Abstract
The isolation of human monoclonal antibodies (hmAb) has emerged as a versatile platform in a wide variety of contexts ranging from vaccinology to therapeutics. In particular, the presence of high titers of circulating auto-antibodies is implicated in the pathology and outcome of autoimmune diseases. Therefore, the molecular characterization of these hmAb provides an avenue to understanding the pathogenesis of autoimmune diseases. Additionally, the phenotype of the auto-reactive B cells may have direct relevance for therapeutic intervention. In this report, we describe a high-throughput single-cell assay, microengraving, for the screening, characterization and isolation of anti-citrullinated protein antibodies (ACPA) from peripheral blood mononuclear cells (PBMC) of rheumatoid arthritis (RA) patients. Stimulated B cells are profiled at the single-cell level in a large array of sub-nanoliter nanowells (~105), assessing both the phenotype of the cells and their ability to secrete cyclic-citrullinated peptide (CCP)-specific antibodies. Single B cells secreting ACPA are retrieved by automated micromanipulation, and amplification of the immunoglobulin (Ig) heavy and light chains is performed prior to recombinant expression. The methodology offers a simple, rapid and low-cost platform for isolation of auto-reactive antibodies from low numbers of input cells and can be easily adapted for isolation and characterization of auto-reactive antibodies in other autoimmune diseases.
1. Introduction
In the last couple of decades, the ability to isolate fully human monoclonal antibodies (hmAb) from B cells has emerged as a versatile platform for many applications including the production of therapeutic antibodies1, revealing molecular insights into the nature of antigen driven antibody affinity maturation2, structural vaccinology3,4, recognition of conserved viral epitopes5, and elucidating fundamental mechanisms of B cell immunology in autoimmune diseases6.
Donor-derived hmAb are isolated by immortalization of primary B cells employing traditional methods like the hybridoma technology7 or in vitro infection with Epstein-Barr virus8, or by utilizing more recent methodologies like genetic reprogramming of memory B cells9. The advantage of these approaches is that upon immortalization the cells serve as production factories for the secretion of the native hmAb. The drawbacks however are that immortalization efficiencies are not high and the cells still need to be screened in a second step to isolate antigen-specific clones. Alternately, primary B cells can be directly interrogated for their antigen specificity using either flow-cytometry or microwell arrays10-12, and single antigen-specific B cells can be isolated for reverse transcription, gene amplification, cloning and recombinant expression of the hmAb13,14. The advantages of these approaches are that they are easier to implement, rapid and facilitate screening up front. Secondly, with regards to the micro/nanowell arrays, the ability to work with small sample sizes like tissue resident B cells, and the ability to screen both memory B cells and antibody-secreting plasmablasts and plasma cells are added advantages. A limitation of these approaches, however is that they rely on recombinant antibody expression.
The isolation of auto-antibodies, antibodies directed against self-antigens, holds promise as a mechanism to delineate the molecular basis of autoimmune diseases15. Auto-antibodies that are highly specific for cellular antigens can be detected both in the sera and target organs of patients with organ-specific autoimmune diseases such as rheumatoid arthritis (RA), type I diabetes and thyroiditis16. In RA patients, the presence of these auto-antibodies like the anti-citrullinated protein antibodies (ACPA) has diagnostic and prognostic significance17-19. In line with other similar autoimmune diseases, it has also been demonstrated, that the ACPA may contribute to development of inflammatory arthritis20,21. Consistent with this finding, therapeutic regimens that utilize antibody-mediated depletion of B cells in autoimmune diseases, may provide clinical benefit22,23. Thus, in addition to the molecular characterization of ACPA, determining the phenotype of auto-reactive B cells is essential for the development of clinical strategies that rely on B cell depletion24,25.
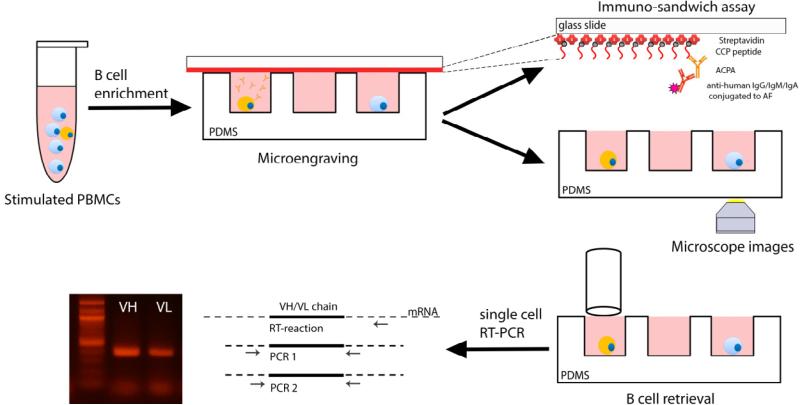
Here, we describe a novel high throughput technology, that allows for the combined screening of the phenotype and antigen specificity of ACPA secreted from single B cells. In this approach, PBMC are briefly stimulated ex vivo with recombinant human interleukin-21 (rhIL-21) and soluble CD40 ligand (sCD40L) to facilitate the generation of antibody secreting cells (ASC), as described previously26. The enriched B cell population is then loaded onto a microfabricated nanowell array (~105 individual nanowells per array) with sub-nanoliter volumes (125 pL) to isolate individual cells. The nanowell array is interrogated for cyclic-citrullinated peptide (CCP)17 specific immunoglobulin (Ig) secretion by using a functionalized glass slide. In combination with automated fluorescence microscopy, CCP-specific live B cells are identified and retrieved by micromanipulation. Subsequently, single cell RT-PCR is performed to amplify Ig variable heavy and light chain (VH:VL) genes from the retrieved B cells. The results outline a workflow to obtain paired Ig VH and VL gene amplification by screening, identification and isolation of CCP-specific memory B cells from RA patients PBMC (Figure 1). This methodology provides a fast, efficient and economical platform for isolation of antigen-specific antibodies.
