Abstract
Commitment to division requires that cells sense, interpret, and respond appropriately to multiple signals. In most eukaryotes, cells commit to division in G1 prior to DNA replication. Beyond a point, known as Start in yeast and the restriction point in mammals, cells will proceed through the cell cycle despite changes in upstream signals. In metazoans, misregulated G1 control can lead to developmental problems or disease, so it is important to understand how cells decipher the myriad external and internal signals that contribute to the fundamental all-or-none decision to divide. Extensive study of G1 control in the budding yeast S. cerevisiae and mammalian culture systems has revealed highly similar networks regulating commitment. However, protein sequences of functional orthologs often indicate a total lack of conservation suggesting significant evolution of G1 control. Here, we review recent studies defining the conserved and diverged features of G1 control and highlight systems-level features that may be common to other biological regulatory networks.
Although both Start and the restriction point govern passage into S phase, their physiological input signals are quite different. A pre-Start cell exposed to mating pheromone immediately arrests in G1 while a post-Start cell proceeds once more through the cell cycle [1–3]. Similarly, beyond the mammalian restriction point, cells complete division irrespective of changes to some growth factor-dependent signals [4,5]. Based on how they were defined in yeast and mammalian cells, Start and the restriction point have very distinct physiologies which might place different requirements on their regulatory networks [6].
Conservation and Evolution of G1 Control Networks
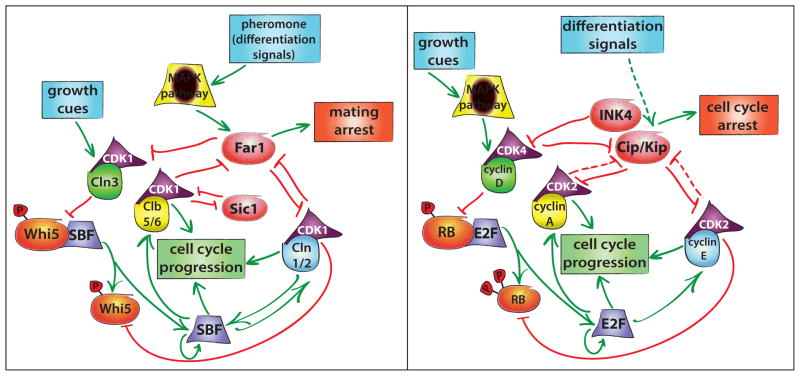
The core G1 control network appears highly similar in budding yeast and mammals (Figure 1) [7]. In both organisms, signaling leads to an increase in cyclin-dependent kinase (CDK) activity via cyclin synthesis, which is largely responsible for promoting progression into S phase [8,9]. Prior to CDK activation, budding yeast spend variable amounts of time in G1, with smaller cells generally taking longer to reach Start [10]. This size-dependent progression functions primarily in daughter cells and requires that growth be coupled to the cell cycle. One likely coupling mechanism would rely on increasing levels of a cell cycle-regulating “sizer” protein whose rate of synthesis is proportional to the overall protein production rate [11]. A good sizer candidate in budding yeast is the G1-S activator CLN3, which drives progression through Start in a dosage-dependent manner [12,13]. The levels of Cln3 are sensitive to both cellular growth rate and metabolic state [14,15]. Cln3 binds and activates CDK1, the sole yeast cyclin-dependent kinase required for cell cycle progression [16]. Cln3-CDK1 phosphorylates and initiates the inactivation of the transcriptional inhibitor Whi5, promoting its disassociation from the transcription factor SBF (Swi4/Swi6). This results in weak transcriptional activation of two downstream G1 cyclins, CLN1 and CLN2 [17,18]. Cln1 and Cln2 promote further inactivation of Whi5 and simultaneous activation of SBF and MBF (Mbp1/Swi6), which drive the cell cycle-dependent expression of over 200 genes including the S-phase cyclins that initiate DNA synthesis [19–22]. The SBF component Swi4 is also an SBF target, suggesting an additional positive feedback loop [23]. While an exact mechanism has yet to be elucidated, rising Cln3 levels may relay synthesis rate information to the G1 control network, with downstream positive feedback setting the threshold for cell cycle commitment.
