Abstract
The ultimate success of cell division relies on the accurate partitioning of the genetic material. Errors in this process occur in nearly all tumors and are the leading cause of miscarriages and congenital birth defects in humans. Two cell divisions, mitosis and meiosis, use common as well as unique mechanisms to ensure faithful chromosome segregation. In mitosis, alternating rounds of DNA replication and chromosome segregation preserves the chromosome complement of the progenitor cell. In contrast, during meiosis two consecutive rounds of nuclear division, meiosis I and meiosis II, follow a single round of DNA replication to reduce the chromosome complement by half. Meiosis likely evolved through changes to the mitotic cell division program. This review will focus on the recent findings describing the modifications that transform mitosis into meiosis.
Maintenance of cellular and organismal fitness requires the proper partitioning of genetic material during cell division. During mitosis, alternating rounds of DNA replication and chromosome segregation preserve the chromosome complement of the progenitor cell (Figure 1A). During meiosis, two consecutive rounds of nuclear division, meiosis I and meiosis II, follow a single round of DNA replication to reduce the chromosome complement by half (Figure 1B). During meiosis I homologous chromosomes are segregated, during meiosis II as during mitosis, sister chromatids split. The specialized meiotic chromosome segregation pattern likely evolved through changes to the mitotic cell division program. This review will focus on recent findings describing the modifications that transform mitosis into meiosis I.
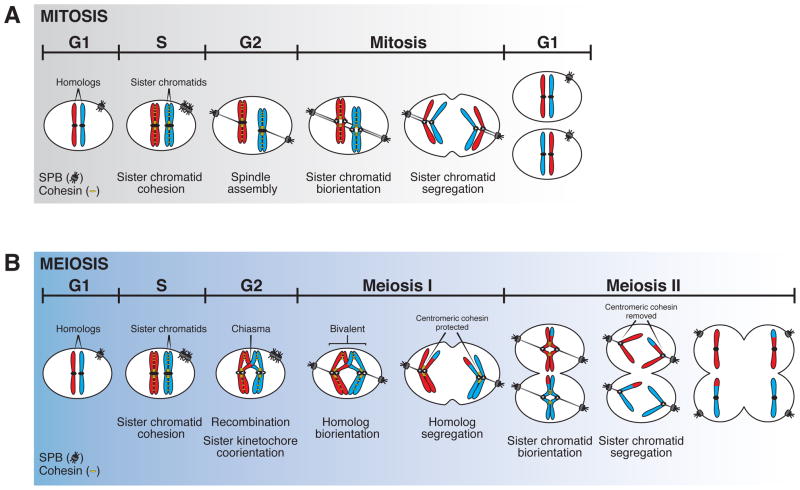
Figure 1. Mitosis and meiosis – a comparison.
A. During mitotic cell division, cohesin complexes (yellow) are loaded onto chromosomes during DNA replication in S phase, and provide a physical linkage between sister chromatids. During metaphase sister chromatids attach to spindle microtubules (gray lines) from opposite poles, a process known as biorientation. During anaphase sister chromatids segregate to opposite poles. SPB = spindle pole body (centrosome in yeast)
B. During meiotic cell division, cohesin complexes (yellow) are loaded onto chromosomes concurrent with DNA replication in pre-meiotic S phase. During meiotic prophase I, homologous chromosomes become physically linked through recombination to form chiasmata. Prior to entry into meiosis I, sister kinetochores become cooriented such that each pair of sister kinetochores attaches to microtubules emanating from a single pole, resulting in biorientation of homologous chromosomes. A pair of homologous chromosomes that is linked by chiasmata is called a bivalent. During meiosis I, arm cohesin is removed leading to segregation of homologous chromosomes away from each other, while centromeric cohesin is protected from removal. In meiosis II, centromeric cohesin is removed and sister chromatids are partitioned as in mitosis.
The basic principles of chromosome segregation
To accurately segregate chromosomes, each pair of sister chromatids (during mitosis and meiosis II) or each pair of homologs (during meiosis I) must attach to microtubules emanating from opposite spindle poles (Figure 1). But how does the cell “know” that attachment has occurred in a manner so that segregation will have the desired outcome of equal partitioning of the genetic material? The key is – Tension (Figure 2). Physical linkages between sister chromatid pairs or homologous chromosome pairs provide resistance to the pulling forces of microtubules. ONLY when chromosomes have attached to the spindle from opposite poles, known as biorientation, will this lead to the exertion of force on the microtubule-chromosome interface, the kinetochore (Figure 2). In other words, linkages between chromosome pairs provide resistance to the pole-ward pulling forces of microtubules, thus generating tension.