Figure 1.
Outline of the methodology for the combined screening of the antigen specificity and phenotype of single auto-reactive B cells. B cells obtained by enrichment from CCP+ donor derived PBMCs are loaded onto a nanowell array. Microengraving is used to determine CCP reactivity and automated fluorescence microscopy is employed to determine the phenotype of the B cells, on-chip. Post data-analysis, the memory B cells secreting CCP-specific hmAb are retrieved by micromanipulation. Single cell RT-PCR is performed to enable the amplification of the Ig VH and VL regions.
The protocol described here, demonstrated in the context of ACPA, can be readily adapted to the screening of hmAb against any antigen of interest.
2. Methods
Human Subjects Statement
All work outlined in this report was performed according to protocols approved by the Institutional Review Boards at the University of Houston (12495-EX) and the Baylor College of Medicine (H30360)
2.1. Detection of antigen-specific antibodies
As outlined previously, the detection of antigen-specific hmAb from single B cells has been most commonly accomplished via the use of flow cytometry. Labeled antibodies directed against B-cell phenotypic markers like CD19 and CD20 are combined with screens for either soluble antigen or even whole cells displaying antigen5,10. The sorted single cells are then cloned for recombinant expression and assayed for their antigen specificity. Similarly, high-throughput cloning of single B cells has been employed to isolate panels of hmAb via recombinant expression, and the antigen specificity of these hmAb is determined in a second step using ELISA6.
More recently, microfabricated arrays have been described for detection of antigen specificity of single B cells by either direct interrogation of surface-bound B cell receptor (BCR) 27 or Ca2+ mobilization28, or by detecting the secreted hmAb (by antibody secreting cells (ASC) or stimulated B cells)11. We outline a protocol here for the detection of secreted hmAb by activated single B cells using microengraving.
2.1.1. Detection of ACPA in RA patient sera
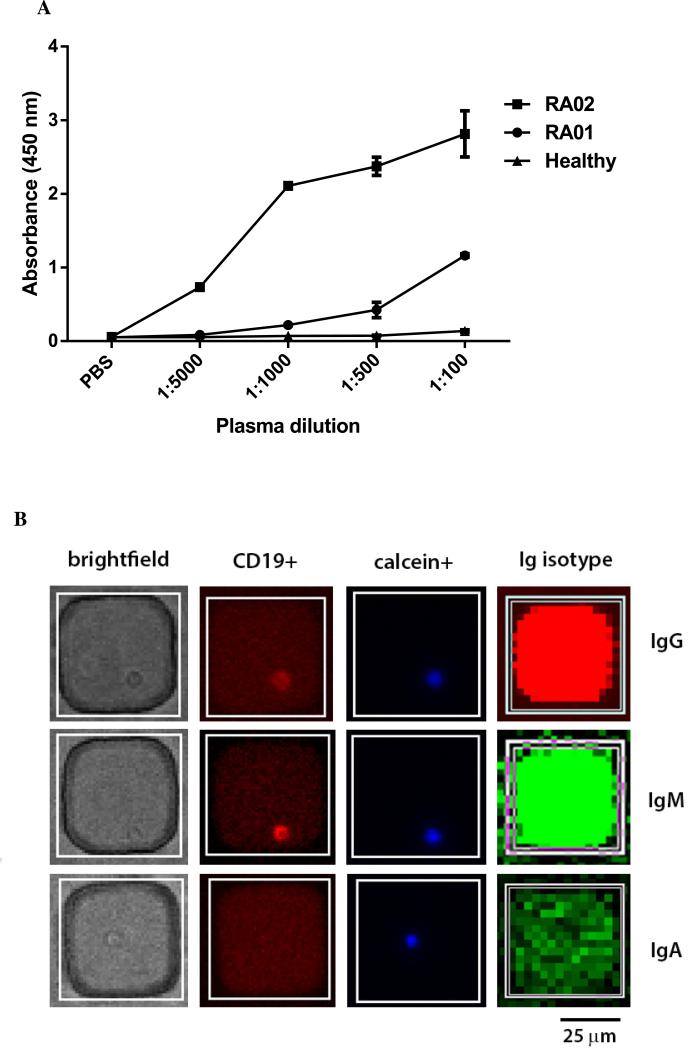
A commercial ELISA-based test, QuantaLite CCP (Innova Diagnostic, San Diego, CA), is used in the clinic for the diagnosis of CCP+ RA. We have adapted this ELISA to enable us to pre-screen RA patients with moderate to high-titers of ACPA (Figure 2A). Briefly, streptavidin-coated microplates are used to capture biotinylated CCP and subsequently incubated with the plasma samples. Detection is accomplished using a secondary horseradish peroxidase (HRP)-conjugated antibody. In our example, since RA02 plasma has higher ACPA titers, we chose this sample for screening ACPA from activated B cells (Figure 2A).
