HLA-A | NM_002116 | 0.75 | 0.89 | 1.91 | 1.07 | 1.50 | 0/4 |
| HLA-G | NM_002127 | 0.68 | 0.73 | 1.03 | 1.02 | 1.02 | 1/4 |
| HLA-E | NM_005516 | 0.71 | 1.38 | 0.83 | 0.97 | 1.14 | 0/4 |
| HS3ST3B1 | NM_006041 | 0.51 | 0.79 | 0.72 | 0.96 | 0.46 | 3/4 |
| KRTAP22-1 | NM_181620 | 0.44 | 0.92 | 1.33 | 1.11 | 0.97 | 0/4 |
| LIFR | NM_002310 | 0.59 | 1.22 | 1.07 | 1.27 | 1.13 | 0/4 |
| LOC347541 | XM_293407 | 0.51 | 1.17 | 1.45 | 1.60 | 1.85 | 0/4 |
| LRIG1 | NM_015541 | 0.49 | 0.84 | 1.05 | 1.18 | 0.90 | 0/4 |
| MICA | NM_000247 | 0.68 | 1.22 | 0.83 | 1.53 | 2.08 | 0/4 |
| MICB | NM_005931 | 0.69 | 0.96 | 1.21 | 0.81 | 0.61 | 1/4 |
| NAGLU | NM_000263 | 0.54 | 1.06 | 1.46 | 1.73 | 1.83 | 0/4 |
| PCDHA2 | NM_018905 | 0.35 | 0.78 | 0.94 | 0.96 | 1.02 | 1/4 |
| PVRL4 | NM_030916 | 0.48 | 1.17 | 1.17 | 1.55 | 1.02 | 0/4 |
| SGCA | NM_000023 | 0.52 | 0.90 | 1.28 | 1.80 | 0.79 | 1/4 |
| ULBP2 | NM_025217 | 0.62 | 1.19 | 1.19 | 0.98 | 1.71 | 0/4 |
HS3ST3B1 specifically carries out the final sulfation steps in the synthesis of heparan sulfate (HS)/heparin glycosaminoglycans (GAGs). HS GAGs are long, unbranched, anionic polysaccharides that are sulfated at N, 2-O, 3-O, and 6-O positions to generate diverse structures with unique protein binding properties(38). HS GAGs are conjugated to a subset of proteins, are found on cell surfaces and the extracellular matrix, and provide docking sites for basic domains on soluble proteins such as chemokines, FGF, and Wnt ligand family members, thereby “presenting” them to cell surface receptors(39-42). An interaction of HS with 2DL4 is in accordance with previously reported specific binding of NKp46, NKp44, and NKp30 to HS, indicating that the sulfated polysaccharide is engaging various NK cell receptors or in some cases influencing their ligand engagement (43-46).
Direct binding of KIR2DL4-Ig to PC3 cells was consistently down-regulated either by pooled siHS3ST3B1 (IFC: Fig. 1C; flow cytometry: Fig. 2A) or three of four individual siRNAs from the same pool (Fig. 2B). The down-regulation of KIR2DL4-Ig binding was specific, since transfection with the siHS3ST3B1 pool did not suppress binding of either KIR2DL1-Ig or LIR1-Ig; both receptors can bind their ligands on the PC3 cell surface in a HS-independent manner (Fig. 2C). We previously reported that the interaction of HS with NKp46 is sensitive to desulfation of the HS (43, 44). Accordingly, the binding of NKp46-Ig to PC3 cells was also down-regulated following siHS3ST3B1 transfection (Fig. 2C).
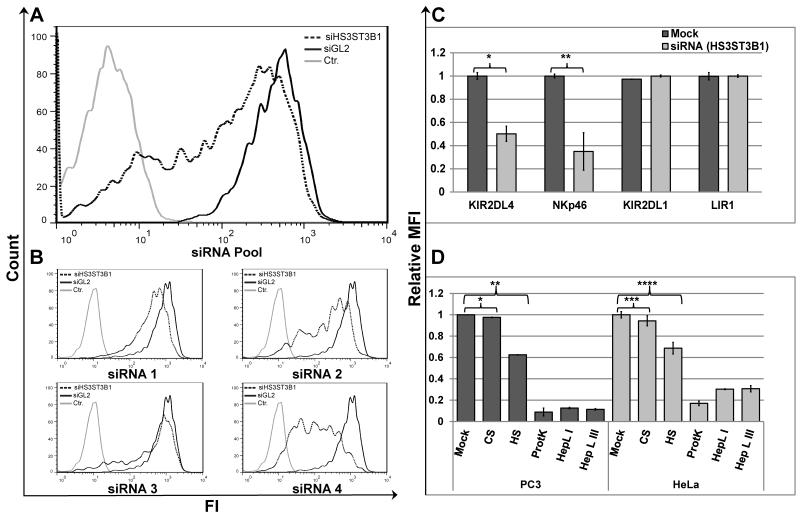
Figure 2. HS3ST3B1-specific siRNA, soluble HS/heparin competition, and HS-specific cleavage prevents binding of KIR2DL4-Ig to PC3 and HeLa cells.
(A) KIR2DL4-Ig staining of PC3 cells transfected with SMARTPool siRNA targeting luciferase (siGL2; bold line) or HS3ST3B1 specific siRNA SMARTPool (dotted line); hFc-Ig staining of control siRNA transfected cells was included as a negative control (grey line). (B) KIR2DL4-Ig staining of PC3 cells transfected with SMARTPool siRNA targeting luciferase (siGL2; bold line) or individual siRNAs from the corresponding SmartPool targeting HS3ST3B1(dotted line); hFc-Ig staining of control siRNA transfected cells was included as a negative control (grey line). Data represent one of n=4 independent experiments for (A) and (B). (C) Comparative staining of KIR2DL4-Ig, NKp46-D2-Ig, KIR2DL1-Ig or LIR1-Ig on PC3 cells transfected with SMARTPool siRNA targeting luciferase (siGL2; Mock) or HS3ST3B1 specific siRNA SMARTPool (siHS3ST3B1). Statistics by T-Test: *p-value 0.005; **p-value 0.01. (D) Comparative staining of PC3 (black bars) or HeLa (grey bars) cells with KIR2DL4-Ig fusion protein under various treatment conditions. Cells were pre-incubated in either DPBS supplemented with 0.5% BSA alone (mock), 5μg/ml of either Heparin LMW (HS) or Chondroitin Sulfate A (CS), or with Proteinase K (ProtK), Heparin Lyase I (HepL I) or Heparin Lyase III (HepL III). KIR2DL4-Ig was used alone (mock and enzyme treated cells) or pre-incubated with either HS or CS for blocking of KIR2DL4-Ig binding. Data represent normalized mean ± s.d. of n=3 independent experiments for (C) and (D). P-values were calculated using T-Test: * p-value and ** p-value ≤ 0.001; *** p-value and **** p-value ≤ 0.01.
