Abstract
PALB2 (Partner And Localizer of BRCA2) binds to and colocalizes with BRCA2 in DNA repair. Germline mutations in PALB2 have been identified in approximately 1–2% of familial breast cancer and 3–4% of familial pancreatic cancer cases. The goal of this study was to evaluate the prevalence of PALB2 mutations in women with breast cancer without BRCA1/2 mutations who also had a personal or family history of pancreatic cancer. PALB2 mutation analysis was performed in 94 non-BRCA1/2 breast cancer patients with a personal or family history of pancreatic cancer. Two truncating PALB2 mutations, c.3549C>CA and c.2962C>CT, were identified resulting in a mutation prevalence of 2.1%. The proband found to carry the c.3549C>CA PALB2 mutation had a mother diagnosed with both breast and pancreatic cancer; this relative was subsequently confirmed to carry the identical mutation. The proband with the c.2962C>CT mutation had a father and paternal aunt diagnosed with pancreatic cancer; neither relative was available for testing. Two novel PALB2 missense variants were also found, one of which was deemed potentially deleterious. The prevalence rate of PALB2 mutations in a non-BRCA1/2 breast cancer population specifically selected for a family history of pancreatic cancer does not appear to be significantly increased compared to that observed in other breast cancer populations studied thus far. Further evaluation is needed to determine the prevalence of PALB2 mutations and the clinical utility of such testing in those individuals affected with both breast and pancreatic cancers.
Keywords: BRCA2, Breast cancer, PALB2, Pancreatic cancer
Introduction
Breast cancer is a common disease, affecting over one million women worldwide every year [1]. Though the vast majority of breast cancers are sporadic in nature, approximately 5–10% of breast cancers are due to autosomal dominant inheritance of a specific genetic mutation [2, 3]. BRCA1 and BRCA2 are the most widely known and best understood genes among the current panel of breast cancer susceptibility genes. However, mutations in BRCA1 and BRCA2 account for no more than half of all hereditary breast cancers [4, 5]. Thus, the need to identify the contribution of other breast cancer susceptibility genes is crucial.
PALB2 (Partner And Localizer of BRCA2) is a recently discovered, moderate-risk breast cancer susceptibility gene [6, 7]. PALB2 (FANCN) and BRCA2 (FANCD1) are Fanconi anemia (FA) genes that function in the FA-Breast Cancer (BRCA) DNA repair pathway. Biallelic mutations in PALB2 or BRCA2 result in the development of Fanconi anemia [8]. The PALB2 gene product functions as a tumor suppressor and interacts closely with both BRCA1 and BRCA2 during double-strand DNA repair. PALB2 acts as a physical link between BRCA1 and BRCA2 to form a “BRCA complex” which then binds with RAD51 to initiate homologous recombination [9–12]. It is this close functional interaction with known high-risk breast cancer genes, particularly BRCA2, that ultimately helped to identify PALB2 as a candidate breast cancer susceptibility gene [13].
Though PALB2 gene mutations are rare among the general population, studies estimate a prevalence of 1–2% in patients with familial breast cancer [6, 14]. PALB2 gene mutation carriers appear to have a two- to four-fold relative risk of developing breast cancer; however, these risk estimates may be even higher in those patients with a family history of breast cancer [6, 15]. There is additional data to suggest that some PALB2-associated breast cancers display a more aggressive tumor phenotype, including triple-negative disease, higher tumor grade and higher Ki67 expression [16]. In fact, nearly 40% of the PALB2-associated breast cancers identified to date display a triple-negative phenotype, regardless of the specific PALB2 mutation [12].
In addition, PALB2 mutations have recently been shown to be associated with an increased risk of familial pancreatic cancer. Tischkowitz et al. [17] screened a cohort of 254 pancreatic cancer patients unselected for family history and found a PALB2 mutation prevalence of <1%. However, Jones et al. [18] sequenced PALB2 in 96 patients with familial pancreatic cancer and found an overall prevalence of 3.1%. Slater et al. [19] found a similar prevalence of PALB2 mutations (3.7%) in a panel of 81 European patients with familial pancreatic cancer. Thus, PALB2 gene mutations appear to be more prevalent in those patients with a family history of pancreatic cancer.