Figure 1.
Despite a lack of sequence homology, G1 control networks are similar in both yeast (left panel) and mammals (right panel). Proteins and signals with similar functions are similarly shaped/colored. Upstream growth cues activate G1 cyclins, which drive progression into S phase via the activation of a positive feedback loop. Differentiation signals, including the pheromone-activated MAPK pathway in yeast, activate proteins that inhibit cyclin-CDK activity, leading to the increased stability of a low CDK activity cellular state.
The core G1 signaling network is similar in mammals, where growth factor stimulation leads to an increase in cyclin D, the upstream activator of G1 progression [24]. Cyclin D, functionally analogous to the yeast Cln3 protein, activates CDKs 4 and 6 to phosphorylate and initiate inactivation of the pocket proteins p107, p130, and the retinoblastoma (Rb) protein [25]. Inactivation of Rb leads to partial activation of the transcription factors E2F1-3, which then activate the transcription of downstream cyclins E and A that likely complete Rb inactivation and initiate DNA replication [25,26]. In both networks, activating signals proceed through an upstream cyclin that initiates a positive feedback loop of downstream cyclins.
While the core cell cycle control networks are broadly similar in both mammalian and yeast systems, many functionally analogous proteins share no sequence homology. For example, despite a lack of sequence similarity, Whi5 and Rb act as inhibitors of cell cycle progression through recruitment of histone deacetylases and are directly inhibited by CDK activity [13,27]. Similarly, SBF shares no sequence homology with E2F while performing an analogous function. In addition, known homologs such as CDK1 appear to have taken on a new role in G1 control in yeast while performing mitotic functions in mammals, which use CDKs 2, 4, and 6 to regulate cell cycle entry [28]. While network structure and the physiological function of integrating multiple signals into a decision to divide are conserved, there has been significant evolution of G1 control and direct inferences from one organism to another should be carefully considered. Nonetheless, the high degree of network similarity suggests that similar molecular mechanisms might be used to generate appropriate input-output relationships governing physiological function.
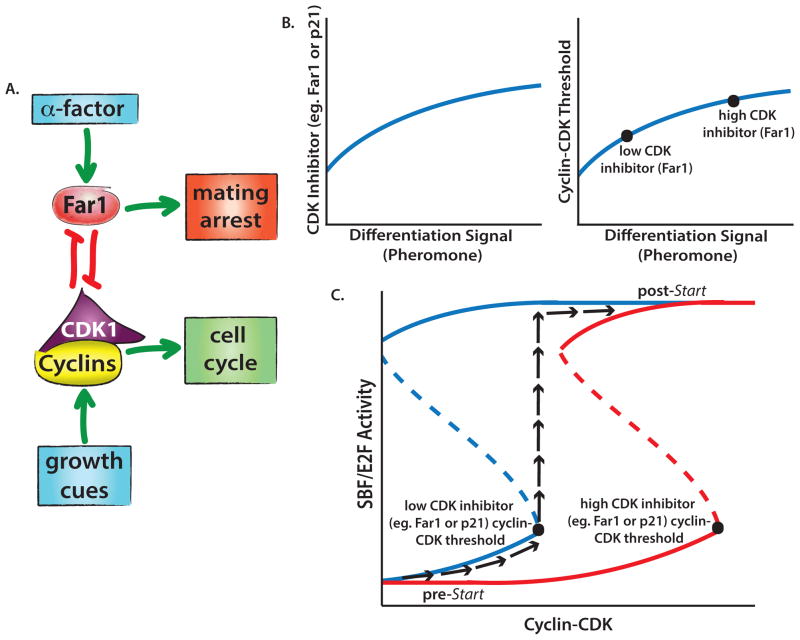
In both mammals and yeast, input signals of diverse origin increase CDK activity to reach a threshold level driving commitment. In this model, when the transcriptional inhibitor Whi5 or Rb is sufficiently inactivated, an all-or-none response occurs that prevents reversion to a pre-committed state [29–31]. Such an all-or-none response, where a higher level of activation is required to initiate commitment than to maintain the downstream state, suggests that the underlying regulatory system undergoes a saddle-node bifurcation (Figure 2) [32]. During such a transition, the system is initially bistable, but the low-CDK state is destabilized by an increasing input signal leading to rapid cell cycle entry [30]. Loss of bistability via saddle-node bifurcation is encountered at other irreversible cellular transitions, such as oocyte maturation in Xenopus laevis, where progesterone treatment of cells above a threshold promotes meiosis [33]. The transition from interphase to meiosis is established through ultrasensitivity of the MAP kinase cascade within a positive feedback loop that drives maturation [34]. Thus, cells employ bistable regulatory networks based on positive feedback to generate all-or-none threshold responses [35–37].