A number of siRNAs were identified that significantly enhanced KIR2DL4-Ig binding to PC3 cells in the primary screen. These included a siRNA pool targeting Sulfatase 1, which selectively removes 6-O-sulfate groups from HS (SULF1; NM_015170). Sulfatase 1-targeted siRNA enhanced the binding of KIR2DL4-Ig by a score of 1.42. Accordingly, transfection of SULF1 cDNA into SKOV-3 cells specifically reduced KIR2DL4-Ig binding, as compared to LIR1 or KIR2DL1 binding (no change), and similarly impacted NKp46D2-Ig binding (Supplemental Fig. 3). Taken together, we conclude that our genome-wide siRNA screen has identified HS GAGs as potential structures recognized by KIR2DL4 as ligands or co-ligands.
Characterization of the KIR2DL4-Ig and HS interaction
To verify whether KIR2DL4 can interact with membrane-associated HS on tumor cell lines, we pre-incubated KIR2DL4-Ig with HS or another GAG, chondroitin sulfate A (CS) and used the mixtures to stain PC3 and HeLa cells. Indeed, pre-incubation of KIR2DL4-Ig with HS, but not with CS, significantly suppressed cell surface staining (Fig. 2D). We next manipulated membrane-associated HS on the tumor cells by pretreating them with heparin lyase I or heparin lyase III (to cleave HS) or by pretreating with proteinase K (to cleave HS Proteoglycans (HSPGs)) prior to staining with KIR2DL4-Ig. As shown in Figure 2D, pretreatment of both PC3 and HeLa cell lines each of these enzymes nearly abolished surface staining by KIR2DL4-Ig.
Next we examined the residues in KIR2DL4 that may be involved in HS binding. Since HS is a negatively charged macromolecule that can interact with basic residues in proteins, we postulated that a region with a high positive surface potential could be a candidate site for HS binding on KIR2DL4. The structure of the KIR2DL4 extracellular domains was modeled by comparison with the 3D crystal structure of NKp46, and a Coulombic surface was then determined for the KIR2DL4 model (Figure 3A (upper left panel)). The map revealed a widespread and pronounced basic patch (blue) within the membrane distal D0 Ig-like domain, that we postulated as the binding site for HS on the surface of KIR2DL4. In contrast, strong and broad basic patches were not observed on the extracellular domains of KIR2DL1 or LIR1 (Fig. 3A, lower panels), which do not interact with HS (44, 46). To test our predictions, we produced soluble forms of the individual Ig-like domains of KIR2DL4 (see Methods), designated KIR2DL4-D0-Ig and KIR2DL4-D2-Ig, and tested their interactions with HS as compared to full length KIR2DL4-Ig using a BIAcore device (Figure 3B-D). As predicted from the modeling analysis, KIR2DL4-Ig and KIR2DL4-D0-Ig displayed characteristic high affinity binding kinetics to HS, but no significant interaction with HS could be detected for KIR2DL4-D2-Ig. The equilibrium dissociation constant (KD) values for KIR2DL4-Ig and KIR2DL4-D0-Ig were nearly identical (24.5nM, χ2 = 0.094 and 27.6nM, χ2 = 0.838, respectively).
We next assessed the binding of the recombinant full-length and individual domain KIR2DL4 fusion proteins to CHO-K1 cells and a mutant variant of this cell line, called CHO-677, which is deficient in HS biosynthesis, but produces high levels of CS (26). Both KIR2DL4-Ig and KIR2DL4-D0-Ig robustly stained parental CHO-K1 cells, as compared to residual staining of CHO-677 cells (Fig. 3E-F). In contrast, KIR2DL4-D2-Ig manifested only residual staining, which was similar for both cell lines (Fig. 3G). Furthermore, we compared staining of CHO-K1 and CHO-677 with the KIR2DL4-Ig, KIR2DL4-D2-Ig and KIR2DL4-D0-Ig, which had been pre-incubated with soluble HS or with various anti-KIR2DL4 mAbs. Soluble HS significantly suppressed the binding of KIR2DL4-Ig and KIR2DL4-D0-Ig to CHO-K1 cells, but did not affect their residual binding to CHO-677 cells (Fig. 3H). This was in contrast to an inability of soluble HS to impact KIR2DL4-D2-Ig binding to either cell line (Fig. 3H).
We further assessed specific antibody binding sites on full KIR2DL4 and the individual Ig-like domains (data not shown). The clone 33 mAb recognized only the full KIR2DL4-Ig protein but not the single domains, suggesting that the binding epitope encompasses both domains. In contrast, mAb 53.1 recognized recombinant full KIR2DL4-Ig and KIR2DL4-D2-Ig but not the KIR2DL4-D0-Ig. Blocking of KIR2DL4 fusion proteins with clone 33 mAb did not affect the binding to CHO-K1 or CHO-677 (Fig. 3H). In contrast, blocking of KIR2DL4 fusion proteins with clone 53.1 mAb effectively prevented the binding of KIR2DL4-Ig to CHO-K1, although it did not affect the binding of the KIR2DL4-D0-Ig (Fig. 3H). This finding could be explained by an allosteric interference of the HS binding epitope on D0 upon binding of 53.1 mAb to D2. In this scenario, interference with the HS interaction is only possible upon interaction with full KIR2DL4, but it is not detectable when the domains are tested separately.
Soluble HS increases IFN-γ production by NK cells stimulated with anti-KIR2DL4 mAbs
We and others have shown that the NCRs (NKp30, NKp44, and NKp46) can also directly bind HSPG associated and soluble HS and that Ab-mediated engagement of NCRs in the presence of soluble HS can potentiate IFN-γ production by NK cells (43-49). Since HS can also interact with KIR2DL4, we hypothesized that KIR2DL4-mediated NK cell responses could be similarly influenced by soluble HS.