Taken together, these studies raise the question as to whether PALB2 gene mutations are more prevalent in families with both breast and pancreatic cancers. We hypothesized that a higher prevalence of PALB2 mutations might exist in this population given: (1) the close functional relationship that PALB2 shares with BRCA2; (2) the known association of PALB2 mutations with both breast and pancreatic cancers; and (3) the well-recognized BRCA2 phenotype of familial breast and pancreatic cancers [20, 21]. Thus, the goal of this study was to determine the prevalence of PALB2 mutations in a population of BRCA1/2-negative breast cancer patients specifically selected for a personal and/or family history of pancreatic cancer.
Methods
Subject selection
Eligible subjects were identified through three university-affiliated cancer risk programs. All subjects were required to have a personal history of breast cancer (invasive or DCIS) and negative testing for both BRCA1 and 2 mutations (Table 1). Other specific eligibility criteria included: (1) Personal history of breast cancer at age ≤50 with family history of one or more cases of pancreatic cancer in first or second degree relatives in the same lineage of the family; or (2) Personal history of breast cancer at age ≤65 with family history of one or more cases of breast cancer diagnosed at age ≤65 and one or more cases of pancreatic cancer in first or second degree relatives in the same lineage of the family; or (3) Personal history of breast cancer at age ≤65 with family history of two or more cases of pancreatic cancer in first or second degree relatives in the same lineage of the family; or (4) Personal history of breast cancer at age ≤65 with personal history of pancreatic cancer; or (5) Personal history of bilateral breast cancer with family history of one or more cases of pancreatic cancer in first or second degree relatives in the same lineage of the family. Cancer diagnoses for probands were confirmed by pathology reports; cancer diagnoses for relatives were determined only through proband interviews, though pathologic confirmation was occasionally possible.
Table 1.
Characteristics of study subjects
| N (%) | |
|---|---|
| Total number of subjects | 94 |
| Median age at first breast cancer diagnosis (range) | 41 (31–66) |
| Subjects with multiple breast cancers | 28 (29.8%) |
| Subjects with both breast and pancreatic cancers | 3 (3.2%) |
| Family history of breast cancer in eligible lineagea | |
| 1 family member | 38 (40.4%) |
| ≥2 family members | 16 (17.0%) |
| Family member with bilateral breast cancer | 4 (4.3%) |
| Family history of pancreatic cancer in eligible lineageb | |
| 1 family member | 82 (87.2%) |
| ≥2 family members | 11 (11.7%) |
Family members include both 1st and 2nd degree relatives
One subject had only a personal history of pancreatic cancer (and no family history)
Data collection
Clinical information relevant to eligible patients was abstracted from medical records and included: age at breast cancer diagnosis, breast cancer treatment and outcome. Information regarding breast cancer pathology was abstracted from pathology reports and included histology, estrogen receptor (ER), progesterone receptor (PR) and HER2 status, grade and nodal status. Information regarding family history of cancers was also collected from both lineages and included: types of cancers, age at cancer diagnoses, and family ethnicity.
Laboratory methods
Blood or saliva was collected from each study subject for DNA analysis. DNA was extracted from blood using a Qiagen DNA extraction kit and from saliva using the Oragene DNA extraction procedure. Patients were tested for PALB2 alterations using Capillary Exon Grouping Analysis (cEGAN). Exon grouping analysis is based on Conformation-Specific Capillary Electrophoresis [22, 23]. All coding exons and surrounding intronic sequences were amplified with 19 primer pairs and analyzed on ABI-3730XL instruments (Applied Biosystems, Foster City, California). Polymerase chain reaction fragments with aberrant mobility were sequenced for the first 12 exons. Exon 13 was directly sequenced in all patients. This method permits detection of mutations and polymorphisms in PALB2 coding regions as well as splice-site mutations. BRCA1 and 2 gene sequencing for all cases was performed through Myriad Genetics Inc.
Statistical methods
The 90% confidence interval (CI) of the prevalence of germline PALB2 mutations was computed using the method of exact binomial confidence interval [24].
All patients provided informed consent for tissue analysis and medical record review. This study was approved by the institutional review boards of Dana Farber/Harvard Cancer Center and University of Pennsylvania Cancer Center.
