Abstract
The repair of DNA double-strand breaks by recombination is key to the maintenance of genome integrity in all living organisms. Recombination can however generate mutations and chromosomal rearrangements, making the regulation and the choice of specific pathways of great importance. In addition to end-joining through non-homologous recombination pathways, DNA breaks are repaired by two homology-dependent pathways that can be distinguished by their dependence or not on strand invasion catalysed by the RAD51 recombinase. Working with the plant Arabidopsis thaliana, we present here an unexpected role in recombination for the Arabidopsis RAD51 paralogues XRCC2, RAD51B and RAD51D in the RAD51-independent single-strand annealing pathway. The roles of these proteins are seen in spontaneous and in DSB-induced recombination at a tandem direct repeat recombination tester locus, both of which are unaffected by the absence of RAD51. Individual roles of these proteins are suggested by the strikingly different severities of the phenotypes of the individual mutants, with the xrcc2 mutant being the most affected, and this is confirmed by epistasis analyses using multiple knockouts. Notwithstanding their clearly established importance for RAD51-dependent homologous recombination, XRCC2, RAD51B and RAD51D thus also participate in Single-Strand Annealing recombination.
Author Summary
The repair of DNA double-strand breaks by recombination is key to the maintenance of genome integrity in all living organisms. Recombination can however generate mutations and chromosomal rearrangements, making the regulation and the choice of specific pathways of great importance. Through modulation of the activity of the recombinase RAD51, the RAD51 paralogue proteins play key roles in the regulation of recombination. Considerable advances have been made in understanding of the RAD51 paralogue proteins and their roles in mediating RAD51-mediated homologous recombination, however very little is known of possible roles that they may have in other recombination pathways. Working with the plant, Arabidopsis thaliana, we show here major roles for three RAD51 paralogues in RAD51-independent single-strand annealing recombination. Notwithstanding their clearly established importance for RAD51-dependent homologous recombination, XRCC2, RAD51B and RAD51D thus also participate in Single-Strand Annealing recombination.
Introduction
DNA double-strand breaks (DSB) are produced by ionizing radiation, free radicals derived from metabolism, DNA crosslinking reagents and during DNA replication [1], [2]. DSB can lead to mutations and rearrangements and/or loss of chromosomes, causing tumorigenesis or cell death. DSB must be repaired to maintain genome integrity, and this is carried out by end-joining through non-homologous recombination or by homologous recombination, which implicates DNA sequence homology of the recombining molecules (for reviews, see [3], [4]). The pathways that utilize homology for repair can be distinguished by their dependence or not on strand-invasion catalysed by the RAD51 recombinase (or DMC1 in meiosis): gene conversion homologous recombination (HR) is RAD51-dependent while single-strand annealing (SSA) is RAD51-independent [3].
RAD51-dependent HR is an error-free DSB repair mechanism involving the use of a homologous template for restoration of the original sequence. It involves resection of the 5′-ended DNA strands at the DSB, generating 3′ single-stranded DNA overhangs that are bound by replication protein A (RPA). Assisted by mediator proteins, RAD51 displaces RPA and forms a helical nucleofilament on the exposed single-stranded DNA (ssDNA) flanking the DSB. This nucleofilament performs the homology search and catalyses invasion of the homologous template DNA, following which the invading 3′ ends are extended through DNA synthesis. The joint recombination intermediate is resolved to separate the recombining DNA molecules and thus restore chromosome integrity (for a review, see [3]).
In addition to RAD51 and the meiosis-specific DMC1, a number of RAD51 paralogue proteins have been described in a variety of organisms. These share 20% to 30% homology with RAD51 and presumably arose by gene duplication and evolved new functions [5]. They clearly play key roles in DNA repair through HR, but their exact functions are not fully understood (for reviews, see [6]–[8]).
Two S. cerevisiae RAD51 paralogues, RAD55 and RAD57, form a heterodimeric complex which associates with the RAD51 nucleoprotein filament, stabilising it against disruption by the SRS2 antirecombinase [9]. Recent work has characterized novel yeast RAD51 paralogues: Shu1, Shu2, Csm2 and Psy3, components of the “suppresses sgs1 hydroxyurea sensitivity” (SHU or PCSS) complex which also promotes RAD51 filament assembly and its stability through counteracting the antirecombination activity of the SRS2 and SGS1 helicases [10]–[17]. Fission yeast has homologues of Shu1, Shu2 and Psy3 (Rlp1, Sws1 and Rdl1) and SWS1 and SWSAP1 are members of a human SHU complex [11], [12], [18].
Five RAD51 paralogues have been identified in animals and plants: RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 (for reviews, see [7], [19], [20]). Animal cells defective in any of the RAD51 paralogues are hypersensitive to DNA cross-linking agents, such as Cisplatin and Mitomycin C, and show spontaneous chromosomal aberrations [21]–[27]. Mouse xrcc2, rad51b, rad51c and rad51d mutants are embryonic lethal [28]–[31]. In contrast, all five RAD51 paralogues Arabidopsis mutants grow and develop normally and rad51c and xrcc3 mutant plants are sterile due to recombination defects [32], [33].