Prepare PBST by adding 0.05% Tween 20 (Sigma-Alrdich, St. Louis, MO) to 1X PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Sodium Phosphate dibasic, 2 mM Potassium Phosphate monobasic, pH 7.4).
Coat 96-well polystyrene MaxiSorp plates (NUNC, Rochester, NY) with 100 μL/well of 50 μg/mL streptavidin (Sigma-Aldrich, St. Louis, MO). Seal the plates with Parafilm M (Pechiney, Chicago, IL), and incubate overnight at 4°C. Wash 5 times with PBST and block with at least 200 μL/well of 10% bovine serum albumin (BSA) in PBST.
Incubate at room temperature (RT) for 2 h.
Wash 5 times with PBST and incubate with 100 μL/well of 5 μg/mL biotinylated CCP (Anaspec, Fremont, CA). Seal the plates and incubate at RT for 1 h. Wash 5 times with PBST.
Add 100 μL/well of the human plasma samples to be tested, at the dilutions 1:100, 1:500, 1:1000, and 1:5000 (each in duplicate). Add 100 μL of 1X PBS as negative control. Seal the plates and incubate at RT for 45 min.
Wash 5 times with PBST. Add 100 μL/well of goat anti-human IgG/IgA HRP-conjugated antibody (Innova Diagnostic) and incubate for 1 h at RT. Wash 5 times with PBST.
Add 100 μL/well of TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Thermo, Waltham, MA) and incubate at RT for 10 min until the blue color develops.
Quantify the absorbance at 450 nm using the InfiniteM300 Microplate Reader (Tecan, Männedorf, Switzerland).
Figure 2.
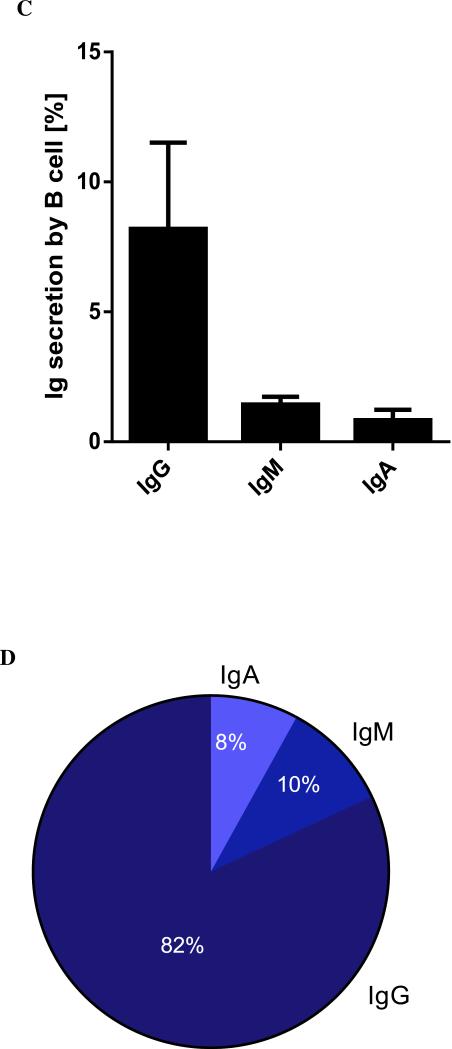
Characterization of ACPA. (A) CCP reactivity of donor plasma as determined by ELISA. Briefly, microtiter plates are coated with streptavidin, incubated first with biotinylated-CCP and then with plasma at different dilutions. Detection is enabled by incubation with HRP conjugated to anti-human IgG/IgA, followed by colorimetric readout. (B) Micrographs of matched images of single B cells to the isotype of the antibody secreted, detected by microengraving. (C) The frequency of immunoglobulin (IgG, IgM and IgA) secretion from three independent sets of stimulated, enriched B cells isolated from the PBMC of RA− donors, determined by microengraving. (D) The distribution of immunoglobulin isotype of these same samples.
2.2. Optimization of memory B cell stimulation
In order to convert B cells into antibody-secreting cells in vitro, a number of different protocols employing cytokines, TLR agonists, CD40L and antibodies have been reported29-31. Each of these methods induces secretion from slightly different subsets of B cells. In our experiments, the use of sCD40L, rhIL-21 and anti-APO1 enables secretion from the highest frequency of B cells using minimal culture times. sCD40L engages with CD40 expressed on the cell surface of B cells to mimic T cell-mediated activation32 and IL-21 is known to promote the differentiation to antibody-secreting cells26,33. Since activation also induces cell death, anti-APO1 is used to rescue B cells from Fas-induced apoptosis34.
We have used PBMC isolated by Ficoll-Paque density gradient centrifugation of fresh whole blood. After 4 days of stimulation, the frequency of antibody-secreting B cells is assessed by microengraving. The percentage of activated B cells is calculated as the number of live B cells secreting Ig as a fraction of the total number of CD19 positive, live B cells present in the nanowell array.
These results indicate that 6-13% of B cells secrete antibodies, with IgG as a predominant isotype, consistent with selective stimulation of memory B cells (Figure 2C-D).
Prepare RPMI-PLGH media: 500 mL RPMI-1640 (Cellgro, Manassas,VA), Penicillin-streptomycin (Cellgro) 50,000 U/50mg + L-glutamine 2mM (Cellgro) + HEPES 10mM (Sigma).