Results
A total of 94 cases from independent families were selected for PALB2 sequence analysis. None of the subjects had a BRCA mutation or variant of uncertain significance. Characteristics of the 94 study subjects are summarized in Table 1. Median age at breast cancer diagnosis was 41 years, with 29.8% of subjects being diagnosed with multiple breast cancers. Three subjects had a personal history of both breast and pancreatic cancers. All subjects except one had at least one first- or second-degree family member with pancreatic cancer, and 11.7% of subjects had two relatives with pancreatic cancer. (One patient had a personal history of breast and pancreatic cancer, but no family history of pancreatic cancer). The majority of the subjects in our study were of Northern European descent (62/94, 65.9%).
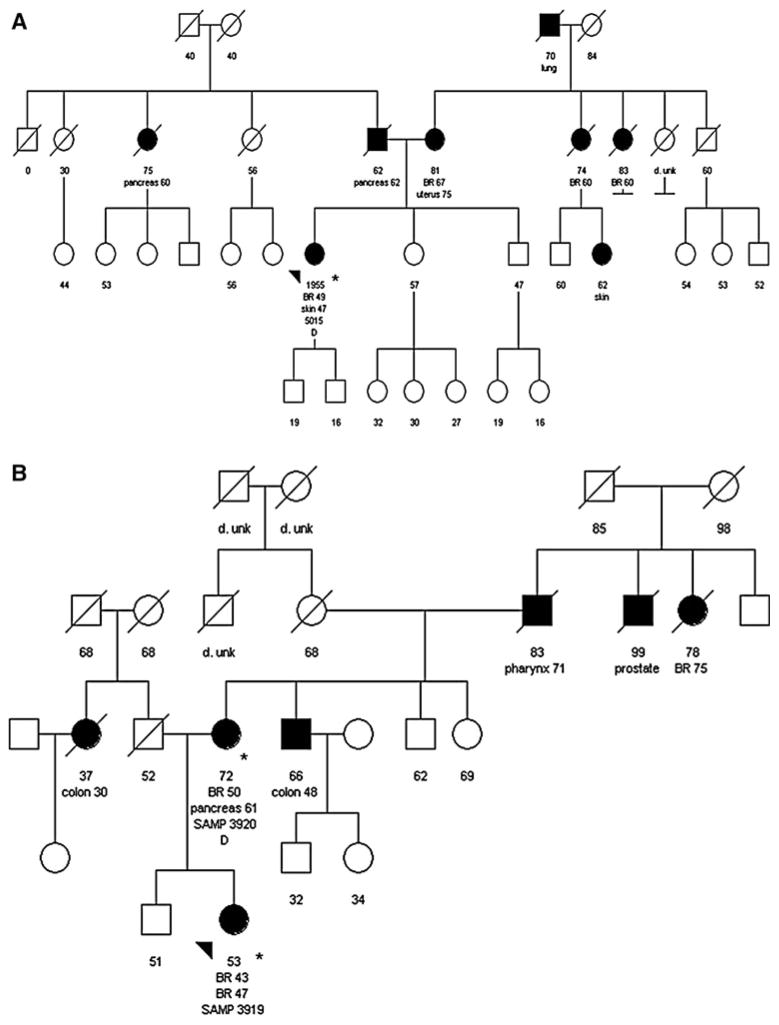
Truncating PALB2 mutations were identified in 2/94 subjects (2.1%; 90% CI = 0.40%, 6.5%). Both subjects were of Northern European/Caucasian descent. In the first subject, PALB2 sequence analysis revealed a nonsense mutation in exon 9, c.2962C>CT, which results in a stop codon at amino acid 988. This subject was diagnosed with pre-menopausal breast cancer at age 49. Pathology review showed a 1.5 cm, poorly differentiated, invasive ductal breast carcinoma that was negative for both estrogen receptor (ER) and Her2 over-expression. The subject also had a basal cell carcinoma of the skin at age 47. The family history was remarkable for a father with pancreatic cancer at age 62 and a paternal aunt with pancreatic cancer at age 60 (Fig. 1A). Of note, the subject’s mother and two maternal aunts were diagnosed with post-menopausal breast cancer. The subject’s mother was also affected with uterine cancer at age 75. Tissue from family members was unavailable for analysis.
