Abstract
Background
Carboetomidate is a pyrrole etomidate analog that is 3 orders of magnitude less potent an inhibitor of in vitro cortisol synthesis than etomidate and does not inhibit in vivo steroid production. Although carboetomidate's reduced functional effect on steroid synthesis is thought to reflect lower binding affinity to 11β-hydroxylase, differential binding to this enzyme has never been experimentally demonstrated. In the current study, we tested the hypothesis that carboetomidate and etomidate bind with differential affinity to 11β-hydroxylase by comparing their abilities to inhibit photoaffinity labeling of purified enzyme by a photoactivatable etomidate analog and to modify the enzyme's absorption spectrum in a way that is indicative of ligand binding. In addition, we made a preliminary exploration of the manner in which etomidate and carboetomidate might differentially interact with this site using spectroscopic methods as well as molecular modeling techniques to better understand the structural basis for their selectivity.
Methods
The ability of azi-etomidate to inhibit cortisol synthesis was tested by assessing its ability to inhibit cortisol synthesis by H295R cells. The binding affinities of etomidate and carboetomidate to 11β-hydroxylase were compared by assessing their abilities to (1) inhibit photoincorporation of the photolabile etomidate analog [3H]azi-etomidate into the enzyme and (2) modify the absorption spectrum of the enzyme's heme group. In silico docking studies of etomidate, carboetomidate, and azi-etomidate binding to 11β-hydroxylase were performed using the computer software GOLD.
Results
Similar to etomidate, azi-etomidate potently inhibits in vitro cortisol synthesis. Etomidate inhibited [3H]azi-etomidate photolabeling of 11β-hydroxylase in a concentration-dependent manner. At a concentration of 40 μM, etomidate reduced photoincorporation of [3H] azi-etomidate by 96% ± 1% whereas carboetomidate had no experimentally detectable effect. On addition of etomidate to 11β-hydroxylase, a type 2 difference spectrum was produced indicative of etomidate complexation with the enzyme's heme iron; carboetomidate had no effect whereas azi-etomidate produced a reverse type 1 spectrum. Computer modeling studies predicted that etomidate, carboetomidate, and azi-etomidate can fit into the heme-containing pocket that forms 11β-hydroxylase's active site and pose with their carbonyl oxygens interacting with the heme iron and their phenyl rings stacking with phenylalanine-80. However, additional unique poses were identified for etomidate and azi-etomidate that likely account for their higher affinities.
Conclusions
Carboetomidate's reduced ability to suppress in vitro and in vivo steroid synthesis as compared with etomidate reflects its lower binding affinity to 11β-hydroxylase and may be attributed to carboetomidate's inability to form a coordination bond with the heme iron located at the enzyme's active site.
Etomidate is an imidazole-based sedative hypnotic that has minimal effects on respiratory and cardiovascular function.1–3 Unfortunately, it potently suppresses the synthesis of adrenocortical steroids that are necessary for a wide range of biological functions including glucose homeostasis, electrolyte, and water balance, and the stress response to trauma and infection.4–6 This side effect restricts etomidate use to single bolus administration for the induction of anesthesia and requires that other drugs be used to maintain anesthesia during surgery.7,8
Etomidate is thought to inhibit adrenocortical steroid synthesis by binding with high affinity to the active site of the cytochrome P450 (CYP) enzyme 11β-hydroxylase.9 Based on the hypothesis that such high affinity binding requires an interaction (coordination bond) between the basic nitrogen in etomidate's imidazole ring and the heme iron at 11β-hydroxylase's active site, we developed carboetomidate as a pyrrole etomidate analog and predicted that it would not bind with high affinity to 11β-hydroxylase;10 carboetomidate is structurally identical to etomidate (and is a potent hypnotic) but lacks the basic nitrogen necessary to form a coordination bond (Fig. 1). Functional studies generally support this hypothesis as carboetomidate is 3 orders of magnitude less potent an inhibitor of cortisol synthesis in vitro10 and does not suppress adrenocorticotropic hormone-stimulated steroid production in vivo.10,11
Figure 1.
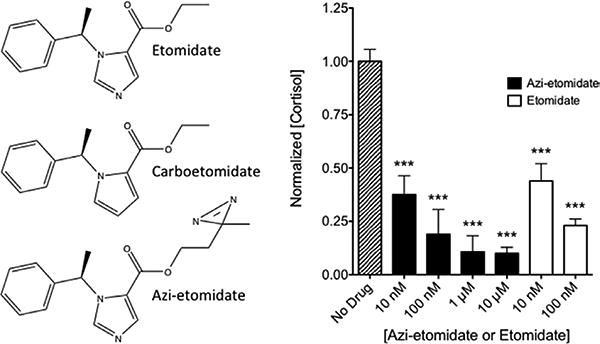
A, Chemical structures of etomidate, carboetomidate, and azi-etomidate. B, Inhibition of cortisol synthesis by azi-etomidate and etomidate. At all concentrations studied, azi-etomidate and etomidate reduced cortisol concentrations in wells containing H295R cells 24 hours after simulation with forskolin. Control (no drug) cortisol concentrations were 1.86 ± 0.10 ng/mL. ***P < 0.001 versus control value; 1-way analysis of variance followed by a Dunnett multiple comparison post-test. Each column shows the mean ± SD from 4 to 8 wells.
