Abstract
The chronic high-level alcohol consumption seen in alcoholism leads to dramatic effects on the hippocampus, including decreased white matter, loss of oligodendrocytes and other glial cells, and inhibition of neurogenesis. Examining gene expression in post mortem hippocampal tissue from 20 alcoholics and 19 controls allowed us to detect differentially expressed genes that may play a role in the risk for alcoholism or whose expression is modified by chronic consumption of alcohol. We identified 639 named genes whose expression significantly differed between alcoholics and controls at a False Discovery Rate (FDR) ≤ 0.20; 52% of these genes differed by at least 1.2-fold. Differentially expressed genes included the glucocorticoid receptor and the related gene FK506 binding protein 5 (FKBP5), UDP glycosyltransferase 8 (UGT8), urea transporter (SLC14A1), zinc transporter (SLC39A10), Interleukin 1 receptor type 1 (IL1R1), thioredoxin interacting protein (TXNIP), and many metallothioneins. Pathways related to inflammation, hypoxia, and stress showed activation, and pathways that play roles in neurogenesis and myelination showed decreases. The cortisol pathway dysregulation and increased inflammation identified here are seen in other stress-related conditions such as depression and post-traumatic stress disorder and most likely play a role in addiction. Many of the detrimental effects on the hippocampus appear to be mediated through NF-κB signaling. Twenty-four of the differentially regulated genes were previously identified by genome-wide association studies of alcohol use disorders; this raises the potential interest of genes not normally associated with alcoholism, such as suppression of tumorigenicity 18 (ST18), BCL2-associated athanogene 3 (BAG3), and von Willebrand factor (VWF).
Keywords: alcoholism, stress, inflammation, cortisol, hippocampus, gene expression, GWAS, NF-κ
Introduction
Alcohol dependence (alcoholism) is a complex disorder with a 40–60% genetic contribution to risk (Edenberg & Foroud, 2006; Heath et al., 1997; McGue, 1999). Although several genes in which variants affect the risk for alcohol dependence have been identified (Rietschel & Treutlein, 2012), their overall effect accounts for only a small portion of the vulnerability to alcohol dependence. Many studies are underpowered, and determining which modest association results are true positives can be difficult. Studies of gene expression in the human brain can reveal differences between alcoholics and controls that might be either risk factors or sequelae of excessive drinking; in either case, this increases the likelihood that such genes are relevant to the disease.
Prior studies have compared gene expression between alcoholics and controls using human post mortem brains (Flatscher-Bader et al., 2010; Flatscher-Bader et al., 2005; Iwamoto et al., 2004; Kryger & Wilce, 2010; Lewohl et al., 2000; Liu et al., 2007; Liu et al., 2006; Mayfield et al., 2002; Sokolov et al., 2003; Zhou et al., 2011b). Others have examined brain regions from animal models (Edenberg et al., 2005; Kerns et al., 2005; Kimpel et al., 2007; McBride et al., 2010; Mulligan et al., 2008; Mulligan et al., 2006; Saito et al., 2004; Tabakoff et al., 2008; Wolen et al., 2012; Worst et al., 2005). The human studies have examined superior frontal cortex (Lewohl et al., 2000; Liu et al., 2007; Liu et al., 2006), frontal cortex (Liu et al., 2007), prefrontal cortex (Flatscher-Bader et al., 2005; Iwamoto et al., 2004), temporal cortex (Sokolov et al., 2003), nucleus accumbens and ventral tegmental area (Flatscher-Bader et al., 2010; Flatscher-Bader et al., 2005), basolateral amygdala (Kryger & Wilce, 2010), and hippocampus (Zhou et al., 2011b). These studies have found down-regulation of myelin-related genes (Liu et al., 2006; Mayfield et al., 2002) and mitochondrial dysfunction (Liu et al., 2007; Sokolov et al., 2003), and dysregulation of genes involved in ubiquitination (Liu et al., 2006; Sokolov et al., 2003) and apoptosis and cell survival (Liu et al., 2004; Liu et al., 2007; Liu et al., 2006).
The hippocampus is a key region related to learning, for which neurogenesis is required (Winocur et al., 2006). Chronic, excessive consumption of alcohol leads to dramatic effects on the hippocampus. Hippocampal size is decreased with chronic drinking (Agartz et al., 1999; Laakso et al., 2000), and abstinence leads to a recovery of this volume loss (Crews & Nixon, 2009). The decrease in hippocampal size is due to a combination of neurodegeneration and decreased neurogenesis (Crews & Nixon, 2009; Morris et al., 2010; Richardson et al., 2009). While neurodegeneration is noted in alcoholism, post mortem studies of the hippocampus have found glial cell loss but no neuronal loss. A post mortem study of the hippocampus found a loss of white matter, including oligodendrocytes, but with no significant loss of neurons (Harding et al., 1997). Alcoholics who had been abstinent before death did not show a significant loss of white matter, implying that recovery from this loss is possible (Harding et al., 1997). A second post mortem examination of the hippocampus showed a 37% loss of glial cells (astrocytes, oligodendrocytes, and to a lesser extent microglia) in alcoholics (Korbo, 1999). Part of the neurodegeneration in brain is related to ethanol-induced inflammation through the Toll-like receptors and induction of the NF-κB pathway (Alfonso-Loeches et al., 2012; Crews & Nixon, 2009; Qin & Crews, 2012). Neuroinflammation may also play a part in the addiction process because alcohol and stress induce innate immune genes via the NF-κB pathway that lead to changes in behavior that mimic addiction (Blednov et al., 2011; Blednov et al., 2012; Crews et al., 2011; Mayfield et al., 2013). Inflammation has been seen to block neurogenesis through the NF-κB pathway in depression (Koo et al., 2010), and neurogenesis can be restored by blocking inflammation (Monje et al., 2003).
To obtain a global picture of changes in gene expression in the hippocampi of alcoholics, we conducted a microarray study of post mortem hippocampi from 20 alcoholics and 19 controls. We report the differences in gene expression between alcoholics and controls and the pathways affected. We compare our results with genes identified in other human brain expression studies and in genome-wide association studies (GWAS) for alcohol dependence or phenotypes associated with alcohol use disorders to look for genes in common and the pathways they delineate.
Materials and Methods
Hippocampal tissue from 20 alcoholics and 19 controls, all of European background (6 females in each group), was obtained from the New South Wales Tissue Resource Centre at the University of Sydney, Australia (Sheedy et al., 2008). Supplemental Table S1 describes the samples used. Total RNA was extracted using TRIzol® Reagent (Invitrogen; Carlsbad, CA) following a modified protocol with twice as much TRIzol® per gram of tissue (Edenberg et al., 2005). RNA was further purified using the Qiagen RNeasy mini-kit (Qiagen; Valencia, CA). Quality of the RNA, determined using the Agilent Bioanalyzer (Agilent; Santa Clara, CA), did not significantly differ between the 2 groups (mean RIN 6.8, SD 1).
RNA was labeled and hybridized to Affymetrix Gene 1.0 ST arrays, following the standard WT protocol (GeneChip® Whole Transcript [WT] Sense Target Labeling Assay, rev. 5, www.affymetrix.com). Samples were processed in 2 groups, balanced by phenotype and sex. Arrays were scanned and data were imported into Partek Genomics Suite version 6.2 (Partek, Inc.; St. Louis, MO).
Robust Multichip Average signals (RMA) (Irizarry et al., 2003) were generated for the core probe sets using the RMA background correction. Quantile normalization and summarization was done by Median Polish analysis using the Partek Genomics Suite. Summarized signals for each probe set were log2 transformed. These data are deposited in the NCBI Gene Expression Omnibus under series number GSE44456. The log2 transformed signals were used for principal components analysis, hierarchical clustering, and signal histograms to determine if there were any outlier arrays; none were found. We have previously shown that removing probe sets not reliably detected above background in any experimental condition improves analysis by reducing the multiple testing burden (McClintick & Edenberg, 2006). The signal histogram (not shown) indicated that probe sets with log2 values < 4 were at background level. Therefore, probe sets with mean log2 values < 4.0 in both alcoholics and controls were removed. The remaining probe sets were analyzed using a 3-way ANOVA with the factors of phenotype (control/alcoholic), sex (male/female), and processing batch (for potential technical variations). Interaction between sex and phenotype was not significant after correcting for multiple testing (Storey & Tibshirani, 2003) and was removed from the analysis. Fold changes were calculated using the untransformed RMA signals. False discovery rates (FDR) were calculated using q-value (Storey & Tibshirani, 2003).
We collected lists of differentially expressed genes from 10 other gene expression studies of post mortem brain tissue comparing alcoholics to controls (Flatscher-Bader et al., 2010; Flatscher-Bader et al., 2005; Iwamoto et al., 2004; Kryger & Wilce, 2010; Lewohl et al., 2000; Liu et al., 2007; Liu et al., 2006; Mayfield et al., 2002; Sokolov et al., 2003; Zhou et al., 2011b). Similarly, we assembled lists of genes identified in 12 recent GWAS studies of risk for alcoholism or related traits (Bierut et al., 2010; Edenberg et al., 2010; Foroud et al., 2007; Hack et al., 2011; Johnson et al., 2011; Kendler et al., 2011; Lind et al., 2010; Treutlein et al., 2009; Wang et al., 2012; Xuei et al., 2006; Zlojutro et al., 2011; Zuo et al., 2012). We annotated the list of differentially expressed genes from our study (Supplemental Table S2) to show these overlaps. We also created a list of genes identified by 2 or more studies (including the present one) in Supplemental Table S4; these will be referred to as “multiply-identified genes” in the rest of the text.