Fig. 1. Pedigrees of the families with PALB2 mutations.
A depicts the family carrying the c.2962C>CT mutation. B depicts the family carrying the c.3549C>CA mutation. Individuals with cancer are shown in black. The arrow indicates the proband carrying the PALB2 mutation. Cancer type (BR breast cancer), age at diagnosis, and age of death are shown below each individual. Asterisks indicate those patients for whom cancer diagnoses were confirmed by pathology report
In the second subject found to have a mutation, PALB2 sequence analysis revealed a nonsense mutation in exon 13, c.3549C>CA, which results in a stop codon at amino acid 1183. This subject was initially diagnosed with pre-menopausal breast cancer at age 43. Pathology review showed a 2.5 cm invasive ductal carcinoma, ER-positive, Her2 status unknown. Despite treatment with mastectomy, chemotherapy and tamoxifen, the breast cancer recurred with distant metastases at age 47. The family history was remarkable for a mother with breast cancer at age 50 and pancreatic cancer at age 61 (Fig. 1B). Other notable cancers within the pedigree included a maternal uncle with colon cancer at age 48, a maternal grandfather with pharyngeal cancer at age 71, and a paternal aunt with colon cancer at age 30. PALB2 sequence analysis of the mother’s blood revealed the same PALB2 mutation identified in the proband. Tissue from other family members was unavailable for analysis.
In addition, sequence analysis revealed thirteen different PALB2 variants and polymorphisms (Table 2). Among these, two novel missense variants were identified: 1222T>TC (p.Y408YH) and 3128G>GC(p.G1043GA). Both of the subjects found to have a novel missense variant were of Northern European descent. Pathogenicity of the novel missense variants was analyzed using three in silico tests (PolyPhen, SIFT and PMut). Based on these results, the variant 1222T>TC was predicted to be a neutral change by two of three tests (SIFT, PMut). However, the variant 3128G>GC was predicted to have potential deleterious effects. This variant codes for 1043G>AG, which is located in WD repeat 4 (1010–1052). Interaction with BRCA2 occurs through a hydrophobic pocket at the crossover between WD repeats 4 and 5 (1053–1057). Thus, it is quite possible that this alteration may abrogate the interaction of PALB2 and BRCA2 [25]. The subject with the 3128G>GC variant was diagnosed with pre-menopausal breast cancer at age 40. Pathology review showed a 2.3 cm, grade 3, invasive ductal carcinoma which was ER-positive and Her 2-negative. Extensive lymphatic invasion was noted in addition to three positive axillary lymph nodes. The family history was remarkable for a paternal grandfather with pancreatic cancer. No family members were available for genetic testing. In addition a novel synonymous variant was also found, 1881G>GT (p. V627VV). Consistent with previously reported synonymous variants, this variant is predicted to be non-deleterious and pathogenicity was therefore not assessed.
Table 2.
Identified PALB2 variants
| Variant type | PALB2 cDNA change | PALB2 protein change | Number observed | Prediction programs
|
||
|---|---|---|---|---|---|---|
| PolyPhen | SIFT | PMut | ||||
| Novel variants | ||||||
| Mis | c.3128G > GC | p.1043G > AG | 1 | Possibly damaging | Affect protein function | Pathological |
| Mis | c.1222T > TC | p.Y408Y > YH | 1 | Possibly damaging | Tolerated | Neutral |
| Syn | c.1881G > GT | p.V627VV | 1 | N/A | N/A | N/A |
| Known variants | ||||||
| Mis | c.1010T > TCa | p.L337LS | 5 | Possibly damaging | Affect protein function | – |
| Mis | c.1676A > AGa | p.Q559QR | 16 | Benign | Tolerated | – |
| Mis | c.2014G > GCa | p.E672EQ | 2 | Benign | Affect protein function | – |
| Mis | c.2590C > CTa | p.P864PS | 3 | Probably Damaging | Tolerated | – |
| Mis | c.2794G > GAb | p.V932VM | 1 | Benign | Tolerated | – |
| Mis | c.2816T > TGa | p.L939LW | 1 | Possibly damaging | Affect protein function | – |
| Mis | c.2993G > GAa | p.G998GE | 1 | Probably damaging | Affect protein function | – |
| Syn | c.1194G > GAb | p.V398VV | 1 | N/A | Tolerated | N/A |
| Syn | c.1572A > AGb | p.S524SS | 1 | N/A | Tolerated | N/A |
| Syn | c.3300T > TGb | p.T1100TT | 2 | N/A | Tolerated | N/A |
Mis Missense variant, Syn synonymous variant, PolyPhen Polymorphism Phenotyping prediction (http://genetics.bwh.harvard.edu/pph), SIFT Sorting Intolerant From Tolerant (http://blocks.fhcrc.org/sift/SIFT_BLink_submit.html), PMut (http://mmb2.pcb.ub.es:8080/PMut)
Previously reported [26]
Previously reported [6]
N/A Not applicable
– Information not available