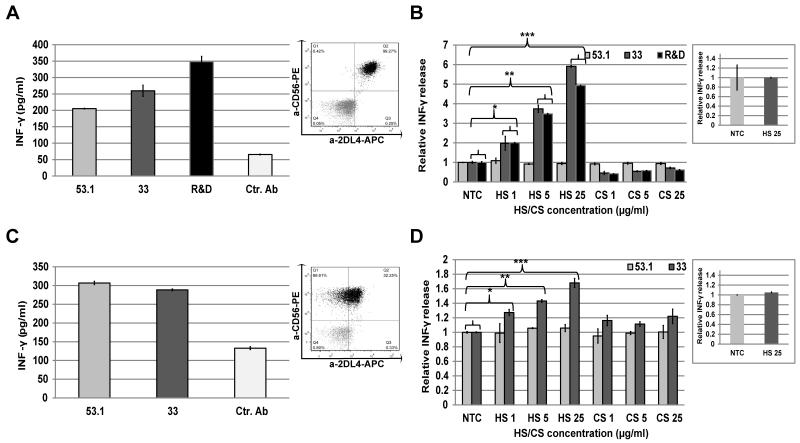
We first tested IFN-γ secretion from the KHYG1-2DL4 cell line and from primary human NK cells obtained from a 10A allele donor, capable of expressing KIR2DL4 on the NK cell surface (4, 8, 9). KHYG1-KIR2DL4 and primary NK cells cultured in IL-2-containing medium demonstrated high surface KIR2DL4 expression (Fig. 4A and 4C; insets). Following incubation with increasing concentrations of either HS or CS, these NK cells were plated in wells that were pre-coated with anti-KIR2DL4 mAbs and IFN-γ secretion was assayed by standard ELISA 16 hours later. Several plate-bound anti-KIR2DL4 mAbs elicited IFN-γ secretion from both the KHYG1-2DL4 cells (mAbs 53.1, 33 and 181703) and primary human NK cells (mAbs 53.1 and 33) (Fig. 4A and 4C). IFN-γ production was significantly enhanced in KHYG1-KIR2DL4 cells supplemented with HS when KIR2DL4 was engaged with anti-KIR2DL4 mAbs 33 and 181703 (Fig. 4B). Similarly, HS enhanced IFN-γ secretion by primary NK cells engaged with anti-KIR2DL4 33 mAb (Fig. 4D). The potentiation induced by HS was concentration-dependent in both NK cell systems (Fig. 4B and 4D). Importantly, HS did not potentiate IFN-γ secretion in response to 53.1 mAb, which competes with HS binding to KIR2DL4 (Fig. 4B and 4D). To test the specificity of the stimulatory impact of HS on IFN-γ secretion by NK cells, we employed CS, which did not affect mAb-induced secretion of IFN-γ (Fig. 4B and 4D). The HS-induced enhancement of IFN-γ secretion required non-competitive KIR2DL4 engagement, since the presence of HS alone along with control anti 2B4 mAb did not alter basal IFN-γ secretion by NK cells (Fig. 4B and 4D, insets).
Figure 4. Impact of HS/heparin on KIR2DL4-mediated IFN-γ responses by NK cells.
A, C. IFN-γ response by KHYG-1 overexpressing KIR2DL4 (A) or primary NK cells from a 10A donor (C) that were stimulated overnight with plate-bound anti-KIR2DL4 mAb (clones 53.1, 33 or R&D 181703) or control mAb (2B4; Ctr.). IFN-γ in culture supernatant was assayed by ELISA. Insert dot-plot panels show FACS analysis of KIR2DL4 expression performed by combined staining with APC-conjugated anti-KIR2DL4 mAb (clone 33) and PE-conjugated anti-CD56a mAb (clone HCD56). B, D. KHYG-1 or Primary NK cell activation by plate-bound mAbs in the presence of various concentrations of soluble HS or CS (1, 5 and 25 μg/ml accordingly) as compared to PBS alone (NTC). NK cells were KHYG-1 (B) or Primary NK (pNK) (D); Insert panels show ELISA analysis of NK cell activation by plate-bound control mAb (2B4) in the presence of maximal concentration of soluble HS or PBS (NTC); IFN-γ in culture supernatant was assayed by ELISA. Data represent mean ((A) and (C)) ± s.d. and normalized to NTC mean ((B),(D) and inserts) ± s.d of n=3 independent experiments. P-values were calculated using T-Test (B): *p-value ≤ 0.03; **p-value and ***p-value ≤ 0.001; (D): *p-value ≤ 0.1; **p-value and ***p-value ≤ 0.008; P-values of NTC vs. CS are > 0.5 for both KHYG and pNK (all concentrations).
Impact of HS on KIR2DL4 localization and function in the HEK293T model
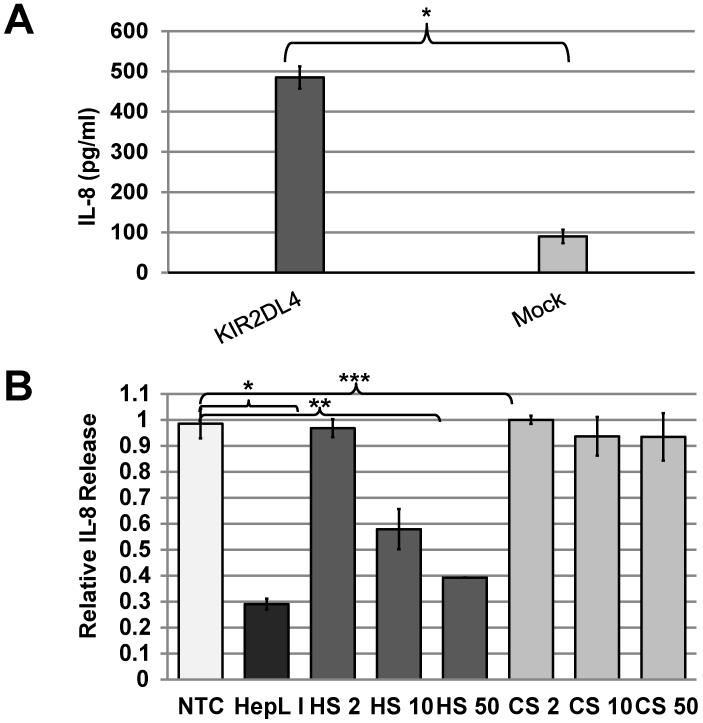
The immune receptors 2B4, TLR4, and KIR2DL4 can stimulate production of IL-8 when expressed in HEK293T cells. While IL-8 secretion in HEK293T cells requires antibody engagement of 2B4 or TLR4, KIR2DL4 expression alone causes spontaneous IL-8 production in HEK293T (14, 50, 51). HEK293T cells naturally lack detectable expression of ligands for 2B4 and TLR4. While HEK293T also do not express HLA-G, KIR2DL4-Ig stained these cells (Supplemental Fig. 1), suggesting that HEK293T cells express an endogenous non HLA-G ligand(s) for KIR2DL4.
