Abstract
The cosmopolitan genus Fraxinus, which comprises about 40 species of temperate trees and shrubs occupying various habitats in the Northern Hemisphere, represents a useful model to study speciation in long-lived angiosperms. We used nuclear external transcribed spacers (nETS), phantastica gene sequences, and two chloroplast loci (trnH-psbA and rpl32-trnL) in combination with previously published and newly obtained nITS sequences to produce a time-calibrated multi-locus phylogeny of the genus. We then inferred the biogeographic history and evolution of floral morphology. An early dispersal event could be inferred from North America to Asia during the Oligocene, leading to the diversification of the section Melioides sensus lato. Another intercontinental dispersal originating from the Eurasian section of Fraxinus could be dated from the Miocene and resulted in the speciation of F. nigra in North America. In addition, vicariance was inferred to account for the distribution of the other Old World species (sections Sciadanthus, Fraxinus and Ornus). Geographic speciation likely involving dispersal and vicariance could also be inferred from the phylogenetic grouping of geographically close taxa. Molecular dating suggested that the initial divergence of the taxonomical sections occurred during the middle and late Eocene and Oligocene periods, whereas diversification within sections occurred mostly during the late Oligocene and Miocene, which is consistent with the climate warming and accompanying large distributional changes observed during these periods. These various results underline the importance of dispersal and vicariance in promoting geographic speciation and diversification in Fraxinus. Similarities in life history, reproductive and demographic attributes as well as geographical distribution patterns suggest that many other temperate trees should exhibit similar speciation patterns. On the other hand, the observed parallel evolution and reversions in floral morphology would imply a major influence of environmental pressure. The phylogeny obtained and its biogeographical implications should facilitate future studies on the evolution of complex adaptive characters, such as habitat preference, and their possible roles in promoting divergent evolution in trees.
Introduction
Vicariance and isolation have been suggested as important drivers of diversification in trees, especially for those distributed over wide areas that have previously been subjected to fragmentation, or experienced major distributional changes (e.g. [1]). Colonization of diverse habitats could also promote divergent evolution. For example, divergent evolution could be associated with transitions from insect to wind pollination, which could lead to simpler flower morphology [2]. A change in pollination mode might also affect the genetic structure via changes in effective population size [3] and the rate of speciation.
Members of the genus Fraxinus, known as ash trees, comprise 43 recognized species [4], and are widely distributed in the northern temperate zone from Eurasia to North America. They display a range of morphologies (e.g., shrubs to trees; simple to compound leaves) and are also found in diverse habitats, from semi-deserts to subtropical or temperate environments, and from sea level to subalpine altitudes. Fraxinus shows great diversity in floral morphology, with taxa with complete, incomplete and naked flowers (i.e., no corolla or calyx), with two or four petals, and in the case of F. excelsior, with male, female or hermaphrodite flowers on the same tree [5]. Given this diversity of locations, habitats, and morphologies, the cosmopolitan Fraxinus should represent an informative model to investigate speciation patterns in long-lived angiosperm species in relation to divergent evolution caused by vicariance or adaptation to different habitats.
Evolutionary inferences are valuable only if conducted in the proper phylogenetic context and in the light of a sound knowledge of the biogeographic history [6], [7]. Several studies have investigated the phylogeny of Fraxinus using nuclear internal transcribed spacer (nITS) [4], [8] and chlroroplastic DNA (cpDNA) sequences [4], [9]. The monophyly of this genus in the tribe Oleeae has been confirmed [10] and six sections (Dipetaleae, Fraxinus, Melioides, Ornus, Pauciflorae, and Sciadanthus) have been delimited on the basis of molecular (reciprocal monophyly) and morphological characters (flower and samara morphology) [4] (Table 1). However, three Fraxinus species (F. cuspidata, F. spaethiana, and F. chiisanensis) remained unclassified (incertae sedis) [4]. The species in the different sections usually form cohesive continental groups (the sections Dipetaleae, Melioides, and Pauciflorae in North America; and the sections Fraxinus, Ornus, and Sciadanthus in Eurasia).
Table 1. Different classification schemes of the genus Fraxinus, including the revisions proposed in this study, and geographic distributions of species.
