Abstract
Compounds able to interfere with amino acid biosynthesis have the potential to inhibit cell growth. In both prokaryotic and eukaryotic microorganisms, unless an ornithine cyclodeaminase is present, the activity of δ1-pyrroline-5-carboxylate (P5C) reductase is mandatory to proline production, and the enzyme inhibition should result in amino acid starvation, blocking in turn protein synthesis. The ability of some substituted derivatives of aminomethylenebisphosphonic acid and its analogues to interfere with the activity of the enzyme from the human pathogen Streptococcus pyogenes was investigated. Several compounds were able to suppress activity in the micromolar range of concentrations, with a mechanism of uncompetitive type with respect to the substrate P5C and non-competitive with respect to the electron donor NAD(P)H. The actual occurrence of enzyme inhibition in vivo was supported by the effects of the most active derivatives upon bacterial growth and free amino acid content.
Keywords: Amino acid metabolism, Antibiotics, P5C reductase, Proline, Streptococcus sp
Introduction
The development of bacterial resistance to current antibiotic therapies is an increasing threat for human health (Chen et al. 2009). Many of therapeutics that are currently in use came from a small set of molecular scaffolds, whose functional lifetime has been extended by synthetic tailoring. Because of the emergence and diffusion of multi-drug resistance, the discovery of new scaffolds is mandatory (Fischbach and Walsh 2009). As an alternative, new antibiotic targets should be identified (Pathania and Brown 2008). In this perspective, although they were exploited to date mainly as active principles for weed control (Tan et al. 2006), inhibitors of enzymes that catalyse key reactions in amino acid metabolism could represent promising new leads for the control of pathogenic microorganisms. In several instances, the inhibition of selected enzymes in amino acid biosynthesis has been indeed found to exert remarkable activity against bacteria (Harth and Horwitz 2003; Hutton et al. 2007; Liu et al. 2008; Ziebart et al. 2010).
From this point of view, little attention has been paid to date to proline synthesis. Proline plays an important role in protein structure, uniquely contributing to protein folding and stability (Ge and Pan 2009). Moreover, in a wide variety of microorganisms, a rapid and reversible increase in the intracellular concentration of free proline has been shown in response to either osmotic or temperature stress, implying a role in stress tolerance and osmoregulation (Empadinhas and Da Costa 2008; Takagi 2008). The ability of changing cellular osmolarity seems essential to cope with fluctuating external water potential, salinity and temperature, and survive in harsh environments (Höper et al. 2005). Some evidence also suggested that the ability of metabolising proline might function as a virulence factor for certain pathogenic bacteria (Nakajima et al 2008). In other cases, the same may occur indirectly: if unable to produce compatible osmolytes, the bacterial cell cannot achieve osmoadaptation in body fluids. As a consequence, the expression of certain virulence determinants (such as the pyelonephritis-associated pilus in Escherichia coli; Culham et al. 1998) is affected.
The development of effective inhibitors of proline synthesis may be hampered by the occurrence in bacteria of redundant biosynthetic pathways. Proline biosynthesis can in fact proceed from either glutamate or arginine, usually the glutamate route being the main pathway (Cunin et al. 1986; Aral and Kamoun 1997). Moreover, a stereospecific and irreversible conversion of l-ornithine to l-proline may be accomplished in a single step by the enzyme ornithine cyclodeaminase [OCD, EC 4.3.1.12] (Goodman et al. 2004). In the presence of two pathways, the inhibition of either of the enzymes that catalyze the rate-limiting steps would be ineffective, because proline starvation would not be achieved and the cell would be allowed to recover. However, because OCD has been identified only in a small set of soil and plant-associated bacteria, and the glutamate and the ornithine pathways share the last reaction catalyzed by a δ1-pyrroline-5-carboxylate (P5C) reductase [EC 1.5.1.2], this goal might be achieved through the development of specific inhibitors for the latter enzyme. Since this dual anabolic route is shared also by plants (Lehmann et al. 2010), P5C reductase inhibitors might find application in crop protection as well. We previously synthesized and screened several derivatives of aminomethylenebisphosphonic acid (compound 01) for the ability to inhibit plant P5C reductase (Forlani et al. 2007). A group of phenyl derivatives were found effective in the micromolar to millimolar range, and indeed showed phytotoxic properties (Forlani et al. 2008a). Here, we report on the effectiveness of some derivatives and analogues of compound 01 against the bacterial P5C reductase, assessed at both the enzyme and the whole cell level.
Materials and methods
Enzyme purification
Escherichia coli BL21(DE3) pLysS cells, made competent by the calcium chloride method, were transformed with the pMCSG7 vector bearing the M1 GAS Streptococcus pyogenes P5C reductase gene (Nocek et al. 2005). Transformants were selected at 37°C on LB plates containing 100 mg l−1 ampicillin and 25 mg l−1 chloramphenicol. Freshly grown cultures in liquid LB medium (0.6 OD600) were induced at 24°C with 1 mM IPTG. Cells were harvested by centrifugation 4 h after induction, and stored at −20°C. Pellets (about 2 g) were thawed and extracted in a mortar with 2 g g−1 alumina. All the subsequent operations were carried out at 4°C. The homogenate was resuspended in 20 ml g−1 of 50 mM Na phosphate buffer, pH 7.5, containing 200 mM NaCl and 0.5 mM DTT. Following clarification at 4,000g for 5 min, the extract was centrifuged at 18,000g for 15 min. The supernatant was immediately loaded at a constant flow of 10 ml h−1 onto a His-Select™ Nickel Affinity Gel (Sigma P6611) column (0.5 cm diameter, 2 ml bed-volume) equilibrated with extraction buffer. After extensive washing, the column was eluted stepwise with buffer containing increasing concentrations of imidazole, harvesting 1-ml fractions. The presence and the purity of the heterologous protein were determined by polyacrylamide gel electrophoresis under denaturing conditions. Pure fractions were combined, adjusted to a protein concentration of 0.5 mg ml−1, filter sterilized (0.2 μm) and stored on ice. Under these conditions, the enzyme was remarkably stable, with more than 90% of the initial activity still retained after 6 month storage.
