Abstract
Background
The human protozoan parasites Leishmania are prototrophic for pyrimidines with the ability of both de novo biosynthesis and uptake of pyrimidines.
Methodology/Principal Findings
Five independent L. infantum mutants were selected for resistance to the pyrimidine analogue 5-fluorouracil (5-FU) in the hope to better understand the metabolism of pyrimidine in Leishmania. Analysis of the 5-FU mutants by comparative genomic hybridization and whole genome sequencing revealed in selected mutants the amplification of DHFR-TS and a deletion of part of chromosome 10. Point mutations in uracil phosphorybosyl transferase (UPRT), thymidine kinase (TK) and uridine phosphorylase (UP) were also observed in three individual resistant mutants. Transfection experiments confirmed that these point mutations were responsible for 5-FU resistance. Transport studies revealed that one resistant mutant was defective for uracil and 5-FU import.
Conclusion/Significance
This study provided further insights in pyrimidine metabolism in Leishmania and confirmed that multiple mutations can co-exist and lead to resistance in Leishmania.
Author Summary
The human protozoan parasites Leishmania present the ability of both de novo biosynthesis and uptake of pyrimidines. The pyrimidine pathway is not well understood in these parasites. In the hope to better understand the pyrimidine pathway in Leishmania, five independent L. infantum mutants were selected for resistance to the pyrimidine analogue 5-fluorouracil (5-FU). Analysis of the 5-FU mutants by comparative genomic hybridization and whole genome sequencing revealed the amplification of the main target enzyme DHFR-TS, and point mutations in three important metabolic enzymes. Transfection experiments confirmed that these point mutations were responsible for 5-FU resistance. Transport studies also revealed that one resistant mutant was defective for uracil and 5-FU import. Overall, this study provided further insights in pyrimidine metabolism in Leishmania and confirmed that multiple mutations can co-exist and lead to resistance in these protozoa.
Introduction
The protozoan parasites Leishmania are distributed worldwide and cause different symptoms including cutaneous, mucocutaneous or visceral leishmaniasis, the latter potentially fatal if left untreated [1], [2]. Treatments include pentavalent antimonials, amphotericin B, paromomycin or miltefosine [3], [4] but these drugs have severe shortcomings including toxicity, high cost, and resistance development that need to be addressed in the near future for a better control of this parasitic diseases [5]. With 350 million people at risk, the impact of leishmaniasis on global heath is non-negligible and the search for new drugs or new formulations along with the development of effective vaccines is urgent.
Several lines of evidences have suggested that the pyrimidine pathway would represent a viable target for drug intervention in protozoan parasites, at least in the related parasite Trypanosoma brucei brucei [6] but also in Leishmania [7] although recent data would suggest that the pyrimidine pathway may not offer potential for therapeutic intervention in Leishmania [8]. Leishmania spp. are able to synthetize UMP [9], although they seem to prefer to import pyrimidines from their environment [10]. While purine transporters have been well studied in Leishmania and trypanosomes (reviewed in [11]), our knowledge of the pyrimidine import machinery is considerably less detailed in these parasites. High affinity transporters for uracil have been reported in both Leishmania and Trypanosoma b. brucei [6], [7], [12], although the gene responsible for this transport activity has not been identified in Leishmania. Leishmania and Trypanosome parasites synthesize pyrimidine nucleotides via both de novo and salvage pathways so they don't need preformed pyrimidine for their growth [8], [13].
One often used strategy to gain insight in a metabolic pathway in Leishmania is to study resistance mechanisms to an antimetabolite. For example, the folate/pterin metabolism and transport in Leishmania was largely derived from studies of mutants selected for resistance to the antifolate methotrexate (MTX) [14], [15]. Indeed, Leishmania parasites are auxotroph for folates and need to import these essential molecules from their environment to meet their folate requirements [14], [16]. Studies of MTX resistance allowed the characterization of plasma membrane transporters of the folate/biopterin transporter family (FBT), a distant family within the major facilitator superfamily [17], [18], [19], [20]. Similarly, studies of sinefugin resistant mutants have allowed the discovery of the AdoMetT1 transporter, a member of the FBT family transporting S-adenosylmethionine [21]. We thus selected Leishmania cells for resistance to the pyrimidine analogue 5-fluorouracil (5-FU), a antineoplastic compound displaying a strong antileishmanial activity [22]. After its entry in mammalian cells, 5FU is converted into 5-fluorodeoxyuridine 5′-monophosphate (5-FdUMP) and 5-fluorouridine 5′-monophosphate (5-FUMP), both of which can be further phosphorylated and incorporated into DNA and RNA, respectively. Thus, the antiproliferative property of 5FU ultimately results in the inhibition of DNA replication and inhibition of the processing and maturation of rRNA, tRNAm snRNA and mRNA precursors, leading to cell death. The enzyme thymidylate synthase (TS) which catalyzes the methylation of deoxyuridine monophosphate (dUMP) to deoxythymidine monophsophate (dTMP) is thought to be the main target of 5-FdUMP (for a review see [23]) but other 5FU targets are being revealed including spliceosomal snRNAs [24] and the RNA exosome component hRrp6 [25]. In order to increase our understanding of pyrimidine metabolism in Leishmania, a genomic analysis of in vitro generated 5FU resistant mutants was performed.
Materials and Methods
Cell lines and culture
The Leishmania infantum WT (strain MHOM/MA/67/ITMAP-263) and 5-fluorouracil (5FU)-resistant strains (Lin5FU500.1, Lin5FU500.2, Lin5FU500.3, Lin5FU500.4 and Lin5FU500.5) were cultured as promastigotes at 25°C in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum and 5 µg/ml hemin. The five Lin5FU mutants were derived from the sensitive WT strain by passaging them in increasing drug concentrations at steps 50, 100, 200, 400 and 500 µM 5-FU (Sigma-Aldrich, St. Louis, MO, USA). Revertants were obtained by culturing the resistant cell lines in absence of 5-FU for 30 passages. Cell growth was monitored by measuring the absorbance of culture aliquots (200 µl) at 600 nm in a multiwell scanning spectrophotometer (Multiskan, Thermo Scientific, Waltham, MA, USA). EC50 values were determined by measuring the absorbance of culture aliquots (200 µl) grown in the presence of various concentrations of drugs at 600 nm in a multiwell scanning spectrophotometer (Multiskan, Thermo Scientific, Waltham, MA, USA). EC50 represents the concentration of drug that inhibits 50% of the growth.
Materials, chemicals and reagents
All restriction enzymes used in this study were acquired from New England Biolabs. Synthetic oligonucleotides for PCR and cloning experiments were purchased from Integrated DNA Technologies. Cyanine fluorescent labelled nucleotides required for microarray probes preparation were from GE Healthcare. Transport assays were performed with [3H]-labelled isotopes purchased from either PerkinElmer (uracil) or Moravek Biochemicals (5-fluorouracil).
DNA manipulations
For Southern blot and PCR analyses, genomic DNAs from parasite cells were isolated using the DNAzol reagent (Invitrogen, Carlsbad, CA, USA) as recommended by the manufacturer. Southern blots, probe labeling, hybridization, and washing conditions were done following standard protocols [26].
For single nucleotide polymorphisms (SNPs) validation, the complete coding regions of genes LinJ.10.1370, LinJ.10.1430 and LinJ.10.1440 were PCR amplified (see Table S1 in Supplementary Material, primers denoted “PCR amplification”) using genomic DNAs derived from the WT 263 strain and each of the five 5-fluorouracil resistant mutants. Southern probes to assay gene deletion and/or amplification events in 5-FU resistant mutants were obtained by PCR, using genes LinJ.10.1380, LinJ.10.1390 and LinJ.10.1420 as targets on WT genomic DNA with the appropriate set of primers (Table S1, Supplementary Material, primers denoted “Southern”). DHFR-TS (LinJ.06.0890) containing amplicons were detected by Southern blot using a PCR amplified probe derived from gene LinJ.06.0910 (Table S1).