In the present study, we sought to test the hypothesis that etomidate and carboetomidate bind with different affinities to 11β-hydroxylase by comparing their abilities to inhibit photoincorporation of a photolabile etomidate analog into 11β-hydroxylase and to modify the absorption spectrum of 11β-hydroxylase in a way that is indicative of ligand binding to the enzyme's heme group. In addition, we made a preliminary exploration of the manner in which etomidate and carboetomidate might differentially interact with this site using molecular modeling techniques to better understand the structural basis for their selectivity.
Methods
[3H]azi-etomidate (12 Ci/mmol in methanol) was synthesized as previously described.12 Etomidate was obtained from Bachem (Torrance, CA), and carboetomidate was synthesized by Aberjonia Laboratories (Beverly, MA) as previously described.10 They were prepared as stock solutions in methanol. Buffer A contained: 50 mM K2HPO4, 20% glycerol, 500 mM Na acetate, 0.1 mM dithiothreitol, 0.1 mM EDTA, 1% Na cholate, and 1% Tween 20, at pH 7.4. Gel-counting buffer consisted of 90% toluene plus 10% tissue solubilizer (TS-2, from Research Products International, Mount Prospect, IL) containing 2.8 g/L of 2,5-diphenyloxa-zole and 0.28 g/L of 1,4-bis(5-phenyloxazol-2-yl)benzene. IRB approval for these studies was not sought because neither animals nor humans were used.
Inhibition of In Vitro Cortisol Synthesis
The abilities of azi-etomidate and etomidate to inhibit cortisol synthesis were assessed using the adrenocortical cell line H295R as previously described. Cortisol concentrations in culture plate wells were measured 24 hours after forskolin stimulation using a commercially available enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, MN, no. KGE008).10
Preparation and Purification of Human 11β-Hydroxylase
Human 11β-hydroxylase was expressed in Escherichia coli as a mature form with N- and C-terminal modifications as described.13 The 11β-hydroxylase expression plasmid was introduced into E coli strain BL21(DE3)pLys along with a GroES/GroEL expression vector pGro12.
Expression and purification were performed as described by Zöllner et al.13 The protein was purified to apparent homogeneity. Its ultraviolet/visible absorption spectrum in the substrate-free form showed an absorption maximum at 392 nm indicating a high-spin form of this P450 enzyme.
Photolabeling of 11β-Hydroxylase with [3H] Azi-Etomidate in the Presence of Etomidate or Carboetomidate
[3H]azi-etomidate (10 nM) along with either etomidate or carboetomidate from the stock solutions was added to the wells of a glass 96-well plate and the methanol evaporated. The ligands were dissolved in 136 μL of buffer A, and 14 μL of human 11β-hydroxylase (2 mg/mL) in buffer A was added. Samples were then irradiated at 365 nm for 45 minutes on ice to induce photoincorporation of the radioactive ligand into the enzyme as previously described.12 After irradiation, samples were loaded on separate lanes of an 8% sodium dodecyl sulfate gel and run overnight. A single protein band at approximately 50 kDa was identified by Coomassie blue staining and excised for scintillation counting in the gel-counting buffer after destaining. Blank sections of the gel were also treated similarly, and the counts in these blanks were subtracted from those containing 11β-hydroxylase.
Spectroscopic Studies of Etomidate and Carboetomidate Interactions with 11β-Hydroxylase
Spectrophotometric analysis was performed using a model V-630 spectrophotometer from Jasco (Easton, MD). Difference spectroscopy was performed in tandem cuvettes as generally described by Schenkman.14 Each tandem cuvette contained 2 chambers. The first chamber of the “test” cuvette was filled with 2 μM of 11β-hydroxylase in buffer, whereas the second chamber was filled only with buffer. For reference, a second tandem cuvette was used. The chambers in the reference cuvette were identically filled with 11β-hydroxylase and buffer. Etomidate or carboetomidate (40 μM final concentration from dimethyl sulfoxide stocks) was added to the first chamber of the test cuvette and to the second chamber of the reference cuvette. An equal volume of dimethyl sulfoxide was added to the remaining 2 chambers of the cuvettes. Difference spectra were recorded from 370 to 450 nm. The buffer was composed of 50 mM potassium phosphate (pH 7.4), 20% glycerol, 0.5% sodium cholate, and 0.05% Tween 20.
Absorption spectra of etomidate and carboetomidate were recorded in quartz cuvettes. The final concentration of each compound was 40 μM in Milli-Q water (Millipore Corp., Billerica, MA). These spectra were recorded from 230 to 300 nm.
In Silico Docking Studies of Etomidate and Carboetomidate Binding to 11β-Hydroxylase
Ligands were docked to the homology model developed by Roumen et al.15 and based primarily on the known structure of CYP101 (pdb: 2CPP)16 with the structure of CYP2C5 (pdb: 1NR6)17 inserted where there are gaps in the amino acid alignment between 11β-hydroxylase and CYP101 (B′–C loop, F-G-H region, J-K loop, K-L loop). The first 50 N-terminal residues, the membrane-binding region, were excluded because there is no known structural template. In addition to sequence alignment, they carefully considered where the steroid might dock in relation to its proximity to iron for the oxidation reaction.