Enzyme assay
The physiological, forward reaction of P5C reductase was measured by following the P5C-dependent oxidation of NAD(P)H. Unless otherwise specified, the assay mixture contained 100 mM HEPES-KOH buffer, pH 7.5, 1 mM MgCl2, 1 mM l-P5C and 0.4 mM NADH, in a final volume of 1 ml. A limiting amount of enzyme (0.60 nkat under standard assay conditions, corresponding to 25 ng protein, freshly water-diluted from the pure enzyme preparation) was added to the pre-warmed mixture, and the decrease in absorbance at 340 nm was determined at 37°C for up to 5 min by monitoring the sample at 30-s intervals against blanks from which P5C had been omitted. The activity was determined from the initial linear rate, with the assumption of an extinction coefficient of 6,220 M−1 cm−1. dl-P5C was synthesized by the periodate oxidation of δ-allohydroxylysine, purified by cation-exchange chromatography on a Dowex AG50 (200–400 mesh) column, and quantified by either the o-aminobenzaldehyde or the ninhydrin method, as described (Williams and Frank 1975). Final preparations in HCl 1 M were stored in the dark at 4°C. Proper dilutions were neutralized with the same volume of KOH 1 M just before being added to the reaction mixture. The indicated final concentration always refers to the l-isomer. The protein concentration was determined by the method of Bradford (1976), using bovine serum albumin as the standard.
Enzyme inhibition and kinetic analysis
N-Substituted derivatives and analogues of compound 01 (Fig. 1) were synthesized and purified as described previously (Forlani et al. 2008a, and supporting information available therein). P5C reductase inhibition was evaluated by adding to the reaction mixture an appropriate water dilution of a 1–5 mM freshly prepared solution of a given compound, brought to pH 7.5–8.0 with KOH, to obtain a final concentration of 0.2 mM. Results were expressed as percent of the activity measured in parallel untreated controls, and are mean ± SE over at least three independent replications. Active compounds were further tested in the micromolar range, from 0.1 to 100 μM. At least three measurements were performed for each dose. The concentration causing 50% inhibition of P5C reductase activity (IC50) was estimated utilizing the linear regression equation of enzyme activity values, expressed as a percentage of untreated controls, plotted against the logarithm of inhibitor concentration. At least three concentrations in the rectilinear part of the resulting sigmoid curve were considered. Confidence limits were computed according to Snedecor and Cochran (1989). IC50 values and confidence intervals were confirmed also by the probit method (Finney 1971).
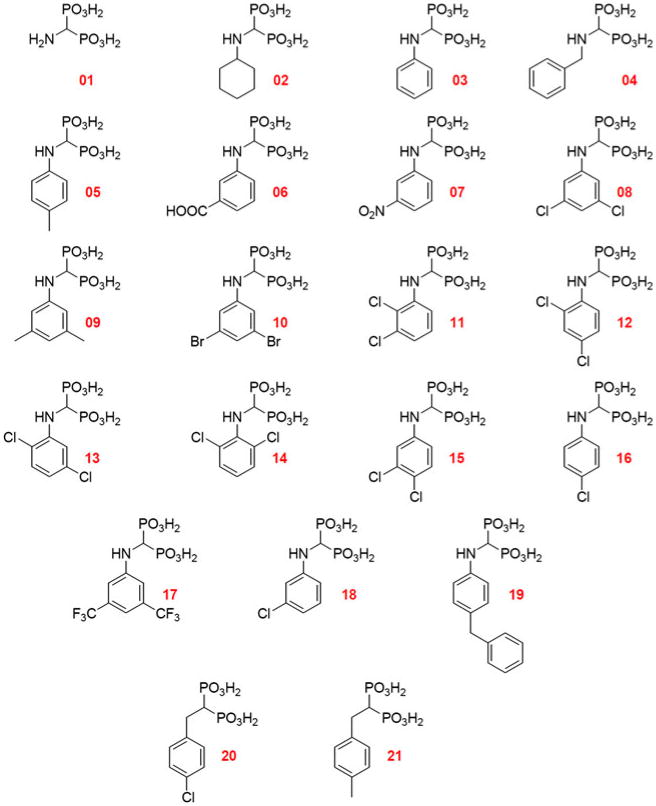
Fig. 1. Derivatives of compound 01 evaluated as possible inhibitors of P5C reductase from Streptococcus pyogenes.
For kinetic evaluations, the enzyme was assayed in the presence of increasing concentrations of a given compound at varying those of the substrates. The concentration for the variable substrate ranged from 80 to 268 μM NADH and from 125 to 500 μM l-P5C. At least eight doses were evaluated for each substrate, at no less than triplicate. In the case of P5C (uncompetitive inhibition), KI values were estimated from Lineweaver–Burk plots of activity, on the basis of the corresponding lowering in the apparent KM value; three inhibitor concentrations, ranging from 0.5 to 1.5-fold the IC50 value, were tested. For NADH, three inhibitor concentrations, ranging between 0.6 and 1.8-fold the IC50 value, were tested. However, because of a noncompetitive type of inhibition, KI values were estimated from Dixon plots of activity by evaluating the effect of six inhibitor levels, ranging from 0.2 to 1.0-fold the IC50 value, in the presence of four substrate concentrations. To confirm the inhibitory mechanism, NADPH was also tested as the variable substrate, at concentrations ranging from 12 to 144 μM. All reported data are presented as mean-s ± SEM (standard error of the mean) over results obtained with different inhibitor or substrate concentrations, respectively.