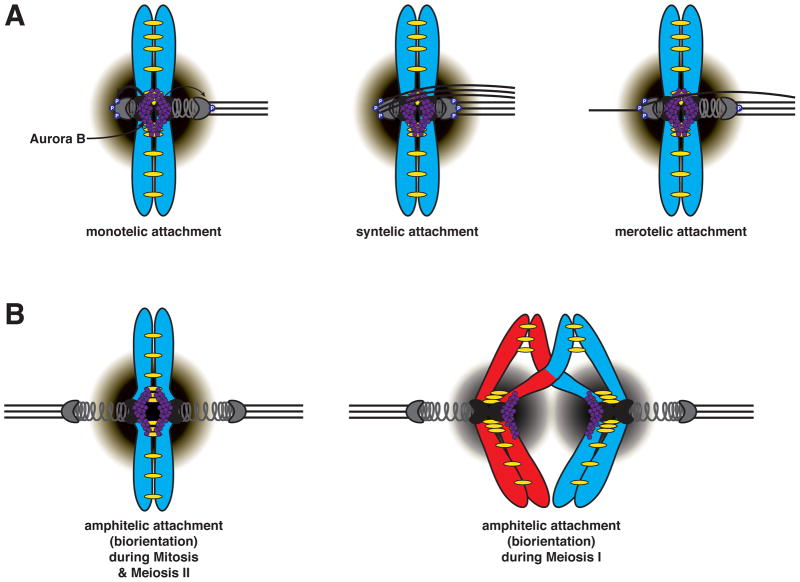
Figure 2. Aurora B selectively destabilizes microtubule attachments that fail to generate tension.
A. The pulling forces exerted by spindle microtubules attached to the kinetochore are resisted by linkages between sister chromatids, mediated by the cohesin complex (yellow). This results in tension across the kinetochores of sister chromatids. Microtubule attachments that fail to generate tension are selectively destabilized by the kinase Aurora B, which is concentrated at the inner centromere (purple circles). Phosphorylation of outer kinetochore components by Aurora B severs microtubule attachments. Thus, non-tension generating attachments are destabilized due to outer kinetochore substrates being in close proximity to Aurora B. Monotelic attachment (left) is the state when only one of the two sister kinetochores is attached to kinetochore microtubules. Syntelic attachment (middle) is a condition in which both sister kinetochores become attached to microtubules from the same pole. Merotelic attachment (right) is when a single kinetochore becomes attached to microtubules from both spindle poles. Purple gradient represents Aurora B phosphorylation gradient. P represents Aurora B-dependent phosphorylation of outer kinetochore components.
B. Tension promotes stable microtubule-kinetochore interactions through spatial separation of Aurora B from kinetochore substrates. Correct amphitelic (bioriented) attachments result in stable microtubule-kinetochore interactions by positioning/pulling the kinetochore substrates away from Aurora B. Biorientation is promoted by Aurora B in both mitosis and meiosis. In mitosis and meiosis II (left), tension occurs across the kinetochores of sister chromatids, while in meiosis I (right), tension occurs across the kinetochores of homologous chromosomes.
Who links sister chromatids and homologs? Linkages between each pair of sister chromatids are established as they are replicated, through an evolutionarily conserved protein complex called cohesin, whose core subunits in mitosis include Smc1, Smc3, Scc3 and the kleisin subunit Scc1/Mcd1 (also known as Rad21) (reviewed in Onn et al., 2008). In meiosis, homologous chromosomes are not linked during S phase but undergo an extensive pairing process during prophase I that culminates in the exchange of genetic material known as homologous recombination. Homologous recombination generates crossovers, which together with cohesin molecules distal to the crossover link homologous chromosomes (Figure 2).
Biorientation is ensured by a surveillance mechanism, that “senses” when microtubule-kinetochore attachments are not under tension. The highly conserved protein kinase and member of the four-subunit chromosomal passenger complex (CPC) Aurora B destabilizes microtubule attachments that fail to generate tension by phosphorylating outer kinetochore components causing them to release the microtubule (Figure 2) (reviewed in Watanabe, 2012). Unattached kinetochores in turn generate an “anaphase halt signal” through the spindle assembly checkpoint (SAC). The SAC inhibits the activity of the ubiquitin ligase APC/C-Cdc20; the inducer of chromosome segregation (reviewed in Murray, 2011; Santaguida and Musacchio, 2009). APC/C-Cdc20 activates a protease known as Separase by degrading its inhibitor Securin. Active Separase cleaves the cohesin subunit Scc1/Mcd1/Rad21 thereby dissolving the linkages between sister chromatids. Anaphase chromosome movement then ensues.