Molecular docking calculations were performed with the program GOLD 4.1 (Cambridge Crystallographic Data Center, Cambridge, United Kingdom; http://www.ccdc.cam.ac.uk/products/life_sciences/gold/) using the default settings. The program was run under the slow mode for improved accuracy, and for efficiency, the ligand was confined to a sphere of 20 Å radius centered on the heme iron. Each ligand was docked for a maximum of 25 poses. The ligands to be docked were prepared using the PRODRG server (http://davapc1.bioch.dundee.ac.uk/prodrg/). The homology model (presented earlier) was loaded into the Hermes program, and hydrogen atoms were added and included in the calculation. The algorithm exhaustively searches the entire rotational and translational space of the ligand with respect to the protein. It considers hydrogen bonding and van der Waals interactions between the ligand and the protein, as well as the ligand's internal van der Waals and intramolecular hydrogen bonding interactions. All the calculations were performed in the absence of water molecules.
Results
Azi-Etomidate Inhibits In Vitro Cortisol Synthesis at Nanomolar Concentrations
Azi-etomidate reduced forskolin-stimulated cortisol concentrations in culture plate wells 37% ± 9%, 19% ± 12%, 11% ± 8%, and 10% ± 3% to (n = 4 replicates, mean ± SD) of the control concentrations measured in the absence of hypnotic at respective concentrations of 10 nM, 100 nM, 1 μM, and 10 μM (Fig. 1B). For comparison, Figure 1B also shows the quantitatively similar inhibitory actions of etomidate determined in the same cell batch (44% ± 8% and 23% ± 3% at 10 and 100 nM, respectively; n = 4–8, mean ± SD).
Etomidate Inhibits [3H]Azi-Etomidate Photolabeling of 11β-Hydroxylase Whereas Carboetomidate Does Not
Because of the predicted nanomolar affinity of etomidate and the experimental need to maintain the 11β-hydroxylase concentration in the micromolar range (ligand depletion conditions), we compared the ability of 40 μM etomidate and carboetomidate (near its aqueous solubility limit) to protect against photoincorporation of 10 nM [3H]azi-etomidate (Table 1). Etomidate's inhibition of [3H]azi-etomidate photolabeling was nearly complete, with just 4.1% ± 1.1% of the photolabeling remaining (n = 3 replicates, mean ± SD, P < 0.0001, 1-way analysis of variance with Dunnett multiple comparison post-test versus control). Conversely, carboetomidate had no experimentally detectable inhibitory effect, as 102% ± 8.2% (n = 3 replicates, mean ± SD) of the photolabeling remained.
Table 1. Protection Against [3H]Azi-Etomidate Photolabeling of 11β-Hydroxylasea.
| Competing ligand | Normalized photoincorporation | ||
|---|---|---|---|
|
| |||
| Average (95% CI) | ±SD | n | |
| Control | 100 (92–109) | 7.5 | 3 |
| Carboetomidateb | 102 (2.8–5.3) | 8.2 | 3 |
| Etomidateb | 4.1 (92–110) | 1.1 | 3 |
n = number of replicates in the experiment.
a6 μM 11β-hydroxylase.
b40 μM drug.
Spectroscopy of the 11β-Hydroxylase Interacting with Ligands
Etomidate, carboetomidate, and azi-etomidate showed absorption maxima at 243, 272.5, and 220 nm, respectively. The difference spectrum of 11β-hydroxylase titrated with etomidate displayed a maximum at 424.5 nm and a minimum at 410 nm (Fig. 2). This is consistent with a type 2 spectral interaction, indicating that the hypnotic binds to the heme iron to form a low-spin complex.14,18,19 The difference spectrum of 11β-hydroxylase titrated with azi-etomidate displayed a maximum at 420 nm and a minimum at 388 nm, consistent with a reverse type 1 spectrum (sometimes also termed a modified type 2 spectrum) and also suggesting an interaction between the ligand and the heme iron. In contrast to etomidate and azi-etomidate, carboetomidate induced essentially no spectral change.
Figure 2.
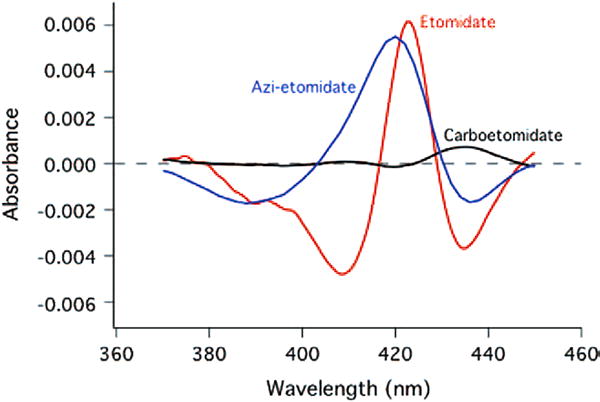
Spectral change induced by addition of etomidate (red curve) or carboetomidate (black curve), both at 40 μM, to purified human 11β-hydroxylase.
In Silico Docking Within the 11β-Hydroxylase Active Site
The docking algorithm that we used seeks different arrangements of the ligand in the binding pocket (called poses) that are consistent with the geometry and surface composition of the binding pocket. Because of the complexity of force fields within proteins, it is has been optimized for the prediction of ligand-binding positions rather than the prediction of binding affinities. Thus, our exploration was designed to ask whether the specific homology model might aid our understanding of the interactions underlying the clear specificity of action of these 2 ligands on 11β-hydroxylase. Our strategy had 2 parts. First, we sought poses that were unique to etomidate and thus might be the ones responsible for its observed higher affinity as an inhibitor. Second, we analyzed situations in which the 2 ligands adopted identical (i.e., isosteric or superimposable) poses to assess the role of imidazole in contributing additional binding energy.