Figure 2.
A. Simplified scheme of the decision-making yeast network regulating Start. B. Increased duration of α-factor exposure leads to the accumulation of Far1, which increases the amount of cyclin-CDK required to drive yeast into the cell cycle. Increasing Far1 reflects the corresponding temporal integration of pheromone pathway activity. C. Cell cycle commitment is a bistable process. Once a threshold level of cyclin-CDK is reached, cells rapidly transition to the committed state. The black arrows illustrate one potential path. The dotted line represents an unstable intermediate state. Increased Far1 concentrations favor the low SBF activity state and shift the curve toward the right indicating that higher cyclin-CDK concentrations are required to commit a cell to division.
Redefining Commitment with Single-Cell Analysis
Our general framework for viewing commitment is that input signals raise cyclin-CDK levels until they traverse a threshold beyond which positive feedback becomes self-sustaining. However, additional pathways can act to modulate this threshold (Figure 1) [38]. In Xenopus, the meiotic progesterone threshold can be tuned by GSK-3β activity [34]. Threshold tuning has also been observed in Xenopus mitosis, where inhibitory phosphorylation of CDK1 must be overcome by Cdc25 phosphatase activity to permit mitotic entry. DNA checkpoint signaling that opposes Cdc25 activity raises the threshold concentration of cyclin B required to progress into mitosis [39]. Thus, traversing a tunable threshold is a common theme among switch-like transitions.
The measurement of commitment thresholds has been greatly improved by recent technical advances. More accurate measurements are providing mechanistic insight into how these thresholds are tuned. While population-based studies have poor threshold resolution due to imperfect cell cycle synchrony, the combination of time-lapse imaging with microfluidic devices, which precisely control the extracellular environment, has become a powerful tool for quantitatively measuring commitment thresholds [40]. This combination of microfluidics and imaging has been successfully applied to yeast Start. Traversal of Start is driven by CDK activity, which can be measured in live cells via the CDK-dependent nuclear export of the transcriptional inhibitor Whi5 [29,41]. Start corresponds precisely to the abrupt activation of the G1 cyclin-positive feedback loop, which occurs when about 50% of a cell’s Whi5 has left the nucleus [42]. There is surprisingly little cell-to-cell variation in the threshold, as the percentage of nuclear Whi5 at the time of pheromone addition predicts cell fate with 97% accuracy. Interestingly, the threshold level of Whi5 is tuned by the addition of mating pheromone, and it was found that the threshold for exiting a pheromone-arrested state is higher (~65% of nuclear Whi5 must be exported). This study illustrates the potential for using microfluidic platforms to accurately determine thresholds in a variety of physiological contexts [43].