| Sections based on [8] | Sections based on [4] | Sections based on this study | Species | Synonyms used in this study | Distribution |
| Ornus | Ornus | Ornus | ornus L. | Mediterranean area, N Africa and SW Asia | |
| Ornaster | apertisquamifera Hara | Japan | |||
| bungeana DC | China | ||||
| floribunda Wall. | retusa Champ. ex Benth.var. henryana | C & E Asia (from Afghanistan to Japan) | |||
| griffithii G. B. Clarke | E Asia (from NE India to Japan and Indonesia) | ||||
| lanuginosa Koidz. var. lanuginosa and var. serrata (Nakai) Hara | Japan | ||||
| malacophylla Hemsl. | China, Thailand | ||||
| paxiana Lingelsh. | sikkimensis (Lingelsh.) Handel-Mazzetti | Himalayas, China | |||
| raibocarpa Regel | C Asia (Turkestan mountains, Iran, Pakistan, Afghanistan) | ||||
| sieboldiana Blume | mariesii Hook. f. | China, Japan, Korea | |||
| trifoliolata W. W. Smith | China | ||||
| baroniana Diels | China | ||||
| chinensis Roxb. | China, Japan, Korea, Vietnam | ||||
| longicuspis Sieb. & Zucc. | Japan | ||||
| micrantha Lingelsh. | C Asia (Punjab to Nepal, Himalayas) | ||||
| Bumelioides | Dipetalae | Dipetalae | anomala Torr. ex S. Wats. | SW USA, N Mexico | |
| dipetala Hook. et Arn. | trifoliata (Torr.) Lewis & Epling | SW USA, N Mexico (Baja California) | |||
| quadrangulata Michx. | E & C USA, C Canada | ||||
| Fraxinus | Fraxinus | angustifolia Vahl. ssp. angustifolia | monophylla Desf. | SW Europe | |
| angustifolia Vahl. ssp. oxycarpa (M.Bieb. ex Willd.) Franco & Rocha Afonso | oxycarpa Willd., pallisiae A. J. Willmott, obliqua Tausch | SE Europe | |||
| angustifolia Vahl. ssp. syriaca (Boiss.) Yalt. | potamophila Herder, holotricha Koehne, sogdiana Bunge, syriaca Boiss. | W & E Asia (Turkey to Pakistan and Russia) and Algeria | |||
| excelsior L. | turkestanica Carrière | C & N Europe | |||
| mandshurica Rupr. | E Asia (China, Japan, Korea, E Russia) | ||||
| nigra Marsh. | E USA, E Canada | ||||
| Melioides | platypoda Oliv. | E Asia | |||
| Melioides | Melioides | americana L. | biltmoreana Beadle | E USA, E Canada | |
| berlandieriana A. DC. | SW USA, NE Mexico | ||||
| caroliniana Mill. | SE USA | ||||
| latifolia Benth. | W USA | ||||
| papillosa Lingelsh. | SW USA (SE Arizona, SW New Mexico, Texas), Mexico (W Chihuahua, NE Sonora) | ||||
| pennsylvanica Marsh. | richardii Bosc | C & E USA, Canada | |||
| profunda (Bush) Bush | tomentosa Michx. f. | E USA | |||
| texensis A. Gray | S USA (Texas) | ||||
| uhdei (Wenzig) Lingelsh. | Guatemala, Honduras, Mexico, USA (Hawaii, Puerto Rico) | ||||
| velutina Torr | SW USA, N Mexico | ||||
| Dipetalae | incertae sedis | cuspidata Torr. | SW USA, Mexico | ||
| N.A. | chiisanensis Nakai | Korea | |||
| spaethiana Lingelsh. | Japan | ||||
| Pauciflorae | Pauciflorae | dubia (Willd. ex Schult & Schult f.) P. S. Green & M. Nee | Mexico, Guatemala | ||
| gooddingii Little | SW USA (Arizona), N Mexico | ||||
| greggii A. Gray | SW USA, Mexico | ||||
| purpusii Brandegee | Mexico, Guatemala | ||||
| rufescens Lingelsh. | Mexico | ||||
| Sciadanthus | Sciadanthus | xanthoxyloides (G. Don) DC. | N Africa (Algeria, Morocco) to Asia (Afghanistan to China) | ||
| hubeiensis S. Z. Qu, C. B. Shang & P. L. Su | hoopiensis | China |
N.A.: not available, taxa that were not included in the considered study.
In addition to cases of incongruence, previous phylogenetic studies failed to achieve a complete resolution of relationships at the species level within the Fraxinus genus and suggested that some morphological species may not be monophyletic [4], [8]. These findings are suggestive of incomplete lineage sorting [11], [12] or reticulate evolution [13], [14], which could make phylogenetic inferences difficult. The occurrence of natural hybridization between ash taxa in parapatric or sympatric contact [15], [16] may add to this difficulty and illustrates that reproductive isolation is a slow process that may affect speciation patterns and speed in Fraxinus, as noted for other temperate and boreal trees (e.g. [13], [17]).
The evolution of floral morphology has been investigated in Fraxinus by mapping floral characters onto phylogenies. A trend toward sequential simplification of floral structure associated with the loss of corolla and calyx was reported, but a reversal of this was also observed [8]. The same trend toward simplification as well as reversal was also inferred using a more complete phylogeny [4], but there were conflicting findings between these two studies owing to incongruences between the estimated phylogenies. Against this background, improving the resolution of the phylogeny of Fraxinus by considering more loci should improve our understanding of the evolution of floral morphology by enabling better estimates of the frequencies of reversals or multiple losses of floral components, and how these floral changes are linked to the biogeographic and speciation history, as well as the colonization of new habitats.
The objectives of this study were to reconstruct the complete phylogeny of Fraxinus from multiple loci for an accurate assessment of the relationships among the sections of the genus, infer the biogeographic history and speciation patterns of this cosmopolitan angiosperm group, and better understand the long-term drivers of speciation in temperate tree taxa. A more robust phylogeny should also improve our understanding of the evolution of phenotypic characters such as floral morphology, and facilitate the distinction between parallel and convergent evolution from a common ancestry. An ancillary objective was to compare the phylogenies obtained from structurally different loci (the repeated nuclear ribosomal regions, cpDNA intergenic regions, and a low-copy nuclear gene) in order to detect possible sources of locus-specific conflicts that could blur phylogenetic inference.