Overview of Poses: Etomidate Versus Carboetomidate
The structural differences between etomidate and carboetomidate (Fig. 1) are minor and cause essentially no change in volume. It is not surprising that the docking routine showed that both agents were able to fit in the heme-containing pocket of 11β-hydroxylase. Thus, steric constraints do not seem to play a role in specificity. The docking routine found 11 different poses for etomidate and 6 different poses for carboetomidate within the heme-containing cavity of the 11β-hydroxylase homology model. For etomidate, these 11 poses were in 2 categories based on their interaction with the enzyme's heme iron. In 5 poses, the iron interacted with etomidate's imidazole nitrogen, whereas in the other 6 poses, the iron interacted with etomidate's carbonyl group. For carboetomidate, only the latter (i.e., carbonyl–iron) interactions were found.
Heme–Imidazole Interactions
Of the 5 predicted etomidate poses in which the iron interacted with the imidazole nitrogen, in 4 cases the imidazole ring was approximately perpendicular to the heme ring, and in 1, it lay nearly parallel to it. One representative of the 5 poses is shown in Figure 3. The docking algorithm predicted nitrogen–iron distances of 1.7 to 2.0 Å, which are comparable with, but generally shorter than, those found in a number of moderately high resolution CYP structures. For example, in the CYP 2B4 structure, 3 different imidazole-containing ligands all exhibit a nitrogen–iron distance of 2.1 Å (pdb, resolution [Å ]: 1SUO, 1.9; 2BDM, 2.3; 2Q6N, 2.5).20–22 In a much higher resolution structure of human oxyhemoglobin (2DN1, 1.25 Å), the imidazole of His-92 is 2.06 Å from the iron.23 In addition to the nitrogen–iron interaction, a common finding was that the aromatic ring of etomidate was almost perfectly aligned with that of Phe-80 on the surface of the pocket. The carbon atoms of the rings were in register (dotted red lines in Fig. 3), and the distance between opposed carbons were in the range expected for strong van der Waals interactions, varying systematically around the ring from 3.4 to 3.8 Å. Such distances may be compared with values of 3.5 to 3.8 Å for the ring stacking of α-naphthoflavone with Phe-226 in cytochrome P4501A2.24
Figure 3.
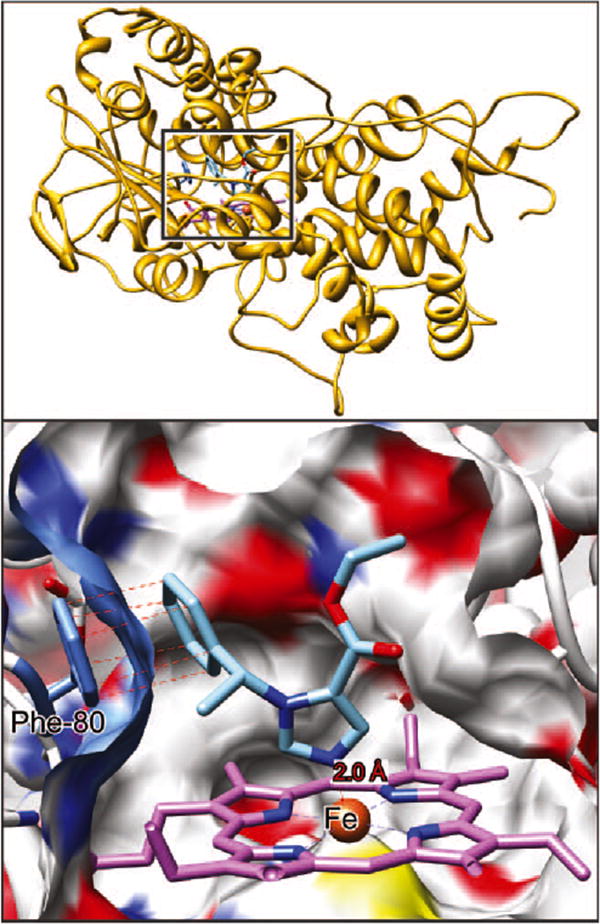
Etomidate docked in the substrate-binding pocket of 11β-hydroxylase. A cross-section through the surface of the binding cavity is shown. The surface is colored according to the atoms behind it; white is carbon, blue nitrogen, red oxygen, and yellow sulfur. Etomidate is depicted in stick representation with blue nitrogens, red oxygens, and sky blue carbons. In the heme system, purple is carbon, blue is nitrogen, and then rust is heme. The heme iron atom is coordinated to Cys-400 below the ring and etomidate imidazole nitrogen above the ring. Top right insert from Figure 1 for reference.
The carbonyl group had only a single interaction with the protein that was shorter than 5 Å, being 4.4 Å from the side chain of Phe-437. The ester oxygen faired slightly better, being 3.6 Å from Phe-437 and 3.7 Å from Thr-268's hydroxyl. Although this latter distance is too long for hydrogen bonding, it should be remembered that we did not allow the protein's structure to react to the presence of the ligand.
The single pose with the imidazole ring parallel to the heme ring (not shown) still maintained the aromatic ring stacking with Phe-80, albeit somewhat weaker with distances ranging from 3.0 to 4.3 Å. The iron–imidazole distance was long (3.2 Å), but this lack of coordination is possibly offset by multiple interactions with the iron because the imidazole ring atoms and the carbonyl and ether oxygens were all within 2.6 to 4.0 Å of the iron.