Time-lapse imaging with fluorescent reporters has not yet been applied to the restriction point where there are many positive feedback loops and other signaling events that could be causal for commitment [6,44,45]. In mammalian cells, low levels of E2F1-3 activate expression of E2F1, forming a positive feedback loop that inactivates its key negative regulator Rb [30,46]. The E2F target cyclin E binds CDK2 to further phosphorylate and complete inactivation of Rb [26,47], producing a second positive feedback loop. A third positive feedback loop involves the E2F target Skp2, the F-box protein specifically targeting the CDK inhibitor p27 for degradation [48–50]. While these positive feedback loops may contribute to the commitment process, recent studies using single-cell time-lapse imaging of cycling normal human fibroblasts concluded that the mammalian restriction point occurs four to six hours before significantly detectable Rb phosphorylation, suggesting that upstream events could define commitment [24,45,51,52]. Moreover, studies in transformed cells reveal a lack of proper restriction point control in response to serum removal [51,53,54]. Much like in yeast, single-cell time-lapse imaging experiments using live cell reporters in primary mammalian cells may precisely pinpoint the molecular events of the restriction point and hopefully resolve the controversy surrounding commitment timing.
Commitment regulation may be modified in distinct cell types and we currently lack understanding of restriction point control within a growing animal tissue. For example, stem cell division control may be distinct because these cells do not rely on a MAPK-dependent mitogenic pathway to enter the cell cycle, and they exhibit a short G1 phase prior to differentiation (Figure 3) [55] [56–58]. We know what markers are expressed in differentiated cells, but we do not yet understand when exactly the decision to differentiate is made nor the causal molecular mechanism [59]. By reconstructing the physiological environment of stem cells using microfluidics and studying activation timing of the molecular pathways underlying differentiation, one might be able to generate a predictive framework for how these cells make decisions.
Figure 3.
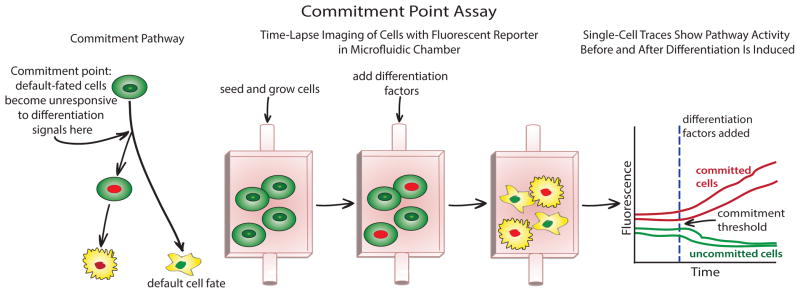
General experimental scheme for using time-lapse imaging, fluorescent reporters, and microfluidics to analyze commitment within a predictive framework. For pathways that employ irreversible transitions, following a cell before and after the transition allows the determination of molecular events committing a cell to a downstream fate if a threshold can be determined using a fluorescent reporter (see text).
Prediction is essential to analyzing cell cycle commitment because it provides a quantitative measure of how well a cellular decision is understood. In this context, commitment points can be analyzed by exposing cells to an abrupt step change in their extracellular environment and then observing the outcome of a binary cellular decision, such as differentiation into two distinct cell types or the distinction between cells committed to division and those responsive to growth factor signaling (Figure 3). It is important to only use information gathered prior to the step change to infer the cellular state because allowing the use of subsequent downstream information would produce trivial predictions: e.g., the expression of any of 1000 differentiation-regulated genes predicts cell fate with 100% accuracy. However, expression of these proteins will not be highly predictive of the decision because they occur downstream of the commitment point. Nevertheless, such trivial predictions are the current state-of-the-art in the stem cell field indicating that the adoption of a predictive framework would represent a large methodological step forward because the most informative measurements will reveal the core of the cell’s decision-making network.