Materials and Methods
Plant material
We collected 292 samples (1 to 28 individuals per species) for 40 Fraxinus species (from a total of 43 sensu [4]) that represent almost the entire genus, from arboreta (n = 198), herbaria (n = 17) or the field (n = 87) (see Table S1 for fieldwork information and vouchers). The sampling did not require any specific permits, given that it was carried out on government-owned sites and did not involve endangered or protected species. In the present study, we retained the classification and synonymies of Wallander [4] because of her exhaustive sampling of the genus (Table 1). Outgroups from the Oleaceae were selected in accordance with Wallander and Albert [10] (Forsythia×intermedia, Jasminum sp., Ligustrum vulgare, Olea europaea, Osmanthus fragrans, Phillyrea angustifolia, and Syringa vulgaris; see Table S1 for details).
The morphological identification of Fraxinus samples can be problematic if the samples are from an area of sympatry in which interspecific hybridization may occur, or because of misidentification due to mislabelling in arboreta or herbaria, or synonymy of taxa. To overcome these issues and render estimated phylogenies more robust, we avoided sampling in areas of sympatry or parapatry, used multiple individuals, and sampled from several sources (including the field, herbaria, and arboreta) (Table S1). Indeed, we expected that, if hybridization or misidentification occurred, only one or a few individuals in a particular species would be affected; as a consequence, most individuals would still be classified in a monophyletic group representative of the species (see Results). Most specimens have been deposited as herbarium vouchers for further morphological confirmation of their identity (see Table S1).
For fresh samples, 50 mg of fresh leaves were dehydrated in an alcohol/acetone 70∶30 solution and stored dry, following a modified version of the protocol of Fernandez-Manjarres et al. [15]. Extraction of DNA was performed for all samples using the DNeasy Plant Mini Kit (Qiagen, Valencia, California, USA) following the manufacturer's instructions.
Sequence datasets
To reconstruct the phylogeny of Fraxinus, we used two cpDNA intergenic regions previously used in Fraxinus (trnH-psbA and rpl32-trnL) [18], and also sequenced the nuclear external transcribed spacer (nETS) regions and the nuclear low-copy gene phantastica. In addition, newly sampled individuals were sequenced for the nuclear ribosomal nITS and added to those already available in GenBank. Of these five loci, three have been used previously in molecular studies of Fraxinus. Sequences of the trnH-psbA and rpl32-trnL loci were tested recently as part of a possible DNA barcoding system for the genus Fraxinus [18], whereas nITS sequences were used in previous phylogenetic analyses of this genus [4], [8].
Chloroplast sequences
Both trnH-psbA and rpl32-trnL sequences were obtained from Arca et al. [18] and concatenated to construct a chloroplast dataset.
Nuclear ribosomal spacers (nETS and nITS)
DNA was amplified from Fraxinus samples using the SST-ETS (GGC WTG TKT GGG TAT GTT GGA T) and 18S-ETS (ACT TAC ACA TGC ATG GCT TAA TCT) primers previously used in Syringa (Oleaceae) [19]. For some samples for which amplification using this primer set was difficult, the primers ETS-F1 (ATG CCT GTT CAT TGG ATG) and ETS-R1 (AAG YCC AVV ACR AGG ARG) were designed de novo. The PCR reaction mix contained two units of Taq DNA polymerase MP-Biomedicals®, 1× buffer, 1.5 mM MgCl2, 0.25 mM dNTPs, 1 µM each primer, 0.1 mg/ml DMSO, and 30 ng of template DNA in a final volume of 25 µl. All reactions were carried out under the following thermal cycle program: 2 min at 94°C, 5 cycles of 15 s at 94°C, 30 s at 57°C, with a touch-down of −1°C per cycle, and 30 s at 72°C, followed by 50 cycles of 15 s at 94°C, 30 s at 52°C, and 30 s at 72°C, and finally 7 min at 72°C. Reactions were performed in a Mastercycler® ep gradient S (Eppendorf, Hamburg, Germany). The PCR quality was checked on 1.5% agarose gels and the amplified PCR products were stored at 4°C.
In addition to Genbank sequences from Wallander [4], we generated nITS sequences for 127 individuals using the primers ITS4 and ITS5 [20], [21], in accordance with the reaction protocol from the same study. For samples for which amplification using either of these primer sets was more difficult, nITS1 and nITS2 were amplified separately using the primer pairs ITS1/ITS6 and ITS7/ITS8 [8]. The PCR quality was checked on 1.5% agarose gels and the amplified PCR products were stored at 4°C until direct sequencing. For nITS, sequences from Genbank and new sequences were combined in a “nITS dataset” that included 43 species.
The phantastica gene
The phantastica gene is a single-copy gene that encodes a transcription factor involved in leaf morphology, and the growth and dorsoventrality of the lateral organs in Antirrhinum [22]. It has also been shown to determine leaflet placement in compound leaves of various angiosperms, notably F. americana [23]. It was retained for use in the present study after the preliminary sequencing of several nuclear regions (e.g. floricaula, frigida, phantastica, adh, waxy, chi, tpi, pgi, waxy, and g3pdh), which revealed that the phantastica gene is sufficiently conserved to be realiably aligned across the tribe Oleeae of Oleaceae, while being sufficiently variable to resolve relationships in the genus Fraxinus (data not shown). Moreover, very few heterozygote positions were identified, none of which revealed evidence of recombination. To our knowledge, this is the first time that phantastica has been used for phylogenetic inference.