Heme–Carbonyl Interactions
In the remaining 6 predicted etomidate poses, the carbonyl group interacted with the heme iron (e.g., Fig. 4). No poses with the ester oxygen interacting with iron were found. There were 2 main categories of pose, each with 3 poses that differed from each other in only minor ways. In 1 set of poses, the phenyl ring of etomidate lay close to Phe-80 in the surface of the binding pocket. In the other set, the phenyl ring was on the opposite side of the pocket near Phe-437. The 6 carboetomidate poses also were in 2 such sets, and they may be considered together with those of etomidate. The carbonyl oxygen–iron distances were all close to 1.9 Å.
Figure 4.
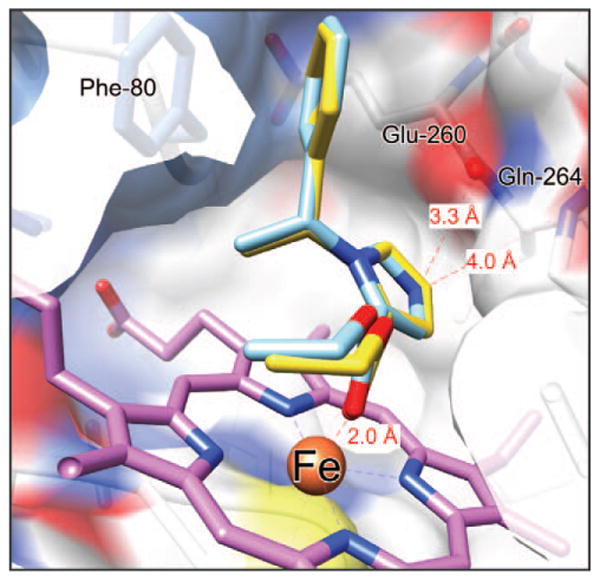
Superimposed structures of etomidate and carboetomidate docked in the substrate-binding pocket of 11β-hydroxylase. Carboetomidate is shown with gold carbons. Both agents adopt very similar poses with the heme iron coordinated to the ester carbonyl oxygen and the phenyl ring proximal to Phe-80.
For the first set of poses mentioned earlier, Figure 4 shows that etomidate and carboetomidate superimpose quite well, no equivalent atom in the 2 molecules being >0.2 Å apart. The carbonyl oxygens interact identically with the iron and the ring stacking with Phe-80 is also identical, with separation of carbons varying from 3.0 to 4.4 Å. Thus, the observed difference in affinity for 11β-hydroxylase is not accounted for by these interactions. However, the imidazole ring's interaction with the protein does provide 2 possible explanations. First, the free nitrogen is 3.3 Å from the backbone carbonyl of Gly-264 and 4.0 Å from the backbone amide nitrogen of Gly-264. Neither of these distances is consistent with strong hydrogen bonding, although some electrostatic interaction must occur. The equivalent carbon is a similar distance from these moieties, but it lacks the electronegativity of the nitrogen. Second, the imidazole ring lies above one of the pyrrole rings of the porphyrin ring system of the heme. The distance of the imidazole ring atoms to the nearest pyrrole ring atoms range from 3.3 to 3.9 Å, consistent with the 2 delocalized π-ring systems undergoing good van der Waals interactions. Because the carboetomidate ring lacks this aromatic quality, it will be less polarizable and should interact more weakly with the porphyrin ring system.
For the second set of poses mentioned earlier, the carbonyl–iron interaction is again very similar. In these poses, the aromatic rings of both agents interact with Phe-437, but they do not ring stack. Instead, they are tilted back so that the agents' aromatic ring is 130° to that of Phe-437 with their carbon atoms separated by 3.6 to 7.1 Å. This weakens the aromatic interaction but allows several other interactions. For example, Ala-263 and Met-180 interact strongly with the agents' aromatic ring. The imidazole nitrogen of etomidate makes a hydrogen bond with the backbone carbonyl of Glu-260 (2.8 Å) and van der Waals contact with the backbone amide of Gly-264 (3.5 Å) and the side chain of Glu-260 (4.7 Å).
An Additional Unique Pose for Azi-Etomidate
In addition to the 2 categories of poses described earlier for etomidate (i.e., heme–imidazole interactions and heme–carbonyl interactions), the docking routine identified an ensamble of azi-etomidate poses in which a diazerine moiety nitrogen interacts with the heme iron (Fig. 5). In these poses, the distance between the azi moiety's nitrogen and the iron ranged from 1.5 to 1.6 Å, and there were no apparent stacking interactions between azi-etomidate's phenyl ring and any of the aromatic residues in or near the binding pocket.
Figure 5.