Prepare R10 media with RPMI-PLGH media supplemented with 10% of heat-inactivated Fetal Bovine Serum (FBS) (Biological, Lawrenceville, GA).
Thaw previously frozen PBMC in R10 media in presence of 20 μg/mL of DNAseI (Sigma). Wash twice with R10 and re-suspend to 3 × 106 cells/mL in R10 media.
Add the stimulation cocktail: 2.5 μg/ml sCD40L (R&D System, Minneapolis, MN), 50 ng/mL rhIL-21 (BD Bioscience, San Jose, CA), and 5 μg/mL anti-APO1 (eBioscience, San Diego, CA) and mix thoroughly.
Incubate at 37°C, 5% CO2 for 4 d.
2.2.2. B cell enrichment
B cell enrichment is performed in order to increase the number of B cells assayed on a single nanowell array. The protocol is performed according to manufacturer's instructions using the EasySep™ Human B Cell Enrichment kit (Stemcell Technologies, Vancouver, Canada). Briefly, 5 × 106 of PBMC are re-suspended in 100μL of recommended media and placed in a round-bottom 96-well plate. Subsequent to the addition of 5 μL of the Human B Cell Enrichment Cocktail, the mixture is incubated for 10 min at RT and 10 μL of the D magnetic particles are added. After a further incubation for 5 min, 135 μL of recommended medium is added and plate is positioned onto the EasyPlate Magnet (Stemcell Technologies) for 10 min. The negatively selected, enriched B cells are carefully removed and transferred to a separate well until use.
2.3. Screening of B cells secreting ACPA by microengraving
Microengraving is a soft-lithographic process for printing protein arrays, where each spot on the array comprises the proteins secreted by a single cell35,36. This technology was first used to isolate hybridomas producing hmAbs and also to identify antigen-specific primary B cells from humans11,35. Here, we describe microengraving for screening, identification and isolation of auto-antibodies (ACPA) secreted from B cells in a rapid and high-throughput manner. The nanowell array is molded into a thin slab of polydimethylsiloxane (PDMS) and is able to isolate large numbers of B cells (~100,000). The array is then placed in contact with a glass slide coated with antigen to locally capture auto-antibodies during a 2h period. Subsequently, the secreted auto-antibodies printed onto the glass slide are revealed using a secondary anti-human IgG/IgM/IgA conjugated to a fluorophore (Alexa Fluor, Molecular Probes, Grand Island, NY) and then imaged on a microarray scanner. The result is a footprint of isolated spots, each corresponding to the individual nanowell containing a single B cell secreting ACPA (Figure 2B). Soluble mouse IgG is used to facilitate the registration of the nanowell array on the printed glass slide. In parallel, the nanowell array is imaged by fluorescence microscopy to enable detection of the B cell surface marker CD19, and the live-cell marker, Calcein Violet (Invitrogen).
After compiling and analyzing the combined data from both the microscopy images and the printed glass slide, quantitative single-cell analysis tables are constructed to determine the location, frequency and phenotype of the B cells secreting ACPA (Figure 2B). This information is then used to guide the retrieval of single cells.
In a routine experiment, the frequency of B cells secreting ACPA detected is 0.02 - 0.10 % of single B cells.
2.3.1. Preparation of nanowell arrays
A “master” template, patterned using photolithography, is used as a mold to print elastomeric PDMS arrays that conform to the dimensions of a standard microscopy slide (25 × 75 mm). Detailed protocols for the design and fabrication of the master are available elsewhere (Figure S1)37,38. Before use, the PDMS nanowell array is sterilized and rendered hydrophilic by brief exposure to air plasma.
Mix thoroughly the Sylgard 184 elastomer kit base and curing agent (Dow Corning, Midland, MI) at 10:1 weight ratio in a disposable cup using a plastic knife.
Degas the mixture in a vacuum chamber for 1 h.
Pour mixture onto the master, seal with a glass slide and let it sit for 30 min.
Transfer the assembly into an oven set to 80°C for 2 h to cure the PDMS and bond it to the glass slide. Then let it cool at RT for 1 h.
Carefully lift off the glass slide from the silicon master containing the PDMS nanowell array, and cover the chip with Scotch tape until use.
2.3.2. Coating of auto-antigen on poly-L-Lysine glass slides
Prepare capture antibody solution by adding 25 μg/mL of Streptavidin (Sigma) and 10 μg/mL of goat anti-mouse Ig (Southern Biotech, Birmingham, AL) in 80 μL of borate buffer (50 mM sodium borate, 50 mM sucrose/trehalose, 80 mM NaCl, pH 9).
Pipet the solution on the poly-L-lysine-coated glass slide and gently place a cover slip to spread uniformly on the slide. Incubate for 1 h at RT in a humidified chamber.
Block with 10 mL of 10% BSA in PBST for 2 hours at RT.
Wash the slide with PBST for 5 minutes and then with PBS for 5 minutes. Rinse quickly with deionized (DI) water and dry the slide in a microarray centrifuge.
Add with 80 μL of 5 μg/mL biotinylated Cyclic-Citrullinated Peptide (Anaspec) and gently place a cover slip to spread uniformly on the slide.
Incubate for 1 hour at RT.
Block with 10 mL of 3% BSA PBST for 10 minutes at RT.