The Phan-F-M13 (TGTAAAACGACGGCCAGGGGTAAGTGGTGGGAGGTTT) and Phan-R-M13 (CAG GAAACAGCTATGACCCGACTCGAGTTGCTGTTC) primers (Basak J., Visva-Bharati Univ., India, unpublished data) were used to amplify an approximately 625-bp-long fragment in the exon. The PCR reaction mix contained two units of Taq DNA polymerase MP-Biomedicals®, 1× buffer, 1.5 mM MgCl2, 0.25 mM dNTPs, 1 µM each primer, 0.1 mg/ml DMSO, and 30 ng of template DNA for a final volume of 25 µl. All reactions involved the following thermal cycle program: 2 min at 94°C, 5 cycles of 15 s at 94°C, 30 s at 62°C, with a touch-down of −1°C per cycle, and 30 s at 72°C, followed by 50 cycles of 15 s at 94°C, 30 s at 57°C, and 30 s at 72°C, and finally 7 min at 72°C. Reactions were performed in a Mastercycler® ep gradient S (Eppendorf, Hamburg, Germany). The PCR quality was checked on 1.5% agarose gels and the amplified PCR products were stored at 4°C until direct sequencing.
DNA sequencing and analyses
All sequencing was carried out at the Génoscope facility (Centre National de Séquençage, Evry, France). The PCR products were purified using exonuclease I and phosphatase, and sequenced using the BigDyeTerminator V3.1 kit (Applied Biosystems) and an ABI 3730XL sequencer. All regions were sequenced for both DNA strands to confirm the accuracy of the sequences. The standard M13 primers [24] were used for phantastica sequencing, as described by Ivanova et al. [25]. All new sequences have been deposited in Genbank under Accession numbers HQ705327 to HQ705604 (nETS), HQ705200 to HQ705326 (nITS), and HM242423 to HM242846 (phantastica).
Sequence alignment and contigs were checked using CodonCode Aligner version 1.6.3 (Codon Code Corporation), and sequences were aligned manually using using Muscle [26] and Se-Al [27]. The datasets (cpDNA, nITS, nETS, and phantastica) were analysed separately, and then concatenated to conduct a “total evidence” analysis. Aligned sequences were used to construct phylogenetic trees based on the maximum likelihood (ML) criterion and Bayesian inference method (BIM). All ML trees were constructed using PhyML [28]. Indels were not considered. Using MrModeltest 2.3 [29], we chose the GTR+I+G model under the AIC criterion, which provides a better estimate of branch lengths than more complex models [30] and does not influence the probabilities of the nodes, even in over-parameterized analysis [31]. We included all individuals, which resulted in a total of 302 samples sequenced for at least one locus (including the outgroups). Bootstrap values were estimated with 1,000 pseudo-replicates.
Regarding the nearly identical nITS sequences reported for F. platypoda and F. mandshurica [4], and given that the two species are in sympatry in Japan, it is possible that these individuals were misidentified and were from the same species, most likely F. mandshurica. To avoid such potential problems related to synonymy, we used many (n = 10) individuals sampled from natural populations in Japan. They all formed a monophyletic group (with the arboretum specimens) for all the different datasets and methods of phylogenetic reconstruction used in the present study (see Figures S2, S3, S4).
All BIM analyses were performed on a reduced dataset using MrBayes [32]. This reduced dataset was created by using one individual per species following Wallander [4], with the selection of individuals located within the ML tree at positions that were reasonable considering their identity for each data partition, that is, grouping with other individuals from the same species. Doing otherwise would have resulted in the need to consider the entire 302 individual sequences, which would have been too time-consuming for Bayesian analysis. For some species, different individuals were included for different sequence datasets, owing to the absence of the previously selected individual in a particular dataset. The newly chosen individual was chosen from the same specific group as the individual it replaced. The GTR+I+Γ model was used in MrBayes, with six substitution types (“nst = 6”) and rate variation across sites modelled using a gamma distribution, with a proportion of sites being invariant (rates = “invgamma”). The Markov chain Monte Carlo search was run with 4 chains for 20 million generations, with trees being sampled every 100 generations. We assessed the convergence of runs and chose to discard the first 2 million generations using Tracer [33].
Estimation of molecular divergence times
The estimation of divergence times was conducted with BEAST v1.7.2 [34] using all taxa available for the four investigated loci (nITS, nETS, phantastica, and concatenated cpDNA) and thus, excluding F. hupehensis. The two oldest fossils that could be reliably attributed to the genus were found in the southeast of North America (Middle Eocene - 39–49 mya, [35], [36], [37]), and were used as a prior for the root node age. Normal prior age distribution was assigned to the root node for the genus, which resulted in a mean age calibration of 44 mya (SD = 3.5 mya); for uncertainty, we considered the entirety of the 95% confidence interval (38.2 to 49.8 mya). In addition, a second fossil, namely pollen representative of the morphology of the extant taxon Fraxinus angustifolia from the Upper Miocene in Europe (12 mya) [38], was used to set the minimum age (uniform distribution) for the rise of European ashes. The GTR+I+Γ model was used, implicating six substitution types with rate variation across sites modelled using a gamma distribution, and the proportion of sites that were invariant was used with base frequencies estimated for all partitions [34]. A random local clock model was further implemented with a Yule process as the tree prior. Markov chain Monte Carlo searches were run for 150 million generations. We assessed the convergence of runs and chose to discard the first 15 million generations using Tracer [33]. Moreover, for more precise inference of the divergence times for the different loci, a prior was set on the ITS substitution rate. In accordance with Kay et al. [39], we selected a truncated normal distribution with a mean of 2.15×10−9 substitutions per site per year and a standard deviation of 3.2×10−9, which is a conservative approach given the relative size of this latter value. These searches achieved sufficient mixing as assessed by the high effective sample size values for most parameters, reaching plateaus for divergence time estimates over generations after burn-in, and repeatability of results over multiple independent runs. The results were combined manually, visualized with FigTree v1.3.1 [40], and the section Dipetalae was set as an outgroup. The rates of diversification were then evaluated using default parameters in the Lineage Through Time (LTT) plot from Tracer v1.5 [33], and subsequent statistics were calculated with R [41] using the package APE [42].