Two-hybrid and co-immunoprecipitation studies have shown that the five RAD51 paralogues form two major complexes: RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) and RAD51C-XRCC3 (CX3), as well as RAD51B-RAD51C (BC) and RAD51D-XRCC2 (DX2) sub-complexes [5], [7], [8], [34]–[40]. RAD51 paralogue complexes act at both early and late stages of the recombinational repair process, although their exact roles remain to be identified [32], [41]–[48]. The early role of RAD51 paralogues in HR is to promote formation and stabilization of RAD51 nucleoprotein filament (reviewed by [6]–[8], [19]), very probably through counteracting disruption of the filament by helicases [9]–[14]. Recent work shows that the BCDX2 complex, and not the CX3 complex, is responsible for RAD51 recruitment at DNA damage sites in human cells [43]. After RAD51-mediated strand invasion, the RAD51 paralogues influence gene conversion tract length [42], [47] and have been linked to Holliday junction (HJ) resolvase activity [45], [46]. In addition, RAD51 paralogues can bind Y-shaped replication-like intermediates and synthetic HJ, in accordance with a role for RAD51 paralogues in repair during DNA replication and in resolution of HR intermediary structures [49], [50].
The second main pathway using homology for repair, single-strand annealing (SSA), promotes recombination between tandemly repeated DNA sequences flanking a DSB. SSA does not involve DNA-strand invasion and has been shown to be independent of RAD51 [51]–[54]. After bidirectional 5′-3′ resection of the DSB ends, the exposed complementary sequences anneal. Subsequent removal of non-homologous 3′-ended ssDNA tails, filling-in of any single-strand gaps and ligation completes the process. The SSA recombination pathway thus leads to deletion of the interstitial DNA sequence lying between the repeats and one of the repeated homologous sequences (for reviews, see [3], [55]).
Little is known about possible involvement of the RAD51 paralogues in RAD51-independent SSA. Yeast Rad55 and Rad57 are not required for SSA in a plasmid assay [51] or spontaneous direct repeat recombination [56], [57] and a recent study has shown that absence of Rad55, Csm2 or Psy3 result in increased SSA recombination at a direct repeat chromosomal locus in yeast [15]. In Arabidopsis, RAD51, RAD51C and XRCC3 are not required for SSA, although a mild reduction in the efficiency of SSA was reported in the rad51c mutant [53].
In this study, we describe an unexpected role in the SSA pathway for Arabidopsis XRCC2, RAD51B and RAD51D, highlighting for the first time a function of these three RAD51 paralogues in RAD51-independent SSA recombination.
Results
XRCC2 is required for SSA recombination
Although XRCC2 is known to be involved in RAD51-dependent homologous recombination in both vertebrates and in plants [6], [7], [19], [43], [44], its potential role in RAD51-independent SSA has not been tested.
SSA recombination was monitored in xrcc2 mutant Arabidopsis thaliana plants using the well-characterised DGU.US recombination reporter locus - consisting of an I-SceI restriction site flanked by 3′ and 5′ truncated copies of the β-glucuronidase gene (GUS) in direct orientation and with an overlap of 557 bp (Figure 1A; [58]). Cleavage of the I-SceI site induces recombination between the flanking GUS sequences and the resulting functional GUS gene is scored histochemically as blue somatic spots. I-SceI induced recombination at this tester locus has been shown not to depend upon RAD51 [53].
Figure 1. I-SceI induced DGU.US recombination depends upon XRCC2.
(A) Schematic map of the recombination substrate DGU.US. (B) β-glucuronidase assay of 14 day-old seedlings grown with or without induction of I-SceI by β-estradiol clearly shows reduced numbers of blue recombinant GUS+ sectors in the xrcc2 mutant. (C) Quantification of recombination events confirms the role of XRCC2. Bars are mean values ± standard errors. * Significant difference (p = 0.036, Mann-Whitney test). (D) Frequency distributions of recombinant spot numbers per plant of 3 independent WT and xrcc2 T2 lines grown in the presence of β-estradiol.
We introduced the GUS recombination reporter locus into xrcc2 mutant and wild-type (WT) plants through crossing and transformed these DGU.US lines with an inducible I-SceI expression cassette (Materials and Methods). Three independent transformants (T2 lines) were selected for each genotype, each with a single insertion site of the I-SceI cassette. Seeds of these lines were plated onto medium containing hygromycin (in order to select plants carrying the I-SceI cassette), in the presence or absence of I-SceI expression inductor (β-estradiol), and numbers of blue GUS+ spots counted after 14 days of growth (Figure 1). Induction of I-SceI expression by β-estradiol treatment in WT plants resulted in a considerable increase of numbers of recombinant blue spots/sectors (Figure 1B and C). In contrast, expression of I-SceI had very little effect on numbers of blue spots in xrcc2 mutant plants, with means of 5.9 spots per plant in the presence of β-estradiol, and 4.1 in its absence. Repetition of these analyses with two other independent I-SceI transformant lines yielded similar results (Figure 1D). XRCC2 thus clearly plays an important role in the SSA recombination pathway.