Repeat step 4 and store in a humidified Petri dish until further use. Note: The glass slide can be stored overnight at 4°C in a humidified chamber.
2.3.3. B cell loading
Oxidize the PDMS nanowell array using standard plasma cleaner for 1 min at high radiofrequency setting and place the array face-down in sterile PBS.
Count the B cells on a hemacytometer by trypan blue exclusion (Sigma). Re-suspend to 5 × 105 cells/mL in 300 μL of R10 media or approximately 0.5-1×106 cell/mL to avoid a large number of cells per well.
Flip the nanowell array face up and aspirate the PBS. Quickly re-immerse the array in 5 mL of R10 and let it stand for 5 min. Note: Ensure that the chip does not dry out.
Aspirate the R10 and load the cells onto the nanowell array by dispensing them drop-wise and letting them settle for 5 min. Check the loading using a standard tissue-culture, inverted microscope and load again if necessary to achieve desired density (~1 cell/well average).
Remove excess cells by rinsing the array with 5 ml of RPMI-PLGH.
2.3.4. Microengraving
Rinse the nanowell array with RPMI-PLGH containing 5 ng/mL soluble mouse IgG.
Remove the excess media from the PDMS nanowell array along the edges until the outline of the microchannels appears. Note: Adequate care needs to be taken to ensure that the cells are not aspirated directly from the nanowells.
Place the PDMS into the hybridization chamber (Agilent Technologies, Santa Clara, CA), and quickly place the coated glass slide face down over the nanowell array. Press gently and close the chamber tightly.
Incubate the assembled chamber in an incubator for 2 h at 37°C/5% CO2 to enable microengraving of the secreted proteins.
Transfer the sandwich (nanowell array + glass slide) in a Petri dish containing pre-warmed RPMI-PLGH media.
Carefully detach the glass slide from nanowell array and wash in 1% milk in PBST for 10 min at RT (for blocking).
Wash with PBST for 5 min and then PBS for 5 min. Rinse the glass slide with DI water and dry by centrifugation.
Prepare 80 μL of detection solution containing 1 μg/mL of anti-human IgG (Jackson Immunoresearch)-Alexa Fluor (AF) 488, anti-human IgA-AF 532 (BD, Franklin Lakes, NJ), anti-human IgM-AF 594 (BD),goat anti-mouse IgG 647 (Invitrogen) in PBS. Deposit on the glass slide and place a cover slip to uniformly coat the slide.
Incubate for 1 h at RT in humidified chamber and protected from dark.
Repeat step 7.
Image the slide on a microarray scanner like the GenePix 4200AL (Molecular Devices, Sunnyvale, CA). Setup the appropriate excitation wavelengths and emission filters to match the fluorescent dyes used in the experiment.
2.3.5. B cell labeling and imaging
Prepare 300 μL of 4 μg/mL anti-CD19-AF 532 (BioLegend, San Diego, CA) and 1 μg/mL Calcein violet-AM in PBS. Deposit on the surface of the array containing the B cells. Incubate with this staining solution for at least 30 min at 37°C/5% CO2 in the dark before imaging. Note: The labeling can also be performed at 4°C. Additional antibodies labeled with orthogonal dyes directed against other phenotypic markers like CD27, CD38 and CD20 can be included in this step.
Acquire images of the nanowell array using a fluorescence microscope such as Axio Observer Z-1 inverted microscope equipped with a motorized stage and Lambda-DG4 illumination system (Zeiss, Jena, Germany).
After imaging, place the nanowell array into a 4-well plate and immobilize it by adding 2% agarose on its glass edges (top and bottom). Immerse in cold PBS and carefully float the coverslip off the array.
Store the nanowell array containing B cells at 4°C until single cell retrieval by micromanipulation.
2.3.6. Data analysis
Data tables that report the phenotype (anti-CD19-AF 532 and calcein violet) and the location of every single B cell on-chip are obtained using image segmentation routines to automatically process the microscopy images.
Separately, the microengraved images are analyzed using appropriate software packages (Genepix Pro 6.1, Molecular Devices) to tabulate the fluorescent intensities of each Ig-positive spot within the array. Standard database matching algorithms are then employed to correlate the two sets of tables to identify the locations of nanowells containing live B cells secreting ACPA.
Once the analysis is complete, the information is exported to the manipulator for B-cell retrieval (Figure S2).
2.3.7. Automated retrieval of antigen-specific B cells
Prepare collection tubes by adding 2 μL of RT buffer 5x (Invitrogen) and 2 μL of sterile nuclease-free water in 0.1 mL PCR tubes (Axygen, Union City, CA).
Place the 4-well plate containing the nanowell array on the motorized stage in the CellCelector microscope (ALS, Jena, Germany).
Prepare the CellCelector micromanipulator by loading the .CSV file containing the locations of ACPA secreting B cells (picking positions) and by calibrating the X, Y and Z positions.
Start the picking and ensure that the desired cells have been retrieved by checking the brightfield images.
Stored the retrieved B cells at −80°C until further use. Note: The frozen cells can be stored at −80°C indefinitely.
2.4. Single cell RT-PCR and amplification
After single B cell isolation, RT-PCR is performed based on previously published protocols with minor modifications39 (Table 1). Lysis of single cells to yield the mRNA is facilitated by freeze-thawing and the addition of detergents. The cDNA is synthetized using a primer mixture complementary to VH or VL chain constant region sequences. Rounds of first and second PCR amplification are performed by using the appropriate primer mixes (Integrated DNA Technologies, Coralville, IA).