DNA constructs and transfections
The genes LinJ.06.0890 (DHFR-TS), LinJ.10.1380, LinJ.10.1390, LinJ.10.1400, LinJ.10.1410, LinJ.10.1420, LinJ.10.1430, LinJ.10.1090, LinJ.21.1450, LinJ.34.1110 and LinJ.34.3040 were amplified from a WT 263 genomic DNA preparation (see Table S1, Supplementary Material, primers denoted “pSP72 αHYGα cloning”). The PCR fragments were first purified on columns (Qiagen, Valencia, CA, USA) according to the manufacturer's recommendations, digested with both XbaI and HindIII then cloned into the Leishmania expression vector pSP72αHYGα (described in [27]) digested with the same enzymes. To check the integrity of all cloned open reading frames, final expression constructs were sequenced before being used in transfection experiments. Transfection and maintenance (hygromycin selection at 600 µg/ml) of these constructs into Leishmania infantum 5-FU resistant promastigotes was performed as previously described [28]. Each transfectant parasite populations were then plated on agar containing drug (hygromycin) for clone isolation and individual clones were further assayed for drug sensitivity.
Microarrays and CGH experiments
The Leishmania DNA oligonucleotides full genome microarray design was described previously [29] as well as prehybridization, hybridization and washing conditions for CGH assays. Genomic DNAs from L. infantum WT (strain MHOM/MA/67/ITMAP-263) and from the five 5-fluorouracil resistant mutants were used as template for probe labelling essentially as described [29]. Normalization and statistical analysis of microarray data were performed in R using the LIMMA 3.12.0 package [30]. Background correction was done using the Edwards method, within-array normalization used loess and inter-array normalization was performed using A quantiles. The entire data set has been deposited in GEO under the accession number series GSE45866.
Whole genome sequencing and data analysis
Genomic DNAs were prepared from mid-log phase clonal cultures of L. infantum 263 WT and from the five 5-FU resistant mutants. Paired-ends sequencing libraries were prepared with the Nextera DNA sample prep kit (each strain tagged with a different index) and libraries were sequenced on an Illumina HiSeq1000 platform with short 101-nucleotide reads. An average genome coverage of over 50-fold was obtained for the five independent mutants as well as the WT strain. This strategy allowed us to identify point mutations when comparing with the reference genome sequence of L. infantum JPCM5 [31]. Sequence reads from each clone were aligned to the L. infantum JPCM5 reference sequence available at TriTrypDB (version 4.0) [32] using the software bwa (bwa aln, version 0.5.9) with default parameters [33]. The maximum number of mismatches was 4, the seed length was 32 and 2 mismatches were allowed within the seed. The detection of single nucleotide polymorphisms (SNPs) was performed using samtools (version 0.1.18), bcftools (distributed with samtools) and vcfutils.pl (distributed with samtools) [34], with a minimum of three reads to call a potential variation prior to further analysis. The quality assessment software samstat (v1.08) was used to generate quality reports [35]. Several python (version 2.4.3) and bash (version 3.2) scripts were created to further analyze the data and for the detection of copy number variations (CNVs). The sequence data for L. infantum 263 WT and the mutant Lin5FU500.1 up to Lin5FU500.5 are available at the EMBL European Nucleotide Archive (http://www.ebi.ac.uk/ena) (study accession ERP001815 and sample accession ERS179382 corresponding to L. infantum 263 WT; and study accession ERP002415, samples ERS226502, ERS226503, ERS226504, ERS226505 and ERS226506 corresponding to the L. infantum 263 mutants Lin5FU500.1 to Lin5FU500.5, respectively). All the putative point mutations detected by whole genome sequencing were verified by PCR amplification and conventional DNA sequencing using primers detailed in supplementary material (Table S1).
Uracil and 5-FU transport assays
Parasite cultures were harvested during their mid-log phase. 1×108 cells were washed and resuspended in transport assay buffer (33 mM HEPES, 98 mM NaCl, 4.6 mM KCl, 0.55 mM CaCl2, 0.07 mM MgSO4, 5.8 mM NaH2PO4, 0.3 mM MgCl2, 23 mM NaHCO3 and 14 mM glucose, pH 7.3) supplemented with 250 nM of [3H] uracil (40.3 Ci mmol−1) (PerkinElmer, Waltham, MA, USA) or [3H] 5-fluorouracil (16.4 Ci mmol−1) (Moravek Biochemicals, Brea, CA, USA). Radioactivity accumulation was measured as previously described [36]. The uptake was normalized to cell numbers and the background transport level was removed by subtracting the accumulation values obtained on ice from each of the test readings.
Results
Comparative phenotypic and genotypic characterisation of 5-fluorouracil mutants
Of the various pyrimidine analogs commercially available, the antimetabolite 5-fluorouracil (5-FU) has strong antileishmanial effects on L. major promastigotes with EC50 values in the low µM [7]. The activity of this drug was tested here against L. infantum WT promastigotes (strain MHOM/MA/67/ITMAP-263) with an in vitro EC50 of 72±0.8 µM to 5-FU (Table 1), being apparently less sensitive than the L. major promastigote strain. Five independent cultures of L. infantum WT parasites were selected by stepwise selection in liquid medium with increasing concentration of the drug up to 500 µM. Cultures were named Lin5FU500.1 up to Lin5FU500.5. Each populations were shown to readily grow in the presence of over 2000 µM 5-FU (Table 1). To evaluate the stability of the resistance phenotype, resistant populations of parasites were sub-cultured for at least 30 passages in absence of 5-FU. Three out of the 5 resistant cultures conserved their high level of resistance to 5-FU (Lin5FU500.3, Lin5FU500.4 and Lin5FU500.5) but an intermediate level at 927±83 µM was observed in Lin5FU500.1, still being 14-fold more resistant than the WT parental strain (Table 1) whereas resistance reverted to WT levels in Lin5FU500.2 (Table 1).
Table 1. Resistance levels to 5-FU and MTX in L. infantum 263 WT and 5-FU resistant mutants.
| EC50 (µM) | |||
| 5FU | MTX | 5FU Rev* | |
| L. infantum 263 WT | 72±0.8 | 114±9 | - |
| L. infantum 5FU500.1 | >2000 | 207±2** | 927±83 |
| L. infantum 5FU500.2 | >2000 | 345±5** | 78±15 |
| L. infantum 5FU500.3 | >2000 | 5±0.3** | >2000 |
| L. infantum 5FU500.4 | >2000 | 106±4 | >2000 |
| L. infantum 5FU500.5 | >2000 | 96±3 | >2000 |
Rev = revertant strain cultured without drug pressure over 30 passages;
(p<0,005).