How Aurora B “knows” whether or not a microtubule-kinetochore attachment is under tension is not yet completely understood. The prevailing model for tension sensing by Aurora B is based on the localization of the protein kinase to the inner kinetochore and the pericentric heterochromatin (also known as the inner centromere). Tension pulls Aurora B substrates located at the outer kinetochore away from the inner centromere such that microtubule attachments are no longer destabilized by the kinase (Liu et al., 2009), Figure 2A). Interestingly, a recent study in budding yeast argues against the Aurora B spatial control model (Campbell and Desai, 2013). A truncation of Sli15/INCENP (a member of the CPC) that cannot localize Aurora B to the inner centromere through known pathways, nevertheless supports proper chromosome segregation (Campbell and Desai, 2013). Clearly, there is more to tension sensing than simply targeting Aurora B to the inner centromere that we must understand to truly comprehend the mechanisms regulating proper chromosome segregation.
Meiosis I – a unique cell division
During the first meiotic division, homologous chromosomes (the maternal and paternal chromosomes) rather than sister chromatids must be partitioned, while leaving in place the means to properly segregate sister chromatids during the second meiotic division. The unique chromosome segregation pattern in meiosis I entails the restructuring of chromosomes during prophase I. Linkages between homologous chromosomes have to be forged and resolved in order to accurately segregate homologous chromosomes. As mentioned above homologous recombination mediates these linkages (Figure 1B). We refer the reader to the following reviews for an in depth summary of the mechanisms of meiotic recombination and of the pathways that prevent DNA replication between the two meiotic divisions (Hochwagen and Amon, 2006; Keeney and Neale, 2006; Longhese et al., 2009; Martinez-Perez and Colaiacovo, 2009). Here we will focus on the two meiosis I-specific cell cycle modifications central to dissolving linkages between homologs and segregating them: the manner in which chromosomes attach to microtubules during meiosis I and how the cohesin complexes are removed from chromosomes.
Segregating homologous chromosomes during meiosis I requires that sister kinetochores attach to microtubules emanating from the same pole (coorientation), rather than opposite poles (biorientation) as they do during mitosis (Figure 1). Furthermore, while all linkages between sister chromatids are lost at the onset of mitotic anaphase, cohesin is lost in a stepwise manner during meiosis (Figure 1B). Loss of cohesin along chromosome arms is necessary to resolve the linkages between homologous chromosomes created by reciprocal recombination, while pericentric cohesin must be maintained to facilitate the segregation of sister chromatids during meiosis II.
Mechanisms of meiosis I sister kinetochore coorientation
In mitosis, the back-to-back configuration of sister kinetochores assists their biorientation by favoring microtubule attachments from opposite poles (Figure 3A). In contrast, during meiosis I, sister kinetochores are fused or are juxtaposed side-by-side, a configuration that facilitates their coorientation by allowing microtubule attachments from the same pole (Figure 3A). Our understanding of the mechanism of sister kinetochore coorientation in meiosis I stems mainly from studies in the budding and fission yeast, both of which contain monocentric chromosomes with a single kinetochore assembly site. However, the centromere size and the corresponding microtubule binding capacity of the kinetochore are different between the two yeasts. While a budding yeast kinetochore assembles onto a defined 125 bp point centromere and associates with a single microtubule, fission yeast, like most eukaryotes, has larger regional centromeres and its kinetochores bind to multiple microtubules (Cleveland et al., 2003; Westermann et al., 2007; Winey et al., 1995). Despite these differences in centromere size and microtubule binding capacity, the conservation of kinetochore proteins across species led to the notion that the repeated assembly of budding yeast-like kinetochores constitutes the larger kinetochores with multiple microtubule binding sites as seen in higher eukaryotes (Joglekar et al., 2009; Zinkowski et al., 1991).
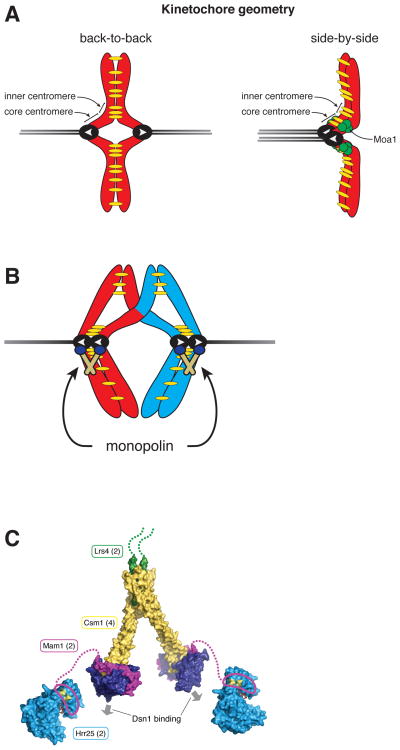
Figure 3. Sister chromatids attach to microtubules emanating from the same spindle pole in meiosis I.