The histochemical GUS assay is an indirect measure of somatic recombination and we thus carried out Southern analyses to demonstrate directly that the decrease of number of GUS+ spots in xrcc2 mutant plants is due to a failure of restoration of the GUS gene. Southern analysis was carried out on SacI-digested genomic DNA of WT and xrcc2 mutant plants (induced or not by β-estradiol). In DGU.US lines, restoration of the GUS gene results in deletion of the repeated sequence, including the inserted I-SceI site. In DNA of WT plants, the reconstituted GUS gene is clearly visible as a band at the expected size (2.5 kb) after induction of I-SceI expression, but not in its absence (Figure 2, lanes 3 and 7). Treatment of the genomic DNA samples with I-SceI in vitro prior to electrophoresis confirms that the 2.5 kb fragment has lost its I-SceI site (Figure 2, lane 9), consistent with elimination of the I-SceI restriction site during the restoration of the marker gene. No restored GUS gene was detected in the xrcc2 mutant line (Figure 2, lanes 8 and 10). This molecular analysis is thus fully consistent with the results of the β-glucuronidase assay and confirms the implication of the XRCC2 protein in the SSA recombination pathway.
Figure 2. Molecular confirmation of recombination in WT, but not xrcc2 mutant plants.

Schematic representation of the GU.US recombination tester locus (A) and Southern analysis (B) of DNA from plants grown in the absence (lanes 1 to 6) or presence of β-estradiol (lanes 7 to 10), digested with SacI (lanes 1,3,4,7,8) or SacI plus I-SceI (lanes 2,5,6,9,10). The blot was hybridized with a GUS-specific probe as indicated in panel (A). The recombined GUS gene has lost its I-SceI site and is seen as a single 2.5 kb SacI fragment only in DNA from WT plants grown in presence of β-estradiol (lanes 7 and 9). Col0: WT plants of Columbia ecotype.
We note also the presence of a 3.2 kb band in the SacI+I-SceI digested DNA from WT plants (Figure 2, lane 9). That this I-SceI resistant band is due to in planta rejoining of I-SceI breaks through end-joining recombination was verified by PCR amplification and DNA sequencing. Approximately 10% of the sequences carried a mutation at the I-SceI restriction site. DNA sequencing showed that these result mostly from small deletions (Figure S1). As previously described [59]–[63], these events can be ascribed to end-joining exploiting the presence of microhomologies either side of the I-SceI cleavage site.
XRCC2 function in spontaneous DGU.US recombination does not depend upon RAD51 activity
Although minor, the DGU.US recombination analyses shown in Figure 1C also showed a difference in numbers of blue spots between WT and xrcc2 plants in the absence of β-estradiol. To check whether this is due to differences in spontaneous recombination rates or to leakiness of the inducible I-SceI cassette (or both), we monitored recombination in xrcc2 mutant and WT plants with the same DGU.US locus (at the same location in genome), but which do not carry I-SceI (Figure 3). This analysis showed a reduction of number of recombinant spots in the absence of the I-SceI cassette, for both WT and xrcc2 plants (from 13.24 to 5.46 and 4.10 to 0.36 spots per plant, respectively; Figures 1C and 3A), confirming the presence of some leakiness in expression of the I-SceI inducible promoter in the absence of β-estradiol. In the absence of the I-SceI cassette, mean numbers of blue spots per plant were however still significantly (15-fold) reduced in xrcc2 mutants (0.36 ; s.e.m = 0.08) compared with WT controls (5.46 ; s.e.m = 0.34 ; Figure 3). An independent repetition of this experiment confirmed these results (Table 1). XRCC2 is thus clearly involved in spontaneous recombination of the DGU.US substrate.
Figure 3. Spontaneous DGU.US recombination is reduced in the xrcc2 mutant.
A significant reduction in spontaneous recombination rate is observed in xrcc2 mutant compared to WT plants. (A) Mean values ± standard errors of the means. *** p<0.0001 (Mann-Whitney test). (B) Frequency distributions.
Table 1. Spontaneous DGU.US recombination in xrcc2 mutant and in wild-type plants.
| Experiment | n | N | m ± SEM | Ratio xrcc2/WT | |
| 1 | WT | 50 | 273 | 5.46±0.34 | |
| xrcc2 | 50 | 18 | 0.36±0.08 | 0.066 | |
| 2 | WT | 50 | 310 | 6.20±0.51 | |
| xrcc2 | 50 | 19 | 0.38±0.09 | 0.061 |
Recombination in the mutants and WT were compared using non-parametric statistical analysis (Mann–Whitney test). Differences between xrcc2 and WT are highly significant (p<0.0001) in both cases. n, the number of plantlets screened; N, the total number of blue spots (recombination events) ; m ± SEM, the mean number of recombination events per plant ± standard error of the mean.
As mentioned above, I-SceI induced recombination at the DGU.US locus has been shown to be RAD51-independent [53]. This has not however been confirmed for spontaneous recombination, for which different mechanisms can be envisaged - single-strand annealing, intermolecular synthesis-dependent strand annealing, break-induced replication [62], [64], [65]. We thus tested the RAD51-dependence of spontaneous recombination at DGU.US by expressing the dominant-negative RAD51-GFP fusion protein [66]. Plants were transformed with the RAD51-GFP fusion protein construct and three T2 lines each with a single insertion (RAD51-GFP plants) were selected and their rad51 mutant phenotype tested by verification of sensitivity to the cross-linking agent, Mitomycin C (MMC) (Figure S2). Wild-type plants are not sensitive to the MMC dose used (2% sensitive plants), in contrast to the segregating RAD51-GFP population, in which 76.9% are sensitive. PCR genotyping confirmed that all of the MMC-sensitive and none of the MMC-resistant T2 plants carry RAD51-GFP. Presence of RAD51-GFP is thus perfectly correlated with MMC-sensitivity, confirming the dominant-negative inhibition of RAD51 by the fusion protein [66]. We then tested spontaneous DGU.US recombination in the RAD51-GFP plants (Figure 4). No significant difference was observed in numbers of GUS+ recombinant spots between control and RAD51-GFP plants (Mann-Whitney test) clearly confirming that spontaneous recombination of the DGU.US substrate does not depend upon RAD51 activity.