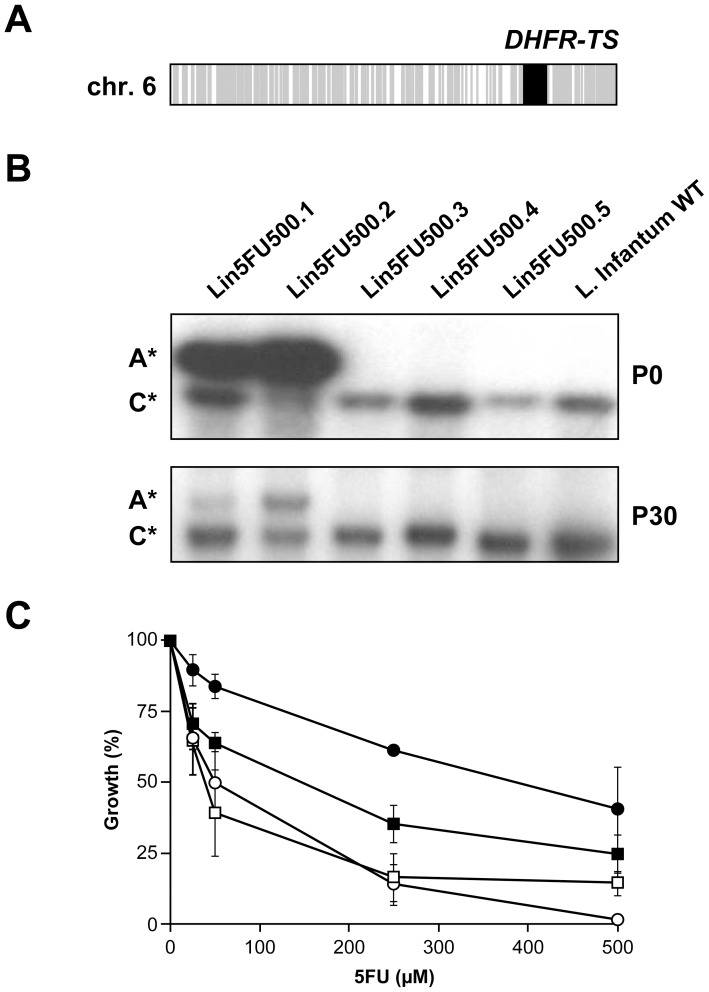
Since unstable resistance in Leishmania is often associated with gene amplification events [37], [38], DNA microarrays covering the whole set of genes encoded by the genome of Leishmania infantum were used to perform comparative genomic hybridization (CGH). The genomic DNA from each 5-FU resistant clone was isolated, labelled with fluorescent dyes and co-hybridized with the WT labelled DNA in order to detect any change in gene copy numbers between the two cell types. The analysis of the CGH results revealed a unique region of about 30 kb on chromosome 6 that was amplified (20-fold compared to the WT level) in Lin5FU500.2 (Fig. 1A, locus indicated in black). This locus was encompassing 6 genes (from LinJ.06.0860 up to LinJ.06.0910) including the gene encoding for the bifunctional enzyme dihydrofolate reductase-thymidylate synthase (DHFR-TS, LinJ.06.0890). This amplicon was found to correspond to an extrachromosomal circle since we could isolate it by standard plasmid preparation (data not shown). Southern blot analysis confirmed that DHFR-TS was amplified in mutant Lin5FU500.2 but also in mutant Lin5FU500.1 (Fig. 1B, panel P0), an amplification surprisingly not detected by CGH. No amplification of the DHFR-TS locus was found in the three other mutants (Fig. 1B, panel P0). A marked decrease in the copy number of the DHFR-TS containing amplicons was observed in both Lin5FU500.1 and Lin5FU500.2 revertant cells grown for 30 passages in absence of 5-FU (Fig. 1B, panel P30). The role of DHFR-TS in 5-FU resistance was tested by transfecting the Leishmania DHFR-TS gene cloned into an expression vector in WT parasites as well as in the revertant strain Lin5FU500.2rev. Transfection of the DHFR-TS construct conferred respectively a 6- and 9- fold increase in EC50 values in L. infantum and in Lin5FU500.2rev when compared to control transfectants (Fig. 1C). Since DHFR-TS gene amplification can also lead to MTX resistance in Leishmania [39], [40], we further tested whether Lin5FU500.1 and Lin5FU500.2 were also cross-resistant to MTX. The Lin5FU500.1 and Lin5FU500.2 resistant parasites were indeed 2- and 3-fold cross-resistant to MTX respectively when compared to the WT strain (Table 1). Mutants Lin5FU500.4 and Lin5FU500.5 were not cross-resistant to MTX but intriguingly, Lin5FU500.3 was 20-fold hypersensitive to MTX (Table 1).
Figure 1. DHFR-TS amplification and resistance to 5-fluorouracil.
(A). Comparative genomic hybridization (CGH) experiments between wild-type and Lin5FU500.2 cells. Grey, equal amount of DNA between the two strains; black, increased copy number of DNA in the mutant Lin5FU500.2. (B). Southern blots of total digested DNAs isolated from WT and each mutant strains at 0 and 30 passages without drug were hybridized to a specific DHFR-TS probe. The DNA was digested to discriminate the chromosomal copy (C*) from the amplified copy (A*). (C). Role of DHFR-TS in 5-FU resistance. Growth curves of wild-type L. infantum parasites transfected with the expression construct pSP72αHYGα- DHFR-TS (black squares) or with the empty vector pSP72αHYGα (white squares) or the Lin5FU500.2 revertant transfected with pSP72αHYGα- DHFR-TS (black circles) or pSP72αHYGα (white circles). Average of at least three independent experiments. Transfection of DHFR-TS led to 5FU resistance that was statistically significant compared to mock transfectants (p<0.05).
Whole-genome sequencing and identification of CNVs and SNPs
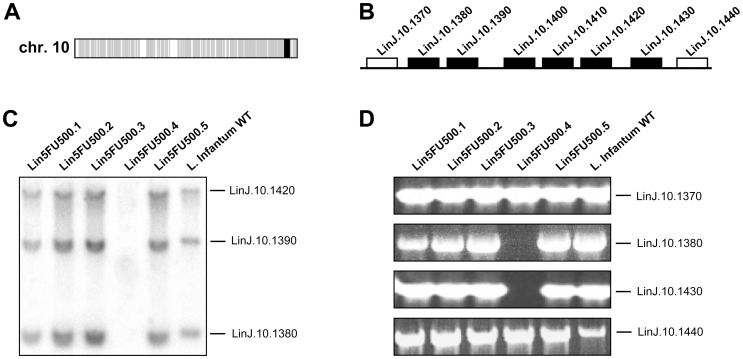
For a more in depth genetic analysis of the 5-FU resistant mutants, we also used a whole genome sequencing (WGS) approach to try to identify and map mutations that could explain the resistance phenotype observed in our panel of 5-FU resistant mutants. This strategy has proven useful to study resistance in Leishmania [41], [42], [43]. Thus, a single clone derived from the WT L. infantum strain and from each of our five resistant populations was sent for sequencing on an Illumina HiSeq1000 system. A total number of 17,437,747 reads was obtained for the WT strain, whereas between 24,501,618 and 39,361,109 reads were obtained for the 5 mutants, leading to an average genome coverage of 50- to 90-fold depending on the strains. Reads depth coverage over the 36 chromosomes of Leishmania was used to predict copy number variations (CNVs), thus revealing either amplifications or deletions at the genome scale. These comparative analyses confirmed the amplification of the DHFR-TS locus on chromosome 6 observed by CGH in the mutant Lin5FU500.2, with an increase in the number of reads of ∼32-fold compared to the WT strain (data not shown). Strangely and similarly to CGH, WGS did not detect any DHFR-TS amplification in the mutant Lin5FU500.1, although southern blot analyses clearly demonstrated the amplification of this locus in this mutant (Fig. 1B). In the mutant Lin5FU500.4, sequence reads analysis revealed a deletion of 6 genes on chromosome 10 with a 64-fold reduction in the number of reads overlapping this locus compared to the WT (data not shown). This sequencing result was in line with CGH analysis (Fig. 2A) where the locus deletion on chromosome 10 had apparently occurred between gene LinJ.10.1370 and gene LinJ.10.1440 in the Lin5FU500.4 mutant (Fig. 2B). Southern blot analyses with specific probes derived from genes localized within this putatively deleted region (LinJ.10.1420, LinJ.10.1390 and LinJ.10.1380) confirmed indeed that this region was deleted in Lin5FU500.4 (Fig. 2C), a result also supported by PCR experiments targeting genes within or outside this locus (Fig. 2D). The WT versions of the six genes part of the deleted locus (LinJ.10.1380 up to LinJ.10.1430, see Fig. 2B) were individually cloned in the expression vector pSP72αHYGα and transfected back into the mutant Lin5FU500.4 to assess their role in 5-FU resistance. None of the transfectants however regained sensitivity to 5-FU (data not shown). Since the deletion on chromosome 10 was close to one of the genes encoding an FBT member (LinJ.10.1450), we decided to test whether this FBT gene would have been responsible for the resistance phenotype observed in the Lin5FU500.4 mutant. The expression of the FBT gene was unchanged in the mutant (results not shown) and transfection of the FBT gene did not change the 5-FU susceptibility in this mutant, however (data not shown).
Figure 2. Locus deletion on chromosome 10 in the Lin5FU500.4 mutant.
(A). Comparative genomic hybridization (CGH) experiments between Lin5FU500.4 and the WT strain. Grey, equal amount of DNA between the strains; black, decrease in the copy number of 5 genes clustered on chromosome 10 was observed in Lin5FU500.4. (B). Schematic representation of the 5 genes part of the locus deleted. The white boxes represent the two genes (LinJ.10.1370 and LinJ.10.1440) flanking the deleted locus on chromosome 10. Black boxes represent the six genes deleted. (C). Southern blot co-hybridized with three different probes (indicated on the right) specific for genes deleted in the Lin5FU500.4 mutant as determined by CGH. (D). PCR amplification of genes outside (LinJ.10.1370 and LinJ.10.1440) or within (LinJ.10.1380 and LinJ.10.1430) the region deleted in Lin5FU500.4 in the various 5FU mutants.
