Abstract
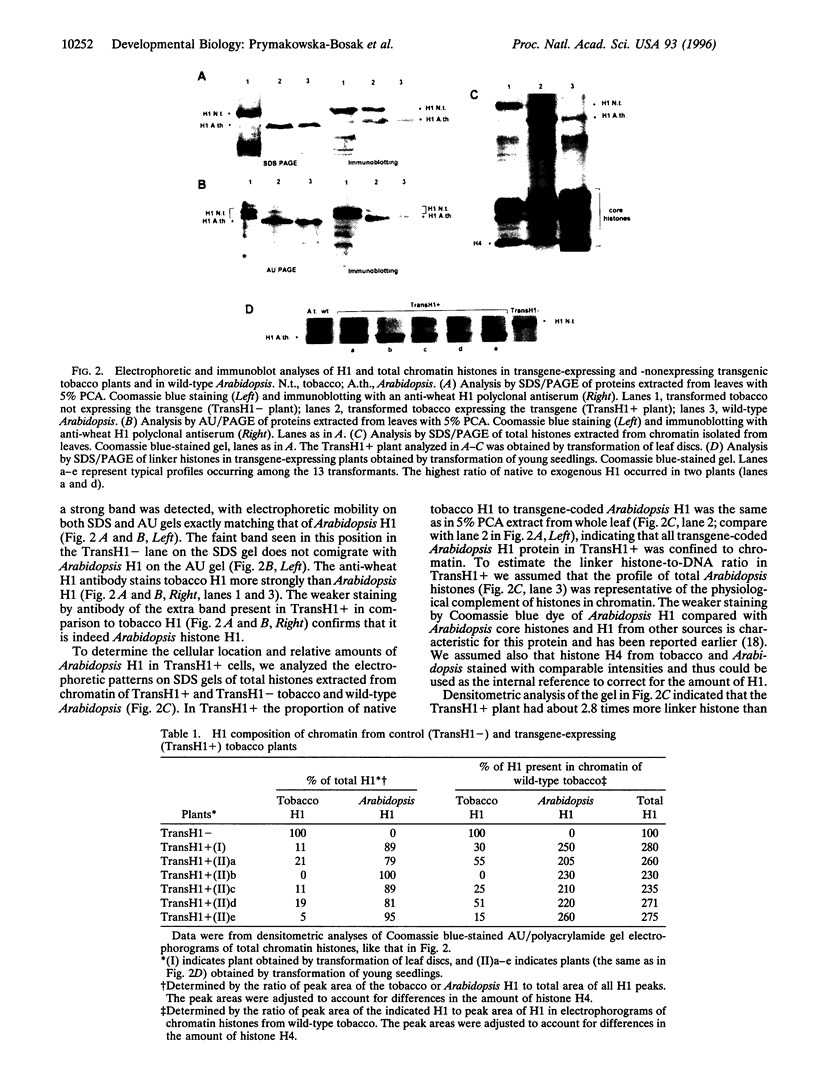
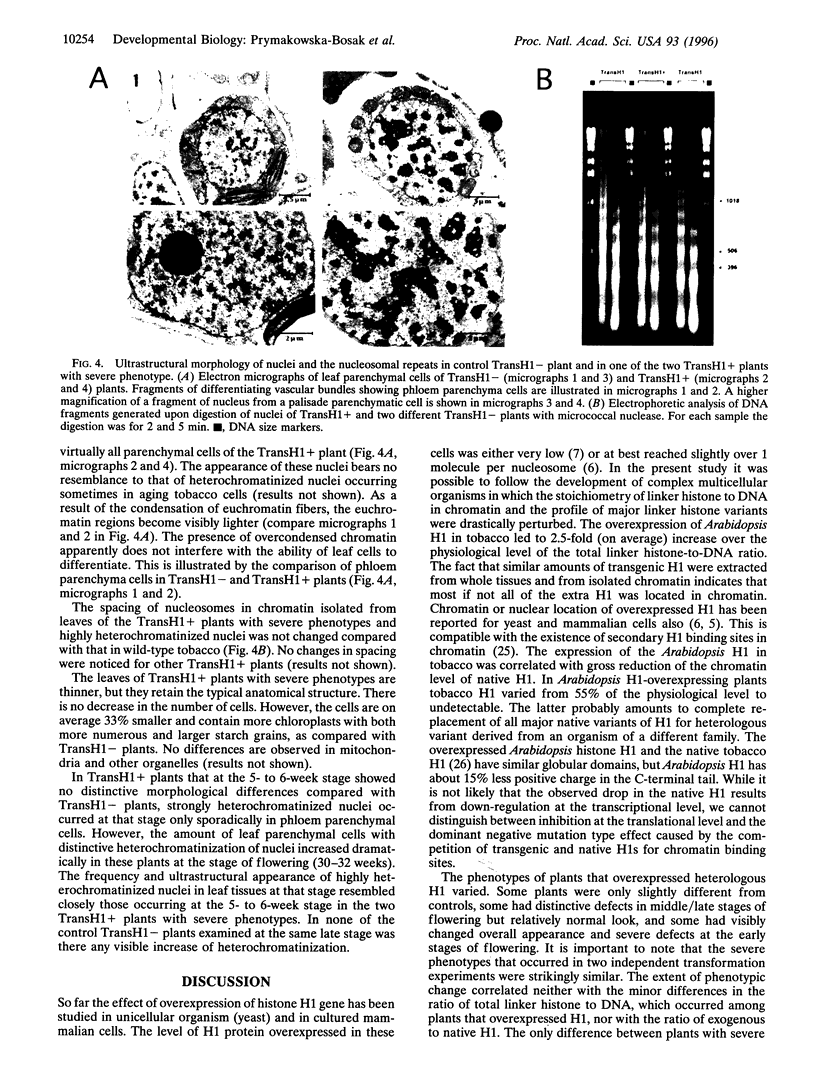
Histone H1, a major structural component of chromatin fiber, is believed to act as a general repressor of transcription. To investigate in vivo the role of this protein in transcription regulation during development of a multicellular organism, we made transgenic tobacco plants that overexpress the gene for Arabidopsis histone H1. In all plants that overexpressed H1 the total H1-to-DNA ratio in chromatin increased 2.3-2.8 times compared with the physiological level. This was accompanied by 50-100% decrease of native tobacco H1. The phenotypic changes in H1-overexpressing plants ranged from mild to severe perturbations in morphological appearance and flowering. No correlation was observed between the extent of phenotypic change and the variation in the amount of overexpressed H1 or the presence or absence of the native tobacco H1. However, the severe phenotypic changes were correlated with early occurrence during plant growth of cells with abnormally heterochromatinized nuclei. Such cells occurred considerably later in plants with milder changes. Surprisingly, the ability of cells with highly heterochromatinized nuclei to fulfill basic physiological functions, including differentiation, was not markedly hampered. The results support the suggestion that chromatin structural changes dependent on H1 stoichiometry and on the profile of major H1 variants have limited regulatory effect on the activity of genes that control basal cellular functions. However, the H1-mediated chromatin changes can be of much greater importance for the regulation of genes involved in control of specific developmental programs.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984 Nov 26;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Lenvik T. R. Arabidopsis thaliana H1 histones. Analysis of two members of a small gene family. Eur J Biochem. 1991 Dec 18;202(3):1029–1039. doi: 10.1111/j.1432-1033.1991.tb16466.x. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Wiebauer K., Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A., Cole R. D. Flanking sequences of Xenopus 5 S RNA genes determine differential inhibition of transcription by H1 histone in vitro. Mitotic phosphorylation of H1 decreases its inhibitory power. J Biol Chem. 1990 Jun 25;265(18):10726–10732. [PubMed] [Google Scholar]