Antimicrobial activity determination
Streptococcus pyogenes Rosenbach, ATCC 19615 strain, was maintained on Trypticase Soy (Oxoid) agar plates containing 5% defibrinated horse blood. A freshly grown single colony was transferred into 10 ml of liquid medium in a 50 ml flask that was incubated overnight at 37°C under shaking (80 rpm). Because of the pathogenic potential, in all instances bacterial suspensions were handled under a biohazard laminar flow hood, taking care to avoid the formation of aerosols leading to environmental contamination. Personnel also wore latex gloves and disposable face mask as an additional safeguard. All contaminated materials/glassware were treated with bleach before being autoclaved and discarded/washed, respectively. The suspension was used to inoculate 15 ml test tubes containing 3 ml of Roche Susceptibility Test medium (Hall et al. 1984) in which proline had been omitted, to an initial density of 0.05 OD600. Growth at 37°C was followed as the increase of absorbance at 600 nm, measured at 1-h intervals with a Novaspec Plus spectrophotometer (Amersham Biosciences) equipped with a tube adapter. Data were used to calculate growth constants (k), and the effect of the presence of a given compound upon bacterial growth was expressed as percent reduction of k. Each treatment was carried out at least in triplicate. The final biomass was evaluated as wet weight increase. The concentration causing 50% inhibition of bacterial growth rate or the final biomass (LC50) was estimated utilizing the linear regression equation of growth values, expressed as a percentage of that for untreated controls, plotted against the logarithm of the inhibitor concentration. Cell viability was assessed by serial dilution with physiological saline and viable count on standard medium. In all cases, absorbance values were consistent with cell density, as estimated by vital count.
Amino acid extraction, separation and quantification
For amino acid analysis, bacterial cells were harvested by centrifugation for 10 min at 4,000g. Pellets were resuspended with 2 ml g−1 of a 3% (w/v) solution of 5-sul-phosalicylic acid, and extracted by sonication with a UP50H ultrasonic processor (Hielscher, Germany) equipped with a Sonotrode MS1 tip, with 6 cycles of 30-s sonication at 70% of maximum power and 2-min cooling on ice. Cell debris was removed by centrifugation at 12,000g at 4°C, and samples (20 μl) were mixed with the same volume of o-phthaldialdehyde solution (0.5 M in 0.5 M sodium borate buffer, pH 10.0, containing 0.5 M β-mercaptoethanol and 10% [v/v] methanol). After exactly 60 s, 20 μl of derivatized samples were injected onto a 4.6 × 250 mm Zorbax ODS column (Rockland Technologies, Newport, DE), and elution proceeded as previously described (Mazzucotelli et al. 2006), monitoring the eluate at 340 nm. This procedure allowed complete resolution of equimolar mixtures of derivatizable amino acids, with detection limit of about 0.1 nmol. Proline and total amino acid content were quantified by the acid ninhydrin method, as described (Williams and Frank 1975). Results were expressed as μmol g−1 cells, and are mean ± SD over at least three replications.
Results and discussion
Phenyl derivatives of compound 01 are potent inhibitors of S. pyogenes P5C reductase
Within the frame of a project aimed at evaluating P5C reductase inhibitors as possible new leads for antibiotics, Streptococcus pyogenes was chosen as the experimental system to take advantage from both the availability of detailed information about the structure of the enzyme (Nocek et al. 2005) and the absence in this species of a gene coding for an OCD. A blast search, performed on the complete genome sequence of the M1 strain (Ferretti et al. 2001; http://www.genome.ou.edu/strep.html) using the Pseudomonas putida KT2440 gene coding for the cyclase as the query, yielded in fact no result. To reduce manipulation of this human pathogen, the P5C reductase gene was cloned (Nocek et al. 2005) and expressed in E. coli. The protein was purified to electrophoretic homogeneity, and the activity characterized throughout in order to ensure proper assay conditions (Petrollino D and Forlani G, manuscript in preparation). When the activity of the enzyme was measured in the presence of various derivatives of compound 01 (Fig. 1) at 0.2 mM, a differential effect was evident (Table 1). Several substituted phenyl derivatives were able to completely suppress activity, while others were substantially ineffective. Active compounds were further tested in the micromolar range, allowing us to calculate the concentrations able to inhibit by 50% the enzymatic activity (Table 1). On the whole, bisphosphonates showed a strikingly higher effectiveness against the bacterial enzyme, with IC50 values about two orders of magnitude lower than those previously found for plant P5C reductase (Fig. S1 in supplemental materials; Forlani et al. 2008a). This is not surprising since the inhibitors, because of the unavailability of the three-dimensional structure of the plant enzyme, had been designed based on the results of a computer-aided docking analysis performed with just the crystal structure of S. pyogenes P5C reductase (Forlani et al. 2007). As to the scaffold, aminobisphosphonates showed higher effectiveness than the corresponding amino-and hydroxyphosphonates (Table S1 in supplemental materials). In addition, the replacement of the phenyl moiety with various pyridyl rings was found detrimental (Table S2 in supplemental materials). Concerning substituents in the phenyl rings, the presence of two halogen moieties yielded the highest inhibitory rates. This is consistent with previous data on the plant enzyme (Forlani et al. 2008a). However, in this case some other analogues showed equipotency, such as compounds 16 and 18, bearing a single chlorine, or compound 19, with a benzyl residue. The latter case is remarkable, since it shows that there is an additional cavity accommodating this bulky group.