Since the CNVs analyses (and CGH) did not revealed any other amplification or deletion events except the ones observed on chromosomes 6 and 10 in our 5-FU resistant mutants, we then analyzed single homozygous nucleotide polymorphisms (SNPs) that were detected by WGS in coding regions (Table 2, n = 29). Seven of the SNPs corresponded to silent mutations and none of the 29 homozygous SNPs were shared between mutants. A selection of seven SNPs within genes encoding the most interesting candidate enzymes possibly involved in 5-FU resistance were PCR amplified from genomic DNAs derived from mutants and PCR products were subjected to direct resequencing for SNP validation. Three SNPs were found to be sequencing errors (indicated as “E” for sequencing error in Table 2) but four were called as real mutations (indicated as “M” for mutation in Table 2). The four validated SNPs were detected in genes encoding respectively for the enzyme thymidine kinase (TK, LinJ.21.1450) in mutant Lin5FU500.3, the uracil phosphoribosyl transferase (UPRT, LinJ.34.1110) as well as a hypothetical protein (LinJ.34.3040) in mutant Lin5FU500.4, and the uridine phosphorylase (UP, LinJ.10.1090) in mutant Lin5FU500.5 (Table 2). With the exception of the hypothetical protein LinJ.34.3040, the other genes have been linked previously to pyrimidine metabolism in kinetoplastids [6], [44] and were thus further investigated along with LinJ.34.3040.
Table 2. Whole genome SNP discovery in 5-FU resistant mutants.
| Strain | Chr | Base in GeneDB | Base in mutant | Gene ID | Gene products | Position in the gene | a.a. change | Individual Sequencing* |
| Lin5FU500.1 | 14 | C | G | LinJ.14.1190 | kinesin K39 | 4557 | H1519Q | ND |
| 15 | T | C | LinJ.15.0490 | hypothetical protein | 8693 | L2898S | ND | |
| 33 | G | T | LinJ.33.2730 | hypothetical protein | 205 | G69C | ND | |
| 35 | A | C | LinJ.35.0500 | proteophosphoglycan ppg3 | 12698 | E4233A | ND | |
| 36 | G | T | LinJ.36.1020 | hypothetical protein | 261 | S87S | ND | |
| Lin5FU500.2 | 3 | C | T | LinJ.03.0260 | hypothetical protein | 3902 | A1301V | ND |
| 3 | C | T | LinJ.03.0260 | hypothetical protein | 4136 | A1379V | ND | |
| 10 | A | C | LinJ.10.1030 | eIF-2B GDP-GTP exchange factor | 1298 | V433G | ND | |
| 15 | A | G | LinJ.15.0490 | hypothetical protein | 2811 | L937L | ND | |
| 35 | T | G | LinJ.35.0490 | proteophosphoglycan ppg4 | 7888 | S2630A | ND | |
| 35 | A | G | LinJ.35.0490 | proteophosphoglycan ppg4 | 8751 | S2917S | ND | |
| 35 | T | G | LinJ.35.4860 | AMP deaminase | 2730 | A910A | ND | |
| Lin5FU500.3 | 6 | C | G | LinJ.06.1360 | hypothetical protein | 1744 | P582A | E |
| 14 | C | G | LinJ.14.0790 | fatty acid elongase | 471 | M157I | ND | |
| 16 | A | G | LinJ.16.1030 | hypothetical protein | 2704 | R902G | ND | |
| 21 | T | G | LinJ.21.1450 | thymidine kinase | 260 | Q87P | M | |
| 35 | T | G | LinJ.35.0520 | proteophosphoglycan ppg4 | 7366 | F2456V | ND | |
| 35 | C | A | LinJ.35.4450 | hypothetical protein | 994 | Q332K | ND | |
| Lin5FU500.4 | 28 | T | C | LinJ.28.2390 | cyclin dependent kinase-binding protein | 1301 | L434P | ND |
| 29 | A | C | LinJ.29.2100 | hypothetical protein | 2673 | T891T | ND | |
| 34 | A | C | LinJ.34.0820 | serine/threonine phosphatase PP1 | 773 | E258A | E | |
| 34 | A | C | LinJ.34.0830 | serine/threonine phosphatase PP1 | 836 | E279A | E | |
| 34 | A | C | LinJ.34.1110 | uracil phosphoribosyl transferase | 434 | K145T | M | |
| 34 | A | C | LinJ.34.2220 | hypothetical protein | 5335 | S1779A | ND | |
| 34 | C | G | LinJ.34.3040 | hypothetical protein | 1907 | S636W | M | |
| Lin5FU500.5 | 10 | A | C | LinJ.10.1090 | uridine phosphorylase | 794 | L265R | M |
| 20 | G | C | LinJ.20.0750 | hypothetical protein | 3132 | Q1044H | ND | |
| 29 | A | G | LinJ.29.1890 | paraflagellar rod protein 1D | 801 | D267D | ND | |
| 35 | A | G | LinJ.35.0490 | proteophosphoglycan ppg4 | 6723 | A2241A | ND |
Experimental validation of SNP variants was performed using PCR-directed sequencing using appropriate pairs of primers (see Table S1, Supplementary Material). Following SNP validation, the WT version of each mutated gene indicated in bold were transfected in the L. infantum 263 WT, Lin5FU500.3, Lin5FU500.4 and Lin5FU500.5 strains. SNPs in italic indicate silent mutations. M, mutation; E, sequencing error; ND, not determined.
To prove the implication of these SNPs in resistance to 5-FU, the WT version of each mutated gene was transfected in the L. infantum WT strain as well as in the three mutants in which they were detected and the EC50 of each transfectant was determined in the presence of 5-FU (Table 3). Transfection of the TK (LinJ.21.1450), the UPRT (LinJ.34.1110), and the UP (LinJ.10.1090) genes reverted resistance in Lin5FU500.3, Lin5FU500.4 and Lin5FU500.5, respectively (Table 3). The phenotype was less strong with LinJ.34.1110 in Lin5FU500.4 but in general each mutation was specific to one mutant. Surprisingly, however, transfection of the UP LinJ.10.1090 gene resensitized both the mutants Lin5FU500.4 and Lin5FU500.5, even though the LinJ.10.1090 gene was only mutated in Lin5FU500.5 (Table 3). The expression of the WT version of gene LinJ.34.3040 coding for a hypothetical protein did not affect the resistance profile to 5-FU in any of the three transfected mutants (data not shown).
Table 3. Resistance to 5-fluorouracil in 5-FU resistant mutants genetically complemented with WT alleles.
| 5FU EC50 (µM) | |
| L. infantum 263 WT+pSP72αHYGα | 29±7 |
| +pSP72αHYGα/LinJ.10.1090 (UP) | 14±3 |
| +pSP72αHYGα/LinJ.21.1450 (TK) | 67±23 |
| +pSP72αHYGα/LinJ.34.1110 (UPRT) | 17±2 |
| L. infantum 5FU500.3+pSP72αHYGα | >2000 |
| +pSP72αHYGα/LinJ.10.1090 | >2000 |
| +pSP72αHYGα/LinJ.21.1450 | 136±13 |
| +pSP72αHYGα/LinJ.34.1110 | >2000 |
| L. infantum 5FU500.4+pSP72αHYGα | >2000 |
| +pSP72αHYGα/LinJ.10.1090 | 31±8 |
| +pSP72αHYGα/LinJ.21.1450 | >2000 |
| +pSP72αHYGα/LinJ.34.1110 | 1140±285 |
| L. infantum 5FU500.5+pSP72αHYGα | >2000 |
| +pSP72αHYGα/LinJ.10.1090 | 22±3 |
| +pSP72αHYGα/LinJ.21.1450 | >2000 |
| +pSP72αHYGα/LinJ.34.1110 | >2000 |
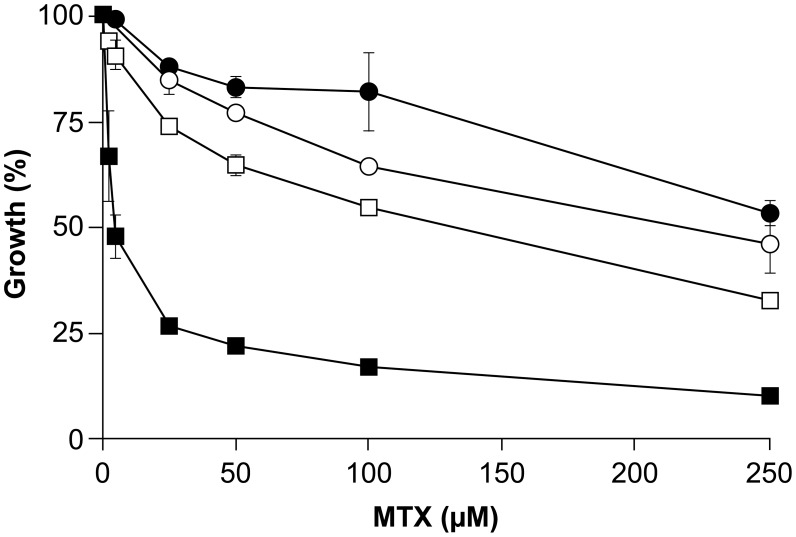
The glutamine (Q) to proline (P) substitution in the TK version found in Lin5FU500.3 is within the active site of the protein (Fig. S1). The Lin5FU500.3 mutant is highly resistant to 5-FU but hypersensitive to MTX (Table 1). Transfection of a WT TK version in the Lin5FU500.3 mutant sensitized parasites to 5-FU (Table 3). We also investigated the MTX sensitivity profile in the Lin5FU500.3 overexpressing TK strain. Interestingly, the overexpression of the TK enzyme in the Lin5FU500.3 mutant abolished the hypersensitivity to MTX in this mutant, thus restoring MTX susceptibility to a level close to that of wild-type cells (Fig. 3).