Figure 4. Spontaneous DGU.US recombination is RAD51-independent.

No significant effect on spontaneous recombination rate was observed in three independent transformants carrying the dominant-negative RAD51-GFP construct. Bars are mean values ± standard errors.
XRCC2, RAD51B and RAD51D have non-epistatic functions in the SSA pathway
XRCC2 is one of five RAD51 paralogue proteins, all of which play important roles in recombination [7]. Given the function of XRCC2 in the RAD51-independent SSA pathway presented above, we also tested for evidence of roles of the other RAD51 paralogues, RAD51B and RAD51D, in this pathway. We thus crossed the DGU.US recombination reporter locus into rad51b and rad51d mutant plants and monitored spontaneous SSA recombination at DGU.US in rad51b and rad51d mutants. Although less pronounced than the 15-fold reduction observed in xrcc2 plants, numbers of spontaneous recombination events are also reduced in rad51b and rad51d mutants (respectively 4.6-fold and 3.4-fold; Figure 5; Table 2) clearly establishing roles for RAD51B and RAD51D in the SSA pathway.
Figure 5. Individual and combined effects of xrcc2, rad51b and rad51d on spontaneous DGU.US recombination.
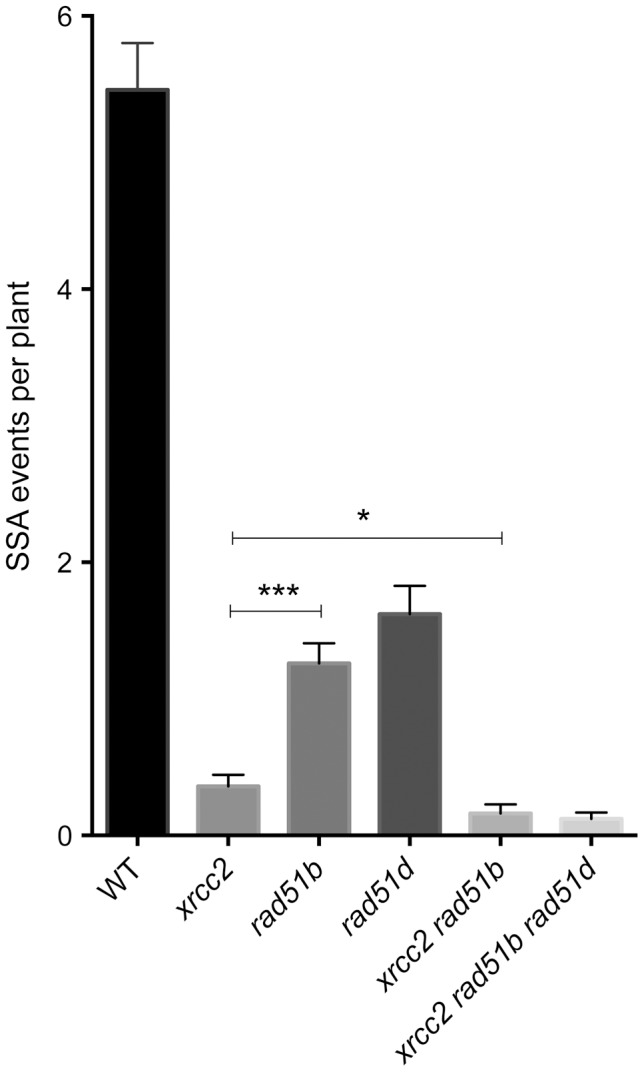
Significant reductions in spontaneous recombination rate are observed in xrcc2, rad51b and rad51d mutants, and the severities of the reductions differ between these single mutants. A further significant reduction is seen in xrcc2 rad51b mutant plants. The triple xrcc2 rad51b rad51d mutant shows a further reduction, but this does not differ significantly from that observed in the xrcc2 rad51b plants. Bars are mean values ± standard errors. * 0.05<p<0.0001; *** p<0.0001 (Mann-Whitney test).
Table 2. Spontaneous DGU.US recombination in wild-type, rad51b, rad51d, double and triple mutants.
| Experiment | n | N | m ± SEM | Ratio mutant/WT | |
| 1 | WT | 50 | 273 | 5.46±0.34 | |
| rad51b | 50 | 63 | 1.26±0.15 | 0.231 | |
| rad51d | 50 | 81 | 1.62±0.21 | 0.297 | |
| xrcc2, rad51b | 50 | 8 | 0.16±0.07 | 0.023 | |
| xrcc2, rad51b, rad51d | 50 | 6 | 0.12±0.05 | 0.022 | |
| 2 | WT | 50 | 310 | 6.20±0.51 | |
| rad51b | 50 | 62 | 1.24±0.18 | 0.200 | |
| rad51d | 50 | 92 | 1.84±0.23 | 0.297 | |
| xrcc2, rad51b | 50 | 6 | 0.12±0.05 | 0.019 | |
| xrcc2, rad51b, rad51d | 50 | 1 | 0.02±0.02 | 0.003 |
Recombination in the mutants and WT was compared using non-parametric statistical analysis (Mann–Whitney test). Differences between each mutant and corresponding WT are highly significant (p<0.0001). n, the number of plantlets screened; N, the total number of blue spots (recombination events); m ± SEM, the mean number of recombination events per plant ± standard error of the mean.