A. For organisms with regional centromeres (e.g. fission yeast) coorientation of sister kinetochores is dictated by the geometry of the sister kinetochores. In mitosis, the back-to-back assembly of kinetochores promotes amphitelic (bioriented) attachments to microtubules from opposite poles (gray lines). In contrast, during meiosis I, cohesin (yellow) at the core centromere establishes a side-by-side kinetochore orientation which promotes attachment of sister kinetochores to microtubules from the same pole. In fission yeast a meiosis-specific protein Moa1 is required for maintenance of cohesin at the core centromere (green circles). White arrows indicate orientation of microtubule-kinetochore attachment.
B. In budding yeast, coorientation of sister kinetochores is mediated by the monopolin complex, which is composed of the meiosis-specific protein Mam1, the casein kinase Hrr25, and two nucleolar proteins Lrs4 and Csm1. Prior to cells entering the meiotic divisions, the polo-like kinase, Cdc5, promotes the release of Lrs4 and Csm1 from the nucleolus and monopolin complex assembly. The monopolin complex then localizes to kinetochores and forces sister chromatids to attach to microtubules emanating from the same spindle pole. In mitosis (and meiosis II), the monopolin complex is absent and sister chromatids attach to microtubules emanating from opposite poles.
C. The crystal structure of the monopolin complex shows the V-shaped complex that is thought to fuse sister kinetochores. Lrs4 = green, Csm1 = yellow and purple, Mam1 = magenta, and Hrr25 = blue. The copy number of each protein in the complex is indicated in parentheses, and arrows indicate the two available Dsn1-binding sites. Adapted from (Corbett & Harrison, 2012).
Centromeres in fission yeast are composed of two domains, a core centromere domain where kinetochore assembly occurs and a neighboring pericentric heterochromatic region, also known as the inner centromere (Figure 3A). It is thought that cohesion at the core centromere induces a side-by-side kinetochore geometry thus promoting coorientation during meiosis I, whereas cohesion at the pericentromeres allows a back-to-back configuration and sister kinetochore biorientation during mitosis and meiosis II (Sakuno et al., 2009). Several observations support this notion: First, during mitosis, where sister kinetochores biorient, Rad21-containing cohesin is enriched at the pericentric heterochromatin but not at the core centromere (Bernard et al., 2001; Nonaka et al., 2002). Second, as occurs in most organisms, Rad21 is substituted by a meiosis-specific kleisin subunit, Rec8. These Rec8-containing cohesin complexes accumulate at the core centromere in addition to the pericentric regions (Watanabe et al., 2001). Replacement of Rec8 with Rad21 in meiosis results in reduced cohesin association with the core centromere and subsequent biorientation of sister kinetochores in meiosis I, while tethering the core centromeres of the two sister chromatids artificially restores sister kinetochore coorientation (Sakuno et al., 2009). The meiosis I-specific protein Moa1 is critical for maintaining Rec8 at the core centromere, but the mechanism whereby this occurs is not yet understood (Kagami et al., 2011; Sakuno et al., 2009; Yokobayashi and Watanabe, 2005). Importantly, Rec8 is also essential for meiosis I sister kinetochore coorientation in plants and nematodes (Chelysheva et al., 2005; Severson et al., 2009; Yu and Dawe, 2000) indicating that the use of a meiosis-specific cohesin is a general feature of meiosis I sister kinetochore coorientation.
Budding yeast does not appear to rely on specialized cohesin to promote sister kinetochore coorientation. Replacing Rec8 with Scc1/Mcd1 does not interfere with sister kinetochore coorientation (Monje-Casas et al., 2007; Toth et al., 2000). Instead, sister kinetochore coorientation is mediated by the monopolin complex (Toth et al., 2000) Figure 3B–C). The use of a different mechanism to promote sister kinetochore coorientation in budding yeast is perhaps not surprising in light of the budding yeast meiosis I kinetochore architecture. Electron microscopy analysis of meiosis I spindles shows that a single microtubule binds to a pair of sister chromatids (Winey et al., 2005), indicating that the two-microtubule binding sites in sister kinetochores are fused into one. Thus, rather than modulating kinetochore orientation as to favor attachment of sister kinetochores to microtubule from the same spindle pole, a meiosis I kinetochore configuration in budding yeast requires the fusion of two microtubule attachment sites into one (Figure 3B).
The fusion of sister kinetochores is accomplished by the monopolin complex (Corbett et al., 2010; Rabitsch et al., 2003). Monopolin is composed of two sub-complexes (Figure 3C). One consists of the meiosis-specific protein Mam1 and the casein kinase Hrr25, the other of Lrs4 and Csm1 (Petronczki et al., 2006; Rabitsch et al., 2003; Toth et al., 2000). The crystal structure of the Lrs4-Csm1 sub-complex has given us insight into the function of the monopolin complex. Lrs4 and Csm1 form a V-shaped structure with globular domains at the two tips comprised of the C-terminus of Csm1 (Corbett et al., 2010) (Figure 3C). These globular domains are thought to bind to the kinetochore component Dsn1. It has been proposed that kinetochore clamping by the monopolin complex occurs by the globular domains of Csm1 binding to Dsn1 proteins in different kinetochores, thus leading to a fusion of the two kinetochores into a single microtubule-binding interface (Corbett and Harrison, 2012). The roles of Mam1 and Hrr25 in the fusion of sister kinetochores is not clear. Mam1 binds to Csm1 as well as to Hrr25 through distinct domains, and Mam1 has been proposed to modulate Hrr25 kinase activity (Corbett and Harrison, 2012).