Figure 3. Role of thymidine kinase (LinJ21.1450) in methotrexate susceptibility.
Growth curves in the presence of methotrexate were determined for L. infantum wild-type cells (lines with circles) and the Lin5FU500.3 mutant (lines with squares), each transfected either with an empty vector (pSP72αHYGα) (black circles and black squares respectively) or with a thymidine kinase expression construct (pSP72αHYGα-LinJ.21.1450) (white circles and white squares respectively). Average of three independent biological replicates. Transfection of TK in the mutant led to MTX susceptibility that was statistically different than the mock control (p<0.05).
Uracil transport activity correlating to drug resistance
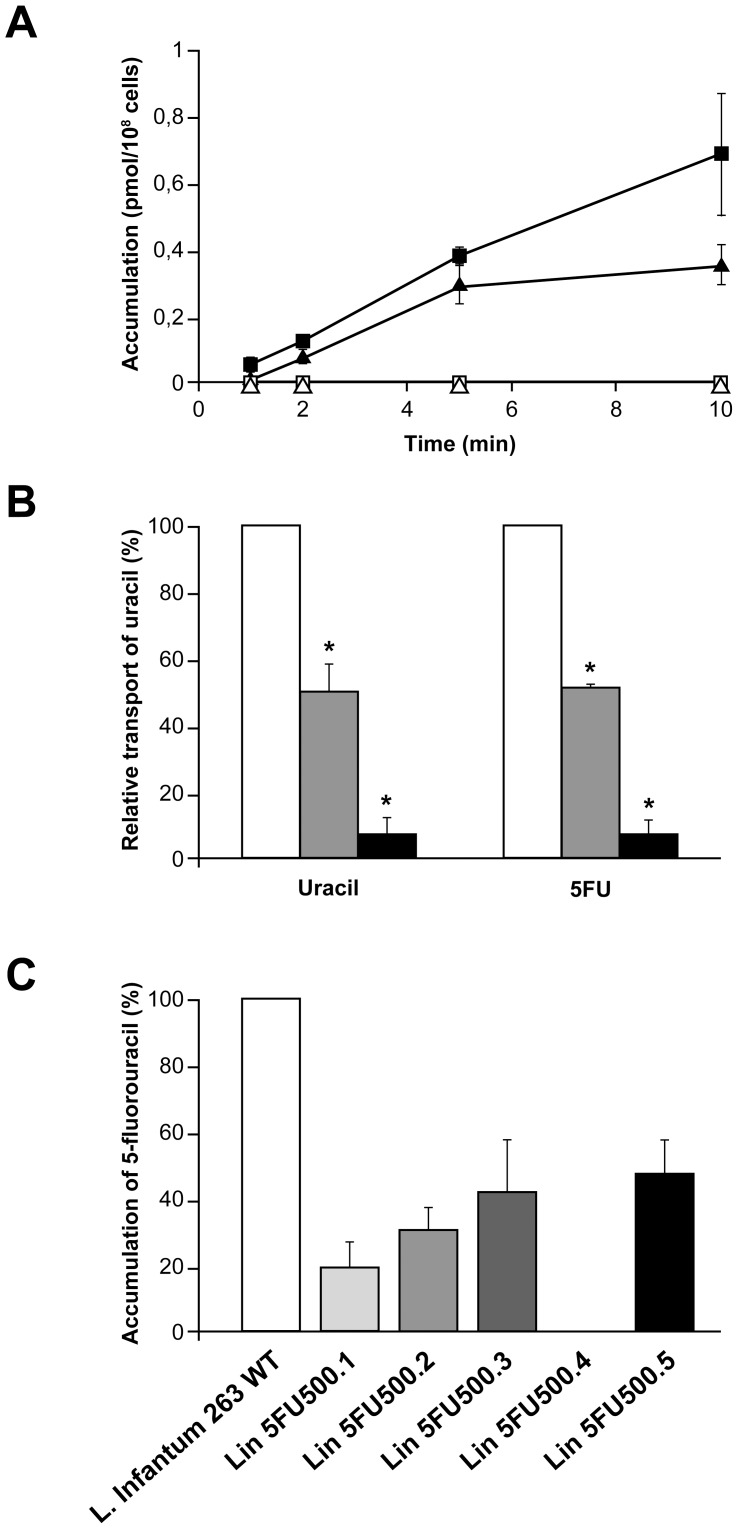
Finally we investigated the ability of L. infantum WT parasites and mutant cells to import uracil and its analogue 5-FU. Transport assays using [3H]-uracil or [3H]-5-FU in the WT strain clearly established that both substrates were transported by this species (Fig. 4A) and most likely by the same transporter. Indeed, the uptake of [3H]-uracil was equally competed with either cold uracil or cold 5-FU (Fig. 4B). In the five mutants tested, we observed a 50–80% decrease in accumulation with the exception of mutant Lin5FU500.4 where no accumulation was observed (Fig. 4A and 4C). This lack of accumulation in Lin5FU500.4 was stable since we could not observe accumulation in revertant parasites grown for 30 passages without drugs (data not shown). Sequence analysis of the mutant did not reveal a candidate mutation that could have identified the uracil transporter. As an alternative for isolating the transporter, we carried out functional cloning where a cosmid bank derived from wild-type L. infantum was transfected in Lin5FU500.4 and spread on hygromycin (the cosmid marker) plates. Individual colonies (n = 4000) were incubated in 96 well plates and screened for 5-FU sensitivity. This approach has been useful to isolate an aquaglyceroporin involved in antimonial transport [45] and for purine transporters in Leishmania [46], [47]. We carried these experiments twice and both times we succeeded in isolating a cosmid rendering Lin5FU500.4 sensitive to 5-FU. Analysis of these transfectants however indicated no transport of uracil but rather each cosmid encoded the uridine phosphorylase LinJ.10.1090 UP gene.
Figure 4. Transport activities of uracil and 5-fluorouracil in L. infantum wild-type cells and 5-FU resistant mutants.
(A). Transport of 5-fluorouracil (black squares) and uracil (black triangles) in L. infantum WT cells and in Lin5FU500.4 mutant cells (5-fluorouracil (white squares) and uracil (white triangles)) after 1, 2, 5 and 10 minutes (B). Transport of [3H]-uracil (white bars) competed with 200× (grey bars) or 2000× ratio (black bars) of cold uracil (left panel) or 5-FU (right panel) after a 10 minutes incubation period. (C). Accumulation of [3H]-5-fluorouracil in L. infantum WT 263 strain and in the 5-FU mutants after 10 minutes. Average of three independent biological replicates. * (p<0.05).