To test this hypothesis, we have transfected HEK293T cells with full-length KIR2DL4 (10A allele) or with empty vector (HEK293T-2DL4 and mock, respectively). Only HEK293T-2DL4 cells secreted high levels of IL-8 (Fig. 5A). This IL-8 secretion was specifically reduced in a concentration-dependent manner upon addition of soluble HS, but not soluble CS (Fig. 5B). These results demonstrate that HS can directly modulate the biological function of KIR2DL4 in the HEK293T-2DL4 model.
Figure 5. Impact of HS/heparin on KIR2DL4-mediated IL-8 production by HEK293T cells.
(A) The expression of KIR2DL4 induces spontaneous IL-8 secretion in HEK293T cells, and (B) IL-8 secretion is inhibited by addition of HS in a concentration-dependent manner (2-50μg), but not by CS (2-50μg) or PBS alone (NTC). . Data represent mean (A) ± s.d. and normalized to NTC mean (B) ± s.d of two out of n=5 independent experiments. P-values were calculated using T-Test: (A) *p-value = 0.002; (B) *p-value ≤ 0.01; **p-value and ***p-value ≤ 0.002.
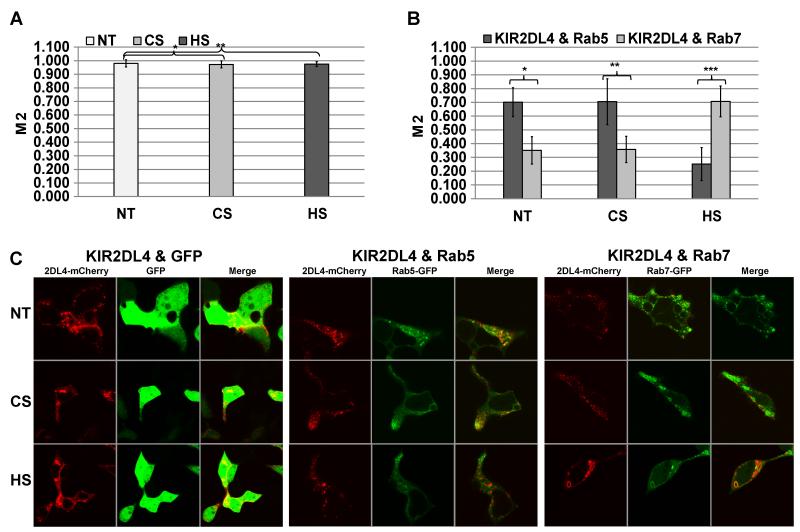
It was previously reported that KIR2DL4 signaling requires internalization of KIR2DL4 to early endosomes (rab5+) but not internalization to late endosomes (rab7+) (14, 52, 53). Based on the negative modulatory effect of HS on IL-8 production in HEK293T-2DL4 cells (Fig. 5), we postulated that exogenous HS may interact with KIR2DL4 and prevent engagement with an HSPG-associated HS, thereby redirecting the internalized receptor to late endosomal (early lysosome) compartment, resulting in decreased IL-8 production. To test this hypothesis, we co-expressed KIR2DL4-mCherry with either rab5-GFP, rab7-GFP or GFP only (as a control) in HEK293T cells. When KIR2DL4-mCherry and GFP were expressed (Fig. 6A), no difference in co-localization was observed following treatment of the cells with HS or CS as compared to no-treatment (NT) control. In contrast, co-localization of KIR2DL4-mCherry with either rab5-GFP or rab7-GFP was significantly affected by HS treatment as compared to CS or NT. Figure 6B summarizes the differential distribution of internalized KIR2DL4-mCherry between rab5+ and rab7+ endosomes following HS or CS treatment. We conclude that HS treatment significantly reduces KIR2DL4-mCherry co-localization with the rab5+ early endosome compartment, while enhancing KIR2DL4-mCherry localization with the rab7+ late endosomal compartment. Figure 6C shows representative images for NT, CS and HS treated HEK293T cells co-expressing KIR2DL4-mCherry and either rab5-GFP, rab7-GFP or GFP alone. The significant impact of soluble HS on shifting rab5 vs. rab7 association of KIR2DL4 indicates that internalized KIR2DL4 is re-directed towards degradation in the late endosome compartment rather than toward signaling in the early endosomes.
Figure 6. HS/heparin treatment promotes KIR2DL4 localization with Rab7+ lysosomes in HEK293T cells.
(A) HEK293T cells expressing KIR2DL4-mCherry and GFP alone (control) were complemented with standard culture medium (NT) or with culture medium containing 1μg/ml of either HS or CS. (B) HEK293T cells expressing KIR2DL4-mCherry and Rab5-GFP or Rab7-GFP (experiment) were complemented with standard culture medium (non-treated; NT) or with culture medium containing 1μg/ml of either HS or CS. M2 coefficient corresponds to the proportion of KIR2DL4-mCherry co-localized with the appropriate GFP-labeled marker. Data represent mean ± s.d. of indicated number of biological replicas (N) for n=4 independent experiments. P-values were calculated using T-test. T-test (control): N=56; *p-value and **p-value ≥ 0.7 (no difference); T-test (experiment): N=112; *p-value, **p-value and ***p-value, ≤ 0.005. (C) Representative images of non-treated (NT) and HS- or CS-treated cells are shown.