The exact mechanism whereby the association of monopolin with sister kinetochores is regulated has not yet been elucidated but factors have been identified that are critical for the process. First, formation of the monopolin complex does not occur until entry into prometaphase I because transcription of MAM1 is not initiated until the completion of recombination. A set of genes critical for entry into meiosis I including meiotic cyclins, the Polo kinase CDC5 and MAM1 are under control of the meiosis-specific transcription factor Ndt80 (Xu et al., 1995). The accumulation of this transcription factor is inhibited by ongoing recombination due to the activity of the recombination checkpoint (Tung et al., 2000). Once this inhibitory signal ceases, Ndt80 is activated by transcriptional and posttranslational means and induces the transcription of factors critical for exit from prophase, progression through the meiotic divisions and spore (gamete) formation. (Okaz et al., 2012; Sourirajan and Lichten, 2008 and reviewed in Winter, 2012). Once MAM1 is expressed, the stable recruitment of the monopolin complex to kinetochores requires the activity of the Polo kinase Cdc5, the meiosis-specific protein Spo13 and the Dbf4-Cdc7 kinase (DDK). Cells lacking any of these factors are defective in sister kinetochore coorientation (Clyne et al., 2003; Katis et al., 2004; Lee and Amon, 2003; Lee et al., 2004; Matos et al., 2008).
Recently it was shown in budding yeast that kinetochores must not be attached to microtubules for the monopolin complex to bind to and fuse sister kinetochores. While the kinetochore retains its microtubule binding capacity during most of the mitotic cell cycle, some outer kinetochore components, such as Ndc80 and Hsk3, associate with kinetochores only as cells enter the meiotic divisions (Miller et al., 2012). When the outer kinetochore is assembled prior to prophase I and cells are allowed to form a meiosis I spindle (by premature expression of meiotic CDKs) microtubule-kinetochore attachments form prior to monopolin association with kinetochores and sister kinetochore coorientation does not occur (Miller et al., 2012). Microtubule-kinetochore attachments may prevent the monopolin complex from binding to the kinetochore. Interestingly, this event appears irreversible. Once premature microtubule-kinetochore attachments have been established in meiotic prophase I, transient microtubule depolymerization fails to restore sister kinetochore coorientation (unpublished observations).
Outer kinetochore disassembly during prophase I may be a conserved aspect of sister kinetochore coorientation. In fission yeast, Ndc80 and its binding partner Nuf2 dissociate from kinetochores during prophase I (Asakawa et al., 2005). Fission yeast mutants that are induced to undergo meiosis in the absence of mating pheromone signaling, do not dissociate Ndc80 from kinetochores during prophase I (Asakawa et al., 2005), and interestingly, split sister chromatids rather than homologs during meiosis I, consistent with defects in coorientation (Yamamoto and Hiraoka, 2003; Yamamoto et al., 2004).
Once sister kinetochores have been cooriented how do they achieve the correct attachment on the meiosis I spindle? Recent work from budding yeast revealed that the conserved kinases Aurora B and Mps1 play unique roles in promoting proper chromosome attachment during meiosis (Meyer et al., 2013). Aurora B first releases kinetochore-microtubule associations during prophase I to allow proper chromosome morphogenesis. Subsequently, during the meiotic divisions, Aurora B promotes biorientation of sister chromatids by destabilizing microtubule attachments that fail to generate tension while Mps1 is required to produce force-generating end-on attachments (Meyer et al., 2013). In some organisms the linkages between homologs, chiasmata, also contribute to the biorientation of homologous chromosomes. It is thought that chiasmata-assisted juxtaposition of sister kinetochores during meiosis I favors the placement of Aurora B underneath the paired kinetochores rather than between them, such that Aurora B is pulled away from its substrates only when the bivalent is bioriented (Sakuno et al., 2011). In fission yeast, sister kinetochores undergo futile biorientation attempts during early prometaphase I, despite the side-by-side kinetochore configuration that favors coorientation. Chiasmata disfavor these erroneous attachments; in their absence sister kinetochore biorientation persists in achiasmatic chromosomes (Dudas et al., 2011; Hirose et al., 2011; Sakuno et al., 2011). Biorientation of achiasmatic chromosomes has also been observed in humans, but not yet in budding yeast, worms and Arabidopsis (Chelysheva et al., 2005; Hassold and Hunt, 2001; Klein et al., 1999; Severson et al., 2009; Watanabe, 2012). Understanding the interplay between coorientation factors, tension-sensing kinases and chiasmata is the critical next step in illuminating how sister kinetochore coorientation is achieved to bring about meiosis I chromosome morphogenesis.