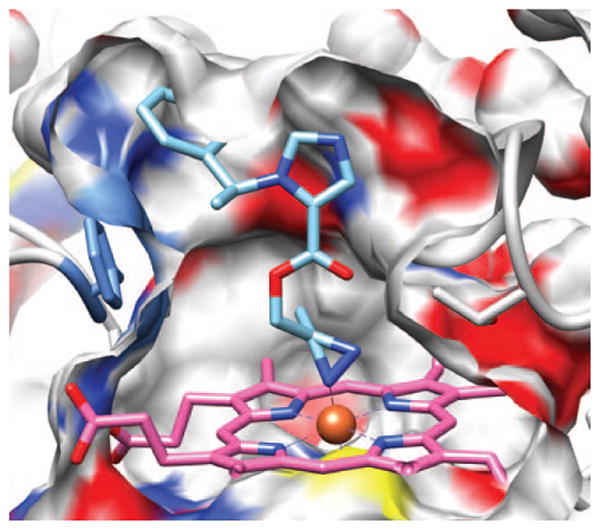
Representative pose showing azi-etomidate docked in the substrate-binding pocket of 11β-hydroxylase with its azi moiety interacting with the enzyme's heme iron. A cross-section through the surface of the binding cavity is shown. The surface is colored according to the atoms behind it; white is carbon, blue nitrogen, red oxygen and yellow sulfur. Azi-etomidate is depicted in stick representation with blue nitrogens, red oxygens, and sky blue carbons.
Discussion
We have shown that etomidate interacts strongly and selectively with purified 11β-hydroxylase, whereas carboetomidate interacts weakly if at all. This conclusion rests on 2 distinct experimental methods. The first method was a kinetic competition assay between photolabeling and ligand binding. Even with competing azi-etomidate concentrations as low as 10 nM, a near-saturated solution of carboetomidate (40 μM) failed to protect effectively against photolabeling, whereas etomidate at the same concentration almost completely inhibited photolabeling. The second method supporting our conclusion was spectroscopic, and it added the additional information that etomidate interacted directly with the heme iron because the spectrum showed the classic type 2 behavior (Fig. 2), whereas carboetomidate did not cause spectral changes. The physiologic implications of carboetomidate's inability to interact with 11β-hydroxylase are of interest because the agent does inhibit in vitro cortisol synthesis defined using a human adrenocortical cell assay, although its potency is 2000-fold less than that of etomidate.10
We sought to gain a qualitative understanding of the origin of the difference in affinities of these 2 agents for 11β-hydroxylase by performing docking studies using a proven homology model of 11β-hydroxylase. Several points should be borne in mind when seeking the origin of difference in interaction strength. First, our understanding of the force fields within proteins is inadequate for precise prediction of ligand–protein interaction energies. Second, for a 1000-fold difference in dissociation constant the difference in free energy of binding is approximately 4 kcal/mol, equivalent to 1 to 2 hydrogen bonds. Third, the free energy for binding contains contributions from the energy of removing the agent from aqueous solution (desolvation) as well as those for insertion into the binding pocket. It seems likely that the free energy of desolvation for carboetomidate would be less than that for etomidate, because the imidazole ring will undergo polar interactions with water that are lost when the >N is replaced by >CH to give a pyrrole ring. The additional hydrophobicity introduced by this substitution would create an entropic driving force for desolvation. This would suggest that the etomidate–11β-hydroxylase interaction itself must be >4 kcal/mol stronger than that of carboetomidate to compensate for desolvation.
Our docking studies revealed that the major difference between the agents was the ability of the imidazole ring to coordinate the sixth position of the porphyrin ring iron atom, an interaction that carboetomidate is incapable of. Theoretical studies on simple models suggest that imidazole can displace water in Fe(H2O)62+ with a favorable free energy of some -20 kcal/mol. This is attributed to its large dipole (3.7 D) and high polarizability25 that lead to favorable charge–dipole and charge-induced dipole interactions. In the pyrrole ring of carboetomidate, the ring dipole is smaller but in the same direction (the negative end pointing toward the iron), and the polarizability is comparable with that of imidazole. Free pyrroles interact via their nitrogen with the iron, but this is not possible in carboetomidate because of steric constraints. Binding to CYP-bound porphyrin–iron complexes is more complex than the aforementioned model system and contains terms from protein conformational changes; nonetheless, for 2 such similar molecules these terms are likely to be similar, and 1 may conclude that the high affinity of etomidate compared with carboetomidate very likely comes from the inability of the >CH in carboetomidate to coordinate strongly with the heme iron.
An alternative interaction with the iron was through the carbonyl oxygen, and both agents adopted similar poses when this interaction was present, so it is unlikely that the carbonyl–iron interaction itself is responsible for the experimentally observed difference in the 2 agents. Furthermore, our photolabeling and spectroscopic experiments indicate that the putative carbonyl–iron interaction is of insufficient strength to significantly stabilize binding of carboetomidate to 11β-hydroxylase.
Results with azi-etomidate were generally similar to etomidate; azi-etomidate is a very potent inhibitor of cortisol synthesis and adopts the same 2 categories of poses as etomidate. However, our docking studies identified an additional set of poses for azi-etomidate in which the photolabel's diazerine moiety interacts with the heme iron. It is unclear whether these poses explain the distinct spectral pattern (i.e., reverse type 1 spectrum) induced by azi-etomidate on purified 11β-hydroxylase. This pattern has been described for other inhibitors of CYP enzymes that do not contain a diazerine, but its precise origin is unknown.26 Future photoaffinity labeling studies aimed at identifying the specific amino acid residues near this azi moiety may provide additional information regarding the orientation(s) of this ligand within the enzyme's active site.