Discussion
Studies of resistance mechanisms to the model drug methotrexate have contributed importantly to our understanding of folate and pterin metabolism and transport in Leishmania (reviewed in [14], [16]). Similarly, studies of resistance mechanisms to the AdoMet analogue sinefungin has led to the isolation of an AdoMet transporter and increased our understanding of one carbon metabolism in Leishmania [21]. One of the main metabolic roles of reduced folates is in the generation of dTMP through the activity of the bifunctional enzyme DHFR-TS. Indeed Leishmania DHFR-TS null mutants are thymidine auxotrophs [48]. In order to further gain insight and link folate and pyrimidine metabolisms, we selected Leishmania cells for resistance to a pyrimidine analogue, 5- fluorouracil (5-FU), as this drug was shown previously to have considerable activities against Leishmania [7]. We selected 5 independent L. infantum mutants highly resistant to 5-FU and analyzed these drug resistant mutants by a combination of comparative genomic hybridization and whole genome sequencing, two approaches that have been useful in studying drug resistance mechanisms in Leishmania [29], [42], [43], [49]. Our analysis has pinpointed several mechanisms of resistance including gene amplification, point mutations in key nucleic acid metabolism enzymes as well as transport defects and is consistent with observations made in 5-FU resistant cancer cells [50], [51], [52].
Thymidylate synthase is the main target of 5-FU in all eukaryotic cells studied [53] including kinetoplastid parasites [6]. It was thus not surprising to observe an extrachromosomal circular amplification of the bifunctional gene DHFR-TS in the Lin5FU500.2 mutant both by CGH (Fig. 1A) and by analyzing sequence reads (data not shown). Growing this mutant in absence of drug led to a marked decrease of the circular amplicon and reversion of the resistance phenotype (Fig. 1B and 1C). Amplification of the DHFR-TS gene also explained the observed MTX cross-resistance in this mutant (Table 1) as MTX targets the Leishmania DHFR enzyme [39]. Growth curves were carried out in the folate rich medium SDM-79. Folate concentration modulates MTX cross-resistance and this may explain the low level of MTX cross-resistance despite a 20-fold amplification of DHFR-TS. Transfection of the DHFR-TS gene confirmed its role in 5-FU resistance (Fig. 1C). Resistance levels reached are lower than the resistant mutants and this may be due to the level of expression of DHFR-TS in transfectants. Southern blot analysis indicated that DHFR-TS was not only amplified in Lin5FU500.2 but also in Lin5FU500.1 (Fig. 1B). Surprisingly, this amplification in Lin5FU500.1 was missed by both CGH and sequencing. This is difficult to explain because we have shown both CGH and sequencing reads depth to be quantitative [43], which is further confirmed by ongoing work with several unrelated resistant strains. The DNA used for Southern blots and sequencing were prepared at different times but usually amplicons are stable in the presence of drugs. Southern blots at varying passages confirmed the stable amplification of DHFR-TS (data not shown). While DHFR-TS amplification seems the only resistance mechanism in Lin5FU500.2, this does not seem to be the case in Lin5FU500.1 since growth in absence of drug led to a decrease in the copy number of the amplicon but only a partial reversion (Table 1, Fig. 1B). Five point mutations (including one silent mutation) are possible candidates for resistance (Table 2) (e.g. the kinesin K39, LinJ.14.1190; the proteophosphoglycan pgp3, LinJ.35.0500; and 3 hypothetical proteins, LinJ.15.0490, LinJ.33.2730 and LinJ.36.1020) and await further additional functional studies.
Sequencing of the genome of the five resistant mutants has also led to the identification of several point mutations in 5-FU resistant parasites, three of which were shown to be involved in 5-FU resistance in three independent mutants. In Lin5FU500.3, we observed a point mutation in the active site of a thymidine kinase (TK, LinJ.21.1450) (Fig. S1). Transfection of the WT copy of the gene in Lin5FU500.3 showed that this is a key mutation involved in 5-FU resistance (Table 3). A mutation in TK would reduce the formation of 5-FdUMP (Fig. 5). However a mutation in TK would reduce the conversion of thymidine into dTMP, hence rending the cell more dependent on the DHFR pathway (Fig. 5), thus making the cell more susceptible to the DHFR inhibitor MTX (Table 1, Fig. 3).
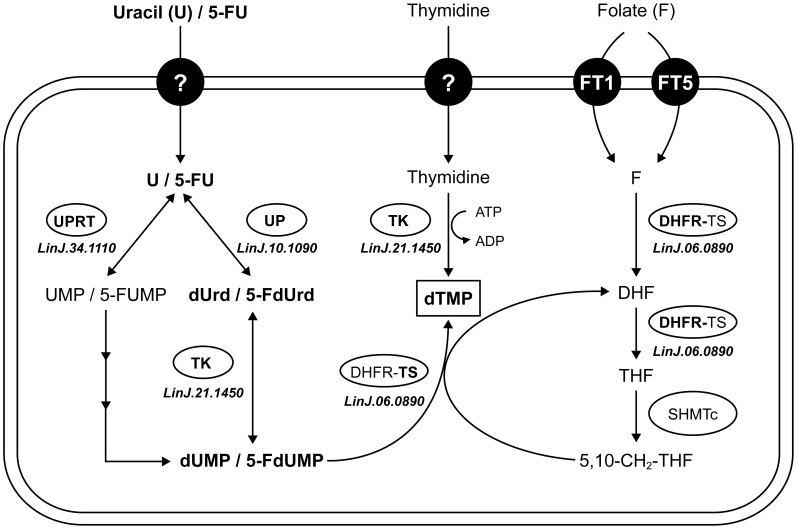
Figure 5. Pyrimidine metabolism and linkages with folate metabolism in Leishmania.
Uracil, after its importation by a non-identified transporter, is metabolized into uridine monophosphate (UMP) by the action of the uracil phosphoribosyl transferase (UPRT, LinJ.34.1110) or into deoxy-uridine (dUrd) by the uridine phosphorylase (UP, LinJ.10.1090). The thymidine kinase (TK, LinJ.21.1450) is involved in synthesis of deoxy-uridine monophosphate (dUMP) but also deoxy-thymidine monophosphate (dTMP). The dUMP produced will lead to dTMP through the action of the bifunctional enzyme dihydrofolate reductase-thymidylate synthase (DHFR-TS). The drug 5-FU is also transported by the uracil transporter and can be metabolized by UPRT, UP and TK. Abbreviations: F, folate; DHF, dihydrofolate; THF, tetrahydrofolate; SHMTc, cytosolic serine hydroxymethyltransferase; U, uracil; 5FU, 5-fluorouracil; 5-FdUrd, 5-fluoro-deoxy-uridine; 5-FUMP, 5-fluoro-uridine monophosphate; 5-FdUMP, 5-fluoro-deoxy-uridine monophosphate.
In mutant Lin5FU500.4 we observed a mutation in the uracil phosphoribosyl transferase (UPRT, LinJ.34.1110). The mutation was located between the flexible loop and the phosphoribosyl-pyrophosphate (PRPP) binding domain in UPRT (Fig. S1). The mutation in UPRT contributes only slightly to 5-FU resistance as suggested by transfection of the wild-type gene in the mutant (Table 3). The main route in Leishmania for 5-FU to become 5-FUMP and being incorporated into RNA is through the UPRT pathway (Fig. 5) and this reduced ability in the mutant may lead to some levels of resistance to 5-FU. However, mutant Lin5FU500.4 has also no measurable accumulation of 5-FU (Fig. 4) and this defect must contribute to resistance. In mutant Lin5FU500.5, the mutation in the uridine phosphorylase (UP, LinJ.10.1090) is key in conferring resistance since transfecting back a wild-type allele completely reverted resistance in Lin5FU500.5. The UP is the main enzyme for activating 5-FU for eventual incorporation into DNA (Fig. 5) and this can explain resistance. The lack of UP would however require an alternate pathway for the generation of dUMP (Fig. 5) and several non-UP pathways have been described in the related parasite T. brucei to lead to the synthesis of dUMP [6]. Transfection of UP (LinJ.10.1090) also rendered Lin5FU500.4 cells more sensitive to 5-FU (Table 3) despite that LinJ.10.1090 is not mutated in Lin5FU500.4. Since Lin5FU500.4 does not transport 5-FU or uracil (Fig. 4) and two independent functional cloning experiments screening for regained sensitivity to 5-FU led to the isolation of cosmids encoding UP (LinJ.10.1090) rather than the uracil transporter, it would suggest that UP is rate limiting and that even in absence of measurable uptake over 10 minutes, slow diffusion of 5FU may be sufficient for UP-mediated increased toxicity.