Temporal Order of Transcription within the G1-to-S Transition
In both mammalian and yeast cells, the temporal order of transcription within the G1-to-S transition has been analyzed [60]. Genes involved in positive feedback loops are transcribed prior to other genes regulated by the same transcription factor. In yeast, the activation of CLN1 and CLN2 precedes other SBF and MBF regulated genes. In mammalian cells, the positive feedback components cyclin E1, cyclin E2, Skp2, and E2F1 are all among the earliest transcribed E2F targets at the G1-to-S transition. Notably, the MBF target NRM1 that inactivates MBF [61] is activated much later than other G1-S genes ensuring enough time for regulon expression prior to its inactivation. That commitment precedes activation of the 5–10% of the genome that is cell cycle regulated in both yeast and mammals suggests the principle that cells tend to make a decision first prior to synthesizing machinery associated with the downstream cell fate.
Time Integration of Signals in a Decision-Making Process
Accurate cellular decisions may be complicated by the fact that within a population of genetically identical cells grown in identical environments, there is substantial cell-to-cell variation in protein concentration. To overcome molecular noise, cells might base their decisions not only on the current strength of an input signal, but on its history [62]. For example, by taking the integral of the signal, cells could formulate a response robust to fluctuations that are removed by time averaging. Integrated responses may be important for p53-dependent regulation in mammalian cells, where p53 may be spontaneously and transiently activated under unstressed conditions [63]. Cells respond differently to transient DNA damage that occurs during normal growth than to damage requiring a full apoptotic response. p53 targets that induce cell cycle arrest, such as p21, respond to persistent but not transient p53 activity perhaps through a temporal integration mechanism yet to be elucidated.
The temporal integration of pathway activity has also been observed in pathways governing cell cycle progression in both yeast and mammals. In human mammary epithelial cells, EGF stimulation leads to the activation of discrete, asynchronous ERK pulses, which increase in frequency with increased concentrations of EGF [64]. That proliferation is proportional to the frequency of ERK pulses suggests that a temporal integration of ERK activity is the relevant input for cell cycle progression [44,50]. In yeast, temporal integration of a signal results in the accumulation of a cell cycle inhibitor rather than an activator. Pheromone activates MAP kinase signaling that arrests the cell cycle by activating transcription of the CDK inhibitor Far1 [65]. The amount of Far1 reflects a temporal integration of pheromone pathway activity, and this corresponds to the amount of cyclin-CDK needed to drive cells into the cell cycle. A similar method of measuring signal dynamics may be employed for the differentiation response in mammalian cells, where the accumulation of cell cycle inhibitors may result from the temporal integration of differentiation signals [66,67].
Conclusion
Cell cycle commitment is an important process, and knowledge of the underlying regulatory networks has been crucial for beginning to uncover the principles governing cellular decisions. Recent advances in single-cell studies have allowed unprecedented temporal resolution of the molecular mechanisms of Start in yeast. Since the Whi5-SBF/MBF-Cln1/2 pathway bears remarkable similarity to the Rb-E2F-cyclin E/A mammalian pathway, we expect that some of the principles underlying commitment in yeast can be usefully applied to the mammalian system. The next few years should be an exciting time for defining how molecular events establish commitment across eukaryotes.
Highlights.
G1 control networks are highly similar in budding yeast and mammals.
Lack of protein sequence conservation suggests significant evolution of G1 control.
Accurate cellular decisions may be based on time-integrated signals.