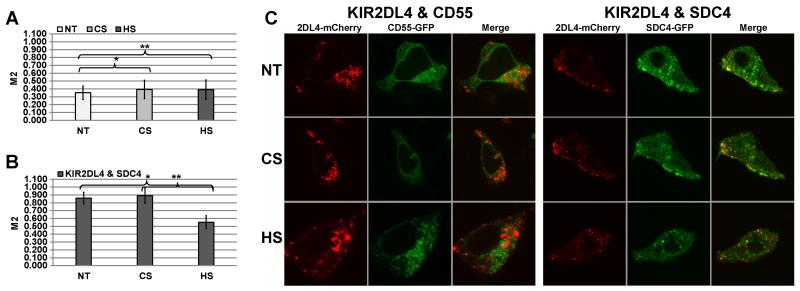
HS can be conjugated to a variety of HSPG, including the most prevalent HSPG in cells - the syndecan family of cell surface proteoglycans (54, 55). Syndecans 1, 2 and 4 are widely expressed in epithelial or fibroblastic cells, while syndecan 3 is mainly restricted to neuronal cells (56, 57). We assessed the syndecan expression profiles by flow cytometry in high KIR2DL4-ligand(s) expressing cell lines (see Supplemental Fig.1) and NK cells (Supplemental Fig. 4). Syndecan 4 (SDC4) exhibited a high expression profile in all cell lines tested. Therefore, we hypothesized that the primary HSPG in HEK293T cells is HS associated with SDC4, and therefore, SDC4 could play an important role in KIR2DL4 localization and function in the HEK293T-2DL4 model. To test this hypothesis we co-expressed KIR2DL4-mCherry with either SDC4-GFP or another heavily glycosylated proteoglycan CD55 (CD55-GFP; reference control) in HEK293T cells. No significant co-localization was observed between KIR2DL4 and CD55, and no impact on localization was detected after either CS or HS treatments (Fig. 7A). In contrast, we observed high co-localization of KIR2DL4 with SCD4, and exogenous HS but not CS treatment significantly reduced this co-localization (Fig. 7B). Figure 7C shows representative images for non-treated, CS and HS treated HEK293T cells co-expressing KIR2DL4-mCherry and either SDC4-GFP or CD55-GFP. We conclude that KIR2DL4 interacts with SDC4 to stimulate IL-8 production in 293T cells and this interaction is HS-dependent (Fig. 3).
Figure 7. KIR2DL4 associates with the heparan sulfate proteoglycan, SDC4 in HEK293T cells.
(A) HEK293T cells expressing KIR2DL4-mCherry and CD55-GFP (control) were complemented with standard culture medium (NT) or with culture medium containing 1μg/ml of either HS or CS. (B) HEK293T cells expressing KIR2DL4-mCherry and SDC4-GFP (experiment) were complemented with standard culture medium (non-treated; NT) or with culture medium containing 1μg/ml of either HS or CS. M2 coefficient corresponds to the proportion of KIR2DL4-mCherry co-localized with the appropriate GFP-labeled marker. Data represent mean ± s.d. of indicated number of biological replicas (N) for n=3 independent experiments. P-values were calculated using T-test. T-test (control): N=37; *p-value and **p-value ≥ 0.9 (no difference); T-test (experiment): N=55; *p-value and **p-value ≤ 0.005. (C) Representative images of non-treated (NT) and HS or CS treated cells are shown.
Discussion
Soluble HLA-G was reported to serve as a ligand for KIR2DL4. Although HLA-G is normally expressed only by fetal-derived trophoblast cells, KIR2DL4 expression is not restricted to the placenta. Indeed, we have observed that several tumor and normal primary cell lines of epithelial and fibroblast origin, which lack expression of HLA-G, are recognized by recombinant KIR2DL4, suggesting that these cells endogenously express alternative KIR2DL4 ligand(s) (Supplemental Figs. 1 and 2).
We performed a whole genome siRNA screen for KIR2DL4 ligand(s) and identified the HS3ST3B1 gene encoding heparan sulfate glucosamine 3-O-sulfotransferase 3B1 as the best scoring hit (Figs. 1, 2). The result is intriguing, since we and others previously published that other NK receptors, namely the NCRs, directly bind HSPG-associated and soluble HS (43-49). Moreover, it was recently shown that heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4 (HS3ST4 gene; Mus musculus) can regulate NK cell functionality (58). Here we showed that HS3ST3B1 (Homo sapiens) is directly involved in NK cell receptors recognition of the target cells via KIR2DL4 (Figs. 2, 3). Following in silico prediction of HS binding site on D0 of KIR2DL4, we showed that HS binding is indeed restricted to the membrane distal D0 domain (Fig. 3) and that KIR2DL4-Ig displayed a characteristic in-vitro binding affinity to HS (KD = 25nM) similar to those of NKp44 and NKp46 (43, 45, 46).
The observation that the three NCRs and KIR2DL4 all recognize HS does not imply that these molecules recognize the same epitope on this complex oligosaccharide. Since the assembly of HS involves up to 23 distinct disaccharides, it has been described as the ‘most information dense biopolymer in nature’ (59, 60). Indeed, we showed that the three NCRs recognize different HS epitopes (43, 44, 47, 48). Nonetheless, the presence of sulfation in HS is essential for the interaction with both NCRs and KIR2DL4 (Figs. 1 and 2), emphasizing the significance of HS3ST3B1 function to the structure of the HSPG-associated HS recognized by all NK cell activating receptors (Fig. 2C for both KIR2DL4 and NKp46). It is important to emphasize that another siRNA targeting the HS 6-O-endo-sulfatase, SULF1, caused increased binding of KIR2DL4-Ig. SULF1 selectively removes 6-O-sulfate groups from HS, further emphasizing the role of HS sulfation in KIR2DL4 recognition of its ligand(s). Accordingly, reduction in staining by KIR2DL4-Ig and NKp46-Ig was further demonstrated in cells transfected with SULF1 cDNA (Supplemental Fig. 3).
Our data also indicate that KIR2DL4 function may be modulated through interactions with HS either on target cells (trans) or on NK cells themselves (cis). HS GAGs are very long, unbranched, anionic polysaccharides, and therefore, the KIR2DL4-HS interaction can most likely take place in the same orientation in either the cis or trans context. To test this we employed HEK293T cells that spontaneously secrete IL-8 following KIR2DL4 expression (14, 51), which was previously suggested to be ligand independent (14, 50, 52). We speculated that the IL-8 secretion resulted from engagement of KIR2DL4 with a non-HLA-G alternative ligand. Indeed, recombinant KIR2DL4-Ig bound to HLA-G-deficient HEK293T cells (Supplemental Fig. 1). In accordance, manipulation of the availability of membrane-associated HS on HEK293T-2DL4 cells significantly reduced IL-8 secretion (Fig. 5B).
We previously demonstrated that HSPG-associated HS is recognized by all NCRs (43-48), presumably by interacting with HS presented in trans on the surface of target cells. Here we showed that PC3 cells expressing HSPG-associated HS can stimulate KIR2DL4+ NK cells as well (Supplemental Fig. 2C), which would be a trans interaction. Additionally, however, our data suggest that KIR2DL4 may also interact with HSPG in cis on the surface of the effector cell to influence receptor function, since HS (soluble or HSPG-associated) directly regulated the localization and function of KIR2DL4 in the HEK293T-2DL4 model (Figs. 5-7). Similarly, soluble HS regulated KIR2DL4 function in NK cells (Fig. 4). It should also be considered, that the high affinity interaction of KIR2DL4 with HS (KD = 25mM) may in fact be adequate to physically engage signaling in an immune synapse upon interaction with HSPG on target cells in trans after exchange from a similar cis interaction with HSPG on the NK cell surface. Therefore we suggest that HS binding function can both directly engage KIR2DL4 to initiate signal transduction and act as an allosteric regulator to modulate the capacity of KIR2DL4 to interact with other ligands in trans. Furthermore, the common use of heparin as a therapeutic agent in patients may in fact be significantly altering the function of the subset of CD56bright NK cells that express KIR2DL4.