Table 1. Effect of phenyl derivatives and analogues of compound 01 upon the catalytic rate of S. pyogenes P5C reductase.
| Compound | Enzyme inhibition at 0.2 mMa | IC50 (μM)b |
|---|---|---|
| 01 | 3.0 ± 2.1 | ND |
| 02 | 8.1 ± 1.5 | ND |
| 03 | 3.0 ± 0.4 | ND |
| 04 | 3.0 ± 2.2 | ND |
| 05 | 33.4 ± 1.5 | >1 mM |
| 06 | 21.6 ± 1.5 | >1 mM |
| 07 | 3.2 ± 0.3 | ND |
| 08 | 91.1 ± 0.3 | 0.22 ± 0.02 |
| 09 | 70.5 ± 6.0 | 25.6 ± 9.6 |
| 10 | 98.4 ± 0.1 | 0.22 ± 0.01 |
| 11 | 95.4 ± 1.3 | 0.87 ± 0.10 |
| 12 | 52.0 ± 1.2 | 12.0 ± 6.1 |
| 13 | 63.9 ± 2.2 | 2.7 ± 0.8 |
| 14 | 96.2 ± 0.1 | 0.88 ± 0.07 |
| 15 | 92.5 ± 0.5 | 0.39 ± 0.02 |
| 16 | 61.5 ± 1.1 | 1.4 ± 0.3 |
| 17 | 71.2 ± 1.0 | 0.78 ± 0.09 |
| 18 | 71.2 ± 2.1 | 0.90 ± 0.13 |
| 19 | 94.5 ± 0.1 | 0.41 ± 0.05 |
| 20 | 33.8 ± 0.7 | >1 mM |
| 21 | 11.5 ± 3.9 | ND |
P5C-dependent NADH oxidation was measured at 37°C for 5 min in the presence of 0.2 mM of a given compound. Results were expressed as percent of the activity measured in parallel, untreated controls. At least three replications were run for each phosphonate, and six for the control. Data are presented as percent inhibition, and are means ± SE over replications
Active compounds (percent inhibition at 0.2 mM > 20%) were further tested in the micromolar range, from 0.1 to 100 μM. The concentrations causing 50% inhibition (IC50) were estimated utilizing the linear regression equation of enzyme activity values, expressed as a percentage of untreated controls, plotted against the logarithm of inhibitor concentration. ND not determined
The mode of action of the most effective inhibitors strengthens the possibility that inhibition may occur and be effective in vivo
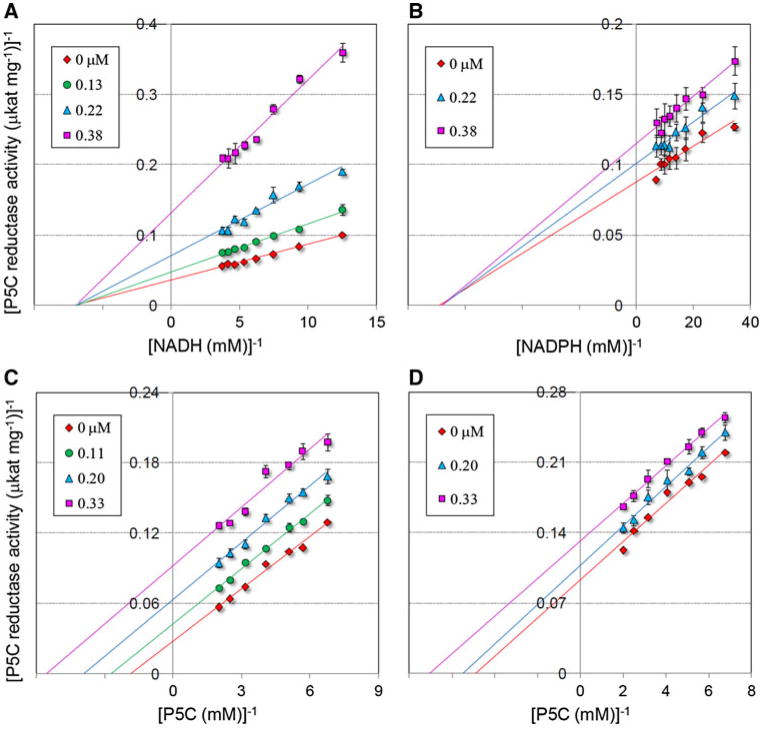
The ability to interfere with the catalytic mechanism of a given enzyme in vitro does not necessarily imply that the treatment with the inhibitor would cause a block of the corresponding metabolic pathway in vivo. This is true for instance in the case of reversible inhibitors acting competitively. The initial suppression of enzyme activity leads to an increase of the intracellular level of the substrate(s), which in turn can relieve the inhibitory effect (Cascante et al. 2002). In order to obtain a better insight into the inhibitory properties of bisphosphonates, a proper kinetic analysis was therefore performed. Results, summarized in Fig. 2, showed that, analogously to the plant enzyme, S. pyogenes P5C reductase is inhibited by compound 08 by a mechanism of noncompetitive type with respect to either NADH or NADPH, and of uncompetitive type with respect to P5C. Similar data were obtained with the other most effective derivative, compound 19 (Fig. S2 in supplemental materials). Replotting of the data allowed calculation of inhibition constants (Table 2) that in all cases were below 1 μM. This set of results has some remarkable implications. Because inhibitors bind only to the P5C-enzyme complex, substrate binding in the P5C reductase active site could be of sequential character [P5C binds before NAD(P)H], as previously hypothesized only on the basis of the crystal structure of the protein (Nocek et al. 2005). This would be an unusual feature, since NAD-dependent reductases usually bind the dinucleotide co-substrate first. It confirms that enzyme inhibition occurs also when NADPH is the electron donor, ruling out the possibility that (because of the higher steric hindrance) its binding may displace the inhibitor from the enzyme. It implies that an intracellular concentration of the inhibitor as low as 10 μM should bring about more than 90%-reduction of P5C reductase activity irrespectively from the increase of P5C level that would be caused as a consequence. All these elements concur in strengthening the possibility that these inhibitors may exert their effect also in vivo.
Fig. 2.