In summary, we have shown that unlike etomidate, carboetomidate neither inhibits photoaffinity labeling of 11β-hydroxylase by azi-etomidate nor alters the enzyme's spectrum in a manner indicative of drug interacting with the enzyme's heme moiety. Using molecular modeling, we identified 11 predicted poses of etomidate in the active site of 11β-hydroxylase. Of these, 6 are shared with carboetomidate. The remaining poses of etomidate, none of which are emulated by carboetomidate, all include an iron–imidazole interaction. Thus, this interaction is most likely the dominant determinant of the high affinity binding of etomidate to 11β-hydroxylase. A secondary motif, the interaction between etomidate's phenyl ring and Phe-80 is a stabilizing factor, but it is not sufficient to confer stability because it is also present in some of the carboetomidate poses.
Acknowledgments
The authors thank Prof. Hermans (Department of Pharmacology and Toxicology, Cardiovascular Research Institute, University Maastricht, the Netherlands) for providing them with the published homology model and Wolfgang Reinle for preparation of the protein. Molecular graphics images were produced using the University of California, San Francisco Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR-01081).
This research was supported by grants to D.E.R. (R01-GM087316 and R21-DA029253 from the National Institutes of Health, Bethesda, MD), R.B. (FOR 13698 from the Deutsche Forschungsgemeinschaft), J.F.C. (K08-GM083216), and by the Department of Anesthesia, Critical Care, and Pain Medicine, Massachusetts General Hospital.
Footnotes
Reprints will not be available from the authors.
See Disclosures at end of article for Author Conflicts of Interest.
Disclosures
Name: Sivananthaperumal Shanmugasundararaj, PhD.
Contribution: This author performed the computer modeling studies.
Attestation: Sivananthaperumal Shanmugasundararaj approved the final manuscript.
Conflicts of Interest: The author has no conflicts of interest to declare.
Name: Xiaojuan Zhou, BS.
Contribution: This author performed the competition experiments.
Attestation: Xiaojuan Zhou approved the final manuscript.
Conflicts of Interest: The author has no conflicts of interest to declare.
Name: Jens Neunzig, MA.
Contribution: This author performed the spectroscopic studies.
Attestation: Jens Neunzig approved the final manuscript.
Conflicts of Interest: The author has no conflicts of interest to declare.
Name: Rita Bernhardt, PhD.
Contribution: This author expressed and purified 11β-hydroxylase.
Attestation: Rita Bernhardt approved the final manuscript.
Conflicts of Interest: The author has no conflicts of interest to declare.
Name: Joseph F. Cotton, MD, PhD.
Contribution: This author helped design the experiment.
Attestation: Joseph F. Cotton approved the final manuscript.
Conficts of Interest: Joseph F. Cotton is a coinventor on a patent application submitted by the Massachusetts General Hospital. He, his department, his laboratory, and his institution could receive royalties relating to the development of carboetomidate or related analogs.
Name: Rile Ge, MD, PhD.
Contribution: This author performed the H295R cell assays.
Attestation: Rile Ge approved the final manuscript.
Conficts of Interest: The author has no conflicts of interest to declare.
Name: Keith W. Miller, DPhil.
Contribution: This author oversaw the binding and computer modeling studies and helped write the manuscript.
Attestation: Keith W. Miller approved the final manuscript and the integrity of the original data and the analysis reported in this manuscript. Keith W. Miller is the archival author.
Conflicts of Interest: Keith W. Miller is a coinventor on a patent application submitted by the Massachusetts General Hospital. He, his department, his laboratory, and his institution could receive royalties relating to the development of carboetomidate or related analogs.
Name: Douglas E. Raines, MD.
Contribution: This author designed the experiments and wrote the manuscript.
Attestation: Douglas E. Raines approved the final manuscript and the integrity of the original data and the analysis reported in this manuscript.
Conflicts of Interest: Douglas E. Raines is a coinventor on a patent application submitted by the Massachusetts General Hospital. He, his department, his laboratory, and his institution could receive royalties relating to the development of carboetomidate or related analogs. Douglas E. Raines is a consultant for and holds an equity position in Annovation BioPharma, a pharmaceutical company that seeks to develop technologies covered by that patent.
This manuscript was handled by: Marcel E. Durieux, MD, PhD.