Discussion
We found PALB2 mutations in two of 94 subjects, a prevalence of 2.1% in this specific breast cancer population. This rate is consistent with the 1–2% prevalence of mutations reported in other familial breast cancer populations [6, 14]. A recent study by Adank et al. [26] evaluated the prevalence of PALB2 mutations in 110 BRCA2-like families, including 45 breast cancer patients with a first or second degree relative with pancreatic cancer. While one truncating PALB2 mutation was found in a male breast cancer patient, none of the 45 subjects with breast cancer and a family history of pancreatic cancer was found to carry a pathogenic mutation. The difference in PALB2 mutation prevalence between our study and that of Adank et al. is likely not significant and may be due to the small sample sizes. While the PALB2 mutation prevalence rate of 2.1% in our population is higher than that reported by Adank et al., it is lower than that found in studies of familial pancreatic cancer (3.1–3.7%) [18, 19].
The c.3549C>CA and c.2962C>CT PALB2 mutations identified in our sample have been previously described in breast cancer and Fanconi anemia patients, respectively [8, 18]. However, our study is the first to specifically identify the c.3549C>CA PALB2 mutation in association with pancreatic cancer. This mutation was identified in the mother of a proband within our sample; the mother was affected with both breast and pancreatic cancer. It is likely that the c.2962C>CT PALB2 mutation is specifically associated with the pancreatic cancer in our proband’s family as well, however tissue from family members affected with pancreatic cancer was unavailable for analysis. With the addition of our data on the family with the c.3549C>CA PALB2 mutation, there are now nine PALB2-related pancreas families identified to date [17–19]. Interestingly, eight of these families include one or more cases of breast cancer, and three families contain individuals with both breast and pancreatic cancer. Within the familial pancreatic cancer population described in the report of Tischkowitz et al. [17] one of nine subjects (11.1%; 90% CI = 0.57%, 42.9%) with both breast and pancreatic cancer carried a PALB2 mutation. We performed PALB2 sequence analysis on four individuals with concomitant breast and pancreatic cancer, namely three probands and one relative (mother) of a proband with breast cancer. Of these, one mutation carrier was found. Though numbers are small, these rates of PALB2 mutations within individuals with both breast and pancreatic cancer is noteworthy and warrants further study of this patient population.
Previous reports have suggested that breast tumors associated with some PALB2 mutations are aggressive in nature [12, 16]. Our data further supports this concept, as both mutation carriers in our sample proved to have aggressive disease. The breast cancer in the first mutation carrier was notable for poor differentiation and triple-negative status, lacking ER, PR and HER2 over-expression. In the second mutation carrier, the breast tumor recurred with distant metastases 4 years after diagnosis despite initial aggressive therapy for ER-positive, node-negative disease. These observations must be interpreted with caution given the small sample size, but are nevertheless intriguing.
In conclusion, we found that PALB2 mutations occur with a prevalence of 2.1% in a population of BRCA1/2-negative breast cancer patients specifically selected for a personal and/or family history of pancreatic cancer. This prevalence rate appears comparable to the rate of PALB2 mutations published in other breast cancer populations. Further study is needed to determine the prevalence of PALB2 mutations and the clinical utility of such testing in those individuals affected with both breast and pancreatic cancers. The benefit of pancreatic cancer screening in patients at increased risk of developing pancreatic cancer, such as those who carry a BRCA2 or PALB2 mutation, is uncertain. Recently, Verna et al. [27] reported that endoscopic ultrasound (EUS) and MRI were able to detect early neoplastic pancreatic lesions in 12% of 51 individuals with a family history of pancreatic cancer. Other studies evaluating the benefit of such screening are ongoing. If pancreatic cancer screening is found to be beneficial in high risk populations, identifying those individuals with PALB2 mutations will likely have even greater clinical significance. Currently, however, routine diagnostic screening for PALB2 mutations in any breast cancer patient is not indicated.