Table 1.
Primer sequences used for reverse-transcription and subsequent amplification of the variable regions from single B cells.
| Description | name | sequence |
|---|---|---|
| cDNA syntesis | ||
| Constant region RT primer mix | CμI | GCAGGAGACGAGGGGGA |
| CγI | AGGG(C/T)GCCAGGGGGAA | |
| CκI | AACAGAGGCAGTTCCAGA | |
| Cλ | AC(C/T)AGTGTGGCCTTGTTGG | |
| Cα | GAGGCTCAGCGGGAAGAC | |
| First PCR | ||
| VH leader sequences | ||
| OH II | VHL-1 | TCACCATGGACTG(C/G)ACCTGGA |
| VHL-2 | CCATGGACACACTTTG(C/T)TCCAC | |
| VHL-3 | CCATG GAR TT(C/T)GGG CTG AGC TGG | |
| VHL-4 | AGAACATGAAACA(C/T)CTGTGGTTCTT | |
| VHL-5 | ATGGGGTCAACCGCCATCCT | |
| VHL-6 | ACAATGTCTGTCTCCTTCCTCAT | |
| OLλ II* | VλL-1 | GGTCCTGGGCCCAGTCTGTGCTG |
| VλL-2 | GGTCCTGGGCCCAGTCTGCC | |
| VλL-3 | ATGGCCTGGA(C/T)C(C/G)CTCTCC | |
| VλL-4/5 | GGTCTCTCTCSCAGC(C/T) TGTGCTG | |
| VλL-6 | GTTCTTGGGCCAATTTTATGCTG | |
| VλL-7 | GGTCCAATTC(C/T)CAGGCTGTGGTG | |
| VλL-8 | GAGTGGATTCTCAGACTGTGGTG | |
| OLκ II | VκL-1/2 | ATGAGG(C/G)TCCC(C/T)GCTCAGCTGCTGG |
| VκL-2 | CTGGGGCTGCTAATGCTCTGG | |
| VκL-3 | TTCCTCCTGCTACTCTGGCTC | |
| VκL-4 | CAGACCCAGGTCTTCATTTCT | |
| Constant regions sequences | ||
| CμII | CAGGAGACGAGGGGGAAAAG | |
| CλII | AGCTCCTCAGAGGAGGG(C/T)GG | |
| CκII | TTTCAACTGCTCATCAGATGGCGG | |
| CγII | GCCAGGGGGAAGAC(C/T)GATG | |
| CαII | GCTCAGCGGGAAGACCTT | |
| Nested PCR | ||
| OH III | VH-1 | TTGCGGCCGCCAGGT(G/C)CAGCTGGT(G/A)CAGTC |
| VH-2 | TTGCGGCCGCCAG(A/G)TCACCTTGAAGGAGTC | |
| VH-3 | TTGCGGCCGC(G/C)AGGTGCAGCTGGTGGAGTC | |
| VH-4 | TTGCGGCCGCCAGGTGCAGCTGCAGGAGTC | |
| VH-5 | TTGCGGCCGCGA(G/A)GTGCAGCTGGTGCAGTC | |
| VH-6 | TTGCGGCCGCCAGGTACAGCTGCAGCAGTC | |
| OLκ III | Vκ-1 | CATAAGATCTCG(A/C)CATCC(A/G)G(A/T)TGACCCAGT |
| Vκ-2 | CACCAGATCTCGAT(A/G)TTGTGATGAC(C/T)CAG | |
| Vκ-3 | ACCAGATCTCGAAAT(T/A)GTG(T/A)TGAC(G/A)CAGTCT | |
| Vκ-4 | CACCAGATCTCGACATCGTGATGACCCAGT | |
| OLλ III | Vλ-1 | TATTAGATCTCCAGTCTGTGCTGACTCAGC |
| Vλ-2 | TATTAGATCTCCAGTCTGCCCTGACTCAGC | |
| Vλ-3 | CACCAGATCTCTCCTATGAGCTGAC(T/A)CAGC | |
| Nested constant regions | CμIII | AGGTCTAGAGAAAAGGGTTGGGGCGGATGC |
| CγIII | AGGTCTAGAGAC(C/G)GATGGGCCCTTGGTGGA | |
| CκIII | TATTCCATGGAAGATGAAGACAGATGGTGC | |
| CλIII | CATTCCATGGGGGAACAGAGTGACCG | |
| CαIII | GACCTTGGGGCTGGTCGGGGA | |
We have successfully amplified immunoglobulin heavy and light chains from auto-reactive single B cells. Typical success rates are ~30-50% for amplification of pairs of variable heavy and light chains (VH:VL).
Thaw the PCR tubes containing single memory B cells and store them on ice.
Add 3 μl of 5% NP-40 (Sigma) and 1 μL of 5 pmol/μL of constant region RT primer mix (Table 1). Place the PCR tubes in a thermocycler (Applied Biosystems, Carlsbad, CA) with heating at 65°C for 3 min, followed by cooling at 25°C for 3 min. Store the tubes on ice.