The loss of measurable accumulation of 5-FU in Lin5FU500.4 was not reverted when growing the mutant in absence of drugs (data not shown). A defect in accumulation can be due to either a decreased uptake or increased efflux. The ABC transporter MDR2 (ABCB2) was shown to be involved in 5-FU resistance in L. amazonensis, most likely by an active extrusion of 5-FU from the parasite cell [22] but in our five resistant mutants, we did not observe any mutation in the MDR2 gene (LinJ.26.2700), nor difference in mRNA levels tested by real time qRT-PCR (data not shown). We have carefully scrutinized the sequencing data of Lin5FU500.4 for either gene deletion or point mutations in proteins with putative transmembrane domains. CGH and WGS analyses detected an 18 kb chromosomal deletion in mutant Lin5FU500.4 on chromosome 10 (Fig. 2). The deleted locus included six genes, from LinJ.10.1380 to LinJ.10.1430. None of the gene products were predicted to have transmembrane domains but all six hypothetical proteins contained a domain of unknown function (DUF) 1861 (GeneDB). DUF1861 containing members are present in Achaea, bacteria and Eukaryota and are the most divergent family of the furanosidase superfamily [54]. Even if their role in 5-FU resistance was not obvious, transfection of the individual WT versions of these genes was nonetheless performed in the mutant Lin5FU500.4 but none did restore sensitivity to 5-FU (data not shown). The mutant Lin5FU500.4 had also a PCR-validated mutated hypothetical gene (LinJ.34.3040, Table 2). This protein has no predicted TM domains but transfection of the WT version of LinJ.34.3040 did not change the resistance profile to 5-FU (data not shown). Members of the equilibrative nucleoside transporter (ENT) were shown to transport purine and pyrimidine [55], [56], [57]. Four members of the ENT family are annotated in the parasite genome (NT1 in 4 copies, NT2, NT3 and NT4) but sequencing and Southern blot analysis have revealed that these genes are neither mutated nor deleted in the mutants (data not shown), supporting a conclusion that the leishmanial uracil transporter is not part of the ENT family [58]. The defect in transport in Lin5FU500.4 may be due to a mutation that we have missed during the analysis of the sequence reads. Additional sequencing and analysis may reveal the identity of this mutation. Alternative in the transport of uracil may depend on more than one gene product and would require either the co-transfection of several genes mutated in the mutant and similarly would complicate its isolation by functional cloning.
In summary, multiple factors contribute to 5-FU resistance in Leishmania. Resistance to 5FU affects mainly the salvage pathway of the parasite, which is the main way to provide pyrimidines for kinetoplastids [59], but gene amplification and transport defect were also associated with resistance. These studies have confirmed the value of studying drug resistance to increase our understanding of pyrimidine metabolism and its interesting connection with folate/antifolate metabolism (DHFR-TS, TK) should be helpful in eventually developing specific inhibitors against Leishmania.
Supporting Information
Sequence alignments of the active sites of thymidine kinase, uridine phosphorylase and uracil phosphoribosyl transferase. The regions of the active sites of the Leishmania TK, UP and UPRT were aligned with the Trypanosoma, mice and human homologues. The highlighted amino acid residues are those in which mutations were found. The highly conserved Q87 in the TK active site is mutated to a P in Lin5FU500.3. Position of the L265R mutation close to the specificity region in the UP enzyme in Lin5FU500.5 mutant. Position of the K145T mutation close to the phosphoribosyl pyrophosphate (PRPP) region in UPRT in Lin5FU500.4.
(TIF)
Primers used in this study. Primer names were given according to the nomenclature of TritrypDB database. Primers were used in PCR reactions either to confirm locus rearrangements or presence or absence of particular genes (PCR amplification); to amplify genes that will be further cloned in plasmids for transfection assays (Cloning); to generate probes for Southern blotting (Southern); or to confirm mutations (SNP validation). Restriction sites are underlined. XbaI, TCTAGA; HindIII, AAGCTT; F, forward primer; R, reverse primer. The size of amplicons is indicated in base pairs (bp).
(RTF)
Funding Statement
JFR was a Training Fellow of the Strategic Training Program in Microbial Resistance, a partnership of the CIHR Institute of Infection and Immunity and the Fonds de Recherche en Santé du Québec. FR was the recipient of a CIHR studentship. JC holds the Canada Research Chair in Medical Genomics. MO holds the Canada Research Chair in Antimicrobial Resistance.This work was funded in part by CIHR operating grant 15501 to MO. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Herwaldt BL (1999) Leishmaniasis. The Lancet 354: 1191–1199. [DOI] [PubMed] [Google Scholar]
- 2. Murray HW, Berman JD, Davies CR, Saravia NG (2005) Advances in leishmaniasis. Lancet 366: 1561–1577. [DOI] [PubMed] [Google Scholar]
- 3. Singh N, Kumar M, Singh RK (2012) Leishmaniasis: current status of available drugs and new potential drug targets. Asian Pac J Trop Med 5: 485–497. [DOI] [PubMed] [Google Scholar]
- 4. Sundar S, Chakravarty J (2013) Leishmaniasis: an update of current pharmacotherapy. Expert Opin Pharmacother 14: 53–63. [DOI] [PubMed] [Google Scholar]
- 5. Croft SL, Sundar S, Fairlamb AH (2006) Drug Resistance in Leishmaniasis. Clinical Microbiology Reviews 19: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ali JAM, Creek DJ, Burgess K, Allison HC, Field MC, et al. (2013) Pyrimidine Salvage in Trypanosoma brucei Bloodstream Forms and the Trypanocidal Action of Halogenated Pyrimidines. Molecular Pharmacology 83: 439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papageorgiou IG, Yakob L, Al Salabi MI, Diallinas G, Soteriadou KP, et al. (2005) Identification of the first pyrimidine nucleobase transporter in Leishmania: similarities with the Trypanosoma brucei U1 transporter and antileishmanial activity of uracil analogues. Parasitology 130: 275–283. [DOI] [PubMed] [Google Scholar]
- 8. Wilson ZN, Gilroy CA, Boitz JM, Ullman B, Yates PA (2012) Genetic dissection of pyrimidine biosynthesis and salvage in Leishmania donovani. J Biol Chem 287: 12759–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutteridge WE, Coombs GH (1977) Biochemistry of parasitic protozoa.: MacMillan Press, London. 172 p. [Google Scholar]
- 10. Hassan HF, Coombs GH (1988) Purine and pyrimidine metabolism in parasitic protozoa. FEMS Microbiol Rev 4: 47–83. [DOI] [PubMed] [Google Scholar]
- 11. de Koning HP, Bridges DJ, Burchmore RJ (2005) Purine and pyrimidine transport in pathogenic protozoa: from biology to therapy. FEMS Microbiol Rev 29: 987–1020. [DOI] [PubMed] [Google Scholar]
- 12. de Koning HP, Jarvis SM (1998) A highly selective, high-affinity transporter for uracil in Trypanosoma brucei brucei: evidence for proton-dependent transport. Biochemistry and Cell Biology 76: 853–858. [DOI] [PubMed] [Google Scholar]
- 13. Ali JA, Tagoe DN, Munday JC, Donachie A, Morrison LJ, et al. (2013) Pyrimidine biosynthesis is not an essential function for Trypanosoma brucei bloodstream forms. PLoS One 8: e58034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nare B, Luba J, Hardy LW, Beverley S (1997) New approaches to Leishmania chemotherapy: pteridine reductase 1 (PTR1) as a target and modulator of antifolate sensitivity. Parasitology 114 Suppl: S101–110. [PubMed] [Google Scholar]
- 15. Ouellette M, Fase-Fowler F, Borst P (1990) The amplified H circle of methotrexate-resistant Leishmania tarentolae contains a novel P-glycoprotein gene. EMBO J 9: 1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouellette M, Drummelsmith J, El-Fadili A, Kundig C, Richard D, et al. (2002) Pterin transport and metabolism in Leishmania and related trypanosomatid parasites. Int J Parasitol 32: 385–398. [DOI] [PubMed] [Google Scholar]
- 17. Cunningham ML, Beverley SM (2001) Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol Biochem Parasitol 113: 199–213. [DOI] [PubMed] [Google Scholar]
- 18. Richard D, Kundig C, Ouellette M (2002) A new type of high affinity folic acid transporter in the protozoan parasite Leishmania and deletion of its gene in methotrexate-resistant cells. J Biol Chem 277: 29460–29467. [DOI] [PubMed] [Google Scholar]