Kinetic analysis of the inhibition of S. pyogenes P5C reductase by compound 08. The enzyme was incubated with a fixed and saturating amount of P5C in the presence of increasing inhibitor concentrations, as indicated, at varying NADH (a) or NADPH (b) level. Lines converging to the x-axis in Lineweaver–Burk plot accounted for an inhibition of non competitive type. A similar analysis was carried out at varying P5C concentration, using either NADH (c) or NADPH (d) as the electron donor. Parallel lines in Lineweaver–Burk plot pointed to an inhibition of uncompetitive type. Unvariable substrate concentration was fixed at 1, 0.4 and 0.2 mM for l-P5C, NADH and NADPH, respectively
Table 2. Inhibition constants of Streptococcus pyogenes P5C reductase activity for two of the most active phenyl derivatives of compound 01.
| Compound 08 (μM) | Compound 19 (μM) | |
|---|---|---|
| KI against l-P5Ca(NADH) | 0.67 ± 0.04 | 0.98 ± 0.07 |
| KI against l-P5Ca(NADPH) | 0.81 ± 0.01 | ND |
| KI against NADHb | 0.11 ± 0.01 | 0.37 ± 0.00 |
| KI against NADPHb | 0.68 ± 0.06 | ND |
ND not determined
Uncompetitive inhibition, KI value estimated from Lineweaver–Burk plots
Noncompetitive inhibition, KI value estimated from Dixon plots
Aminomethylenebisphosphonic acids inhibit the growth of S. pyogenes, and cause an actual reduction of the intracellular pool of free proline
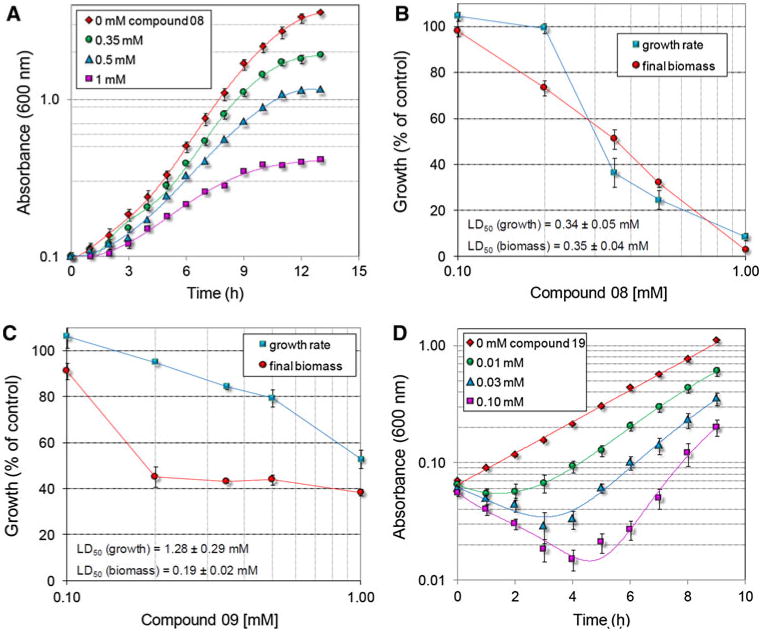
To substantiate this possibility, the growth of bacterial cultures was measured following the addition of increasing concentrations of the most active compounds to the culture medium. When supplied in the same range (10−6 to 10−5 M) in which they had been found to progressively reduce enzyme activity in vitro, they failed to exert cytostatic effects. However, if the concentration was increased over 100 μM, they showed a remarkable ability to inhibit cell proliferation (Fig. 3). The effect was proportional to the dose, and to the relative effectiveness shown by a given compound in vitro. Notably, for all compounds tested but mainly for compound 19 the treatment with low doses, which were unable to reduce growth, caused a remarkable and proportional lag before the attainment of the exponential phase. Such behaviour may be suggestive of the occurrence of detoxifying activiti(es) able to modify the inhibitor molecule. Once induced, they would be able to destroy low amounts of bisphosphonates, allowing the bacterial cell to recover. This would also explain the reason why higher concentrations are required in vivo than in vitro to suppress P5C reductase activity: growth inhibition would take place only when the concentration of the compounds overcomes the detoxifying potential of the bacterial cell. However, other reasons might explain the lower effectiveness in vivo. Because of the high polarity of the phosphonic moiety, these compounds are endowed with a remarkable negative charge that can strongly reduce their uptake into the cell. This hypothesis would be consistent with results obtained in the case of both the phosphonate herbicide glyphosate (Forlani et al. 2008b) and some anticorrosive polyphosphonates (Forlani et al. 2011), for which an almost negligible uptake rate at low external concentrations was found to limit metabolization by the bacterial cell.
Fig. 3.