Centromeric cohesin is maintained until meiosis II
Stepwise dissolution of cohesion ensures proper chromosome segregation in the two consecutive meiotic divisions. During meiosis I, cleavage of cohesin distal to sites of crossovers allows homologs to disjoin (Buonomo et al., 2000) (Figure 1B). Accurate segregation of sister chromatids during meiosis II requires that cohesin around centromeres be protected from cleavage during meiosis I (Figure 1B). A key factor in maintaining centromeric cohesin during meiosis I is the substitution of the kleisin subunit of the cohesin complex, Scc1/Mcd1/Rad21, with the meiosis-specific subunit Rec8 (Klein et al., 1999; Watanabe et al., 2001). In budding and fission yeast, replacing Rec8 with the mitosis-specific kleisin leads to cohesin loss along the entire chromosome at anaphase I onset (Toth et al., 2000; Yokobayashi et al., 2003). It is worth noting that the kleisin subunit is not the only cohesin subunit with meiosis-specific isoforms. For example in fission yeast, Rec11 replaces the Scc3 subunit along chromosome arms where it is required for recombination and arm cohesion (Kitajima et al., 2003).
How is cohesin removed in a stepwise manner during meiosis? During meiosis I, removal of cohesin is mediated by phosphorylation of the kleisin subunit, which promotes its cleavage by Separase. In budding yeast, a number of kinases have been implicated in promoting cleavage of Rec8, including the Polo kinase Cdc5, the casein kinase Hrr25 and DDK, a protein kinase critical for DNA replication (Brar et al., 2006; Katis et al., 2010, Figure 4A). In fission yeast, the casein kinase 1, Hhp2, mediates Rec8 phosphorylation (Ishiguro et al., 2010). In mammalian cells, two pathways exist to remove cohesin from chromosomes. During mitosis, most cohesin dissociates from chromosome arms before metaphase through a cleavage-independent pathway, which involves the phosphorylation of the SA2 (Scc3 homolog) subunit by Polo kinase and only a small pool of cohesin located around centromeres is removed by Separase mediated cleavage (Hauf et al., 2005; Nakajima et al., 2007). Thus, it appears that the removal of cohesin from chromosome arms, either by Separase-dependent or -independent pathways require phosphorylation of its non-Smc subunits by various kinases.
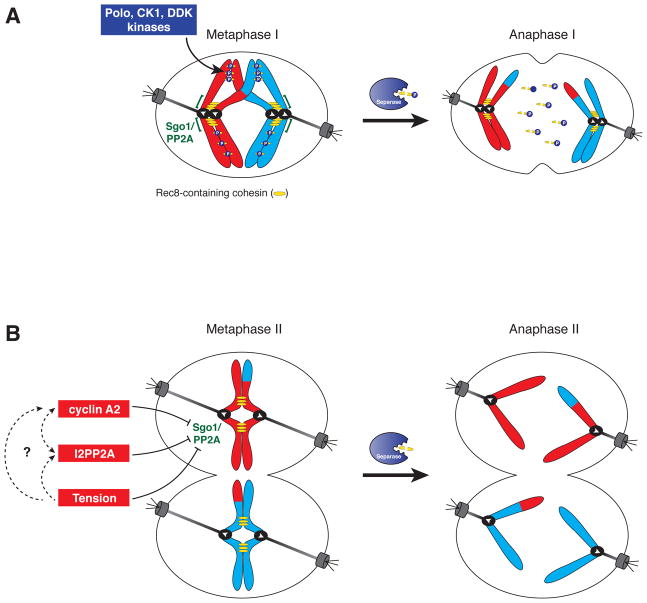
Figure 4. Step-wise cohesin removal during meiosis.
A. Segregation of homologous chromosomes in meiosis I requires the dissolution of arm cohesion, however, cohesin near the centromere must be maintained to promote proper segregation of sister chromatids in meiosis II. Protection of centromeric cohesin is accomplished by preventing phosphorylation of Rec8. This is mediated by Sgo1 (MEI-S332)-dependent recruitment of the protein phosphatase PP2A to centromeric regions (denoted as green brackets) where it antagonizes Rec8 phosphorylation. Rec8 is phosphorylated by a host of kinases including Polo-like kinase, Casein kinase and Dbf4-dependent kinase. Phosphorylated Rec8 (on chromosome arms) is removed by Separase promoting the onset of anaphase I.