- 19. Richard D, Leprohon P, Drummelsmith J, Ouellette M (2004) Growth phase regulation of the main folate transporter of Leishmania infantum and its role in methotrexate resistance. J Biol Chem 279: 54494–54501. [DOI] [PubMed] [Google Scholar]
- 20. Vickers TJ, Beverley SM (2011) Folate metabolic pathways in Leishmania . Essays Biochem 51: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dridi L, Ahmed Ouameur A, Ouellette M (2010) High Affinity S-Adenosylmethionine Plasma Membrane Transporter of Leishmania Is a Member of the Folate Biopterin Transporter (FBT) Family. Journal of Biological Chemistry 285: 19767–19775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katakura K, Fujise H, Takeda K, Kaneko O, Torii M, et al. (2004) Overexpression of LaMDR2, a novel multidrug resistance ATP-binding cassette transporter, causes 5-fluorouracil resistance in Leishmania amazonensis . FEBS Letters 561: 207–212. [DOI] [PubMed] [Google Scholar]
- 23. Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, et al. (2002) Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta 1587: 194–205. [DOI] [PubMed] [Google Scholar]
- 24. Zhao X, Yu YT (2007) Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res 35: 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kammler S, Lykke-Andersen S, Jensen TH (2008) The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol Cancer Res 6: 990–995. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 27. El Fadili A, Kündig C, Ouellette M (2002) Characterization of the folylpolyglutamate synthetase gene and polyglutamylation of folates in the protozoan parasite Leishmania . Molecular and Biochemical Parasitology 124: 63–71. [DOI] [PubMed] [Google Scholar]
- 28. Papadopoulou B, Roy G, Ouellette M (1992) A novel antifolate resistance gene on the amplified H circle of Leishmania . EMBO J 11: 3601–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ubeda JM, Legare D, Raymond F, Ouameur AA, Boisvert S, et al. (2008) Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol 9: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 31. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, et al. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Research 38: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lassmann T, Hayashizaki Y, Daub CO (2011) SAMStat: monitoring biases in next generation sequencing data. Bioinformatics 27: 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papadopoulou B, Roy G, Ouellette M (1993) Frequent amplification of a short chain dehydrogenase gene as part of circular and linear amplicons in methotrexate resistant Leishmania . Nucleic Acids Res 21: 4305–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beverley SM (1991) Gene Amplification in Leishmania . Annual Review of Microbiology 45: 417–444. [DOI] [PubMed] [Google Scholar]
- 38. Borst P, Ouellette M (1995) New Mechanisms of Drug Resistance in Parasitic Protozoa. Annual Review of Microbiology 49: 427–460. [DOI] [PubMed] [Google Scholar]
- 39. Coderre JA, Beverley SM, Schimke RT, Santi DV (1983) Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica . Proceedings of the National Academy of Sciences 80: 2132–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ellenberger TE, Beverley SM (1987) Reductions in methotrexate and folate influx in methotrexate-resistant lines of Leishmania major are independent of R or H region amplification. Journal of Biological Chemistry 262: 13501–13506. [PubMed] [Google Scholar]
- 41. Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, et al. (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21: 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coelho AC, Boisvert S, Mukherjee A, Leprohon P, Corbeil J, et al. (2012) Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl Trop Dis 6: e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mukherjee A, Boisvert S, do Monte-Neto RL, Coelho AC, Raymond F, et al. (2013) Telomeric gene deletion and intrachromosomal amplification in antimony resistant Leishmania . Mol Microbiol [DOI] [PubMed] [Google Scholar]
- 44. Hammond DJ, Gutteridge WE (1982) UMP synthesis in the kinetoplastida. Biochimica et Biophysica Acta (BBA) - General Subjects 718: 1–10. [DOI] [PubMed] [Google Scholar]
- 45. Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M (2005) Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania . Mol Microbiol 57: 1690–1699. [DOI] [PubMed] [Google Scholar]
- 46. Vasudevan G, Carter NS, Drew ME, Beverley SM, Sanchez MA, et al. (1998) Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc Natl Acad Sci U S A 95: 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carter NS, Drew ME, Sanchez M, Vasudevan G, Landfear SM, et al. (2000) Cloning of a novel inosine-guanosine transporter gene from Leishmania donovani by functional rescue of a transport-deficient mutant. J Biol Chem 275: 20935–20941. [DOI] [PubMed] [Google Scholar]
- 48. Cruz A, Beverley SM (1990) Gene replacement in parasitic protozoa. Nature 348: 171–173. [DOI] [PubMed] [Google Scholar]
- 49. Leprohon P, Legare D, Raymond F, Madore E, Hardiman G, et al. (2009) Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum . Nucleic Acids Res 37: 1387–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Longley DB, Harkin DP, Johnston PG (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3: 330–338. [DOI] [PubMed] [Google Scholar]
- 51. Zhang N, Yin Y, Xu SJ, Chen WS (2008) 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules 13: 1551–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Longley DB, Johnston PG (2005) Molecular mechanisms of drug resistance. Journal of Pathology 205: 275–292. [DOI] [PubMed] [Google Scholar]
- 53. Wang W, Marsh S, Cassidy J, McLeod HL (2001) Pharmacogenomic dissection of resistance to thymidylate synthase inhibitors. Cancer Res 61: 5505–5510. [PubMed] [Google Scholar]
- 54. Naumov DG (2012) [Furanosidase superfamily: search of homologues]. Mol Biol (Mosk) 46: 354–360. [PubMed] [Google Scholar]
- 55. Burchmore RJS, Wallace LJM, Candlish D, Al-Salabi MI, Beal PR, et al. (2003) Cloning, Heterologous Expression, and in Situ Characterization of the First High Affinity Nucleobase Transporter from a Protozoan. Journal of Biological Chemistry 278: 23502–23507. [DOI] [PubMed] [Google Scholar]
- 56. Sanchez MA, Tryon R, Pierce S, Vasudevan G, Landfear SM (2004) Functional expression and characterization of a purine nucleobase transporter gene from Leishmania major . Mol Membr Biol 21: 11–18. [DOI] [PubMed] [Google Scholar]
- 57. Henriques C, Sanchez MA, Tryon R, Landfear SM (2003) Molecular and functional characterization of the first nucleobase transporter gene from African trypanosomes. Mol Biochem Parasitol 130: 101–110. [DOI] [PubMed] [Google Scholar]
- 58. Gudin S, Quashie NB, Candlish D, Al-Salabi MI, Jarvis SM, et al. (2006) Trypanosoma brucei: a survey of pyrimidine transport activities. Exp Parasitol 114: 118–125. [DOI] [PubMed] [Google Scholar]
- 59. Fernandes JF, Castellani O (1959) Nucleotide and polynucleotide synthesis in Trypanosoma cruzi. II. In vitro effect of tioguanine and of the aminonucleoside of stylomycin. Exp Parasitol 8: 480–485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments of the active sites of thymidine kinase, uridine phosphorylase and uracil phosphoribosyl transferase. The regions of the active sites of the Leishmania TK, UP and UPRT were aligned with the Trypanosoma, mice and human homologues. The highlighted amino acid residues are those in which mutations were found. The highly conserved Q87 in the TK active site is mutated to a P in Lin5FU500.3. Position of the L265R mutation close to the specificity region in the UP enzyme in Lin5FU500.5 mutant. Position of the K145T mutation close to the phosphoribosyl pyrophosphate (PRPP) region in UPRT in Lin5FU500.4.
(TIF)
Primers used in this study. Primer names were given according to the nomenclature of TritrypDB database. Primers were used in PCR reactions either to confirm locus rearrangements or presence or absence of particular genes (PCR amplification); to amplify genes that will be further cloned in plasmids for transfection assays (Cloning); to generate probes for Southern blotting (Southern); or to confirm mutations (SNP validation). Restriction sites are underlined. XbaI, TCTAGA; HindIII, AAGCTT; F, forward primer; R, reverse primer. The size of amplicons is indicated in base pairs (bp).
(RTF)