To identify transcripts enriched in different cell types we used 3 files from Cahoy et al. (2008): astrocytes (Cahoy Supplemental Table S4), oligodendrocytes (Cahoy Supplemental Table S5), and neurons (Cahoy Supplemental Table S6). These were matched by the official gene symbol (HUGO Gene Nomenclature Committee) to our data set.
Ingenuity Pathway Analysis (IPA, www.Ingenuity.com) was performed using probe sets with an FDR ≤ 0.20 to examine Canonical Pathways. For all of our analyses the Ingenuity knowledge base was used as the reference set to insure all analyses used similar parameters. We analyzed the list of probe sets identified at FDR ≤ 0.20 from our study, the list of multiply identified genes described above, and the cell-type enriched sets of genes described above. We also carried out an IPA Upstream Regulator report to identify transcription factors, cytokines, and chemicals, etc. that are predicted to be activated or inactivated based on the direction of change in their downstream targets; a positive Z-score indicates likely activation and a negative Z-score indicates likely inactivation in alcoholics relative to the controls.
Quantitative Real-Time PCR (qRT-PCR) was used to confirm differences in 4 genes: FKBP5, GRM3, NR3C1, and NR4A2. Primers were selected from Life Technologies™ (Carlsbad, CA) catalog of Taqman® Gene Expression Assays (http://bioinfo.appliedbiosystems.com/genome-database/gene-expression.html). One µg of total RNA from each sample was used for reverse transcription using the High Capacity cDNA Reverse Transcription Kit (Life Technologies™, Carlsbad, CA). Each gene of interest was measured in duplicate using TaqMan® Fast Advanced Master Mix (Life Technologies). Primers for POL2RA (Taqman® primer: Hs00172187_m1) were included in each well as a control. The CT of the POL2RA run in the same well was subtracted from the CT of the target gene to yield the Delta CT (relative expression). The Delta CT from 2 replicates for each sample was used in a 3-way ANOVA using phenotype, sex, and sample ID as factors.
Results
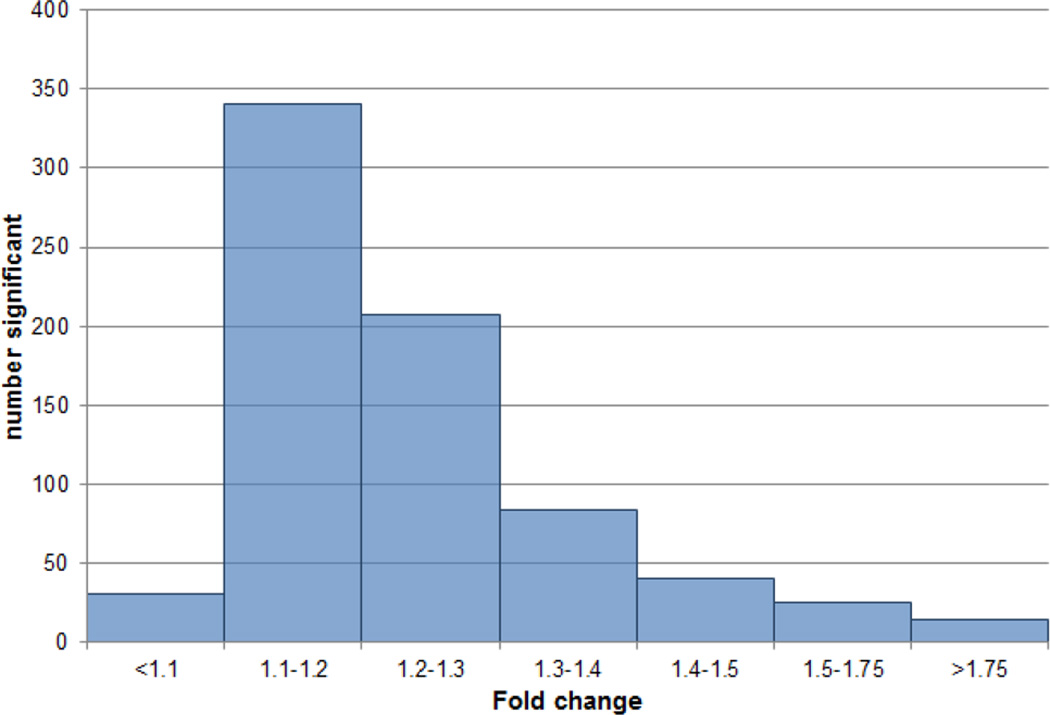
We analyzed RNA extracted from the hippocampi of 20 alcoholics and 19 controls (6 females in each group) using Affymetrix Gene 1.0 ST microarrays. Supplemental Table S1 describes the samples. Subject age and RNA integrity (RIN) did not significantly differ between alcoholics and controls (all p > 0.4). The single factor that most affects microarray measurement of gene expression from post mortem brain tissue is the pH (Atz et al., 2007); pH (mean 6.5, SD 0.3) did not significantly differ between alcoholics and controls. A total of 22,987 probe sets (80% of the core probe sets on the Affymetrix Gene 1.0 ST array) were expressed (detected above background) in at least 1 of the 2 groups (alcoholics or controls). A 3-way ANOVA using factors for phenotype (alcoholic/control), sex, and microarray-processing batch detected 743 probe sets that significantly differed between alcoholics and controls at a False Discovery Rate (FDR) ≤ 0.20. This represented 639 named genes (46 of which were measured twice) plus 58 unnamed probe sets (Supplemental Table S2). Among the significant probe sets, 50% (52% of the named genes) showed absolute fold changes ≥ 1.2 (Figure 1). Slightly over half the changes (53%) reflected lower expression in the alcoholics.
Figure 1. Distribution of fold changes for the 743 transcripts significant at FDR ≤ 0.20.
Large fold changes were found among genes associated with inflammatory and immune response (GO:0006954 and GO:0006955), particularly interleukin receptors (Table 1A). Twenty-one genes involved in hypoxia (GO:0001666) were differentially expressed, with two-thirds of them showing higher expression in the brains of alcoholics (Table 1B). The expression of most genes in the glucocorticoid pathway, including the glucocorticoid receptor (NR3C1) and 2 FK506 binding proteins (FKBP4, FKBP5), differed significantly between alcoholics and controls. NR3C1 expression was 30% lower in alcoholics, whereas FKBP5, which functions as a negative regulator of the pathway, was increased over 2-fold (Table 1C). Genes related to myelination and oligodendrocytes demonstrated decreased expression in the alcoholic hippocampi (Table 1D). Fourteen of 16 significantly changed genes in this group were expressed at lower levels in alcoholics, averaging 74%, whereas only 2 were at higher levels. Eight metallothioneins (MT) with an FDR ≤ 20% were expressed at higher levels in the hippocampus of alcoholics (mean 1.44-fold), and 9 more (20% < FDR ≤ 40%) were also expressed at higher levels in alcoholics (mean 1.2-fold; Table 1E).
Table 1.
Functional categories of selected genes that significantly differ between alcoholics and controls (FDR ≤ 0.20)
| A. Inflammatory / immune
response GO (0006954 & 0006955) | ||||
|---|---|---|---|---|
| Gene symbol | Gene title | Fold | p value | FDR |
| ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 |
−1.55 | 1.1E-02 | 0.21 |
| TAC1 | tachykinin, precursor 1 | −1.53 | 2.9E-02 | 0.27 |
| TNFSF10 | tumor necrosis factor
(ligand) superfamily, member 10 |
−1.37 | 1.5E-02 | 0.23 |
| LIPA | lipase A, lysosomal acid,
cholesterol esterase |
−1.34 | 1.4E-03 | 0.13 |
| HDAC9 | histone deacetylase 9 | −1.31 | 7.8E-03 | 0.19 |
| PXK | PX domain containing
serine/threonine kinase |
−1.31 | 5.8E-03 | 0.18 |
| IKBKAP | inhibitor of kappa light polypeptide
gene enhancer in B-cells, kinase complex-associated protein |
−1.23 | 3.8E-04 | 0.08 |
| SEMA4D | sema domain, immunoglobulin
domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, |
−1.22 | 1.2E-02 | 0.21 |
| KLRG1 | killer cell lectin-like receptor
subfamily G, member 1 |
−1.21 | 5.6E-04 | 0.10 |
| BLNK | B-cell linker | −1.20 | 3.2E-02 | 0.28 |
| OAS1 | 2’,5’-oligoadenylate
synthetase 1, 40/46kDa |
−1.20 | 2.6E-03 | 0.15 |
| PRKRA | protein kinase,
interferon-inducible double stranded RNA dependent activator |
−1.20 | 2.7E-02 | 0.27 |
| PLA2G4C | phospholipase A2, group IVC (cytosolic, calcium-independent) |
−1.18 | 3.6E-02 | 0.29 |
| IGKC | immunoglobulin kappa constant | −1.13 | 1.6E-02 | 0.23 |
| TRAF6 | TNF receptor-associated factor 6 | −1.12 | 4.1E-03 | 0.16 |
| ITCH | itchy E3 ubiquitin protein
ligase homolog (mouse) |
−1.11 | 2.7E-02 | 0.27 |
| ADORA1 | adenosine A1 receptor | −1.11 | 3.4E-03 | 0.16 |
| AKT1 | v-akt murine thymoma viral
oncogene homolog 1 |
1.08 | 7.0E-03 | 0.19 |
| GTPBP1 | GTP binding protein 1 | 1.10 | 3.1E-03 | 0.15 |
| KIR2DL3 | killer cell immunoglobulin-like
receptor, two domains, long cytoplasmic tail, 3 |
1.10 | 2.7E-02 | 0.27 |
| FCGRT | Fc fragment of IgG,
receptor, transporter, alpha |
1.12 | 2.7E-02 | 0.27 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta |
1.13 | 3.7E-02 | 0.29 |
| LTB4R | leukotriene B4 receptor | 1.13 | 6.5E-03 | 0.18 |
| FTH1 | ferritin, heavy polypeptide 1 | 1.13 | 3.0E-02 | 0.28 |