Biogeographic history
The dispersal-extinction-cladogenesis (DEC) model implemented in Lagrange v.20120508 [43] was used to infer the biogeographic history. A geographical range matrix coding each species as present or absent in each of four geographical areas (North America, Asia, Europe, North Africa) was constructed. Species that inhabit more than one geographical area were coded accordingly. Thus, the Lagrange analysis was based on this geographical matrix, and the divergence times calibrated from the BIM consensus tree were obtained from molecular dating with branch lengths equal to mean ages and a root age of 45.4 mya. The dispersal connections between areas were parameterized on the basis of documented land-bridge connections (www.scotese.com) and cooling periods of the Cenozoic [44]. The detailed matrices are shown in Figure S1.
Reconstruction of floral evolution
Floral evolution was traced onto the phylogenetic tree that resulted from the BIM and BEAST analyses of the combined dataset (cpDNA, ETS, ITS, and phantastica) using Mesquite [45]. Parsimony and ML reconstructions were conducted using default parameters in Mesquite [45], with one of the three following states scored for each of the extant species: complete flower (calyx+corolla), incomplete flower (loss of corolla or calyx), or naked flower (no calyx or corolla). Given the lesser ambiguity in the reconstruction of the evolution of floral morphology than that obtained with MP, only the ML reconstruction was mapped onto the phylogenetic tree. Ancestral states of floral characters were estimated using a stochastic model of evolution [46], setting the model to Mk1 (“Markov k-state 1 parameter model”) in Mesquite [45].
Results
The levels of sequence polymorphism and tree resolution are shown in Table 2 for each genomic region analysed. Given that ML trees and BIM trees were largely congruent, only the Bayesian trees are presented, with the ML bootstrap values added onto the branch lengths. ML trees that include all the individuals for ETS, ITS and phantastica datasets are shown in Figures S2, S3 and S4, respectively.
Table 2. Sequence variation and parameters of phylogenetic analyses for the various datasets used to estimate the phylogeny of Fraxinus.
| Sequence dataset | Number of sequences(Fraxinus) | Length | Number of variable sites (%) | Number of informative sites (%) | Number of nodes with support | |
| PP≥0.50 | PP≥0.90 | |||||
| cpDNA | 276 | 1721 | 223 (12.9) | 94 (5.5) | 19 | 6 |
| nITS | 118 | 676 | 170 (25.1) | 142 (21.0) | 34 | 27 |
| nETS | 260 | 480 | 170(35.4) | 112 (23.3) | 26 | 16 |
| phantastica | 225 | 627 | 146 (23.3) | 48 (7.7) | 17 | 15 |
| Combined | - | 3504 | 709 (20.2) | 396 (11.3) | 36 | 32 |
PP≥0.50, number of nodes with posterior probability ≥0.50 in Bayesian analyses; PP≥0.90, number of nodes with posterior probability ≥0.90 in Bayesian analyses. Only internal nodes to the genus Fraxinus were considered for the estimation of statistical support.
Analysis of separate datasets
The trees for each of the loci are shown in Figures S5, S6, S7 and S8. Briefly, the sections were well resolved by cpDNA data, but the BIM tree showed moderate support for the branches leading to taxonomical sections (Fig. S6). On average, the phylogenies obtained with the cpDNA intergenic spacer sequences were not as well supported as those using nuclear DNA sequences. This is likely related to the generally lower sequence divergence observed for the cpDNA loci (Table 2). In the BIM nETS tree (Fig. S7), the sections were well resolved and it showed strong support for the branches underlying the different sections of the genus, like for the BIM nITS tree (Fig. S8). This tree was generally congruent with a previous study [4], except for the placement of F. platypoda and the section Pauciflorae. Finally, the BIM tree for the phantastica dataset (Fig. S9) showed high support for the monophyly of all recognized sections (all PP = 1.0), except for the section Ornus, in which F. micrantha and F. griffithii were not included.
Analysis of the combined dataset (cpDNA, nITS, nETS and phantastica)
When considering only the strongly (PP≥0.90) and moderately (PP≥0.70) supported nodes, the cpDNA tree showed little conflict with the trees produced by the analyses of nuclear DNA loci. Only a few minor incongruences were observed in the placement of some species. For example, F. floribunda and F. griffithii formed a sister clade separate from all other species of the genus. These incongruences may have been due to incomplete lineage sorting, homoplasy, or very low levels of sequence divergence among the cpDNA sequences of Oleaceae. These minor misplacements may also represent artefacts derived from possible misidentification of specimens in arboreta and herbaria.