Epistasis relationships in SSA recombination between the three RAD51 paralogue genes were tested in xrcc2 rad51b double and xrcc2 rad51b rad51d triple mutants. Spontaneous SSA recombination was significantly less efficient in xrcc2 rad51b double mutants that in the corresponding single mutants (p<0.02) (Figure 5). A slight further reduction in numbers of blue spots per plant was observed in the triple xrcc2 rad51b rad51d mutants with respect to the double xrcc2 rad51b mutant, but the difference is not significant.
To confirm at the molecular level the results of the GUS assays, we transformed rad51b, rad51d, xrcc2 rad51b and xrcc2 rad51b rad51d mutant plants with the inducible I-SceI expression cassette. Southern analysis of recombination was carried out on β-estradiol induced T2 plants. As expected, the 2.5 kb fragment of the recombination product is only detected in the WT, confirming the GUS assay data (Figure 6).
Figure 6. Molecular confirmation of recombination defects in rad51b, rad51d, xrcc2 rad51b and xrcc2 rad51b rad51d mutants.
(A) Southern analysis of DNA from rad51b and rad51d mutant plants grown in the presence of β-estradiol, digested with SacI (lanes 1,3,5,7) or SacI plus I-SceI (lanes 2,4,6,8). (B) Southern analysis of DNA from xrcc2 rad51b and xrcc2 rad51b rad51d mutant plants grown in the presence of β-estradiol, digested with SacI. The blots were hybridized with a GUS-specific probe. The recombined GUS gene has lost its I-SceI site and is seen as a 2.5 kb band only in DNA from WT plants grown in presence of β-estradiol (A, lanes 3 and 4; B, lane 2). Col0: WT plants of Columbia ecotype.
Arabidopsis XRCC2, RAD51B and RAD51D thus play roles in SSA recombination pathway and these roles are non-epistatic, at least for XRCC2 and RAD51B.
Discussion
The roles of RAD51 paralogues in RAD51-dependent recombination have been the subject of considerable interest in recent years [6]–[8], [19]. Little is known however of possible roles in RAD51-independent SSA recombination. In Arabidopsis, no effect was found on SSA in xrcc3 mutants and a barely statistically significant reduction observed in rad51c plants [53]. We show here the involvement of three RAD51 paralogues, XRCC2, RAD51B and RAD51D, in RAD51-independent single-strand annealing in Arabidopsis thaliana. XRCC2 plays a major role in this pathway with a striking reduction of I-SceI induced recombination and a 15-fold reduction in the number of spontaneous SSA events in its absence (Figures 1 and 3). Spontaneous SSA is also clearly reduced in rad51b and rad51d mutants (4.6-fold and 3.4-fold reduction respectively; Figure 5; Table 2), although less strongly than in xrcc2 mutants. The differing severity of the phenotypes of the three mutants is suggestive of individual roles for these proteins, and this is supported by epistasis analyses of double and triple mutant plants (Figure 5, Table 2). An alternative to a direct role of these proteins is that the presence of non-functional RAD51 nucleofilaments in these mutants which might block SSA. The lack of effect on SSA of RAD51-GFP (which forms foci at DSBs and is dominant-negative for GC/SDSA recombination) however argues against this interpretation. Data suggesting differing roles for individual RAD51 paralogues, or sub-complexes, can be found in a number of reports. Individual paralogue mutants in DT-40 cells show non-epistatic phenotypes [67] and biochemical analyses show specific roles for the sub-complexes [68]–[70]. In Arabidopsis, absence of XRCC2 and RAD51B, but not RAD51D, increases rates of meiotic crossing-over [44] and RAD51D appears to be the only RAD51 paralogue to be essential for telomere integrity in human cells [71]. A recent report shows opposing effects on cell-cycle regulation of the inhibition of XRCC3 and RAD51C in HeLa cells, with inhibition of XRCC3 eliciting checkpoint defects and inhibition of RAD51C inducing G2/M cell cycle arrest [48].
What can the roles of XRCC2, RAD51B and RAD51D be in the SSA pathway? The main steps of SSA are (1) bidirectional 5′ to 3′ resection of the DSB ends flanking a DSB, (2) annealing of exposed complementary sequences, (3) excision of non-homologous 3′-ended overhangs, (4) DNA synthesis and (5) ligation which restores two continuous strands [72], [73]. A role in the annealing step is suggested by the capacity of the human BCDX2 complex to catalyse annealing between single-strand DNAs in vitro [50]. This study also showed a high affinity of the BCDX2 complex for branched DNA structures, such as Y-shaped DNA, that result from this annealing between tandem repeats during single-strand annealing. Taken together, these results strongly suggest a role of XRCC2, RAD51B and RAD51D in the annealing of the two exposed repeat sequences on either side of the DSB.