Effect of increasing concentrations of P5C reductase inhibitors on S. pyogenes growth. Late-log grown cultures were used to inoculate parallel tubes containing 3 ml of minimal medium to an initial OD600 of 0.1. One hour after the inoculum, a proper amount of a given bisphosphonate was added. The growth was followed for the subsequent 12 h as the increase in absorbance (a, d). After logarithmic transformation of the data, growth constants were calculated over the rectilinear part of the curves, whereas the final biomass (fresh weight) was evaluated following centrifugation 24 h after the inoculum (b, c)
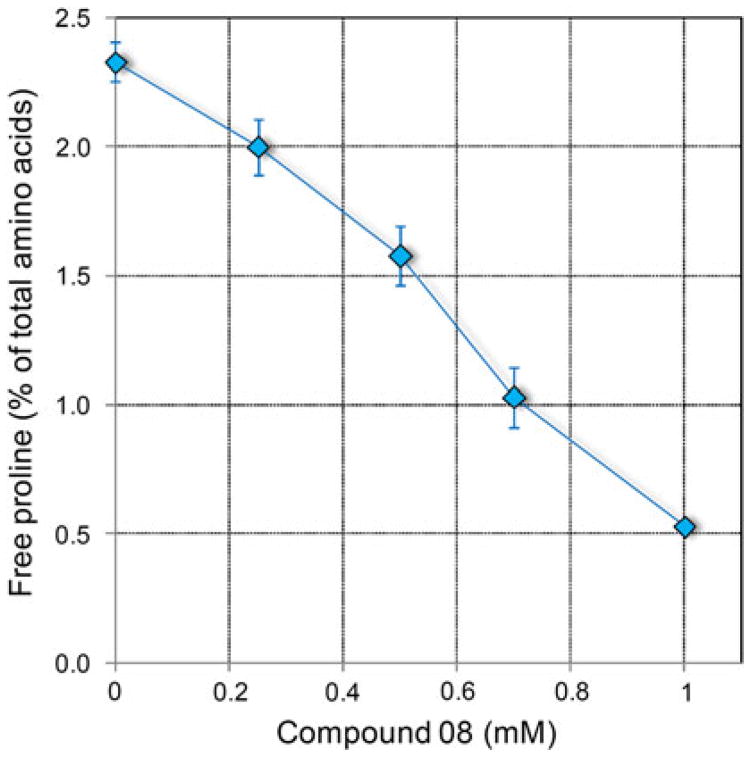
Whatever the reason for the reduced efficacy in vivo, further evidence was required to substantiate an actual occurrence of proline starvation. With this aim, free amino acid content was determined in bacterial cultures treated with inhibitory levels of compound 08. Results, outlined in Table 3, indeed showed a reduction of free proline concentration inside the cell. In addition, the level of the metabolically related intermediate ornithine was severely affected. Therefore, a corresponding increase of free glutamate would be expected, which on the contrary lowered, but if the sum of glutamate and glutamine is considered instead, a mild increase was in fact evident at 500 μM. However, significant fluctuations were noticed also for amino acids of the aspartate family, and for glycine. The metabolic basis of these effects is not plain, and could indicate the occurrence of further phosphonate targets in amino acid metabolism. Therefore, the existence of a causal connection between proline starvation and inhibition of cell proliferation is not certain. Above 500 μM, a general reduction of free amino acid content was found (data not shown), such that made difficult to distinguish specific effects. This notwithstanding, if the percent contribution of proline to total amino acid content was considered instead of its absolute concentration, a consistent pattern was evident (Fig. 4).
Table 3. Free amino acid content in S. pyogenes cells treated with the most active inhibitor of P5C reductase, compound 08.
| aa | Untreated control | 250 μM compound 08 | 500 μM compound 08 | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| μmol (g fw)−1 | % | μmol (g fw)−1 | % | μmol (g fw)−1 | % | |
| Asp | 2.76 ± 0.42 | 12.0 | 1.45 ± 0.01 | 7.2 | 1.48 ± 0.04 | 6.9 |
| Glu | 5.99 ± 0.21 | 26.1 | 2.91 ± 0.14 | 14.4 | 3.97 ± 0.12 | 18.4 |
| Asn | 0.18 ± 0.02 | 0.8 | 0.22 ± 0.01 | 1.1 | 0.23 ± 0.02 | 1.1 |
| Unknown | 3.24 ± 0.06 | 14.1 | 2.25 ± 0.02 | 11.2 | 2.27 ± 0.06 | 10.5 |
| Ser | 0.38 ± 0.03 | 1.7 | 0.41 ± 0.02 | 2.0 | 0.33 ± 0.01 | 1.5 |
| Gln | 3.09 ± 0.19 | 13.4 | 4.56 ± 0.09 | 22.6 | 4.81 ± 0.24 | 22.4 |
| His | 0.92 ± 0.09 | 4.0 | 1.00 ± 0.02 | 5.0 | 0.93 ± 0.13 | 4.3 |
| Arg | 0.21 ± 0.06 | 0.9 | 0.33 ± 0.06 | 1.6 | 0.22 ± 0.03 | 1.0 |
| Gly | 0.59 ± 0.07 | 2.6 | 1.71 ± 0.04 | 8.5 | 1.73 ± 0.10 | 8.0 |
| Thr | 1.20 ± 0.16 | 5.2 | 1.66 ± 0.04 | 8.3 | 1.75 ± 0.08 | 8.1 |
| Ala | 0.12 ± 0.01 | 0.5 | 0.09 ± 0.02 | 0.4 | 0.10 ± 0.01 | 0.5 |
| Tyr | 0.47 ± 0.02 | 2.1 | 0.48 ± 0.02 | 2.4 | 0.48 ± 0.04 | 2.2 |
| Trp + Met | 0.32 ± 0.04 | 1.4 | 0.24 ± 0.06 | 1.2 | 0.36 ± 0.04 | 1.7 |
| Val | 0.70 ± 0.09 | 3.0 | 0.78 ± 0.03 | 3.9 | 0.68 ± 0.02 | 3.1 |
| Phe | 0.61 ± 0.05 | 2.7 | 0.67 ± 0.02 | 3.3 | 0.76 ± 0.05 | 3.5 |
| Iso | 0.24 ± 0.01 | 1.1 | 0.24 ± 0.00 | 1.2 | 0.26 ± 0.02 | 1.2 |
| Leu | 0.41 ± 0.02 | 1.8 | 0.39 ± 0.00 | 2.0 | 0.43 ± 0.03 | 2.0 |
| Orn | 0.22 ± 0.03 | 0.9 | 0.04 ± 0.01 | 0.2 | 0.06 ± 0.02 | 0.3 |
| Lys | 0.68 ± 0.08 | 3.0 | 0.30 ± 0.04 | 1.5 | 0.35 ± 0.03 | 1.6 |
| Pro | 0.63 ± 0.03 | 2.8 | 0.41 ± 0.02 | 2.0 | 0.34 ± 0.01 | 1.6 |
| All | 22.98 ± 0.86 | 100.0 | 20.10 ± 0.38 | 100.0 | 21.52 ± 1.50 | 100.0 |
Amino acid pools were quantified by reverse-phase HPLC following derivatization with oPDA; Pro and total amino acid content were measured by the ninhydrin method. Cys was undetectable. Mean values ± SE over 4 replicates are reported
Fig. 4.