| SMAD1 | SMAD family member 1 | 1.14 | 1.5E-02 | 0.23 |
| MR1 | major histocompatibility complex,
class I-related |
1.15 | 3.2E-02 | 0.28 |
| PROK2 | prokineticin 2 | 1.15 | 3.5E-02 | 0.29 |
| ULBP2 | UL16 binding protein 2 | 1.18 | 2.8E-02 | 0.27 |
| S1PR3 | sphingosine-1-phosphate receptor 3 | 1.20 | 1.4E-02 | 0.22 |
| TGFBR3 | transforming growth factor,
beta receptor III |
1.25 | 1.2E-02 | 0.21 |
| TNFRSF1A | tumor necrosis factor
receptor superfamily, member 1A |
1.25 | 2.6E-03 | 0.15 |
| KIR2DL3 | killer cell immunoglobulin-like
receptor, two domains, long cytoplasmic tail, 3 |
1.27 | 3.1E-02 | 0.28 |
| C1R | complement component 1,
r subcomponent |
1.27 | 7.8E-03 | 0.19 |
| PNP | purine nucleoside phosphorylase | 1.27 | 2.9E-03 | 0.15 |
| TARP | TCR gamma alternate reading
frame protein |
1.39 | 3.0E-04 | 0.08 |
| IFITM2 | interferon induced
transmembrane protein 2 (1–8D) |
1.46 | 1.7E-03 | 0.14 |
| SLC11A1 | solute carrier family 11
(proton-coupled divalent metal ion transporters), member 1 |
1.51 | 2.1E-03 | 0.14 |
| IL4R | interleukin 4 receptor | 1.57 | 2.1E-04 | 0.08 |
| IFITM3 | interferon induced
transmembrane protein 3 (1-8U) |
1.60 | 6.2E-05 | 0.05 |
| IL1R1 | interleukin 1 receptor, type I | 1.71 | 1.7E-05 | 0.03 |
| CD163 | CD163 molecule | 1.80 | 3.7E-03 | 0.16 |
| S100A8 | S100 calcium binding protein A8 | 1.85 | 7.8E-03 | 0.19 |
| IL1RL1 | interleukin 1 receptor-like 1 | 1.87 | 7.9E-04 | 0.11 |
| SERPINA3 | serpin peptidase inhibitor, clade
A (alpha-1 antiproteinase, antitrypsin), member 3 |
2.28 | 4.0E-03 | 0.16 |
| B. Hypoxia GO (0001666) | ||||
| Gene symbol | Gene title | Fold | p value | FDR |
| ITGA2 | integrin, alpha 2 (CD49B, alpha
2 subunit of VLA-2 receptor) |
−1.40 | 1.5E-02 | 0.23 |
| PYGM | phosphorylase, glycogen, muscle | −1.29 | 2.8E-02 | 0.27 |
| VLDLR | very low density lipoprotein receptor | −1.28 | 3.7E-03 | 0.16 |
| PRKCQ | protein kinase C, theta | −1.21 | 2.8E-02 | 0.27 |
| HSP90B1 | heat shock protein 90kDa beta
(Grp94), member 1 |
−1.17 | 1.5E-02 | 0.23 |
| ADAM17 | ADAM metallopeptidase domain 17 | −1.14 | 3.0E-02 | 0.28 |
| BIRC2 | baculoviral IAP repeat-containing 2 | −1.13 | 1.7E-02 | 0.24 |
| EGLN2 | egl nine homolog 2 (C. elegans) | 1.10 | 8.0E-03 | 0.19 |
| PLD2 | phospholipase D2 | 1.11 | 2.9E-02 | 0.28 |
| ECE1 | endothelin converting enzyme 1 | 1.14 | 3.7E-02 | 0.29 |
| HIF1A | hypoxia inducible factor 1,
alpha subunit (basic helix-loop-helix transcription factor) |
1.15 | 2.0E-02 | 0.25 |
| SOD2 | superoxide dismutase 2, mitochondrial | 1.16 | 1.7E-02 | 0.24 |
| SDC2 | syndecan 2 | 1.18 | 2.5E-02 | 0.27 |
| ADM | adrenomedullin | 1.25 | 2.2E-02 | 0.26 |
| SOCS3 | suppressor of cytokine signaling 3 | 1.31 | 7.6E-03 | 0.19 |
| TGFB3 | transforming growth factor, beta 3 | 1.32 | 3.7E-02 | 0.29 |
| DDIT4 | DNA-damage-inducible transcript 4 | 1.39 | 7.9E-03 | 0.19 |
| HIF3A | hypoxia inducible factor 3,
alpha subunit |
1.40 | 1.5E-03 | 0.13 |
| PDLIM1 | PDZ and LIM domain 1 | 1.42 | 1.1E-02 | 0.21 |
| ANGPTL4 | angiopoietin-like 4 | 1.57 | 8.6E-05 | 0.06 |
| EDN1 | endothelin 1 | 1.65 | 3.3E-04 | 0.08 |
| C. HPA Axis | ||||
| Gene symbol | Gene title | Fold | p value | FDR |
| HSPA1A | heat shock 70kDa protein 1A | −1.47 | 7.7E-03 | 1.9E-01 |
| HSP90AA1 | heat shock protein 90kDa
alpha (cytosolic), class A member 1 |
−1.37 | 3.7E-04 | 8.5E-02 |
| HSPA5 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | −1.29 | 1.9E-03 | 1.4E-01 |
| NR3C1 | nuclear receptor subfamily 3, group
C, member 1 (glucocorticoid receptor) |
−1.29 | 1.8E-05 | 2.7E-02 |
| HSPA8 | heat shock 70kDa protein 8 | −1.26 | 7.7E-03 | 1.9E-01 |
| FKBP4 | FK506 binding protein 4, 59kDa | −1.16 | 3.4E-03 | 1.6E-01 |
| FKBP5 | FK506 binding protein 5 | 2.21 | 4.6E-06 | 2.1E-02 |
| D. Myelination | ||||
| Gene symbol | Gene title | Fold | p value | FDR |
| UGT8 | UDP glycosyltransferase 8 | −1.73 | 1.4E-04 | 0.07 |
| ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 |
−1.55 | 1.1E-02 | 0.21 |
| KLK6 | kallikrein-related peptidase 6 | −1.46 | 3.0E-03 | 0.15 |
| MOG | myelin oligodendrocyte glycoprotein | −1.46 | 3.6E-03 | 0.16 |
| TF | transferrin | −1.44 | 6.0E-03 | 0.18 |
| ASPA | aspartoacylase | −1.28 | 6.6E-02 | 0.33 |
| PLP1 | proteolipid protein 1 | −1.26 | 1.6E-03 | 0.14 |
| OMG | oligodendrocyte myelin glycoprotein | −1.25 | 3.9E-03 | 0.16 |
| PLLP | plasmolipin | −1.23 | 1.9E-02 | 0.25 |
| MAG | myelin associated glycoprotein | −1.23 | 8.1E-02 | 0.35 |
| CNP | 2’,3’-cyclic nucleotide
3’ phosphodiesterase |
−1.20 | 4.2E-02 | 0.30 |
| ERBB3 | v-erb-b2 erythroblastic leukemia
viral oncogene homolog 3 (avian) |
−1.20 | 4.3E-02 | 0.31 |
| PMP2 | peripheral myelin protein 2 | −1.18 | 5.8E-02 | 0.33 |
| MYEF2 | myelin expression factor 2 | −1.09 | 3.6E-02 | 0.29 |
| MYT1 | myelin transcription factor 1 | 1.16 | 2.4E-02 | 0.26 |
| MPZL2 | myelin protein zero-like 2 | 1.84 | 1.5E-03 | 0.14 |
| E. Metallothioneins | ||||
| Gene symbol | Gene title | Fold | p value | FDR |
| MT1X | metallothionein 1X | 1.98 | 1.8E-05 | 0.03 |
| MT1M | metallothionein 1M | 1.50 | 1.1E-03 | 0.13 |
| MT1A | metallothionein 1A | 1.48 | 2.6E-04 | 0.08 |
| MT2A | metallothionein 2A | 1.47 | 1.1E-03 | 0.13 |
| MT1G | metallothionein 1G | 1.38 | 2.1E-02 | 0.25 |
| MT1L | metallothionein 1L (gene/pseudogene) | 1.37 | 2.0E-03 | 0.14 |
| MT1JP | metallothionein 1J (pseudogene) | 1.26 | 1.6E-02 | 0.23 |
| MT1P3 | metallothionein 1 pseudogene 3 | 1.25 | 3.3E-03 | 0.16 |
| MT1DP | metallothionein 1D (pseudogene) | 1.24 | 7.1E-03 | 0.19 |
| MT1E | metallothionein 1E | 1.22 | 5.7E-02 | 0.33 |
| MT1B | metallothionein 1B | 1.21 | 1.1E-02 | 0.21 |
| MT1P2 | metallothionein 1 pseudogene 2 | 1.21 | 6.9E-03 | 0.19 |
| MT1F | metallothionein 1F | 1.19 | 1.1E-01 | 0.37 |
| MT3 | metallothionein 3 | 1.16 | 6.1E-02 | 0.33 |
| MT1H | metallothionein 1H | 1.15 | 6.4E-02 | 0.33 |
| MT1IP | metallothionein 1I (pseudogene) | 1.14 | 4.6E-02 | 0.31 |
| MT4 | metallothionein 4 | 1.13 | 7.4E-02 | 0.34 |
Ingenuity Pathways Analysis (IPA) of genes with FDR ≤ 0.20 revealed many canonical pathways that differed between alcoholics and controls (Table 2). Signaling pathways predominated, along with stress or immune responses. Acute phase response signaling, IL-6 signaling, IL-8 signaling, IL-10 signaling, LPS/IL-1 mediated inhibition of RXR function, mTOR signaling, hypoxia signaling, p38 MapK signaling, EIF2 signaling (eukaryotic translation initiation factor 2), and glucocorticoid signaling were up. GADD45 (growth arrest and DNA-damage-inducible) signaling, p38 signaling, and Her2 signaling, were mixed or down. Many of the pathways shared key genes. ATM (ataxia telangiectasia mutated; down 20%) is in 39 of the 60 pathways and AKT1 (v-akt murine thymoma viral oncogene homolog 1; increased 8%) is in 32 of the pathways. TRAF6, PRKD1, MAP2K3, RHOB and RHOC, CREB1, CCND, and the guanine binding proteins GNAI1, GNB2, and GNG5 were each in at least 12 of the pathways.