Biochemical studies have identified two main complexes of the five RAD51 paralogue proteins in animal and plant cells: RAD51B-RAD51C-RAD51D-XRCC2 and RAD51C-XRCC3 [5], [7], [8], [34]–[40]. No self-assembly of individual RAD51 paralogues have been detected. Analysis of epistasis relationships of RAD51 paralogues in chicken DT-40 cells show that rad51b and rad51d are epistatic while xrcc3 rad51d double mutant cells exhibit an additive sensitivity to ionizing radiation [67], consistent with differential actions of two major complexes in cellular response to DNA damage. That the three RAD51 paralogues involved in SSA are components of the BCDX2 complex suggests this complex is the active species in SSA. However, the differing severity of the phenotypes of the xrcc2, rad51b and rad51d (and rad51c; [53]) mutants argues against the implication of the BCDX2 complex as such. The proposed structure of the complex also argues against being the active form in SSA, with protein-protein interaction studies showing that the four proteins are linked in the order: RAD51B-RAD51C-RAD51D-XRCC2 [35]. Absence of RAD51D should thus exclude XRCC2 from the complex, yet SSA in the xrcc2 mutants is significantly more affected than in rad51d (and similarly for rad51b versus rad51c). This argument also applies to the RAD51B-RAD51C and RAD51D-XRCC2 sub-complexes (for reviews, [7], [8]). Our data thus favour individual roles of XRCC2, RAD51B and RAD51D in single-strand annealing recombination.
The yeast RAD51 paralogues Rad55 and Rad57 are not required in SSA recombination in a plasmid-based assay [51] and a chromosomal assay shows that absence of Csm2, Psy3 (also RAD51 paralogues) or Rad55 favours SSA with respect to gene conversion recombination [15]. The description here of roles for XRCC2, RAD51B and RAD51D in the RAD51-independent SSA pathway thus highlights a difference in the roles of Arabidopsis and yeast RAD51 paralogues in the SSA pathway. Such a difference is also seen in the roles of RAD51 paralogues in meiotic recombination with psy3 mutants exhibiting a strong hypo-recombination in yeast [12], while absence of XRCC2 or RAD51B increases meiotic crossing-over in Arabidopsis [44].
In conclusion, we describe here an unexpected role in recombination for the Arabidopsis RAD51 paralogues XRCC2, RAD51B and RAD51D. The roles of these proteins are seen in spontaneous and in DSB-induced recombination at a tandem direct repeat recombination tester locus, both of which are unaffected by the absence of RAD51. Notwithstanding their clearly established importance for RAD51-dependent homologous recombination, these proteins thus also participate in RAD51-independent Single-Strand Annealing recombination.
Materials and Methods
Plant material
The Arabidopsis thaliana xrcc2, rad51b [74] and rad51d [44] mutants used in this work have been previously described. A triple xrcc2/xrcc2 rad51b/rad51b rad51d/rad51d mutant was crossed with the recombination tester DGU.US-1 line [58] and single, double and triple mutants homozygous for the DGU.US substrate were identified in the F2. Wild-type control plants come from the same crosses.
The I-SceI coding sequence [75] was placed under control of β-estradiol in the plasmid pMDC7 [76] by Gateway cloning. The resulting vector was transferred into Agrobacterium tumefaciens, and used to transform the plant lines utilising the floral dip method [77].
Growth conditions
Surface-sterilized seeds were stratified at 4°C for 2 days and grown in vitro on germination medium (0.8% w/v agar, 1% w/v sucrose and half-strength Murashige & Skoog salts (M0255; Duchefa Biochemie, Netherlands)) in a growth cabinet with a 16-h light/8-h dark cycle, at 23°C with 45–60% relative humidity.
The growth medium was supplemented with 170 µM 17-β-estradiol (E2758; Sigma-Aldrich) for induction of I-SceI expression.
Histochemical staining of GUS expression
Fourteen-day old seedlings grown under standard conditions (supplemented or not with 17-β-estradiol) were harvested and incubated in staining buffer (0.2% Triton X-100, 50 mM sodium phosphate buffer (pH 7.2), 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid; Biosynth), dissolved in N,N-dimethylformamide). Plants were infiltrated under vacuum for 15 min and incubated at 37°C overnight. The staining solution was then replaced with 70% ethanol to remove leaf pigments, and the blue spots were counted under a binocular microscope.
Mitomycin C treatment
Seeds were sown on plates containing fresh solid germination medium supplemented with 40 µM Mitomycin C (M0503; Sigma-Aldrich). The plates were then incubated for 20 days (23°C, 16-h light). A plant with 3 or more true leaves is considered resistant.
Plant DNA extraction and Southern analysis
DNA was prepared from seedlings as described previously [78] and 1.5 µg digested with 100 units of SacI or 25 units of I-SceI in a final volume of 200 µl for 15 h. Digested DNA samples were isopropanol precipitated, resuspended in TE, and electrophoresed in 0.8% agarose-TBE gel. Gel was blotted into a positively charged nylon membrane (Hybond-XL, Amersham Biosciences), which was hybridized in 0.5 M phosphate buffer, 7% w/v SDS, 1 mM EDTA (pH 8) and 1% BSA at 65°C. The DNA probes to the GUS gene (a PCR fragment amplified with 5′-TGGATCCCCGGGATCATCTACTTCTG and 5′-AGCCATGCACACTGATACTCTTCACTCC) and the actin gene (a PCR fragment amplified with 5′-GGCTCCTCTTAACCCAAAGG and 5′-TTACCTGCTGGAATGTGCTG) were labelled with [α-32P]dCTP using a random priming labelling kit (Megaprime DNA labelling system, Amersham) according to the manufacturer's instructions. Blots were washed with 0.5% SSC, 0.1% SDS solution at 65°C and imaged with a PhosphoImager (Bio-Rad Personal FX).