Effect of the treatment with increasing concentrations of compounds 08 on free proline concentration in S. pyogenes cells. Data were expressed as percent of total amino acid content, and are mean ± SE over 5 replications obtained in two independent experiments
Conclusions and perspectives
To the best of our knowledge, this is the first paper considering the possibility to affect pathogen growth by proline starvation. This aim was pursued by inhibiting P5C reductase, the enzyme at the converging point of the two main metabolic routes leading to proline biosynthesis in bacteria. Several substituted phenyl derivatives of compound 01 were found to completely suppress enzyme activity in vitro at micromolar concentrations, and the mode of action of the most active compounds strengthened the possibility that a block of proline synthesis may occur in vivo as a consequence. Experiments performed with the human pathogen S. pyogenes indeed showed both a reduction of bacterial growth and a decrease of the intracellular level of free proline. Such effects were found at concentrations largely exceeding those able to inhibit the enzyme in vitro, most likely due to poor membrane permeability. Moreover, the intracellular level of other amino acids than proline was found to be reduced, therefore, suggesting that further targets may occur. It has in fact been reported that bisphosphonates inhibit the activity of several enzymes, including some known antibacterial targets, like 1-deoxy-d-xylulose-5-phosphate reductoisomerase [E.C. 1.1.1.267], undecaprenyl diphosphate synthase [E.C. 2.5.1.31], geranyltranstransferase [E.C. 2.5.1.10] and farnesyltranstransferase [E.C. 2.5.1.29] (Guo et al. 2007). The MEP/DOXP pathway is lacking in S. pyogenes (http://kegg.jp/kegg-bin/show_pathway?spg00900), but the latter enzymes are present, and their inhibition may concur in determining the observed cytostatic effects. Instead of being a drawback, the occurrence of multiple targets would be a positive feature for an antibiotic, as it lowers the risk of resistance selection. In any case, the present work clearly established the possibility of starving cells for proline through the treatment with P5C reductase inhibitors. A structure-activity relationship analysis is currently in progress based upon these results in order to identify new derivatives that may retain high effectiveness against the bacterial enzyme while coupling it with improved permeability across the biological membrane. The attainment of this goal would provide us with new leads and increase the possibility of developing effective therapeutics targeting proline biosynthesis.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the University of Ferrara within the frame of the projects FAR2009-2010 Approcci biotecnologici per un incremento della sostenibilità della produzione agro-zootecnica. Davide Petrollino gratefully acknowledges an applied research fellowship from Spinner Consortium, Emilia Romagna Region (prot. N. 626/09).
Abbreviations
- IC50
Concentration causing 50% inhibition of enzyme activity
- LC50
Concentration causing 50% inhibition of growth rate
- OCD
Ornithine cyclodeaminase
- P5C
δ1-Pyrroline-5-carboxylic acid
Footnotes
Electronic supplementary material: The online version of this article (doi: 10.1007/s00726-011-0970-7) contains supplementary material, which is available to authorized users.
Contributor Information
Giuseppe Forlani, Email: flg@unife.it, Department of Biology and Evolution, University of Ferrara, via L. Borsari 46, 44100 Ferrara, Italy.
Davide Petrollino, Department of Biology and Evolution, University of Ferrara, via L. Borsari 46, 44100 Ferrara, Italy.
Massimo Fusetti, Department of Biology and Evolution, University of Ferrara, via L. Borsari 46, 44100 Ferrara, Italy.
Letizia Romanini, Department of Biology and Evolution, University of Ferrara, via L. Borsari 46, 44100 Ferrara, Italy.
Bogusław Nocek, Biosciences Division, Argonne National Laboratory, Midwest Center for Structural Genomics, 9700 South Cass Avenue, Argonne, IL 60439, USA.
Andrzej Joachimiak, Biosciences Division, Argonne National Laboratory, Midwest Center for Structural Genomics, 9700 South Cass Avenue, Argonne, IL 60439, USA.
Łukasz Berlicki, Department of Bioorganic Chemistry, Faculty of Chemistry, Wrocław University of Technology, Wybrzezże Wyspiańskiego 27, 50-370 Wrocław, Poland.
Paweł Kafarski, Department of Bioorganic Chemistry, Faculty of Chemistry, Wrocław University of Technology, Wybrzezże Wyspiańskiego 27, 50-370 Wrocław, Poland.