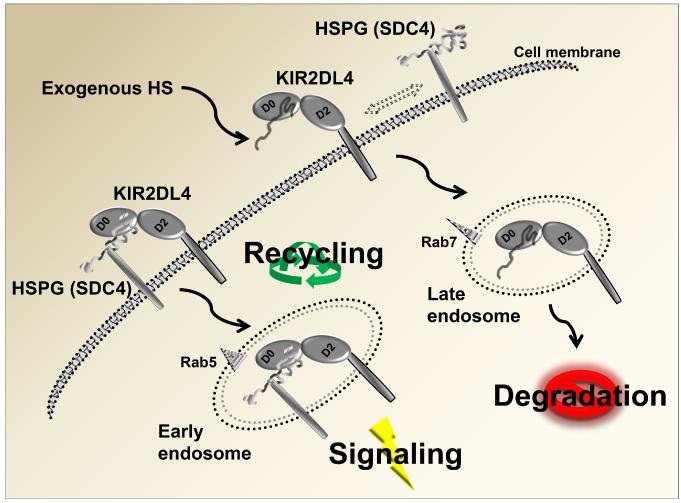
Figure 8 depicts our working model for KIR2DL4 modulation by HSPG in the HEK293T-2DL4 model and could also be applicable in NK cells: HSPG (SDC4)-associated HS binds to membrane-associated KIR2DL4 and directs its endocytosis and trafficking toward the rab5+ early endosomes to mediate signaling and re-cycling. This result is in accordance with data published by Rajagopalan et al. and endorsed in our previous studies (14, 52, 61). We suggest that exogenous HS induces KIR2DL4 to dissociate from HSPGs and traffic to rab7+ late endosomes, where it is destined for degradation (Fig. 8). Furthermore, the intimate co-localization of SDC4 HSPG and KIR2DL4 is directly affected by addition of exogenous HS, thus modulating receptor endocytosis and membrane trafficking (Fig. 7).
Figure 8. Model of KIR2DL4 functional modulation by HS/HSPG.
Interaction of KIR2DL4 with the HSPG syndecan 4 (SDC4) on the cell surface triggers endocytosis to the Rab5+ early endosome compartment to induce spontaneous activation signaling in HEK293T cells. We predict that KIR2DL4 can be recycled back to the cell surface from early endosomes. If cells are exposed to exogenous HS, this displaces SDC4 from interacting and KIR2DL4 is instead endocytosed to Rab7+ late endosomes for subsequent lysosomal degradation.
Most of the cellular HS is derived from the syndecan HSPGs and syndecan 4 (SDC4) in particular (55). SDC4 is widely expressed, in contrast to the other syndecans, which exhibit rather tissue-specific distributions (56). Indeed, we found high expression of the SDC4 in all cell lines tested, including NK cells (Supplemental Fig. 4). It has been suggested that the vesicular co-localization of different SDC4-binding proteins is an indication that SDC4 itself could be involved in endocytosis and membrane trafficking (62, 63), wich is consistent with our results (Figs. 6, 7).
SDC4 can oligomerize (64) and may provide a mechanical link between extracellular ligands (i.e. NK receptors, interacting with HSPG) and the actin cytoskeleton (63, 65, 66) and thus stabilize the formation of the receptor-ligand complex as it was previously reported for FGFR (62, 67, 68). Therefore, the primary impact of the KIR2DL4-HS interaction could be the regulation of the receptor through cis interaction with NK-expressed HSPG rather than a trans interaction with target cell-expressed HSPG. This hypothesis is in accordance with the observation that exogenous HS potentiates IFN-γ secretion in NK cells stimulated with anti-KIR2DL4 mAbs 33 and 181703, but not 53.1, however, presumably because of an allosteric interference with the binding between KIR2DL4 and HS or steric effect (possibly due to the size of IgG), suggesting that the HS-induced potentiation of IFN-γ secretion requires non-competitive KIR2DL4 engagement. We theorize that exogenous HS might have blocked a cis interaction between KIR2DL4 and HSPG, and therefore released the receptor, making it available for more efficient engagement by specific mAbs. This is in accordance with our previous reports of similarly potentiated IFN-γ by NCRs (45-49). Therefore, we suggest that the same mechanism could be applied also for NCRs, i.e. the main function of the NCR-HS interaction is to regulate NCR function through the NK-expressed HSPG and not through the target cell-expressed HSPG. Whether SDC4 or other HSPGs expressed on NK cells are also involved in the regulation of NCR function remains to be further elucidated.
To summarize, our results demonstrate that KIR2DL4 receptor can interact with HS/heparin and HSPGs, and these interactions can modulate function of the receptor to impact NK cell activation. Our discovery of HS/HSPGs as non-HLA-G ligands or co-ligands that can interact with KIR2DL4 provides novel insight that significantly enhances our understanding of how this unique receptor contributes to NK cell responses in humans.
Supplementary Material
Acknowledgements
The authors wish to thank Cell Culture, High Throughput Screening, Biostatistics and Bioinformatics, Molecular Modeling and Flow Cytometry facilities of The Research Institute of Fox Chase Cancer Center.
Special thanks to S M Shahjahan Miah, Alexander W MacFarlane IV, Jennifer Oshinsky and Tatiana Rabinsky, who provided valuable assistance to the undertaking of the research summarized here.
Funding
The present work was supported by the by the United States–Israel Binational Science Foundation Grant 2007282 (AP and KSC), Israel Science Foundation (AP), Israeli Ministry of Science and Technology/German Cancer Research Center program (AP), NIH CA100226 (KSC), CA083859 (KSC), and CA06927 (FCCC).