Table 2. Ingenuity pathway analysis using genes differentially expressed in hippocampi of alcoholics.
Pathways in Section A are common to genes identified in multiple studies. Section B lists pathways identified only in this study.
| Canonical Pathways | p value | Significant genes in the pathway |
|---|---|---|
| A. Pathways common to multiple studies | ||
| Acute Phase Response Signaling |
1.1E-04 |
SOCS3, TCF4, SERPING1,
TNFRSF1A MAP3K1, VWF, SERPINA3, IL1R1, NR3C1 TRAF6, C1R, AKT1, TF, CFB, MAP2K3, OSMR |
| Aldosterone Signaling in Epithelial Cells |
1.1E-04 |
HSPA1A/HSPA1B, HSPH1, SLC12A2,
DNAJA1 HSPA5, HSPA1L, PLCD1, HSPA8, HSPE1 HSP90AA1, HSPB7, DNAJB6, PLCD4, PRKD1 ATM |
| Axonal Guidance Signaling | 2.1E-02 |
PXN, PAPPA, C9orf3, GNAI1, DPYSL5,
SLIT2 ADAMTS9, TUBA1B, PLCD1, SEMA6D, AKT1 GNB2, ADAM10, RTN4, GNG5, ERBB2 SEMA4B, PLCD4, MYL3, FARP2, PRKD1, ATM |
| Cell Cycle: G1/S
Checkpoint Regulation |
4.9E-02 | HDAC9, CCND3, PAK1IP1, CCND1, ATM |
| CXCR4 Signaling | 2.6E-02 |
PXN, AKT1, RHOB, RHOC, GNB2,
GNAI1 GNG5, MYL3, PRKD1, ATM |
| Cyclins and Cell Cycle Regulation |
4.4E-02 |
CCNA2, HDAC9, CCNA1, CCND3,
CCND1 ATM |
| EIF2 Signaling | 2.8E-05 |
RPL24, RPS2, RPL23A,
RPS17/RPS17L RPLP0, RPL7, RPL10A, RPL35, RPS3A, AKT1 RPL7A, RPL39, RPL19, RPL12, RPS5, RPL29 ATM, RPSA |
| Estrogen-mediated S-phase Entry |
4.1E-02 | CCNA2, CCNA1, CCND1 |
| Glioma Invasiveness Signaling | 3.8E-02 | TIMP4, RHOB, TIMP1, RHOC, ATM |
| HGF Signaling | 4.2E-02 |
PXN, AKT1, MAP3K6, MAP3K1,
CCND1 PRKD1, ATM |
| ILK Signaling | 4.0E-02 |
PXN, CDH1, AKT1, RHOB, TNFRSF1A,
RHOC CREB1, ITGB4, CCND1, MYL3, ATM |
| Inhibition of
Matrix Metalloproteases |
7.9E-03 | TIMP4, TIMP1, THBS2, ADAM10, MMP24 |
| mTOR Signaling | 3.5E-03 |
NAPEPLD, DDIT4, RHOC, RPS2,
PRR5L RPS17/RPS17L, PLD1, AKT1, RPS3A, RHOB RPS5, PRKD1, ATM, RPSA |
| p70S6K Signaling | 4.0E-02 |
PLCD1, IL4R, AKT1, GNAI1, PLCD4,
PLD1 PRKD1, ATM |
| Protein Ubiquitination Pathway | 1.8E-02 |
USP28, MED20, HSPA1A/HSPA1B,
HSPH1 USP19, DNAJA1, HSPA5, HSPA1L, HSPA8 TRAF6, UBE2G1, HSPE1, HSP90AA1, HSPB7 DNAJB6 |
| Reelin Signaling in Neurons | 4.2E-02 |
AKT1, ARHGEF2, PAFAH1B1, VLDLR,
ATM APP |
| RhoGDI Signaling | 2.4E-02 |
CDH1, PPP1R12C, RHOB, RHOC,
GNB2 GNAI1, GNG5, ARHGEF17, ARHGEF2, DLC1 MYL3 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis |
3.8E-02 |
SOCS3, TCF4, IL1RL1, TNFRSF1A,
CEBPD IL1R1, CCND1, PLCD1, TRAF6, AKT1, CREB1 MAP2K3, PLCD4, PRKD1, TCF7L2, ATM |
| Signaling by Rho Family GTPases |
7.6E-03 |
SEPT8, PPP1R12C, RHOC, SEPT7,
GNAI1 ARHGEF17, PLD1, CDH1, RHOB, GNB2 GNG5, ARHGEF2, PARD3, MYL3, ATM |
| TR/RXR Activation | 1.9E-02 |
KLF9, AKT1, NXPH2, ACACA,
THRA TBL1XR1, ATM |
| Type II Diabetes
Mellitus Signaling |
3.4E-02 |
SOCS3, AKT1, TNFRSF1A, MAP3K1,
ACSL5 SLC27A3, PRKD1, ATM |
| B. Additional significant pathways | ||
| Activation of IRF by Cytosolic Pattern Recognition Receptors |
1.5E-02 | DHX58, IFIH1, TRAF6, ZBP1, IKBKAP, IFIT2 |
| Acute Myeloid Leukemia Signaling |
1.2E-02 |
TCF4, AKT1, CCNA1, MAP2K3,
CCND1 TCF7L2, ATM |
| Aryl Hydrocarbon
Receptor Signaling |
1.9E-03 |
TGM2, ALDH4A1, CCNA2, ALDH1L1,
CCNA1 CCND3, HSP90AA1, HSPB7, DHFR, CCND1 PTGES3, ATM |
| ATM Signaling | 4.4E-02 | MDM4, GADD45A, CREB1, BLM, ATM |
| Biotin-carboxyl Carrier
Protein Assembly |
5.9E-03 | ACACB, ACACA |
| Cardiac Hypertrophy Signaling | 1.4E-02 |
MAP3K6, RHOC, MAP3K1, GNAI1,
PLCD1 AKT1, RHOB, CREB1, GNB2, GNG5, MAP2K3 PLCD4, MYL3, ATM |
| Colorectal Cancer
Metastasis Signaling |
4.9E-02 |
TCF4, TNFRSF1A, RHOC, CCND1,
MMP24 CDH1, AKT1, MSH2, RHOB, GNB2, GNG5 TCF7L2, ATM |
| Complement System | 2.0E-02 | C1R, SERPING1, CD59, CFB |
| Endometrial Cancer Signaling | 2.5E-02 | CDH1, AKT1, ERBB2, CCND1, ATM |
| eNOS Signaling | 9.8E-03 |
HSPA8, CCNA2, AKT1,
CCNA1 HSPA1A/HSPA1B, HSP90AA1, HSPA5 NOSTRIN, HSPA1L, ATM |
| GADD45 Signaling | 3.4E-03 | GADD45A, CCND3, CCND1, ATM |
| Germ Cell-Sertoli Cell
Junction Signaling |
4.6E-03 |
PXN, CDH1, AKT1, MAP3K6,
RHOB TNFRSF1A, RHOC, MAP3K1, MTMR2 MAP2K3, TUBA1B, ATM |
| Glucocorticoid Receptor Signaling |
4.7E-03 |
HSPA1A/HSPA1B, MAP3K1, HSPA5,
CD163 NR3C1, TAF13, TSC22D3, PTGES3, HSPA1L HSPA8, TRAF6, AKT1, CREB1, FKBP4 HSP90AA1, FKBP5, ATM |
| HER-2 Signaling in Breast Cancer |
1.1E-02 |
AKT1, ITGB4, ERBB2, PARD3,
CCND1 PRKD1, ATM |
| Hereditary Breast Cancer Signaling |
3.4E-02 |
HDAC9, AKT1, MSH2, GADD45A,
BLM CCND1, FANCL, ATM |
| HIF1α Signaling | 4.8E-02 |
EGLN2, AKT1, EDN1, MAPK4,
HSP90AA1 MMP24, ATM |
| HMGB1 Signaling | 3.0E-02 |
AKT1, RHOB, TNFRSF1A, RHOC,
MAP2K3 IL1R1, ATM |
| Huntington’s Disease Signaling | 5.8E-03 |
HDAC9, HSPA1A/HSPA1B, DNM3,
HSPA5 HSPA1L, ZDHHC17, HSPA8, TGM2, DYNC1I2 AKT1, CREB1, GNB2, GNG5, PRKD1, ATM |
| Hypoxia Signaling in the Cardiovascular System |
1.9E-02 |
AKT1, EDN1, UBE2G1, CREB1,
HSP90AA1 ATM |
| IL-1 Signaling | 3.0E-02 |
TRAF6, MAP3K1, GNB2, GNAI1,
GNG5 MAP2K3, IL1R1 |
| IL-10 Signaling | 2.2E-02 | TRAF6, SOCS3, IL4R, IL1RL1, MAP2K3, IL1R1 |
| IL-6 Signaling | 1.3E-02 |
TRAF6, SOCS3, AKT1, TNFRSF1A,
IL1RL1 HSPB7, MAP2K3, IL1R1, ATM |
| IL-8 Signaling | 3.0E-03 |
NAPEPLD, RHOC, GNAI1, CCND1,
PLD1 TRAF6, CDH1, AKT1, CCND3, RHOB, GNB2 GNG5, PRKD1, ATM |
| LPS/IL-1 Mediated Inhibition of RXR Function |
2.7E-02 |
ALDH4A1, TNFRSF1A, IL1RL1,
MAP3K1 IL1R1, FMO5, TRAF6, ALDH1L1, UST, ACSL5 NR5A2, SLC27A3, HS3ST5 |
| LXR/RXR Activation | 4.2E-02 |
SCD, TF, TNFRSF1A, IL1RL1, MYLIP,
ACACA S100A8, IL1R1 |
| Melanoma Signaling | 4.5E-02 | CDH1, AKT1, CCND1, ATM |
| P2Y Purigenic Receptor Signaling Pathway |
6.0E-03 |
PLCD1, AKT1, CREB1, GNB2, GNAI1,
P2RY12 GNG5, PLCD4, PRKD1, ATM |
| p38 MAPK Signaling | 3.6E-02 |
TRAF6, TNFRSF1A, IL1RL1, DUSP10,
CREB1 HSPB7, MAP2K3, IL1R1 |
| Phospholipase C Signaling | 4.0E-02 |
HDAC9, NAPEPLD, RHOC, ARHGEF17,
PLD1 TGM2, RHOB, CREB1, GNB2, GNG5 ARHGEF2, MYL3, PRKD1 |
| Phospholipases | 3.5E-02 | PLCD1, NAPEPLD, PLA1A, PLCD4, PLD1 |
| Protein Kinase A Signaling | 3.9E-02 |
TCF4, PXN, PTPRD, MAP3K1, GNAI1,
TTN PDE8A, PLCD1, DUSP10, CREB1, GNB2 GNG5, DUSP7, PLCD4, MYL3, PDE6B TCF7L2, PRKD1, DUSP16 |
| Role of BRCA1 in DNA Damage Response |
4.4E-02 | MSH2, GADD45A, BLM, FANCL, ATM |
| Role of NFAT in
Cardiac Hypertrophy |
3.3E-02 |
PLCD1, HDAC9, AKT1, MAP3K1,
GNB2 GNAI1, GNG5, MAP2K3, PLCD4, PRKD1, ATM |
| Role of PKR in Interferon Induction and Antiviral Response |