References
- Aral B, Kamoun P. The proline biosynthesis in living organisms. Amino Acids. 1997;13:189–217. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cascante M, Boros LG, Comin-Anduix B, de Atauri P, Centelles JJ, Lee PW. Metabolic control analysis in drug discovery and disease. Nat Biotechnol. 2002;20:243–249. doi: 10.1038/nbt0302-243. [DOI] [PubMed] [Google Scholar]
- Chen LF, Chopra T, Kaye KS. Pathogens resistant to antibacterial agents. Infect Dis Clin North Am. 2009;23:817–845. doi: 10.1016/j.idc.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Culham DE, Dalgado C, Gyles CL, Mamelak D, MacLellan S, Wood JM. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology. 1998;144:91–102. doi: 10.1099/00221287-144-1-91. [DOI] [PubMed] [Google Scholar]
- Cunin R, Glansdorff N, Piérard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empadinhas N, da Costa MS. Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int Microbiol. 2008;11:151–161. [PubMed] [Google Scholar]
- Engel LS. The dilemma of multidrug-resistant Gram-negative bacteria. Am J Med Sci. 2010;340:232–237. doi: 10.1097/MAJ.0b013e3181e939c3. [DOI] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney DJ. Probit analysis. 3rd. Cambridge University Press; Cambridge: 1971. [Google Scholar]
- Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani G, Giberti S, Berlicki Ł, Petrollino D, Kafarski P. Plant P5C reductase as a new target for aminomethylenebisphosphonates. J Agric Food Chem. 2007;55:4340–4347. doi: 10.1021/jf0701032. [DOI] [PubMed] [Google Scholar]
- Forlani G, Occhipinti A, Berlicki Ł, Dzie˛dzioła G, Wieczorek A, Kafarski P. Tailoring the structure of aminophosphonates to target plant P5C reductase. J Agric Food Chem. 2008a;56:3193–3199. doi: 10.1021/jf800029t. [DOI] [PubMed] [Google Scholar]
- Forlani G, Pavan M, Gramek M, Kafarski P, Lipok J. Biochemical bases for a widespread tolerance of cyanobacteria to the phosphonate herbicide glyphosate. Plant Cell Physiol. 2008b;49:443–456. doi: 10.1093/pcp/pcn021. [DOI] [PubMed] [Google Scholar]
- Forlani G, Prearo V, Wieczorek D, Kafarski P, Lipok J. Polyphosphonate degradation by Spirulina spp: a cyanobacterial biofilter for the removal of anticorrosive polyphosphonates from wastewater. Enzyme Microb Tech. 2011;48:299–305. doi: 10.1016/j.enzmictec.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Ge M, Pan XM. The contribution of proline residues to protein stability is associated with isomerization equilibrium in both unfolded and folded states. Extremophiles. 2009;13:481–489. doi: 10.1007/s00792-009-0233-7. [DOI] [PubMed] [Google Scholar]
- Goodman JL, Wang S, Alam S, Ruzicka FJ, Frey PA, Wedekind JE. Ornithine cyclodeaminase: Structure, mechanism of action, and implications for the mu-crystallin family. Biochemistry. 2004;43:13883–13891. doi: 10.1021/bi048207i. [DOI] [PubMed] [Google Scholar]
- Guo RT, Cao R, Liang PH, Ko TP, Chang TS, Hudock MP, Jeng WY, Chen CKM, Zhang Y, Song Y, Kuo CJ, Yin F, Oldfield E, Wang AHJ. Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases. Proc Natl Acad Sci USA. 2007;104:10022–10027. doi: 10.1073/pnas.0702254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydel SE. Extensively drug-resistant tuberculosis: a sign of the times and an impetus for antimicrobial discovery. Pharmaceuticals (Basel) 2010;3:2268–2290. doi: 10.3390/ph3072268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MJ, Kern AC, Middleton RF, Worthington HEC. Roche Susceptibility Test (RST) medium, a defined formulation for susceptibility testing. II. Manufacture, use and stability. J Microbiol Methods. 1984;2:215–220. [Google Scholar]
- Höper D, Völker U, Hecker M. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J Bacteriol. 2005;187:2810–2826. doi: 10.1128/JB.187.8.2810-2826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G, Horwitz MA. Inhibition of Mycobacterium tuberculosis glutamine synthetase as a novel antibiotic strategy against tuberculosis: demonstration of efficacy in vivo. Infect Immun. 2003;71:456–464. doi: 10.1128/IAI.71.1.456-464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton CA, Perugini MA, Gerrard JA. Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol Biosyst. 2007;3:458–465. doi: 10.1039/b705624a. [DOI] [PubMed] [Google Scholar]
- Lehmann S, Funck D, Szabados L, Rentsch D. Proline metabolism and transport in plant development. Amino Acids. 2010;39:949–962. doi: 10.1007/s00726-010-0525-3. [DOI] [PubMed] [Google Scholar]
- Liu JS, Cheng WC, Wang HJ, Chen YC, Wang WC. Structure-based inhibitor discovery of Helicobacter pylori dehydroquinate synthase. Biochem Biophys Res Commun. 2008;373:1–7. doi: 10.1016/j.bbrc.2008.05.070. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Tartari A, Cattivelli L, Forlani G. GABA metabolism during cold acclimation and freezing in barley and wheat. J Exp Bot. 2006;57:3755–3766. doi: 10.1093/jxb/erl141. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Inatsu S, Mizote T, Nagata Y, Aoyama K, Fukuda Y, Nagata K. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29:9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- Nathan C. Antibiotics at the crossroads. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- Nocek B, Chang C, Li H, Lezondra L, Holzle D, Collart F, Joachimiak A. Crystal structures of δ1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J Mol Biol. 2005;354:91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania R, Brown ED. Small and lethal: searching for new antibacterial compounds with novel modes of action. Biochem Cell Biol. 2008;86:111–115. doi: 10.1139/O08-011. [DOI] [PubMed] [Google Scholar]
- Sakoulas G, Moellering RC., Jr Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46:S360–S367. doi: 10.1086/533592. [DOI] [PubMed] [Google Scholar]
- Silber AM, Colli W, Ulrich H, Alves MJ, Pereira CA. Amino acid metabolic routes in Trypanosoma cruzi: possible therapeutic targets against Chagas' disease. Curr Drug Targets Infect Disord. 2005;5:53–64. doi: 10.2174/1568005053174636. [DOI] [PubMed] [Google Scholar]
- Snedecor GW, Cochran WG. Statistical methods. 8th. The Iowa State University Press; Ames: 1898. [Google Scholar]
- Tan S, Evans R, Singh B. Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids. 2006;30:195–204. doi: 10.1007/s00726-005-0254-1. [DOI] [PubMed] [Google Scholar]
- Takagi H. Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol. 2008;81:211–223. doi: 10.1007/s00253-008-1698-5. [DOI] [PubMed] [Google Scholar]
- Williams I, Frank L. Improved chemical synthesis and enzymatic assay of δ1-pyrroline-5-carboxylic acid. Anal Biochem. 1975;64:85–97. doi: 10.1016/0003-2697(75)90408-x. [DOI] [PubMed] [Google Scholar]
- Ziebart KT, Dixon SM, Avila B, El-Badri MH, Guggenheim KG, Kurth MJ, Toney MD. Targeting multiple chorismateutilizing enzymes with a single inhibitor: validation of a three-stage design. J Med Chem. 2010;53:3718–3729. doi: 10.1021/jm100158v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.