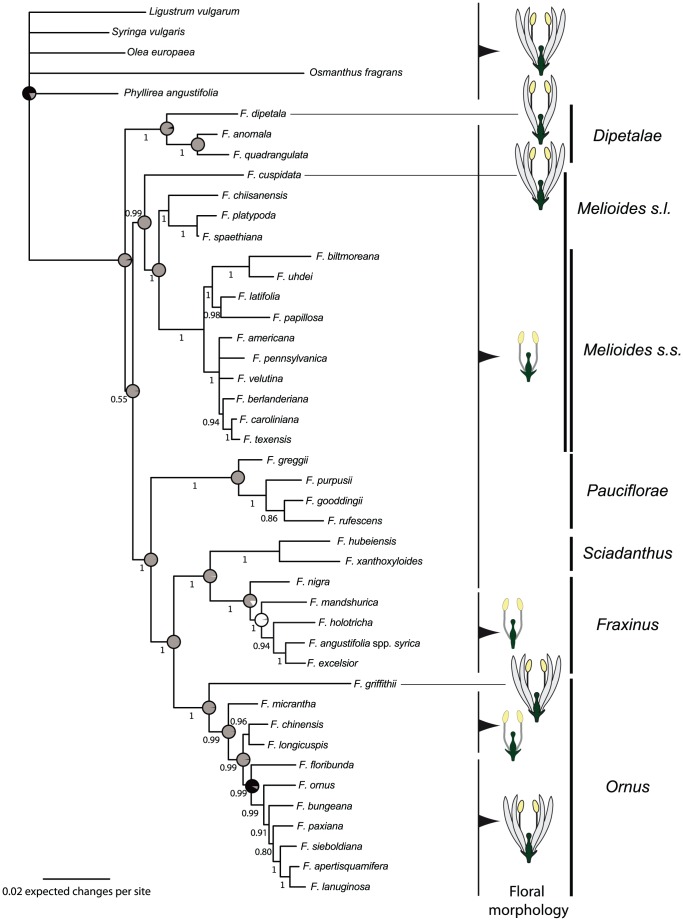
The genus appeared to be divided into three main groups (Fig. 1). The section Dipetalae was monophyletic (PP = 1.0) and a sister group to all other sections of the genus, but this relationship had much lower support (PP = 0.55). All other sections were monophyletic (PP = 1.0), and the section Fraxinus was closely associated with the section Sciadanthus (PP = 1.0); they formed a sister group to the section Ornus (all PP = 1.0). The group formed by these sections had the section Pauciflorae as a sister group (PP = 1.0). The other part of the tree grouped together the species of the section Melioides sensu lato. In this group, F. cuspidata was in a basal position (PP = 1.0). The section Melioides sensu stricto was divided into two groups (PP = 1.0 for both groups). One of these groups included a polytomy (F. americana, F. pennsylvanica, and F. velutina) and a group that comprised F. berlanderiana, F. caroliniana, and F. texensis (PP = 0.94). The other group (PP = 1.0) included F. biltmoreana and F. uhdei in a subgroup (PP = 1.0), with F. latifolia and F. papillosa (PP = 0.98) being found in the other subgroup.
Figure 1. Phylogenetic tree resulting from BIM analysis of the combined dataset (cpDNA, ETS, ITS and phantastica).
Posterior probabilities ≥0.50 are indicated below the branches. Tree nodes that involve floral changes and the relative likelihoods of floral states are indicated by circles: black, complete flower (calyx+corolla); grey, incomplete flower (loss of corolla or calyx); white, naked flower (no calyx or corolla). On the right, schematic floral character states are shown at the tip of the branches. On the right, sections are indicated by vertical bars; the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here shown to be closely related to the incertae sedis species.