Acknowledgments
This work was supported by grants from the Breast Cancer Research Foundation (to KLN, NT, and JG); the Dana Farber/Harvard Cancer Center (CA 006516); the Dana Farber/ Harvard Cancer Center Breast SPORE (CA 089393); and the Mac-Donald Family Foundation (to SMD).
Contributor Information
Erin W. Hofstatter, Email: erin.hofstatter@yale.edu, Section of Medical Oncology, Yale Cancer Center, Yale School of Medicine, 333 Cedar Street, PO Box 208032, New Haven, CT 06520-8032, USA
Susan M. Domchek, Division of Medical Oncology, Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA, USA
Alexander Miron, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA.
Judy Garber, Division of Population Sciences and Adult Oncology, Dana-Farber Cancer Institute, Boston, MA, USA.
Molin Wang, Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute, Boston, MA, USA.
Kathryn Componeschi, Division of Hematology-Oncology, Beth Israel Deaconess Medical Center, Boston, MA, USA.
Leigh Boghossian, Division of Medical Oncology, Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA, USA.
Penelope L. Miron, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA
Katherine L. Nathanson, Division of Medical Oncology, Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA, USA
Nadine Tung, Division of Hematology-Oncology, Beth Israel Deaconess Medical Center, Boston, MA, USA.
References
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2008 doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Buchholz TA, Wazer DE. Molecular biology and genetics of breast cancer development: a clinical perspective. Semin Radiat Oncol. 2002;12:285–295. doi: 10.1053/srao.2002.35248. [DOI] [PubMed] [Google Scholar]
- 3.Tan DSP, Marchio C, Reis-Filho JS. Hereditary breast cancer: from molecular pathology to tailored therapies. J Clin Pathol. 2008;61:1073–1082. doi: 10.1136/jcp.2008.057950. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Ford D, Easton DF, Stratton M. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Amer J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. pii: S0002-9297(07)63848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. 10.1038/ng 1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Ma J, Wu J, et al. PALB2 Links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. 10.1073/ pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirshfield KM, Rebbeck TR, Levine AJ. Germline mutations and polymorphisms in origins of cancers in women. J Oncol. 2010:1–11. doi: 10.1155/2010/297671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res. 2008;10:208–214. doi: 10.1186/bcr2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heikkinen T, Karkkainen H, Aaltonen K, et al. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15:3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 17.Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010 doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 20.The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 21.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–719. doi: 10.1136/jmg.2004.028829. 10.1136/jmg. 2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velasco E, Infante M, Durán M, et al. Heteroduplex analysis by capillary array electrophoresis for rapid mutation detection in large multiexon genes. Nat Protoc. 2007;2(1):237–246. doi: 10.1038/nprot.2006.482. [DOI] [PubMed] [Google Scholar]
- 23.Davies H, Dicks E, Stephens P, et al. High throughput DNA sequence variant detection by conformation sensitive capillary electrophoresis and automated peak comparison. Genomics. 2006;87(3):427–432. doi: 10.1016/j.ygeno.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Mehta CR, Patel NR. A network algorithm for the exact treatment of Fisher’s exact test in rxc contingency tables. J Am Stat Assoc. 1983;78:427–434. [Google Scholar]
- 25.Oliver AW, Swift S, Lord CJ, et al. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adank MA, van Mil SE, Gille JJ, et al. PALB2 analysis in BRCA2-like families. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-1001-1. 10.1007/ s10549-010-1001-1. [DOI] [PubMed] [Google Scholar]
- 27.Verna EC, Hwang C, Stevens PD, et al. Pancreatic cancer screening in a prospective cohort of high-risk patients: a comprehensive strategy of imaging and genetics. Clin Cancer Res. 2010;16:OF1–OF10. doi: 10.1158/1078-0432.CCR-09-3209. [DOI] [PubMed] [Google Scholar]