Add 2 μL of RT buffer 5x, 2 μL of DTT (Invitrogen), 1 μL of 10mM dNTP (Takara, Shiga, Japan) and 0.5 μL of 200 U/μL Superscript III RT (Invitrogen) to each tube, up to a final volume 19.5 μL.
Place the tubes into the thermocycler (Applied Biosystems) and heat at 37°C for 1 h for first strain cDNA synthesis and then heat to 70°C for 10 min to inactivate the enzyme.
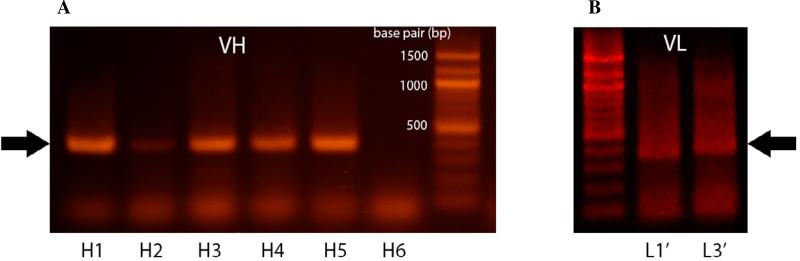
Use the cDNA synthesized in the previous step as template for the first PCR. Take two aliquots of 8 μL cDNA products to amplify VH and VL. To one sample aliquot of 8 μL cDNA product, add 6 μL of PCR buffer 10x, 1.6 μL of 10 mM dNTP (Takara, Shiga, Japan), 0.5 μL of leader sequence primer mixture (containing 20 pmol/μL of each primer), 0.5 μL of constant region primer mixture (20 pmol/μl of each primer), 1 μL of 2.5 U/μL Taq Polymerase (Takara, Shiga, Japan) and 48 μL of H2O. Note: In order to conserve reagents, we apply a tiered approach to the amplification of single cells. The VH regions are amplified first and VL region amplification is only performed for those samples from which we could identify successful VH amplicons on an agarose gel (Figure 3)
Program 3 cycles of pre-amplification at 94°C for 45 s, 45°C for 45 s and 72°C for 1 min 45 s. Next, perform 30 cycles of amplification at 94°C for 45 s, 50°C for 45 s, 72°C for 1 min 45 s and complete by incubation at 72°C for 10 min.
Prepare the nested PCR mix by transferring 3 μL of first PCR product to a new tube and adding 5 μL of PCR buffer 10x, 1.25 μl of 10 mM dNTP, 1 μL of nested PCR primer mixture (20 pmol/μl of each primer), 1 μL of constant region III primer (20 pmol/μL of each primer), 1 μL of 2.5 U/μL Taq Polymerase and 38 μL of H2O.
Run the PCR reaction using the following conditions: 30 cycles of amplification at 94°C for 45 s, 50°C for 45 s, 72°C for 1 min 45 s and final incubation at 72°C for 10 min.
The DNA products are separated using agarose gel electrophoresis. Briefly, 3 μL of PCR product is mixed with 7.5 μL of Tris-acetate-EDTA (TAE) 1x buffer and loaded into 0.8% (wt/vol) agarose gel (Gibco, Grand Island, NY) with 10 μL of 1 kb DNA ladder (NEB, Ipswich, MA) and run at 110 V for 40 min. After staining for 20 min with 0.5 μg/mL ethidium bromide dye (NEB, Ipswich, MA) in TAE buffer, the gel is imaged by using UV light assisted visualization of bands at ~400 bp.
The PCR product is then used as template for further re-amplification to facilitate DNA sequencing and recombinant cloning, essentially as described previously14. Transient transfection into human kidney epithelial cells (HEK293) is used for expression of full-length hmAb and ELISA of the supernatants/purified proteins performed to confirm the antigen specificity and of these antibodies. Subsequent to amplification, around 30-40 % of antigen-specific antibodies selected using microengraving did not clone or express as full-length human IgG1 after transfection (unpublished data).
Figure 3.
Amplification of the VH and VL regions from single B cells. Subsequent to cell lysis and reverse transcription to yield cDNA, two rounds of PCR are employed to amplify the VH and VL regions. The PCR products are run on an agarose gel and visualized by ethidium bromide staining (~400 bp, dark arrow). (A) Lanes H1–H6 and (B) L1’-L3’ show gene amplification for VH and VL products, respectively, from 6 single B cells. Sequential amplification of VH and then the VL is performed to conserve reagents.
3. Conclusions
Here, we describe a technology to screen and isolate auto-antibodies derived from single B cells originating from the PBMC of RA patients. The entire procedure including the stimulation, microengraving, retrieval and amplification of cells can be accomplished in 1 week. The advantages of using microengraving are the ability to detect low numbers of candidate B cells secreting specific antibodies needed to isolate the auto-reactive hmAbs and the ability to determine the phenotype of these auto-reactive B cells.
We anticipate that this technology can be readily adapted for the screening and isolation of other auto-reactive antibodies to help elucidate their molecular contribution in pathology of autoimmune diseases and also determine their phenotype to facilitate therapeutic intervention.