References
- 1.Dubois-Primo J, Bastenier-Geens J, Genicot C, Rucquoi M. A comparative study of etomidate and methohexital as induction agents for analgesic anesthesia. Acta Anaesthesiol Belg. 1976;27(suppl):187–95. [PubMed] [Google Scholar]
- 2.Gooding JM, Corssen G. Effect of etomidate on the cardiovascular system. Anesth Analg. 1977;56:717–9. doi: 10.1213/00000539-197709000-00021. [DOI] [PubMed] [Google Scholar]
- 3.Gooding JM, Weng JT, Smith RA, Berninger GT, Kirby RR. Cardiovascular and pulmonary responses following etomidate induction of anesthesia in patients with demonstrated cardiac disease. Anesth Analg. 1979;58:40–1. doi: 10.1213/00000539-197901000-00016. [DOI] [PubMed] [Google Scholar]
- 4.de Jong FH, Mallios C, Jansen C, Scheck PA, Lamberts SW. Etomidate suppresses adrenocortical function by inhibition of 11 beta-hydroxylation. J Clin Endocrinol Metab. 1984;59:1143–7. doi: 10.1210/jcem-59-6-1143. [DOI] [PubMed] [Google Scholar]
- 5.Wagner RL, White PF. Etomidate inhibits adrenocortical function in surgical patients. Anesthesiology. 1984;61:647–51. doi: 10.1097/00000542-198412000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Wagner RL, White PF, Kan PB, Rosenthal MH, Feldman D. Inhibition of adrenal steroidogenesis by the anesthetic etomidate. N Engl J Med. 1984;310:1415–21. doi: 10.1056/NEJM198405313102202. [DOI] [PubMed] [Google Scholar]
- 7.Absalom A, Pledger D, Kong A. Adrenocortical function in critically ill patients 24 h after a single dose of etomidate. Anaesthesia. 1999;54:861–7. doi: 10.1046/j.1365-2044.1999.01003.x. [DOI] [PubMed] [Google Scholar]
- 8.Bloomfield R, Noble DW. Etomidate and fatal outcome–even a single bolus dose may be detrimental for some patients. Br J Anaesth. 2006;97:116–7. doi: 10.1093/bja/ael124. [DOI] [PubMed] [Google Scholar]
- 9.Hahner S, Stuermer A, Kreissl M, Reiners C, Fassnacht M, Haenscheid H, Beuschlein F, Zink M, Lang K, Allolio B, Schirbel A. [123 I]Iodometomidate for molecular imaging of adrenocortical cytochrome P450 family 11B enzymes. J Clin Endocrinol Metab. 2008;93:2358–65. doi: 10.1210/jc.2008-0050. [DOI] [PubMed] [Google Scholar]
- 10.Cotten JF, Forman SA, Laha JK, Cuny GD, Husain SS, Miller KW, Nguyen HH, Kelly EW, Stewart D, Liu A, Raines DE. Carboetomidate: a pyrrole analog of etomidate designed not to suppress adrenocortical function. Anesthesiology. 2010;112:637–44. doi: 10.1097/ALN.0b013e3181cf40ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotten JF, Le Ge R, Banacos N, Pejo E, Husain SS, Williams JH, Raines DE. Closed-loop continuous infusions of etomidate and etomidate analogs in rats: a comparative study of dosing and the impact on adrenocortical function. Anesthesiology. 2011;115:764–73. doi: 10.1097/ALN.0b013e31821950de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husain SS, Ziebell MR, Ruesch D, Hong F, Arevalo E, Kosterlitz JA, Olsen RW, Forman SA, Cohen JB, Miller KW. 2-(3-Methyl-3H-diaziren-3-yl)ethyl 1-(1-phenylethyl)-1H-imidazole-5-car-boxylate: a derivative of the stereoselective general anesthetic etomidate for photolabeling ligand-gated ion channels. J Med Chem. 2003;46:1257–65. doi: 10.1021/jm020465v. [DOI] [PubMed] [Google Scholar]
- 13.Zöllner A, Kagawa N, Waterman MR, Nonaka Y, Takio K, Shiro Y, Hannemann F, Bernhardt R. Purification and functional characterization of human 11beta hydroxylase expressed in Escherichia coli. FEBS J. 2008;275:799–810. doi: 10.1111/j.1742-4658.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 14.Schenkman JB. Studies on the nature of the type I and type II spectral changes in liver microsomes. Biochemistry. 1970;9:2081–91. doi: 10.1021/bi00812a009. [DOI] [PubMed] [Google Scholar]
- 15.Roumen L, Sanders MP, Pieterse K, Hilbers PA, Plate R, Custers E, de Gooyer M, Smits JF, Beugels I, Emmen J, Ottenheijm HC, Leysen D, Hermans JJ. Construction of 3D models of the CYP11B family as a tool to predict ligand binding characteristics. J Comput Aided Mol Des. 2007;21:455–71. doi: 10.1007/s10822-007-9128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulos TL, Finzel BC, Howard AJ. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 17.Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003;42:9335–45. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- 18.Parker JE, Warrilow AG, Cools HJ, Martel CM, Nes WD, Fraaije BA, Lucas JA, Kelly DE, Kelly SL. Mechanism of binding of prothioconazole to Mycosphaerella graminicola CYP51 differs from that of other azole antifungals. Appl Environ Microbiol. 2011;77:1460–5. doi: 10.1128/AEM.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locuson CW, Hutzler JM, Tracy TS. Visible spectra of type II cytochrome P450-drug complexes: evidence that “incomplete” heme coordination is common. Drug Metab Dispos. 2007;35:614–22. doi: 10.1124/dmd.106.012609. [DOI] [PubMed] [Google Scholar]
- 20.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–81. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Sun L, Muralidhara BK, Kumar S, White MA, Stout CD, Halpert JR. Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Biochemistry. 2007;46:11559–67. doi: 10.1021/bi7011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SY, Yokoyama T, Shibayama N, Shiro Y, Tame JR. 1.25 A resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J Mol Biol. 2006;360:690–701. doi: 10.1016/j.jmb.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Kassimi NE, Doerksen RJ, Thakkar AJ. Polarizabilities of aromatic five-membered rings: azoles. J Phys Chem A. 1995;99:12790–6. [Google Scholar]
- 25.Ricca A, Bauschlicher CW. Theoretical study of the interaction of water and imidazole with iron and nickel dications. J Phys Chem A. 2002;106:3219–23. [Google Scholar]
- 26.Berwanger A, Eyrisch S, Schuster I, Helms V, Bernhardt R. Polyamines: naturally occurring small molecule modulators of electrostatic protein-protein interactions. J Inorg Biochem. 2010;104:118–25. doi: 10.1016/j.jinorgbio.2009.10.007. [DOI] [PubMed] [Google Scholar]