Single-cell analysis has elucidated Start and should be used to study the restriction point.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proceedings of the National Academy of Sciences of the United States of America. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 3.Morgan DO. The Cell Cycle: Principles of Control. London; Sunderland, MA: New Science Press; 2007. [Google Scholar]
- 4.Pardee AB. A restriction point for control of normal animal cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetterberg A, Larsson O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc Natl Acad Sci U S A. 1985;82:5365–5369. doi: 10.1073/pnas.82.16.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **6.Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 Cell Cycle Progression: Distinguishing the Restriction Point from a Nutrient-Sensing Cell Growth Checkpoint(s) Genes & cancer. 2010;1:1124–1131. doi: 10.1177/1947601910392989. This review presents a straightforward summary of work that supports a separate restriction point and cell growth checkpoint in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos Trans R Soc Lond B Biol Sci. 2011;366:3532–3544. doi: 10.1098/rstb.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 9.Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER, Rubin SM, Morgan DO, Loog M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 11.Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Current biology : CB. 2012;22:R350–359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 2009;7:e1000221. doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 2009;7:e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L, Tu BP. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2013;110:7318–7323. doi: 10.1073/pnas.1302490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider BL, Zhang J, Markwardt J, Tokiwa G, Volpe T, Honey S, Futcher B. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start. Mol Cell Biol. 2004;24:10802–10813. doi: 10.1128/MCB.24.24.10802-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 17.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 18.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Ferrezuelo F, Colomina N, Futcher B, Aldea M. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome biology. 2010;11:R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Travesa A, Kuo D, de Bruin RA, Kalashnikova TI, Guaderrama M, Thai K, Aslanian A, Smolka MB, Yates JR, 3rd, Ideker T, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31:1811–1822. doi: 10.1038/emboj.2012.28. In these papers, the authors show that MBF, but not SBF-regulated genes, are specifically activated at the G1-S DNA replication checkpoint demonstrating distinct functions with the G1/S cell cycle regulated genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Bastos de Oliveira FM, Harris MR, Brazauskas P, de Bruin RA, Smolka MB. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 2012;31:1798–1810. doi: 10.1038/emboj.2012.27. In these papers, the authors show that MBF, but not SBF-regulated genes, are specifically activated at the G1-S DNA replication checkpoint demonstrating distinct functions with the G1/S cell cycle regulated genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris MR, Lee D, Farmer S, Lowndes NF, de Bruin RA. Binding specificity of the g1/s transcriptional regulators in budding yeast. PLoS One. 2013;8:e61059. doi: 10.1371/journal.pone.0061059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–4179. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- 25.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 27.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 28.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 29.Charvin G, Oikonomou C, Siggia ED, Cross FR. Origin of irreversibility of cell cycle start in budding yeast. PLoS biology. 2010;8:e1000284. doi: 10.1371/journal.pbio.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nature cell biology. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 31.Yao G, Tan C, West M, Nevins JR, You L. Origin of bistability underlying mammalian cell cycle entry. Mol Syst Biol. 2011;7:485. doi: 10.1038/msb.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyson JJ, Csikasz-Nagy A, Novak B. The dynamics of cell cycle regulation. Bioessays. 2002;24:1095–1109. doi: 10.1002/bies.10191. [DOI] [PubMed] [Google Scholar]
- 33.Justman QA, Serber Z, Ferrell JE, Jr, El-Samad H, Shokat KM. Tuning the activation threshold of a kinase network by nested feedback loops. Science. 2009;324:509–512. doi: 10.1126/science.1169498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 35.Ferrell JE, Xiong W. Bistability in cell signaling: How to make continuous processes discontinuous, and reversible processes irreversible. Chaos. 2001;11:227–236. doi: 10.1063/1.1349894. [DOI] [PubMed] [Google Scholar]
- 36.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novak B, Tyson JJ, Gyorffy B, Csikasz-Nagy A. Irreversible cell-cycle transitions are due to systems-level feedback. Nature cell biology. 2007;9:724–728. doi: 10.1038/ncb0707-724. [DOI] [PubMed] [Google Scholar]