3.9E-02 | TRAF6, AKT1, TNFRSF1A, MAP2K3 |
| Superpathway of D-myo-inositol
(1,4,5)-trisphosphate Metabolism |
4.1E-02 | INPP1, ITPKC, IMPA2 |
| Thrombin Signaling | 8.9E-03 |
RHOC, GNAI1, PLCD1, AKT1, RHOB,
CREB1 GNB2, GNG5, ARHGEF2, PLCD4, MYL3 PRKD1, ATM |
| Thyroid Cancer Signaling | 3.9E-02 | CDH1, TCF4, CCND1, TCF7L2 |
| Xanthine and Xanthosine Salvage |
3.2E-02 | PNP |
| γ-linolenate Biosynthesis
II (Animals) |
1.6E-02 | ACSL5, CYB5R3, SLC27A3 |
To see whether the alcoholics differed in expression of genes enriched in particular cell types, we examined the sets of genes whose expression is known to be enriched in astrocytes, oligodendrocytes, or neurons (Cahoy et al., 2008), noted in Supplemental Table S2. The vast majority of these cell-enriched genes were not differentially expressed: about 95% have FDR > 0.20. However, for those genes that were differentially expressed, the fraction up and down was skewed compared to the overall results. Eighty-three percent of the differentially expressed transcripts enriched in oligodendrocytes were expressed at lower levels in alcoholics (p = 3.9 × 10−9), as were 83% of the differentially expressed transcripts in neurons (p = 2.1 × 10−4), whereas only 53% of the total probe sets were down. The differentially expressed genes expressed in astrocytes demonstrated the opposite trend, with 61% at higher levels in alcoholics (p = 0.003), including hypoxia response genes.
Analyzing upstream regulators can clarify the pathway findings by looking for commonalities in their regulation, i.e. it may be possible to identify sets of differentially expressed genes that are downstream targets of specific transcription factors, cytokines, signaling cascades, and endogenous and exogenous chemicals. Both the glucocorticoid and aldosterone pathways were significantly altered in alcoholic brains, and the upstream effectors analysis indicated that their receptors, NR3C1 and NR3C2, are in an activated state (Supplemental Table S3). Other genes identified as activated include many regulators related to immune function (including cytokines IL1B, IL10, IL11, IL15, IL17A, and EDN1), other regulators, including hypoxia-related gene HIF1A and Endothelial PAS domain-containing protein 1 (EPAS1), and 2 genes that are general indicators of stress, TP53 and TGFB1. The expression of downstream targets for the Wnt/β catenin pathway and the ERBB4 pathways involved in neurogenesis (Lazarov & Marr, 2010), including TCF4 and cyclin D1, provide evidence that both of these pathways were less active in the alcoholics (Supplemental Table S3).
Bioinformatic analysis found 386 genes that were identified in 2 or more studies (GWAS or gene expression, including the present study), which we refer to as multiply identified genes (listed in Supplemental Table S4). One hundred seven of these genes were identified by our study and at least one other (noted in Supplemental Tables S2 & Supplemental Tables S4). Twenty-four of these 107 were identified by at least one of the GWAS (Supplemental Table S2). The 386 multiply identified genes (Supplemental Table S4) were used for Ingenuity analysis, and 81 pathways were significantly altered (p < 0.05; Supplemental Table S5). There were 21 pathways in common between the multiply identified genes and our dataset (section A of Table 2 and of Supplemental Table 5).
We chose 4 genes to test by qRT-PCR, based upon their roles in pathways that are affected. NR3C1 is the glucocorticoid receptor gene, the key transcription factor in the glucocorticoid pathway. FKBP5 (FK506 binding protein 5) is an immunophilin gene important in that pathway that also interacts with 90 kDa heat shock protein and sequesters NR3C1 in the cytosol, increasing glucocorticoid resistance. NR4A2 is a transcription factor in the steroid-thyroid hormone-retinoid receptor superfamily, mutations in which have been related to dopaminergic dysfunction. NR4A2 has been shown to repress inflammatory genes activated by NF-κB (Saijo et al., 2009) in microglia. GRM3 (glutamate receptor, metabotropic 3) was chosen because L-glutamate is the major excitatory neurotransmitter in the central nervous system, and affects most aspects of brain function. All 4 genes showed similar fold-changes in qRT-PCR as they did in the microarrays (Table 3).
Table 3.
Confirmation by qRT-PCR
| Gene Symbol |
RT-PCR p-value |
RT-PCR Fold |
Array p-value |
Array Fold |
|---|---|---|---|---|
| FKBP5 | 2.6E-27 | 1.84 | 4.6E-06 | 2.21 |
| NR3C1 | 8.6E-03 | −1.26 | 1.8E-05 | −1.29 |
| NR4A2 | 3.9E-02 | −1.79 | 3.5E-04 | −1.95 |
| GRM3 | 1.0E-18 | −1.47 | 2.7E-05 | −1.45 |
Primers used FKBP5, Hs01561010_m1; NR3C1, Hs00353740_m1; NR4A2, Hs00428691_m1; GRM3, Hs00168260_m1; POL2RA, Hs00172187_m1 used as control to normalize sample-to-sample variation.
Discussion
This study presents a global picture of differences between alcoholics and controls in gene expression in the post mortem hippocampus. A major theme that emerges from the data is that the hippocampus in alcoholics shows dramatic signs of stress. Genes and pathways (Table 2) involved in stress responses are mostly increased in alcoholics. Metallothioneins, a large number of which are increased in the hippocampus (Table 1E), are increased in many stress conditions (Aschner & West, 2005). EIF2 signaling, which is increased, functions to resolve endoplasmic reticulum (ER) stress; if ER stress cannot be resolved, apoptosis can result (Lerner et al., 2012). TXNIP (1.7-fold higher in alcoholics) can be transcriptionally induced by TGFβ1 and glucocorticoids (Chen et al., 2010; Han et al., 2003), and can link oxidative stress to inflammation via the NLRP3 inflammasome (NLR family, pyrin domain containing 3) (Zhou et al., 2010), an upstream activator of NF-κB signaling that plays a role in the regulation of inflammation, the immune response, and apoptosis.
Signs of hypoxia are present, as evidenced by the increases in Angiopoietin-like 4, EPAS1 (endothelial PAS domain protein 1, also known as HIF2α), HIF3α, and HIF1α (15% increase, FDR 0.26) shown in Supplemental Table S2. Analysis of upstream regulators (Supplemental Table S3) reinforces this, since the pattern of expression of the genes regulated by EPAS1 and HIF1α also indicates that they are activated.