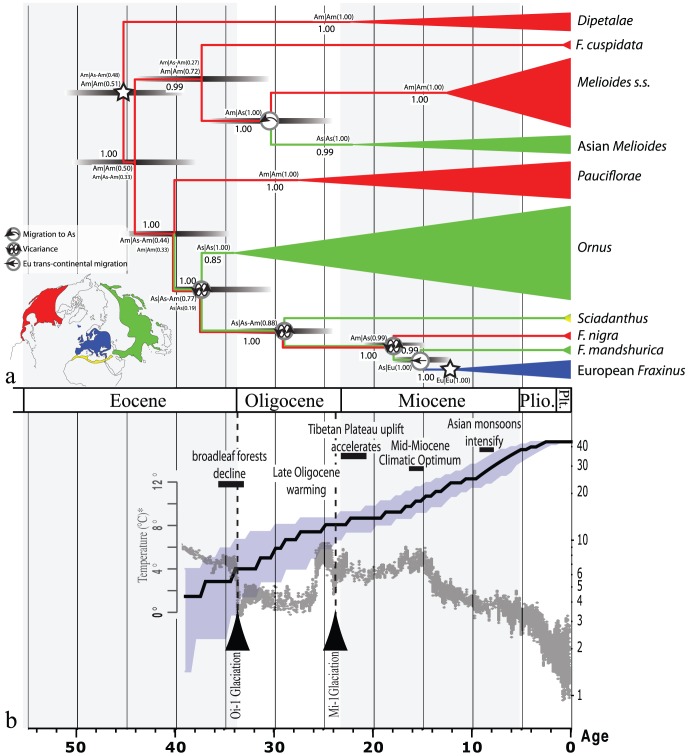
Molecular dating
The divergence and diversification times estimated for each section and major groups of the genus are shown in Table 3 and Figure 2a. A minimum age of 45.4±3.5 mya was obtained for the divergence of the section Dipetalae. The divergence times for four of the six major groups (sections Dipetalae, Melioides sensu lato, Pauciflorae and Ornus) date back to the middle and late Eocene (35–52 mya), whereas divergence was considered to have occurred during the Oligocene and Miocene for two groups (sections Sciadanthus and Fraxinus). The split between the American and Asian lineages of the section Melioides sensus lato was assumed to have also occurred during the Oligocene. The times for diversification (i.e., the date of the node for the last common ancestor) of four of the seven taxonomical groups (sections Dipetalae, Melioides s.s., Fraxinus, and the Asian Melioides species) were shown to have been during the Miocene (5.3–23 mya) (Figure 2a). The sections Ornus and Pauciflorae diversified during the upper and lower Oligocene, respectively. All of the inter-continental migrations that led to divergence events were estimated to have occurred before the Miocene, except in the section Fraxinus, for which the divergence that led to the North American F. nigra from its Eurasian ancestor was postulated to have occurred during the Miocene (Fig. 2a). In general, no clear relationship was found between global temperature changes and the diversification rate over such a large period of time: the cumulative number of divergence events showed a more or less linear increase over time, whereas several large temperature shifts were observed during the past 40 mya (Fig. 2b). The LTT plot showed a decrease in the diversification rates between ≈17 and 25 mya (Fig. 2b), which was not significantly supported by the MCCR test [47] (P = 0.153), but was significant using the γ statistic of Pybus and Harvey [47] (P = 0.049). According to the test for rate variation using the delta-AICrc statistic [48], the most probable model to fit the diversification rates was the yule2rates, with a shift in rate at ≈5 mya. This model assumes that the group has diversified under speciation rate r1 until time st, at which point the speciation rate shifts to a new rate r2. Thus, overall, the diversification rate was quite constant through time, except more recently during the Pleistocene.
Table 3. Estimated times for observed divergence and diversification events in the genus Fraxinus.
| Node description1 | Time period | Age ± S.D. (mya) |
| Divergence of the section Dipetalae | Eocene | 45.4±1.8 |
| Diversification of the section Dipetalae | Oligocene | 24.4±1.8 |
| Divergence of the section Melioides sensu lato | Eocene | 44.2±1.8 |
| Divergence of the section Melioides sensu stricto | Eocene | 30.4±1.8 |
| Diversification of the section Melioides s.s. | Miocene | 12.9±1.5 |
| Diversification of the Asian species of the section Melioides s.l. | Oligocene | 23.7±1.8 |
| Divergence of the section Ornus | Eocene | 38.5±1.7 |
| Diversification of the section Ornus | Oligocene | 34.1±1.7 |
| Divergence of the section Pauciflorae | Eocene | 40.2±1.7 |
| Diversification of the section Pauciflorae | Oligocene | 28.9±1.9 |
| Divergence of the section Sciadanthus | Miocene | 28.7±1.6 |
| Divergence of F. nigra | Oligocene | 18.0±1.3 |
| Divergence of F. mandshurica | Miocene | 15.3±1.1 |
| Diversification of European Fraxinus | Miocene | 12.9±0.8 |
With reference to Fig. 1.
Figure 2. a. Chronogram generated from BEAST analysis and estimations of times of divergence and diversification of sections in Fraxinus (in mya), and showing the results from Lagrange analysis.
Bars at nodes represent the 95% confidence interval; stars indicate the calibration nodes. Map showing the colours used to identify the areas of occurrence of Fraxinus species: red, North America (Am); green, Asia (As); blue, Europe (Eu); yellow, North Africa. The same colours were used on the tree to indicate the reconstructed geographic range across the branches and nodes. For the Lagrange results, a slash indicates the split of areas into two daughter lineages, namely, left/right, where “up” and “down” are the ranges inherited by each descendant branch. The values in brackets represent relative probabilities. When a node has alternative scenarios within 2 log-likelihood units of the optimal reconstruction, the relative probability (fraction of the global likelihood) for the optimal reconstruction is indicated with a smaller font. Circles on nodes indicate migration events or vicariance processes as inferred according to the Lagrange analysis. b. Semilogarithmic Lineage Through Time (LTT) plot (solid line) averaged over 135,000 posterior trees from the BIM analysis: right axis, the solid line represents the cumulative number of lineages and the blue area, the 95% confidence interval; in dark grey, plot of the global deep-sea temperature and its variance, with temperature estimates, geological and biological events reported elsewhere [44]; age in millions of years.