References
- 1.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunology today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 2.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. The Journal of experimental medicine. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–280. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 4.Goodridge JP, Witt CS, Christiansen FT, Warren HS. KIR2DL4 (CD158d) genotype influences expression and function in NK cells. Journal of immunology. 2003;171:1768–1774. doi: 10.4049/jimmunol.171.4.1768. [DOI] [PubMed] [Google Scholar]
- 5.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. Journal of immunology. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 9.Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, Hurley CK, Christiansen FT, Witt CS. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. European journal of immunology. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 10.Yusa S, Catina TL, Campbell KS. KIR2DL5 can inhibit human NK cell activation via recruitment of Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) Journal of immunology. 2004;172:7385–7392. doi: 10.4049/jimmunol.172.12.7385. [DOI] [PubMed] [Google Scholar]
- 11.Vilches C, Rajalingam R, Uhrberg M, Gardiner CM, Young NT, Parham P. KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. Journal of immunology. 2000;164:5797–5804. doi: 10.4049/jimmunol.164.11.5797. [DOI] [PubMed] [Google Scholar]
- 12.Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J. Immunol. 2002;168:5047–5057. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. Journal of immunology. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 14.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS biology. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan S, Fu J, Long EO. Cutting Edge: Induction of IFN- γ production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J. Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 16.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J. Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 17.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2 regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFNγ production. J. Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 18.Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, Bertone S, Moretta A, Moretta L, Mingari MC. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl. Acad. Sci. USA. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 20.Amiot L, Onno M, Renard I, Drenou B, Guillaudeux T, Le Bouteiller P, Fauchet R. HLA-G transcription studies during the different stages of normal and malignant hematopoiesis. Tissue antigens. 1996;48:609–614. doi: 10.1111/j.1399-0039.1996.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 21.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, Avril MF, Dausset J, Guillet JG, Carosella ED. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4510–4515. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyson JE, Erskine R, Whitman MC, Chiu M, Lau JM, Koopman LA, Valter MM, Angelisova P, Horejsi V, Strominger JL. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16180–16185. doi: 10.1073/pnas.212643199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Lozano N, de Pablo R, Puente S, Vilches C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. European journal of immunology. 2003;33:639–644. doi: 10.1002/eji.200323741. [DOI] [PubMed] [Google Scholar]
- 24.Witt CS, Goodridge J, Gerbase-Delima MG, Daher S, Christiansen FT. Maternal KIR repertoire is not associated with recurrent spontaneous abortion. Human reproduction. 2004;19:2653–2657. doi: 10.1093/humrep/deh483. [DOI] [PubMed] [Google Scholar]
- 25.Witt CS, Whiteway JM, Warren HS, Barden A, Rogers M, Martin A, Beilin L, Christiansen FT. Alleles of the KIR2DL4 receptor and their lack of association with pre-eclampsia. European journal of immunology. 2002;32:18–29. doi: 10.1002/1521-4141(200201)32:1<18::AID-IMMU18>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Lidholt K, Weinke JL, Kiser CS, Lugemwa FN, Bame KJ, Cheifetz S, Massague J, Lindahl U, Esko JD. A single mutation affects both N-acetylglucosaminyltransferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:2267–2271. doi: 10.1073/pnas.89.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer biology & therapy. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zilka A, Mendelson M, Rosental B, Hershkovitz O, Porgador A. Generating NK cell receptor-Fc chimera proteins from 293T cells and considerations of appropriate glycosylation. Methods in molecular biology. 2010;612:275–283. doi: 10.1007/978-1-60761-362-6_18. [DOI] [PubMed] [Google Scholar]
- 29.Manders EM, Hoebe R, Strackee J, Vossepoel AM, Aten JA. Largest contour segmentation: a tool for the localization of spots in confocal images. Cytometry. 1996;23:15–21. doi: 10.1002/(SICI)1097-0320(19960101)23:1<15::AID-CYTO3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 30.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. Journal of microscopy. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 31.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. Journal of cell science. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 32.Hoebe RA, van Noorden CJ, Manders EM. Noise effects and filtering in controlled light exposure microscopy. Journal of microscopy. 2010;240:197–206. doi: 10.1111/j.1365-2818.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Canutescu AA, Dunbrack RL., Jr. SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nature protocols. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krivov GG, Shapovalov MV, Dunbrack RL., Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Mochizuki H, Yoshida K, Shibata Y, Kimata K. Tetrasulfated disaccharide unit in heparan sulfate: enzymatic formation and tissue distribution. The Journal of biological chemistry. 2008;283:31237–31245. doi: 10.1074/jbc.M801586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mochizuki H, Yoshida K, Gotoh M, Sugioka S, Kikuchi N, Kwon YD, Tawada A, Maeyama K, Inaba N, Hiruma T, Kimata K, Narimatsu H. Characterization of a heparan sulfate 3-O-sulfotransferase-5, an enzyme synthesizing a tetrasulfated disaccharide. The Journal of biological chemistry. 2003;278:26780–26787. doi: 10.1074/jbc.M301861200. [DOI] [PubMed] [Google Scholar]
- 38.Nakato H, Kimata K. Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim Biophys Acta. 2002;1573:312–318. doi: 10.1016/s0304-4165(02)00398-7. [DOI] [PubMed] [Google Scholar]
- 39.Loo BM, Salmivirta M. Heparin/Heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. The Journal of biological chemistry. 2002;277:32616–32623. doi: 10.1074/jbc.M204961200. [DOI] [PubMed] [Google Scholar]
- 40.Baeg GH, Lin X, Khare N, Baumgartner S, Perrimon N. Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development. 2001;128:87–94. doi: 10.1242/dev.128.1.87. [DOI] [PubMed] [Google Scholar]