There is also evidence of involvement of the hypothalamus-pituitary-adrenal (HPA) axis, specifically the cortisol pathway (Table 1C), and particularly in astrocytes: 37% of the astrocyte-enriched genes that showed increased expression are downstream of the glucocorticoid signaling, and others are downstream of either IL1β or TGFβ1. Pathway and upstream analysis (Table 2, Supplemental Table S3) indicates that the glucocorticoid receptor is activated although its transcript level (NR3C1) is decreased. Cortisol-releasing-hormone (CRH) increases as a result of stress, ethanol abuse, chronic drinking, and the early stage of withdrawal (Armario, 2010; Gianoulakis et al., 2003; Roy et al., 2002), which should activate the glucocorticoid receptor. The HPA and CRH are also activated by alcohol consumption (Clarke & Schumann, 2009), increasing the amount of adrenocorticotropic hormone (ACTH) produced, which in turn stimulates the release of glucocorticoids (Mesotten et al., 2008). Glucocorticoids down-regulate the further release of CRH through a negative feedback loop to the hypothalamus, but increase the production of CRH outside the hypothalamus, e.g. in the central amygdala (Pastor et al., 2008). Dysregulation of the HPA axis is a known problem in alcoholism and other addictions (Armario, 2010; Koob & Kreek, 2007; Sorocco et al., 2006) as well as in at-risk individuals (Sorocco et al., 2006). The increased levels of CRH may lead to increased alcohol consumption as the brain tries to adapt to its increasingly dysregulated state (Koob & Le Moal, 2005). Increased CRH levels also lead to increased sensitivity of stress-induced alcohol consumption (Ciccocioppo et al., 2009; Clarke et al., 2009). Glucocorticoids mediate the development of sensitization to drugs such as ethanol (Roberts et al., 1995) in a feed-forward fashion.
FKBP4 and FKBP5 (over 2-fold higher) and NR3C1 itself are downstream targets of glucocorticoid signaling. FKBP5 functions as a negative regulator of the pathway by lowering the cortisol affinity of the glucocorticoid receptor and keeping it in the cytoplasm, which increases cortisol resistance and short-circuits the glucocorticoid feedback circuit (Binder, 2009; Binder et al., 2008). Mice with chronic exposure to corticosterone (the rodent equivalent of cortisol) develop anxiety and have decreased expression of Nr3c1 and Hsp90 and increased expression of FKBP5 in many tissues (Lee et al., 2010). Increased FKBP5 expression due to known polymorphisms leads to increased risk of affective and anxiety disorders (Binder et al., 2008) and bipolar disorder (Willour et al., 2009).
Our data show decreased myelination (Table 1D) in the hippocampus. Decreased hippocampal volume (Agartz et al., 1999; Laakso et al., 2000; Tyan et al., 2012) and decreases in hippocampal neurogenesis have been observed in alcoholism (Crews & Nixon, 2009; Morris et al., 2010; Richardson et al., 2009). Pathways (WNT/β catenin, reelin signaling in neurons, and ERBB4) and genes (APP, PSEN1, ADAM10, ERBB2, and reelin) that play a role in neurogenesis (Lazarov & Marr, 2010) have decreased activity or expression in the hippocampi of the alcoholics. Both inflammation (Monje et al., 2003) and stress with increased cortisol production (Schoenfeld & Gould, 2012) can inhibit neurogenesis. Chronic cortisol decreases neurogenesis and treatment with the glucocorticoid antagonist mifepristone reverses this reduction (Mayer et al., 2006).
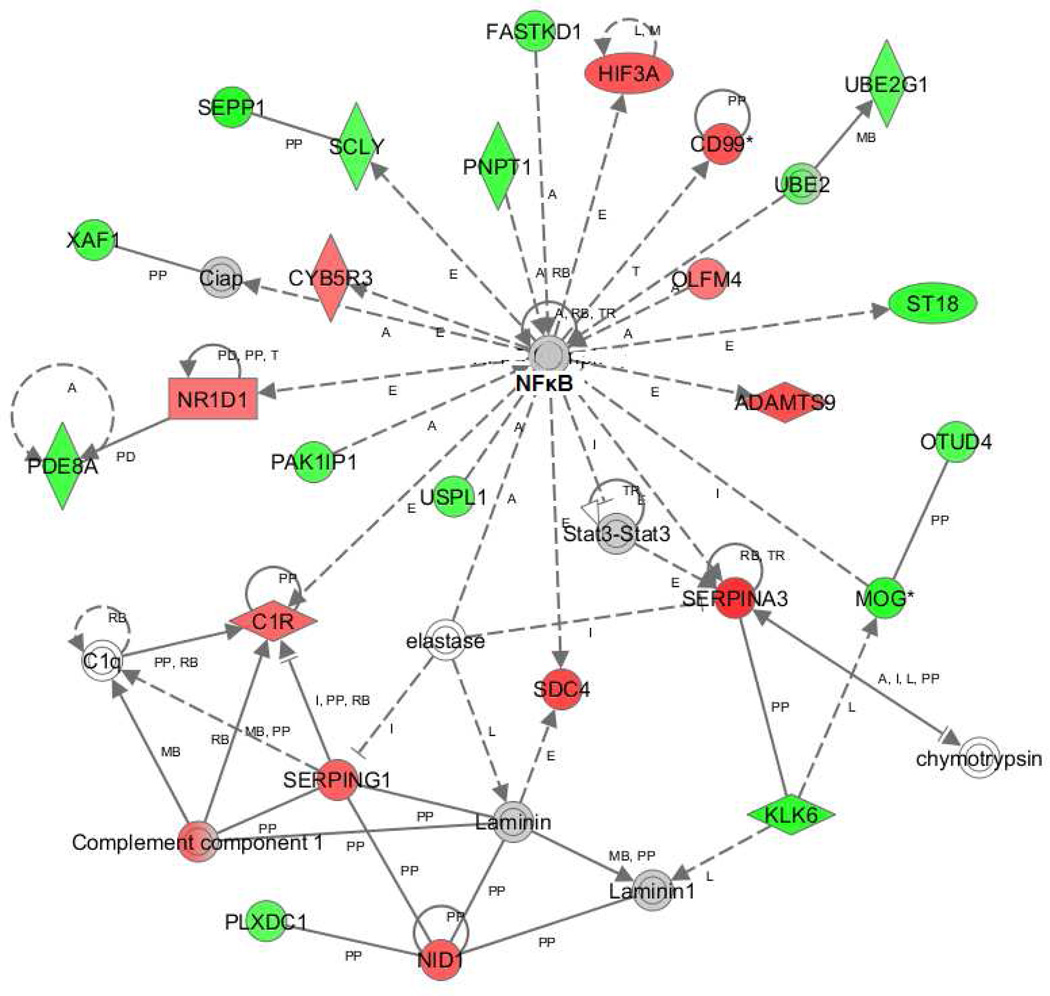
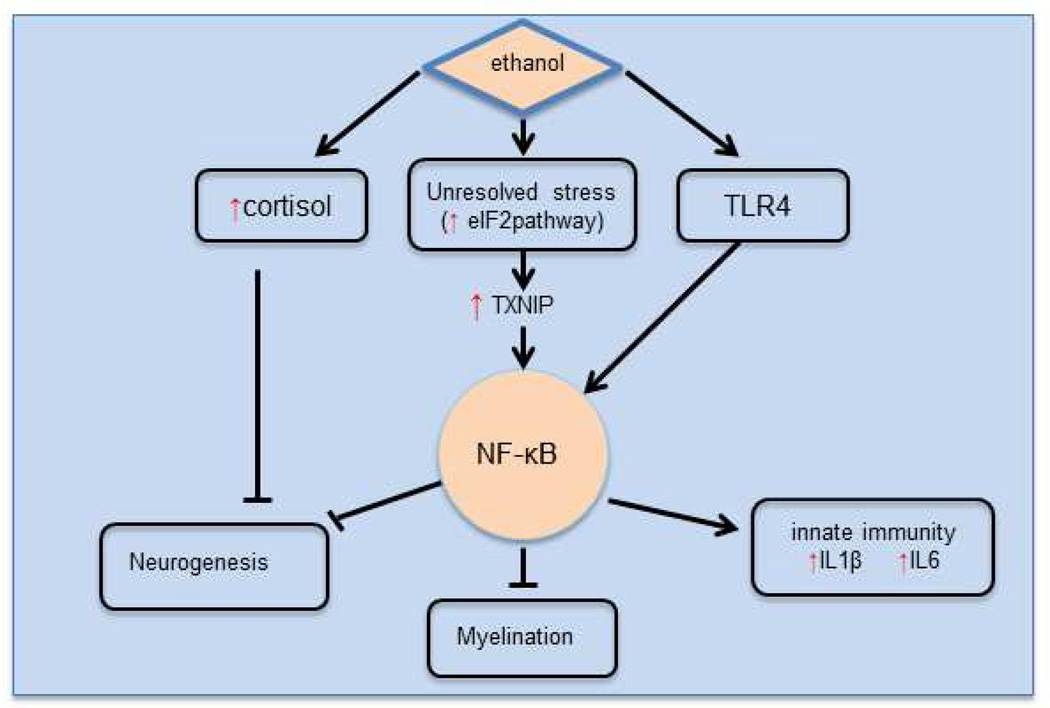
Are these stresses and dysfunctional changes related? Most of these stresses can be linked to NF-κB (Figure 2), which is connected to 25 of the differentially expressed genes in this dataset, including genes related to hypoxia, inflammation, neurogenesis, and myelin. Variations within NFKB1, a subunit of NF-κB, have been associated with alcoholism (Edenberg et al., 2008). One can conceptualize the inter-relationships as in Figure 3. Ethanol activates inflammation via the TLR4 pathway and NF-κB. Increased inflammation, via the toll-like receptor 4 (TLR4), can play a role in the loss of white matter seen in alcoholics (Alfonso-Loeches et al., 2012). Wild-type mice chronically treated with ethanol for 5 months had decreased expression of several myelin-related genes in multiple brain regions, and also a reduced number of oligodendrocytes, but Tlr4 knockout mice similarly treated did not show decreased expression of the myelin genes. The ER stress we have found, if unresolved, can also increase inflammation via TXNIP (strongly increased) and NF-κB.