Supporting Information
I-SceI induced mutations in end-joining products of DGU.US repair. The unmodified sequence surrounding the I-SceI cut site of the DGU.US recombination tester locus is shown at the top of the alignment, with the I-SceI restriction site boxed and the cut-sites for each strand arrowed. Mutations are highlighted by gray boxes and the size of deletions (bp) is indicated at right. Flanking microhomologies presumably involved in the end-joining of I-SceI induced DSB are underlined.
(PDF)
Sensitivity to Mitomycin C in T2 Rad51-GFP plants. WT and three independent Rad51-GFP T2 transformants were tested for their sensitivity to the cross-linking agent MMC. The dominant-negative effect of the RAD51-GFP allele is clearly visible in the 3∶1 segregating MMC hypersensitivity of the plantlets. (A) photos of the plantlets and (B) quantitation of sensitive versus resistant plants.
(PDF)
Acknowledgments
We thank Holger Puchta for providing the I-SceI plasmid and DGU.US recombination tester. Members of the recombination group are thanked for their help and discussions.
Funding Statement
HS was the recipient of a PhD fellowship financed by Institut National des Sciences Biologiques (INSB/CNRS) and the Conseil régional d'Auvergne (PAVIRMA project). This work was financed by a European Union research grant (FP7-KBBE-2008-227190), the Centre National de la Recherche Scientifique, the Université Blaise Pascal, the Université d'Auvergne, and the Institut National de la Santé et de la Recherche Médicale. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, et al. (2000) The importance of repairing stalled replication forks. Nature 404: 37–41. [DOI] [PubMed] [Google Scholar]
- 2. Whitaker SJ (1992) DNA damage by drugs and radiation: what is important and how is it measured? European journal of cancer 28: 273–276. [DOI] [PubMed] [Google Scholar]
- 3. Heyer W-D, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annual review of genetics 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waterworth WM, Drury GE, Bray CM, West CE (2011) Repairing breaks in the plant genome: the importance of keeping it together. The New phytologist 192: 805–822. [DOI] [PubMed] [Google Scholar]
- 5. Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, et al. (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes & development 15: 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krejci L, Altmannova V, Spirek M, Zhao X (2012) Homologous recombination and its regulation. Nucleic acids research 40: 5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suwaki N, Klare K, Tarsounas M (2011) RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Seminars in cell & developmental biology 22: 898–905. [DOI] [PubMed] [Google Scholar]
- 8. Thacker J (2005) The RAD51 gene family, genetic instability and cancer. Cancer letters 219: 125–135. [DOI] [PubMed] [Google Scholar]
- 9. Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, et al. (2011) Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernstein KA, Reid RJD, Sunjevaric I, Demuth K, Burgess RC, et al. (2011) The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Molecular biology of the cell 22: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, et al. (2006) Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J 25: 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, et al. (2013) A new protein complex promoting the assembly of Rad51 filaments. Nature communications 4: 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shor E, Weinstein J, Rothstein R (2005) A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mankouri HW, Ngo HP, Hickson ID (2007) Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell 18: 4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godin S, Wier A, Kabbinavar F, Bratton-Palmer DS, Ghodke H, et al. (2013) The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic acids research 41: 4525–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. She Z, Gao ZQ, Liu Y, Wang WJ, Liu GF, et al. (2012) Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS Lett 586: 2306–2312. [DOI] [PubMed] [Google Scholar]
- 17. Tao Y, Li X, Liu Y, Ruan J, Qi S, et al. (2012) Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. J Biol Chem 287: 20231–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu T, Wan L, Wu Y, Chen J, Huang J (2011) hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem 286: 41758–41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bleuyard J-Y, Gallego ME, White CI (2006) Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA repair 5: 1–12. [DOI] [PubMed] [Google Scholar]
- 20. Karpenshif Y, Bernstein KA (2012) From yeast to mammals: recent advances in genetic control of homologous recombination. DNA Repair (Amst) 11: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Schärer OD, van Buul PPW, et al. (2002) Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic acids research 30: 2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, et al. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Molecular and cellular biology 21: 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, et al. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Molecular and cellular biology 20: 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson RD, Liu N, Jasin M (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401: 397–399. [DOI] [PubMed] [Google Scholar]
- 25. Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & development 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Molecular cell 1: 783–793. [DOI] [PubMed] [Google Scholar]
- 27. Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, et al. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proceedings of the National Academy of Sciences of the United States of America 92: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deans B, Griffin CS, Maconochie M, Thacker J (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. The EMBO journal 19: 6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuznetsov SG, Haines DC, Martin BK, Sharan SK (2009) Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res 69: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pittman DL, Schimenti JC (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 26: 167–173. [DOI] [PubMed] [Google Scholar]
- 31. Shu Z, Smith S, Wang L, Rice MC, Kmiec EB (1999) Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Molecular and cellular biology 19: 8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bleuyard J-Y, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. The EMBO journal 23: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W, Yang X, Lin Z, Timofejeva L, Xiao R, et al. (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant physiology 138: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu N, Schild D, Thelen MP, Thompson LH (2002) Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic acids research 30: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller KA, Sawicka D, Barsky D, Albala JS (2004) Domain mapping of the Rad51 paralog protein complexes. Nucleic acids research 32: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, et al. (2002) RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. The Journal of biological chemistry 277: 8406–8411. [DOI] [PubMed] [Google Scholar]