- 41.Lortat-Jacob H. The molecular basis and functional implications of chemokine interactions with heparan sulphate. Curr Opin Struct Biol. 2009;19:543–548. doi: 10.1016/j.sbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Laguri C, Arenzana-Seisdedos F, Lortat-Jacob H. Relationships between glycosaminoglycan and receptor binding sites in chemokines-the CXCL12 example. Carbohydr Res. 2008;343:2018–2023. doi: 10.1016/j.carres.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 43.Hecht ML, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, Schauer S, Porgador A, Seeberger PH. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. Journal of proteome research. 2009;8:712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 44.Bloushtain N, Qimron U, Bar-Ilan A, Hershkovitz O, Gazit R, Fima E, Korc M, Vlodavsky I, Bovin NV, Porgador A. Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. Journal of immunology. 2004;173:2392–2401. doi: 10.4049/jimmunol.173.4.2392. [DOI] [PubMed] [Google Scholar]
- 45.Zilka A, Landau G, Hershkovitz O, Bloushtain N, Bar-Ilan A, Benchetrit F, Fima E, van Kuppevelt TH, Gallagher JT, Elgavish S, Porgador A. Characterization of the heparin/heparan sulfate binding site of the natural cytotoxicity receptor NKp46. Biochemistry. 2005;44:14477–14485. doi: 10.1021/bi051241s. [DOI] [PubMed] [Google Scholar]
- 46.Hershkovitz O, Jivov S, Bloushtain N, Zilka A, Landau G, Bar-Ilan A, Lichtenstein RG, Campbell KS, van Kuppevelt TH, Porgador A. Characterization of the recognition of tumor cells by the natural cytotoxicity receptor, NKp44. Biochemistry. 2007;46:7426–7436. doi: 10.1021/bi7000455. [DOI] [PubMed] [Google Scholar]
- 47.Ito K, Higai K, Shinoda C, Sakurai M, Yanai K, Azuma Y, Matsumoto K. Unlike natural killer (NK) p30, natural cytotoxicity receptor NKp44 binds to multimeric alpha2,3-NeuNAc-containing N-glycans. Biological & pharmaceutical bulletin. 2012;35:594–600. doi: 10.1248/bpb.35.594. [DOI] [PubMed] [Google Scholar]
- 48.Ito K, Higai K, Sakurai M, Shinoda C, Yanai K, Azuma Y, Matsumoto K. Binding of natural cytotoxicity receptor NKp46 to sulfate- and alpha2,3-NeuAc-containing glycans and its mutagenesis. Biochemical and biophysical research communications. 2011;406:377–382. doi: 10.1016/j.bbrc.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 49.Jarahian M, Watzl C, Issa Y, Altevogt P, Momburg F. Blockade of natural killer cell-mediated lysis by NCAM140 expressed on tumor cells. International journal of cancer. Journal international du cancer. 2007;120:2625–2634. doi: 10.1002/ijc.22579. [DOI] [PubMed] [Google Scholar]
- 50.Yang H, Young DW, Gusovsky F, Chow JC. Cellular events mediated by lipopolysaccharide-stimulated toll-like receptor 4. MD-2 is required for activation of mitogen-activated protein kinases and Elk-1. The Journal of biological chemistry. 2000;275:20861–20866. doi: 10.1074/jbc.M002896200. [DOI] [PubMed] [Google Scholar]
- 51.Miah SM, Hughes TL, Campbell KS. KIR2DL4 differentially signals downstream functions in human NK cells through distinct structural modules. Journal of immunology. 2008;180:2922–2932. doi: 10.4049/jimmunol.180.5.2922. [DOI] [PubMed] [Google Scholar]
- 52.Rajagopalan S. Endosomal signaling and a novel pathway defined by the natural killer receptor KIR2DL4 (CD158d) Traffic. 2010;11:1381–1390. doi: 10.1111/j.1600-0854.2010.01112.x. [DOI] [PubMed] [Google Scholar]
- 53.Rajagopalan S, Moyle MW, Joosten I, Long EO. DNA-PKcs controls an endosomal signaling pathway for a proinflammatory response by natural killer cells. Science signaling. 2010;3:ra14. doi: 10.1126/scisignal.2000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dreyfuss JL, Regatieri CV, Jarrouge TR, Cavalheiro RP, Sampaio LO, Nader HB. Heparan sulfate proteoglycans: structure, protein interactions and cell signaling. Anais da Academia Brasileira de Ciencias. 2009;81:409–429. doi: 10.1590/s0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- 55.Gallo RL, Ono M, Povsic T, Page C, Eriksson E, Klagsbrun M, Bernfield M. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11035–11039. doi: 10.1073/pnas.91.23.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim CW, Goldberger OA, Gallo RL, Bernfield M. Members of the syndecan family of heparan sulfate proteoglycans are expressed in distinct cell-, tissue-, and development-specific patterns. Molecular biology of the cell. 1994;5:797–805. doi: 10.1091/mbc.5.7.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods A, Couchman JR. Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Molecular biology of the cell. 1994;5:183–192. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, Simonet T, Horard B, Jamet K, Cervera L, Mendez-Bermudez A, Poncet D, Grataroli R, de Rodenbeeke CT, Salvati E, Rizzo A, Zizza P, Ricoul M, Cognet C, Kuilman T, Duret H, Lepinasse F, Marvel J, Verhoeyen E, Cosset FL, Peeper D, Smyth MJ, Londono-Vallejo A, Sabatier L, Picco V, Pages G, Scoazec JY, Stoppacciaro A, Leonetti C, Vivier E, Gilson E. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nature cell biology. 2013 doi: 10.1038/ncb2774. [DOI] [PubMed] [Google Scholar]
- 59.Sasisekharan R, Venkataraman G. Heparin and heparan sulfate: biosynthesis, structure and function. Current opinion in chemical biology. 2000;4:626–631. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 60.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. The Journal of clinical investigation. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miah SM, Purdy AK, Rodin NB, MacFarlane A. W. t., Oshinsky J, Alvarez-Arias DA, Campbell KS. Ubiquitylation of an internalized killer cell Ig-like receptor by Triad3A disrupts sustained NF-kappaB signaling. Journal of immunology. 2011;186:2959–2969. doi: 10.4049/jimmunol.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elfenbein A, Lanahan A, Zhou TX, Yamasaki A, Tkachenko E, Matsuda M, Simons M. Syndecan 4 regulates FGFR1 signaling in endothelial cells by directing macropinocytosis. Science signaling. 2012;5:ra36. doi: 10.1126/scisignal.2002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bass MD, Humphries MJ. Cytoplasmic interactions of syndecan-4 orchestrate adhesion receptor and growth factor receptor signalling. The Biochemical journal. 2002;368:1–15. doi: 10.1042/BJ20021228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carey DJ. Syndecans: multifunctional cell-surface co-receptors. The Biochemical journal. 1997;327(Pt 1):1–16. doi: 10.1042/bj3270001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans. Nature reviews. Molecular cell biology. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reproductive biology and endocrinology: RB&E. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsia E, Richardson TP, Nugent MA. Nuclear localization of basic fibroblast growth factor is mediated by heparan sulfate proteoglycans through protein kinase C signaling. Journal of cellular biochemistry. 2003;88:1214–1225. doi: 10.1002/jcb.10470. [DOI] [PubMed] [Google Scholar]
- 68.Richardson TP, Trinkaus-Randall V, Nugent MA. Regulation of basic fibroblast growth factor binding and activity by cell density and heparan sulfate. The Journal of biological chemistry. 1999;274:13534–13540. doi: 10.1074/jbc.274.19.13534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.