- Juan L. J., Utley R. T., Adams C. C., Vettese-Dadey M., Workman J. L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994 Dec 15;13(24):6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linder C., Thoma F. Histone H1 expressed in Saccharomyces cerevisiae binds to chromatin and affects survival, growth, transcription, and plasmid stability but does not change nucleosomal spacing. Mol Cell Biol. 1994 Apr;14(4):2822–2835. doi: 10.1128/mcb.14.4.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini L., Vaeck M., van Montagu M. Conserved epitopes on plant H1 histones recognized by monoclonal antibodies. Eur J Biochem. 1989 Jan 2;178(3):779–787. doi: 10.1111/j.1432-1033.1989.tb14509.x. [DOI] [PubMed] [Google Scholar]
- McArthur M., Thomas J. O. A preference of histone H1 for methylated DNA. EMBO J. 1996 Apr 1;15(7):1705–1714. [PMC free article] [PubMed] [Google Scholar]
- Miloshev G., Venkov P., van Holde K., Zlatanova J. Low levels of exogenous histone H1 in yeast cause cell death. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11567–11570. doi: 10.1073/pnas.91.24.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto J. J. Immunoblotting. Methods Cell Biol. 1993;37:105–117. [PubMed] [Google Scholar]
- Paro R. Mechanisms of heritable gene repression during development of Drosophila. Curr Opin Cell Biol. 1993 Dec;5(6):999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers A., Muyldermans S., Wyns L. The interaction of histone H5 and its globular domain with core particles, depleted chromatosomes, polynucleosomes, and a DNA decamer. J Biol Chem. 1991 Jan 25;266(3):1502–1508. [PubMed] [Google Scholar]
- Shen X., Yu L., Weir J. W., Gorovsky M. A. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995 Jul 14;82(1):47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Sirotkin A. M., Edelmann W., Cheng G., Klein-Szanto A., Kucherlapati R., Skoultchi A. I. Mice develop normally without the H1(0) linker histone. Proc Natl Acad Sci U S A. 1995 Jul 3;92(14):6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker S. A modification of the acetic acid-urea system for use in microslab polyacrylamide gel electrophoresis. Anal Biochem. 1980 Nov 1;108(2):263–265. doi: 10.1016/0003-2697(80)90579-5. [DOI] [PubMed] [Google Scholar]
- Sun J. M., Ali Z., Lurz R., Ruiz-Carrillo A. Replacement of histone H1 by H5 in vivo does not change the nucleosome repeat length of chromatin but increases its stability. EMBO J. 1990 May;9(5):1651–1658. doi: 10.1002/j.1460-2075.1990.tb08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Haizel T., Adam E., Nagy F. Molecular characterization and expression of a tobacco histone H1 cDNA. Plant Mol Biol. 1995 Feb;27(3):597–605. doi: 10.1007/BF00019325. [DOI] [PubMed] [Google Scholar]
- Timmermans M. C., Maliga P., Vieira J., Messing J. The pFF plasmids: cassettes utilising CaMV sequences for expression of foreign genes in plants. J Biotechnol. 1990 Jun;14(3-4):333–344. doi: 10.1016/0168-1656(90)90117-t. [DOI] [PubMed] [Google Scholar]