B. Sgo1/PP2A is localized to centromeric regions during metaphase II, however centromeric cohesin is removed during anaphase II onset by Separase, indicating that centromere localized Sgo1/PP2A is unable to protect cohesin during meiosis II. Multiple negative regulators for Sgo1/PP2A have been reported. In mouse oocytes, both cyclin A2 and I2PP2A localize to centromeres and have been implicated as Sgo1/PP2A inhibitors. Additionally tension has been proposed to inhibit Sgo1/PP2A by direct inhibition of PP2A activity via conformational change and/or by tension-dependent spatial separation of Sgo1-PP2A from Rec8-cohesin.
Given that Rec8 phosphorylation is a prerequisite for cohesin removal from chromosomes during anaphase I, it is not surprising that the protection of centromeric cohesin is accomplished by preventing Rec8 phosphorylation. This occurs, at least in part, by MEI-S332/ Shugoshin-dependent recruitment of the protein phosphatase PP2A to centromeric regions where it antagonizes Rec8 phosphorylation (Katis et al., 2004; Kerrebrock et al., 1995; Kitajima et al., 2004; Kitajima et al., 2006; Riedel et al., 2006) Figure 4A). In fission yeast and in humans, recruitment of Shugoshin to pericentric heterochromatin requires phosphorylation of histone H2A on a conserved residue by the protein kinase Bub1 (Kawashima et al., 2010).
How is centromeric cohesin rendered resistant to cleavage in meiosis I, but is sensitive to cleavage in meiosis II? A simple explanation is that Sgo1-PP2A is removed prior to anaphase II, thus leaving the centromeric pool of cohesin sensitive to cleavage. However, both in budding yeast and in mouse oocytes, PP2A associates with centromeric regions during meiosis I as well as meiosis II (Chambon et al., 2013; Katis et al., 2010). In addition, a recent report in budding yeast showed that premature microtubule-kinetochore interactions in prophase I lead to de-protection of centromeric cohesin in meiosis I without compromising Sgo1-PP2A localization (Miller et al., 2012). These findings indicate that localization of the protective machinery per se is not sufficient to promote centromeric protection of cohesin and suggest that PP2A must be inactivated in meiosis II by other mechanisms.
One way by which de-protection of centromeric cohesin could occur is through tension generated by centromeric cohesion counteracting the microtubule pulling forces. Tension could modulate the activity of the cohesin protective machinery through tension-dependent deformation of PP2A resulting in inhibition of catalytic activity (Grinthal et al., 2010), and/or tension-dependent spatial separation of centromeric cohesin from PP2A (Lee et al., 2008). It will be interesting to distinguish whether microtubule-kinetochore interactions directly modulate PP2A activity or whether they indirectly interfere with PP2A function by providing pericentric access to an inhibitor of PP2A. However, it is important to note that in budding yeast, sister kinetochores come under tension in metaphase I in mutants defective in monopolin function, yet centromeric cohesin is not removed prematurely (Toth et al., 2000). Therefore, tension between sister kinetochores alone cannot explain how centromeric cohesin becomes de-protected.
It is also possible that PP2A is inactivated during meiosis II by factors that specifically interact with the phosphatase after anaphase I onset. Recently, meiosis II-specific modulators of PP2A activity have been reported in mouse oocytes. An inhibitor of PP2A, I2PP2A, colocalizes with PP2A and centromeric cohesin in metaphase II and is required for removal of centromeric cohesin during anaphase II (Chambon et al., 2013). Cyclin A2 has also been implicated in regulating the stepwise loss of cohesin. Cyclin A2 associates with centromeric regions at all stages of meiosis, except for anaphase I. Importantly, the cyclin is required for centromeric cohesin removal during anaphase II, whereas stable cyclin A2-expressing oocytes prematurely loose centromeric cohesin in meiosis I (Touati et al., 2012). Deciphering how microtubule kinetochore attachments and/or tension modulate the activity of the cohesin protective machinery and whether they do so by affecting the activity of PP2A regulators will be critical questions to address for the future.