- 38.Buchler NE, Cross FR. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sha W, Moore J, Chen K, Lassaletta AD, Yi CS, Tyson JJ, Sible JC. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci U S A. 2003;100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS One. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrell JE., Jr Simple rules for complex processes: new lessons from the budding yeast cell cycle. Molecular cell. 2011;43:497–500. doi: 10.1016/j.molcel.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. Uses microfluidics in conjunction with single-cell time-lapse imaging to analyze a commitment point (Start) at unprecedented temporal resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- *44.Zwang Y, Sas-Chen A, Drier Y, Shay T, Avraham R, Lauriola M, Shema E, Lidor-Nili E, Jacob-Hirsch J, Amariglio N, et al. Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Molecular cell. 2011;42:524–535. doi: 10.1016/j.molcel.2011.04.017. Cells require growth factor at two distinct short windows in G1. In the first window, an EGF pulse induces metabolic and p53-regulated genes including CDK inhibitors, while the second window down-regulates p53 activity and sets a threshold for cell cycle entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hitomi M, Yang K, Guo Y, Fretthold J, Harwalkar J, Stacey DW. p27Kip1 and cyclin dependent kinase 2 regulate passage through the restriction point. Cell cycle. 2006;5:2281–2289. doi: 10.4161/cc.5.19.3318. [DOI] [PubMed] [Google Scholar]
- 46.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- *47.Merrick KA, Wohlbold L, Zhang C, Allen JJ, Horiuchi D, Huskey NE, Goga A, Shokat KM, Fisher RP. Switching Cdk2 on or off with small molecules to reveal requirements in human cell proliferation. Mol Cell. 2011;42:624–636. doi: 10.1016/j.molcel.2011.03.031. The authors utilize an analog-sensitive CDK2 to reveal its specific functions in cell proliferation, including restriction point control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yung Y, Walker JL, Roberts JM, Assoian RK. A Skp2 autoinduction loop and restriction point control. J Cell Biol. 2007;178:741–747. doi: 10.1083/jcb.200703034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coats S, Flanagan WM, Nourse J, Roberts JM. Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- 50.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & development. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 51.Larsson O, Zetterberg A. Existence of a commitment program for mitosis in early G1 in tumour cells. Cell Prolif. 1995;28:33–43. doi: 10.1111/j.1365-2184.1995.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 52.Martinsson HS, Starborg M, Erlandsson F, Zetterberg A. Single cell analysis of G1 check points-the relationship between the restriction point and phosphorylation of pRb. Experimental cell research. 2005;305:383–391. doi: 10.1016/j.yexcr.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 53.Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 54.Zetterberg A, Larsson O. Coordination between cell growth and cell cycle transit in animal cells. Cold Spring Harb Symp Quant Biol. 1991;56:137–147. doi: 10.1101/sqb.1991.056.01.018. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Austin RH. Applications of Microfluidics in Stem Cell Biology. Bionanoscience. 2012;2:277–286. doi: 10.1007/s12668-012-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fluckiger AC, Marcy G, Marchand M, Negre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24:547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 58.White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **59.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. Studies embryonic stem cell differentiation in single cells to understand the pluripotency gene network in the context of differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *60.Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Molecular cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. By analyzing gene expression data from yeast and mammalian cells, the authors determine that in both systems G1-S positive feedback elements are activated first and drive commitment prior to the activation of several hundred other G1-S genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, Russell P, Wittenberg C. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Molecular cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- *62.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. Time-lapse imaging of single cells suggests that cells respond to integrated p53 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **64.Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2012;49:249–261. doi: 10.1016/j.molcel.2012.11.002. ERK is activated in asynchronous pulses whose frequency and duration is determined by extracellular EGF concentrations, suggesting that cells formulate a proliferative response based on the integration of ERK signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *65.Doncic A, Skotheim JM. Feedforward Regulation Ensures Stability and Rapid Reversibility of a Cellular State. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.04.014. The authors show that the coherent feed-forward regulation of the CDK inhibitor Far1 results in a stable yet rapidly reversible pheromone arrest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wainwright LJ, Lasorella A, Iavarone A. Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc Natl Acad Sci U S A. 2001;98:9396–9400. doi: 10.1073/pnas.161288698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18:851–855. doi: 10.1101/gad.1205304. [DOI] [PubMed] [Google Scholar]