Figure 2. Ingenuity Pathway Analysis network with NF-κB as central hub.
Red: genes with increased expression; green: genes with decreased expression; gray: gene in dataset but was not significantly changed; white: not in the data set used for analysis.
Figure 3. Key pathways affected by ethanol.
Ethanol intake increases cortisol and activates NF-κB via Toll-like receptor 4 (TLR4). NF-κB activation increases innate immune activity. Hippocampal neurogenesis is inhibited via NFκB. NR4A2 represses NF-κB transactivation of other genes. When stress cannot be resolved by the eIF2 pathway, transcription of TXNIP is increased which also increases NF-κB transactivation. Red and Green vertical arrows indicate pathways, genes, or signaling molecules that have increased/decreased expression or activity in the hippocampus of alcoholics.
One goal of examining gene regulation in the brain is to inform the analyses of genes that may influence risk for alcoholism. Toward that end, we compiled data from 10 previously published gene expression studies (Flatscher-Bader et al., 2010; Flatscher-Bader et al., 2005; Iwamoto et al., 2004; Kryger & Wilce, 2010; Lewohl et al., 2000; Liu et al., 2007; Liu et al., 2006; Mayfield et al., 2002; Sokolov et al., 2003; Zhou et al., 2011b), from this study, and from 12 GWAS for risk of alcoholism or alcoholic traits (Bierut et al., 2010; Edenberg et al., 2010; Foroud et al., 2007; Hack et al., 2011; Johnson et al., 2011; Kendler et al., 2011; Lind et al., 2010; Treutlein et al., 2009; Wang et al., 2012; Xuei et al., 2006; Zlojutro et al., 2011; Zuo et al., 2012). There were 386 genes identified by at least 2 of these collected studies (Supplemental Table S4). Five genes were identified in 4 studies, and are thus strong candidates for further study: selenoprotein P (SEPP1), heterochromatin protein 1 binding protein 3 (HP1BP3), transferrin (TF), EGF-like repeats and discoidin I-like domains 3 (EDIL3), and contactin associated proteinlike 2 (CNTNAP2). SEPP1 binds selenium and has antioxidant activity and is down-regulated by both inflammatory cytokines like IL1β (Dreher et al., 1997) and glucocorticoids (Rock & Moos, 2009); it is decreased in the hippocampi of alcoholics (Supplementary Table S2). Transferrin is an iron transporter and is also a negative acute phase response protein; it is also decreased. HP1BP3 has been identified as a biomarker for postpartum depression (Guintivano et al., 2013). EDIL3 can stimulate cerebral angiogenesis (Fan et al., 2008) and was down-regulated in mouse embryos exposed to ethanol (Zhou et al., 2011a). CNTNAP2 is an extremely large protein in the neurexin family, polymorphisms in which were recently found to be associated with depression and schizophrenia in a Han Chinese population (Ji et al., 2013). Several pathways identified using this list of genes overlap with the pathways identified by our study (Supplemental Table S5, Section A) which include stress-related pathways EIF2 and mTOR signaling. IPA also identified NF-κB as significantly altered for this group of multiply identified genes.
Twenty-four of the genes identified by our study were previously identified by GWAS (Supplemental Table S2, GWAS column). This list includes several genes with large fold changes, such as SLC39A10 (a zinc transporter), suppression of tumorigenicity 18 (ST18), protein tyrosine phosphatase receptor type D (PTPRD), BCL2-associated athanogene 3 (BAG3), and von Willebrand factor (VWF). Although these genes might not be thought of as related to alcoholism, their differential expression in alcoholic brains, together with their genetic connection, suggests they might be. The IPA analysis of the 107 genes in our study that were identified in other studies indicated that 38 of these genes are related to cell death, including ST18 and BAG3.
This study demonstrates many differences in gene expression between the hippocampi of alcoholics and controls, and highlights interrelated insults to the hippocampus: stress, hypoxia, inflammation, and excess cortisol (Figures 2, 3). These may play roles in the demyelination, loss of glial cells, and decreased neurogenesis seen with chronic alcohol abuse. NF-κB appears to be a key player in these processes (Figure 3). Some of these differences in gene expression may be due to genetic variations that precede the addiction process and may play an active role in the addiction process. Others may be the result of years of excessive alcohol consumption, and still others may be altered due to the interaction of genetic variation with excessive alcohol consumption. A post mortem study such as this cannot distinguish among these possibilities. The modifications seen here in gene expression in these pathways could be part of the allostatic change suggested by Koob & Kreek (2007). In the hippocampus, resetting the cortisol pathway may be one way to break this chain of events. Decreased neurogenesis and increased inflammation are also seen in major depressive illness (Koo et al., 2010), but antidepressant treatment has had mixed results in the treatment of alcoholism per se (Kranzler et al., 2012). Animal and human post mortem research indicate the innate immune function induced by TLRs and NF-κB signaling creates negative affect and stress, which with repeated cycles of ethanol abuse leads to addiction (Crews et al., 2011). This study demonstrates that this increase in the innate immune system and NF-κB signaling is still present after years of chronic drinking. With multiple stressors increasing NF-κB signaling, it may take a multi-pronged approach to normalize the brain of chronic drinkers.
Supplementary Material
Acknowledgments
COGA (principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, and L. Bierut) includes 10 different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy); Howard University (R. Taylor); and Virginia Commonwealth University (D. Dick). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and we also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Brain tissues were received from the New South Wales Tissue Resource Centre, which is supported by the National Health and Medical Research Council of Australia, The University of Sydney, Prince of Wales Medical Research Institute, Neuroscience Institute of Schizophrenia and Allied Disorders, National Institute of Alcohol Abuse and Alcoholism (Grant R01 AA12725), and NSW Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agartz I, Momenan R, Rawlings RR, Kerich MJ, Hommer DW. Hippocampal volume in patients with alcohol dependence. Archives of General Psychiatry. 1999;56:356–363. doi: 10.1001/archpsyc.56.4.356. [DOI] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual M, Gomez-Pinedo U, Pascual-Lucas M, Renau-Piqueras J, Guerri C. Toll-like receptor 4 participates in the myelin disruptions associated with chronic alcohol abuse. Glia. 2012;60:948–964. doi: 10.1002/glia.22327. [DOI] [PubMed] [Google Scholar]
- Armario A. Activation of the hypothalamic-pituitary-adrenal axis by addictive drugs: different pathways, common outcome. Trends in Pharmacological Sciences. 2010;31:318–325. doi: 10.1016/j.tips.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Aschner M, West AK. The role of MT in neurological disorders. Journal of Alzheimer’s Disease. 2005;8:139–145. doi: 10.3233/jad-2005-8206. discussion 209–215. [DOI] [PubMed] [Google Scholar]
- Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, Overman K, Stein R, Tomita H, Potkin S, et al. Methodological considerations for gene expression profiling of human brain. Journal of Neuroscience Methods. 2007;163:295–309. doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain, Behavior, and Immunity. 2011;25(Suppl 1):S92–S105. doi: 10.1016/j.bbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA. Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addiction Biology. 2012;17:108–120. doi: 10.1111/j.1369-1600.2010.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yoshihara E, Son A, Matsuo Y, Masutani H, Sugie K, Maeda M, Yodoi J. Differential roles of Annexin A1 (ANXA1/lipocortin-1/lipomodulin) and thioredoxin binding protein-2 (TBP-2/VDUP1/TXNIP) in glucocorticoid signaling of HTLV-I-transformed T cells. Immunology Letters. 2010;131:11–18. doi: 10.1016/j.imlet.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Ciccocioppo R, Gehlert DR, Ryabinin A, Kaur S, Cippitelli A, Thorsell A, Lê AD, Hipskind PA, Hamdouchi C, Lu J, et al. Stress-related neuropeptides and alcoholism: CRH, NPY, and beyond. Alcohol. 2009;43:491–498. doi: 10.1016/j.alcohol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Krause K, Li T, Schumann G. An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addiction Biology. 2009;14:366–370. doi: 10.1111/j.1369-1600.2009.00151.x. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Schumann G. Gene-environment interactions resulting in risk alcohol drinking behaviour are mediated by CRF and CRF1. Pharmacology, Biochemistry, and Behavior. 2009;93:230–236. doi: 10.1016/j.pbb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Crews FT, Nixon K. Mechanisms of neurodegeneration and regeneration in alcoholism. Alcohol and Alcoholism. 2009;44:115–127. doi: 10.1093/alcalc/agn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Zou J, Qin L. Induction of innate immune genes in brain create the neurobiology of addiction. Brain, Behavior, and Immunity. 2011;25(Suppl 1):S4–S12. doi: 10.1016/j.bbi.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher I, Jakobs TC, Köhrle J. Cloning and characterization of the human selenoprotein P promoter. Response of selenoprotein P expression to cytokines in liver cells. The Journal of Biological Chemistry. 1997;272:29364–29371. doi: 10.1074/jbc.272.46.29364. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addiction Biology. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcoholism: Clinical and Experimental Research. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes, Brain, and Behavior. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, et al. Association of NFKB1, which encodes a subunit of the transcription factor NF-kappaB, with alcohol dependence. Human Molecular Genetics. 2008;17:963–970. doi: 10.1093/hmg/ddm368. [DOI] [PubMed] [Google Scholar]