Conclusions and Perspectives
Meiosis I chromosome morphogenesis lies at the heart of germ cell differentiation. Understanding this process and determining how the developmental program inducing the germ cell fate reprograms the canonical cell cycle machinery to bring about the unique meiosis I division is central to understanding this defining feature of sexual reproduction. As errors in meiotic chromosome segregation are the leading cause of congenital birth defects and miscarriages in humans (Hassold and Hunt, 2001), further insight into the mechanistic basis by which cells properly orchestrate this process will also have important implications for human health and fertility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell. 2005;16:2325–2338. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- **.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. doi: 10.1038/nature12057. This paper demonstrates that tension-sensing by Aurora B can be uncoupled from survivin-based centromere localization in budding yeast by using an engineered truncation of INCENP that eliminates its interaction with survivin while retaining Aurora B activation domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chambon JP, Touati SA, Berneau S, Cladiere D, Hebras C, Groeme R, McDougall A, Wassmann K. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr Biol. 2013;23:485–490. doi: 10.1016/j.cub.2013.02.004. This paper identifies I2PP2A as an inhibitor of the cohesin protector PP2A and shows that in mouse oocytes I2PP2A is essential for faithful sister chromatid segregation by mediating de-protection of centromeric cohesin in meiosis II. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- **.Corbett KD, Harrison SC. Molecular architecture of the yeast monopolin complex. Cell Rep. 2012;1:583–589. doi: 10.1016/j.celrep.2012.05.012. This paper determines the stoichiometry and overall architecture of the monopolin complex of budding yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas A, Ahmad S, Gregan J. Sgo1 is required for co-segregation of sister chromatids during achiasmate meiosis I. Cell Cycle. 2011;10:951–955. doi: 10.4161/cc.10.6.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci U S A. 2010;107:2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Hirose Y, Suzuki R, Ohba T, Hinohara Y, Matsuhara H, Yoshida M, Itabashi Y, Murakami H, Yamamoto A. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 2011;7:e1001329. doi: 10.1371/journal.pgen.1001329. This paper shows that in fission yeast chiasmata contribute to the elimination of inappropriate sister chromatid bi-orientation during meiosis I. This finding indicates that crossovers play a crucial role in promoting proper microtubule-kinetochore attachments during meiosis I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 2011;12:1189–1195. doi: 10.1038/embor.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. This paper defines the mechanism whereby the cohesin protector Sgo1 is recruited to centromeres in fission yeast. It occurs through Bub1 kinase-dependent phosphorylation of the conserved serine 121 of histone H2A. [DOI] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Lee BH, Kiburz BM, Amon A. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr Biol. 2004;14:2168–2182. doi: 10.1016/j.cub.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- **.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. By measuring localized phosphorylation dynamics in living mammalian cells, this paper shows that Aurora B-dependent substrate phosphorylation at the kinetochore depends on the substrate’s distance from the kinase located at the inner centromere. This suggests that tension can be sensed by increased spatial separation of Aurora B from kinetochore substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009;8:1127–1138. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Colaiacovo MP. Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- *.Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339:1071–1074. doi: 10.1126/science.1232518. Through live cell microscopy of budding yeast cells, this paper examines the stages of chromosome-microtubule interactions and their regulation by Aurora B and Mps1 kinases during meiosis I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Miller MP, Unal E, Brar GA, Amon A. Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife. 2012;1:e00117. doi: 10.7554/eLife.00117. This paper shows that premature interactions of kinetochores with microtubules transform meiosis I into a mitosis-like division by disrupting two key meiosis I events: coorientation of sister kinetochores and maintenance of centromeric cohesin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. A brief history of error. Nat Cell Biol. 2011;13:1178–1182. doi: 10.1038/ncb2348. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci. 2007;120:4188–4196. doi: 10.1242/jcs.011528. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, Zachariae W. Meiotic Prophase Requires Proteolysis of M Phase Regulators Mediated by the Meiosis-Specific APC/C(Ama1) Cell. 2012;151:603–618. doi: 10.1016/j.cell.2012.08.044. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- **.Sakuno T, Tanaka K, Hauf S, Watanabe Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev Cell. 2011;21:534–545. doi: 10.1016/j.devcel.2011.08.012. This paper indicates that chiasmata-dependent stabilization of correct microtubule-kinetochore attachments in meiosis I is achieved by configuring kinetochores to the outer edge of the bivalent, while bringing Aurora B inward. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- **.Touati SA, Cladiere D, Lister LM, Leontiou I, Chambon JP, Rattani A, Bottger F, Stemmann O, Nasmyth K, Herbert M, et al. Cyclin A2 is required for sister chromatid segregation, but not separase control, in mouse oocyte meiosis. Cell reports. 2012;2:1077–1087. doi: 10.1016/j.celrep.2012.10.002. This study reveals that in mouse oocytes cyclin A2 is localized to kinetochores throughout meiosis II and is required for separase-dependent sister chromatid separation in meiosis II. [DOI] [PubMed] [Google Scholar]
- Tung KS, Hong EJ, Roeder GS. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc Natl Acad Sci U S A. 2000;97:12187–12192. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13:370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Morgan GP, Straight PD, Giddings TH, Jr, Mastronarde DN. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol Biol Cell. 2005;16:1178–1188. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2012;76:1–15. doi: 10.1128/MMBR.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. Embo J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto TG, Chikashige Y, Ozoe F, Kawamukai M, Hiraoka Y. Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J Cell Sci. 2004;117:3875–3886. doi: 10.1242/jcs.01248. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Dawe RK. Functional redundancy in the maize meiotic kinetochore. J Cell Biol. 2000;151:131–142. doi: 10.1083/jcb.151.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]