- 37. Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, et al. (2005) Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells. Plant molecular biology 57: 819–833. [DOI] [PubMed] [Google Scholar]
- 38. Osakabe K, Yoshioka T, Ichikawa H, Toki S (2002) Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana. Plant molecular biology 50: 71–81. [DOI] [PubMed] [Google Scholar]
- 39. Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. The Journal of biological chemistry 275: 16443–16449. [DOI] [PubMed] [Google Scholar]
- 40. Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, et al. (2002) Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic acids research 30: 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Badie S, Liao C, Thanasoula M, Barber P, Hill MA, et al. (2009) RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. The Journal of cell biology 185: 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA (2002) XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Molecular cell 10: 387–395. [DOI] [PubMed] [Google Scholar]
- 43. Chun J, Buechelmaier ES, Powell SN (2013) Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Molecular and cellular biology 33: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Da Ines O, Degroote F, Amiard S, Goubely C, Gallego ME, et al. (2013) Effects of XRCC2 and RAD51B mutations on somatic and meiotic recombination in Arabidopsis thaliana. The Plant Journal 74: 959–970. [DOI] [PubMed] [Google Scholar]
- 45. Liu Y, Masson J-Y, Shah R, O'Regan P, West SC (2004) RAD51C is required for Holliday junction processing in mammalian cells. Science 303: 243–246. [DOI] [PubMed] [Google Scholar]
- 46. Liu Y, Tarsounas M, O'Regan P, West SC (2007) Role of RAD51C and XRCC3 in genetic recombination and DNA repair. The Journal of biological chemistry 282: 1973–1979. [DOI] [PubMed] [Google Scholar]
- 47. Nagaraju G, Hartlerode A, Kwok A, Chandramouly G, Scully R (2009) XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Molecular and cellular biology 29: 4283–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rodrigue A, Coulombe Y, Jacquet K, Gagne JP, Roques C, et al. (2013) The RAD51 paralogs ensure cellular protection against mitotic defects and aneuploidy. J Cell Sci 126: 348–359. [DOI] [PubMed] [Google Scholar]
- 49. Compton SA, Ozgur S, Griffith JD (2010) Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. J Biol Chem 285: 13349–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, et al. (2004) Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic acids research 32: 2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, et al. (2008) Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res 36: 4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roth N, Klimesch J, Dukowic-Schulze S, Pacher M, Mannuss A, et al. (2012) The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. The Plant journal 72: 781–790. [DOI] [PubMed] [Google Scholar]
- 54. Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M (2004) Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annual review of genetics 38: 233–271. [DOI] [PubMed] [Google Scholar]
- 56. McDonald JP, Rothstein R (1994) Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics 137: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mozlin AM, Fung CW, Symington LS (2008) Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics 178: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Orel N, Kyryk A, Puchta H (2003) Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. The Plant Journal 35: 604–612. [DOI] [PubMed] [Google Scholar]
- 59. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, et al. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic acids research 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heacock M, Spangler E, Riha K, Puizina J, Shippen DE (2004) Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. The EMBO journal 23: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miller JC, Tan S, Qiao G, Barlow KA, Wang J, et al. (2011) A TALE nuclease architecture for efficient genome editing. Nature biotechnology 29: 143–148. [DOI] [PubMed] [Google Scholar]
- 62. Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. Journal of experimental botany 56: 1–14. [DOI] [PubMed] [Google Scholar]
- 63. Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. The EMBO journal 17: 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Malkova A, Ivanov EL, Haber JE (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A 93: 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, et al. (2009) The Structural Maintenance of Chromosomes 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, et al. (2013) Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role. PLoS genetics 9: e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, et al. (2005) Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic acids research 33: 4544–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ (2003) Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem 278: 2469–2478. [DOI] [PubMed] [Google Scholar]
- 69. Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, et al. (2002) Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem 277: 14315–14320. [DOI] [PubMed] [Google Scholar]
- 70. Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, et al. (2001) Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, et al. (2004) Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117: 337–347. [DOI] [PubMed] [Google Scholar]
- 72. Lin FL, Sperle K, Sternberg N (1984) Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Molec Cell Biol 4: 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prado F, Aguilera A (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics 139: 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bleuyard J-Y, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. The Plant Journal 41: 533–545. [DOI] [PubMed] [Google Scholar]
- 75. Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, et al. (2012) In planta gene targeting. Proceedings of the National Academy of Sciences of the United States of America 109: 7535–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant physiology 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 78. Gallego ME, White CI (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 98: 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
I-SceI induced mutations in end-joining products of DGU.US repair. The unmodified sequence surrounding the I-SceI cut site of the DGU.US recombination tester locus is shown at the top of the alignment, with the I-SceI restriction site boxed and the cut-sites for each strand arrowed. Mutations are highlighted by gray boxes and the size of deletions (bp) is indicated at right. Flanking microhomologies presumably involved in the end-joining of I-SceI induced DSB are underlined.
(PDF)
Sensitivity to Mitomycin C in T2 Rad51-GFP plants. WT and three independent Rad51-GFP T2 transformants were tested for their sensitivity to the cross-linking agent MMC. The dominant-negative effect of the RAD51-GFP allele is clearly visible in the 3∶1 segregating MMC hypersensitivity of the plantlets. (A) photos of the plantlets and (B) quantitation of sensitive versus resistant plants.
(PDF)