Supplementary Material
Acknowledgments
This publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA174385 and the University of Houston New Faculty Award (I102560). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Vandana Kaul and Balakrishnan Ramesh for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187:4229–4235. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- 3.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein F, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuels J, Ng YS, Coupillaud C, Paget D, Meffre E. Impaired early B cell tolerance in patients with rheumatoid arthritis. J Exp Med. 2005;201:1659–1667. doi: 10.1084/jem.20042321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, et al. Human antibodies for immunotherapy development generated via a human B cell hybridoma technology. Proc Natl Acad Sci U S A. 2006;103:3557–3562. doi: 10.1073/pnas.0511285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickell PM. Immortalization of human B-lymphocytes by epstein-barr virus. Methods Mol Biol. 1992;8:213–218. doi: 10.1385/0-89603-191-8:213. [DOI] [PubMed] [Google Scholar]
- 9.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradshaw EM, et al. Concurrent detection of secreted products from human lymphocytes by microengraving: cytokines and antigen-reactive antibodies. Clin Immunol. 2008;129:10–18. doi: 10.1016/j.clim.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin A, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. 2009;15:1088–1092. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- 13.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkon K, Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4:491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppu S, Ronkainen MS, Kulmala P, Akerblom HK, Knip M. GAD65 antibody isotypes and epitope recognition during the prediabetic process in siblings of children with type I diabetes. Clin Exp Immunol. 2004;136:120–128. doi: 10.1111/j.1365-2249.2004.02416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Venrooij WJ, van Beers JJ, Pruijn GJ. Anti-CCP Antibody, a Marker for the Early Detection of Rheumatoid Arthritis. Ann N Y Acad Sci. 2008;1143:268–285. doi: 10.1196/annals.1443.013. [DOI] [PubMed] [Google Scholar]
- 18.Leslie D, Lipsky P, Notkins AL. Autoantibodies as predictors of disease. J Clin Invest. 2001;108:1417–1422. doi: 10.1172/JCI14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioan-Facsinay A, et al. Anti-cyclic citrullinated peptide antibodies are a collection of anti-citrullinated protein antibodies and contain overlapping and non-overlapping reactivities. Ann Rheum Dis. 2011;70:188–193. doi: 10.1136/ard.2010.131102. [DOI] [PubMed] [Google Scholar]
- 20.Sokolove J, et al. Autoantibody epitope spreading in the pre-clinical phase predicts progression to rheumatoid arthritis. PLoS One. 2012;7:e35296. doi: 10.1371/journal.pone.0035296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McInnes IB, Schett G. The pa thogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 22.Koumakis E, Wipff J, Avouac J, Kahan A, Allanore Y. Severe refractory rheumatoid arthritis successfully treated with combination rituximab and anti-tumor necrosis factor-alpha-blocking agents. J Rheumatol. 2009;36:2125–2126. doi: 10.3899/jrheum.090160. [DOI] [PubMed] [Google Scholar]
- 23.Keystone E, et al. Improvement in patient-reported outcomes in a rituximab trial in patients with severe rheumatoid arthritis refractory to anti-tumor necrosis factor therapy. Arthritis Rheum. 2008;59:785–793. doi: 10.1002/art.23715. [DOI] [PubMed] [Google Scholar]
- 24.Tedder TF. CD19: a promising B cell target for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5:572–577. doi: 10.1038/nrrheum.2009.184. [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Benoist C, Mathis D. Rituximab specifically depletes short-lived autoreactive plasma cells in a mouse model of inflammatory arthritis. Proc Natl Acad Sci U S A. 2010;107:4658–4663. doi: 10.1073/pnas.1001074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ettinger R, et al. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 27.Tajiri K, et al. Cell-microarray analysis of antigen-specific B-cells: single cell analysis of antigen receptor expression and specificity. Cytometry A. 2007;71:961–967. doi: 10.1002/cyto.a.20471. [DOI] [PubMed] [Google Scholar]
- 28.Ozawa T, et al. MAC-CCD system: a novel lymphocyte microwell-array chip system equipped with CCD scanner to generate human monoclonal antibodies against influenza virus. Lab Chip. 2009;9:158–163. doi: 10.1039/b810438g. [DOI] [PubMed] [Google Scholar]
- 29.Huggins J, et al. CpG DNA activation and plasma-cell differentiation of CD27- naive human B cells. Blood. 2007;109:1611–1619. doi: 10.1182/blood-2006-03-008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arpin C, et al. Generation of memory B cells and plasma cells in vitro. Science. 1995;268:720–722. doi: 10.1126/science.7537388. [DOI] [PubMed] [Google Scholar]
- 31.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 32.van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 33.Good KL, Bryant VL, Tangye SG. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J Immunol. 2006;177:5236–5247. doi: 10.4049/jimmunol.177.8.5236. [DOI] [PubMed] [Google Scholar]
- 34.Mehta DS, et al. IL-21 induces the apoptosis of resting and activated primary B cells. J Immunol. 2003;170:4111–4118. doi: 10.4049/jimmunol.170.8.4111. [DOI] [PubMed] [Google Scholar]
- 35.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 36.Varadarajan N, et al. Rapid, efficient functional characterization and recovery of HIV-specific human CD8+ T cells using microengraving. Proc Natl Acad Sci U S A. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liadi I, Roszik J, Romain G, Cooper LJ, Varadarajan N. Quantitative high-throughput single-cell cytotoxicity assay for T cells. J Vis Exp. 2013:e50058. doi: 10.3791/50058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat Protoc. 2009;4:767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Stollar BD. Human immunoglobulin variable region gene analysis by single cell RTPCR. J Immunol Methods. 2000;244:217–225. doi: 10.1016/s0022-1759(00)00260-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.