- Fan Y, Zhu W, Yang M, Zhu Y, Shen F, Hao Q, Young WL, Yang GY, Chen Y. Del-1 gene transfer induces cerebral angiogenesis in mice. Brain Research. 2008;1219:1–7. doi: 10.1016/j.brainres.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcoholism: Clinical and Experimental Research. 2010;34:1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. Journal of Neurochemistry. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, Nurnberger J, Schuckit M, Carr L, Porjesz B, et al. Association of alcohol craving with alpha-synuclein (SNCA) Alcoholism: Clinical and Experimental Research. 2007;31:537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Gianoulakis C, Dai X, Brown T. Effect of chronic alcohol consumption on the activity of the hypothalamic-pituitary-adrenal axis and pituitary beta-endorphin as a function of alcohol intake, age, and gender. Alcoholism: Clinical and Experimental Research. 2003;27:410–423. doi: 10.1097/01.ALC.0000056614.96137.B8. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Arad M, Gould TD, Payne JL, Kaminsky ZA. Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Molecular Psychiatry. 2013 doi: 10.1038/mp.2013.62. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack LM, Kalsi G, Aliev F, Kuo PH, Prescott CA, Patterson DG, Walsh D, Dick DM, Riley BP, Kendler KS. Limited associations of dopamine system genes with alcohol dependence and related traits in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Alcoholism: Clinical and Experimental Research. 2011;35:376–385. doi: 10.1111/j.1530-0277.2010.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Jeon JH, Ju HR, Jung U, Kim KY, Yoo HS, Lee YH, Song KS, Hwang HM, Na YS, et al. VDUP1 upregulated by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–4046. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- Harding AJ, Wong A, Svoboda M, Kril JJ, Halliday GM. Chronic alcohol consumption does not cause hippocampal neuron loss in humans. Hippocampus. 1997;7:78–87. doi: 10.1002/(SICI)1098-1063(1997)7:1<78::AID-HIPO8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychological Medicine. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neuroscience Research. 2004;49:379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ji W, Li T, Pan Y, Tao H, Ju K, Wen Z, Fu Y, An Z, Zhao Q, Wang T, et al. CNTNAP2 is significantly associated with schizophrenia and major depression in the Han Chinese population. Psychiatry Research. 2013;207:225–228. doi: 10.1016/j.psychres.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Walther D, Uhl GR. Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PLoS One. 2011;6:e19210. doi: 10.1371/journal.pone.0019210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcoholism: Clinical and Experimental Research. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. The Journal of Neuroscience. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JW, Russo SJ, Ferguson D, Nestler EJ, Duman RS. Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2669–2674. doi: 10.1073/pnas.0910658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. The American Journal of Psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Plasticity of reward neurocircuitry and the 'dark side' of drug addiction. Nature Neuroscience. 2005;8:1442–1444. doi: 10.1038/nn1105-1442. [DOI] [PubMed] [Google Scholar]
- Korbo L. Glial cell loss in the hippocampus of alcoholics. Alcoholism: Clinical and Experimental Research. 1999;23:164–168. [PubMed] [Google Scholar]
- Kranzler HR, Feinn R, Armeli S, Tennen H. Comparison of alcoholism subtypes as moderators of the response to sertraline treatment. Alcoholism: Clinical and Experimental Research. 2012;36:509–516. doi: 10.1111/j.1530-0277.2011.01609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryger R, Wilce PA. The effects of alcoholism on the human basolateral amygdala. Neuroscience. 2010;167:361–371. doi: 10.1016/j.neuroscience.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Laakso MP, Vaurio O, Savolainen L, Repo E, Soininen H, Aronen HJ, Tiihonen J. A volumetric MRI study of the hippocampus in type 1 and 2 alcoholism. Behavioural Brain Research. 2000;109:177–186. doi: 10.1016/s0166-4328(99)00172-2. [DOI] [PubMed] [Google Scholar]
- Lazarov O, Marr RA. Neurogenesis and Alzheimer's disease: at the crossroads. Experimental Neurology. 2010;223:267–281. doi: 10.1016/j.expneurol.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Yang X, Purcell RH, Harvey A, Willour VL, Huo Y, Rongione M, Wand GS, Potash JB. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metabolism. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcoholism: Clinical and Experimental Research. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Research and Human Genetics : the official journal of the International Society for Twin Studies. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. Journal of Neurochemistry. 2004;90:1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcoholism: Clinical and Experimental Research. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Mayer JL, Klumpers L, Maslam S, de Kloet ER, Joëls M, Lucassen PJ. Brief treatment with the glucocorticoid receptor antagonist mifepristone normalises the corticosterone-induced reduction of adult hippocampal neurogenesis. Journal of Neuroendocrinology. 2006;18:629–631. doi: 10.1111/j.1365-2826.2006.01455.x. [DOI] [PubMed] [Google Scholar]
- Mayfield J, Ferguson L, Harris RA. Neuroimmune signaling: a key component of alcohol abuse. Current Opinion in Neurobiology. 2013 doi: 10.1016/j.conb.2013.01.024. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. Journal of Neurochemistry. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M. Phenotyping alcoholism. Alcoholism: Clinical and Experimental Research. 1999;23:757–758. doi: 10.1111/j.1530-0277.1999.tb04180.x. [DOI] [PubMed] [Google Scholar]
- Mesotten D, Vanhorebeek I, Van den Berghe G. The altered adrenal axis and treatment with glucocorticoids during critical illness. Nature Clinical Practice. Endocrinology & Metabolism. 2008;4:496–505. doi: 10.1038/ncpendmet0921. [DOI] [PubMed] [Google Scholar]
- Monje ML, Toda H, Palmer TD. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Morris SA, Eaves DW, Smith AR, Nixon K. Alcohol inhibition of neurogenesis: a mechanism of hippocampal neurodegeneration in an adolescent alcohol abuse model. Hippocampus. 2010;20:596–607. doi: 10.1002/hipo.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Boehm SL, 2nd, Owen JA, Levin PS, Berman AE, Blednov YA, Crabbe JC, Williams RW, Miles MF, et al. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes, Brain, and Behavior. 2008;7:677–689. doi: 10.1111/j.1601-183X.2008.00405.x. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor R, McKinnon CS, Scibelli AC, Burkhart-Kasch S, Reed C, Ryabinin AE, Coste SC, Stenzel-Poore MP, Phillips TJ. Corticotropin-releasing factor-1 receptor involvement in behavioral neuroadaptation to ethanol: a urocortin1-independent mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9070–9075. doi: 10.1073/pnas.0710181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Crews FT. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. Journal of Neuroinflammation. 2012;9:130. doi: 10.1186/1742-2094-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson HN, Chan SH, Crawford EF, Lee YK, Funk CK, Koob GF, Mandyam CD. Permanent impairment of birth and survival of cortical and hippocampal proliferating cells following excessive drinking during alcohol dependence. Neurobiology of Disease. 2009;36:1–10. doi: 10.1016/j.nbd.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J. The genetics of alcohol dependence. Annals of the New York Academy of Sciences. 2012;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Lessov CN, Phillips TJ. Critical role for glucocorticoid receptors in stress- and ethanol-induced locomotor sensitization. The Journal of Pharmacology and Experimental Therapeutics. 1995;275:790–797. [PubMed] [Google Scholar]
- Rock C, Moos PJ. Selenoprotein P regulation by the glucocorticoid receptor. Biometals. 2009;22:995–1009. doi: 10.1007/s10534-009-9251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Mittal N, Zhang H, Pandey SC. Modulation of cellular expression of glucocorticoid receptor and glucocorticoid response element-DNA binding in rat brain during alcohol drinking and withdrawal. The Journal of Pharmacology and Experimental Therapeutics. 2002;301:774–784. doi: 10.1124/jpet.301.2.774. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Szakall I, Toth R, Kovacs KM, Oros M, Prasad VV, Blumenberg M, Vadasz C. Mouse striatal transcriptome analysis: effects of oral self-administration of alcohol. Alcohol. 2004;32:223–241. doi: 10.1016/j.alcohol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Schoenfeld TJ, Gould E. Stress, stress hormones, and adult neurogenesis. Experimental Neurology. 2012;233:12–21. doi: 10.1016/j.expneurol.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy D, Garrick T, Dedova I, Hunt C, Miller R, Sundqvist N, Harper C. An Australian Brain Bank: a critical investment with a high return! Cell and Tissue Banking. 2008;9:205–216. doi: 10.1007/s10561-008-9076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. Journal of Neuroscience Research. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Sorocco KH, Lovallo WR, Vincent AS, Collins FL. Blunted hypothalamic-pituitary-adrenocortical axis responsivity to stress in persons with a family history of alcoholism. International Journal of Psychophysiology. 2006;59:210–217. doi: 10.1016/j.ijpsycho.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mammalian genome : official journal of the International Mammalian Genome Society. 2008;19:352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Archives of General Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyan SH, Shih AY, Walsh JJ, Maruyama H, Sarsoza F, Ku L, Eggert S, Hof PR, Koo EH, Dickstein DL. Amyloid precursor protein (APP) regulates synaptic structure and function. Molecular and Cellular Neurosciences. 2012;51:43–52. doi: 10.1016/j.mcn.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Molecular Psychiatry. 2012 doi: 10.1038/mp.2012.143. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Chen H, Toolan J, Belmonte P, Cutler DJ, Goes FS, Zandi PP, Lee RS, MacKinnon DF, Mondimore FM, et al. Family-based association of FKBP5 in bipolar disorder. Molecular Psychiatry. 2009;14:261–268. doi: 10.1038/sj.mp.4002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winocur G, Wojtowicz JM, Sekeres M, Snyder JS, Wang S. Inhibition of neurogenesis interferes with hippocampus-dependent memory function. Hippocampus. 2006;16:296–304. doi: 10.1002/hipo.20163. [DOI] [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW, Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS One. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. Journal of Neuroscience Research. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr, et al. Association of the kappa-opioid system with alcohol dependence. Molecular Psychiatry. 2006;11:1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, Edenberg HJ, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011a;12:124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nature Immunology. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, et al. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. American Journal of American Genetics. Part B, Neuropsychiatry Genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156B:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, et al. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37:557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.