Biogeographical reconstruction
The results of the Lagrange analyses mapped on the dated phylogeny and a graphical representation of major historical events are shown in Fig. 2a and Fig. 3, respectively. The North American origin of the genus was confirmed (relative probability = 0.51), despite the fact that an ancient vicariance effect between North America and Asia was also retrieved with marginally lower probability (relative probability = 0.48). The analysis inferred colonization events in the section Melioides (relative probability = 1.0), resulting in lineages leading to the species F. spaethiana, F. platypoda, and F. chiisanensis. The geographical history of the other Eurasian sections was more complex, with two divergence events in Asia resulting from vicariance, one leading to the section Ornus (relative probability = 0.77) and the other to the section Sciadanthus, represented only by F. xanthoxyloides in our analysis (relative probability = 0.88). Two additional divergence events were inferred in the section Fraxinus. Whereas one was suggestive of allopatric divergence that resulted in the lineage leading to F. nigra (relative probability = 0.99), the other event implicating the colonization of Europe resulted in the differentiation of three species (F. excelsior, F. angustifolia and F. holotricha) (relative probability = 1.0).
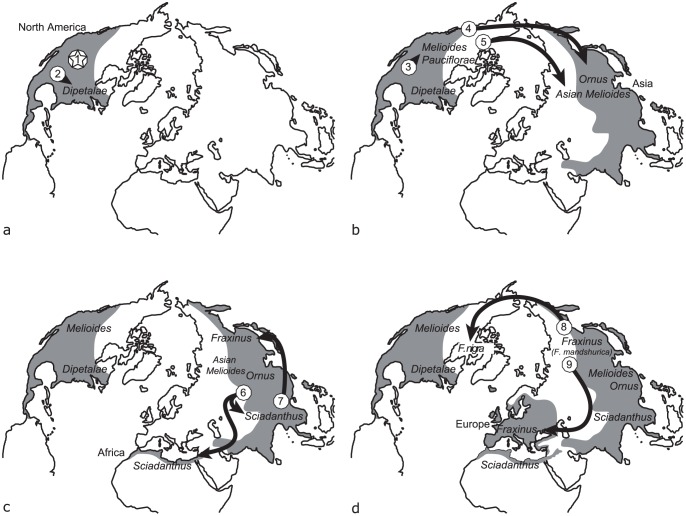
Figure 3. Map illustrating major events in the biogeographic history of the genus Fraxinus.
(a) The genus likely appeared in North America (1), the section Dipetalae diverging early (2). (b) Diversifications leading to the sections Melioides and later Pauciflorae (3) in North America were followed by migrations to Asia, leading to the sections Ornus (4) and to the Asian species of the section Melioides (5). (c) The section Sciadanthus expanded in Eurasia and Africa (6), (d) followed by the divergence of the Asian section Fraxinus (7) and the migration of a lineage leading to F. nigra in North America (8). More recently, the differentiation of F. mandshurica in Asia and transcontinental expansion of the lineage towards Europe (9) resulted in the observed geographical diversity of the section Fraxinus throughout Eurasia.
Reconstruction of evolution of floral morphology
Due to the congruence of results using either the BIM or the BEAST tree, the reconstruction of the evolution of floral morphology is reported in details from the BIM analysis (Fig. 1) while the BEAST tree is reported in Figure S9. The BIM reconstruction that showed the highest likelihood (−log L = 24.68) indicated that there was an ancient state of incomplete flowers at the time of the common ancestor of the genus, with one loss of floral pieces in the section Fraxinus, and four reversions from incomplete flowers to complete flowers. There was uncertainty relative to whether a reversion in F. nigra or a loss of calyx in Eurasian species occurred, depending on whether a naked or incomplete flower was present at the root of the sections Sciadanthus and Fraxinus. However, despite the relatively low number of modifications, the floral morphology was not uniform within taxonomical sections. For example, even though the section Ornus mostly comprises species with complete flowers, F. chinensis, F. micrantha, and F. longicuspis harbour flowers without a corolla (Fig. 1). In the same way, F. cuspidata displays complete flowers in the section Melioides sensu lato, which is a group otherwise characterised by flowers without a corolla; likewise, F. dipetala displays flowers with only two petals, which is a unique feature in the genus (Fig. 1). Moreover, the extensive sharing of floral morphology among different sections of the genus without any particular correlation with the taxonomic relationships (e.g. incomplete flowers, lacking a corolla but with a calyx, were observed in almost all species of sections Dipetalae, Melioides sensu lato, Pauciflorae and Sciadanthus) clearly indicates that floral morphology has not evolved along a single, linear trajectory and that its evolutionary history shows little congruence with the phylogeny of the genus.
Discussion
The phylogeny of the genus Fraxinus
The phylogenetic tree that resulted from the analysis of the combined dataset had higher resolution than the trees based on analyses of separate datasets and the previous nITS trees of Wallander [4] and Jeandroz et al. [8]. It resulted in a notable improvement in delineation of the relationships among the different sections of the genus. Despite the same placement as in Wallander [4] and Jeandroz et al. [8], the support for the position of the section Dipetalae at the root of the tree was only moderate. The section Fraxinus, which previously formed a polytomy with the sections Sciadanthus and Melioides [8], was closely related to the section Sciadanthus and, to a lesser extent, to the section Ornus. While the section Pauciflorae was not considered by Jeandroz et al. [8], our results agree with the previous placement of this section as the sister group to the Old World sections [4].
In all analyses, F. platypoda appeared to be closely related to F. spaethiana, which was previously considered as an incertae sedis species [4]. This close relationship was initially suggested by Nakaike [49] and Wei and Green [50] on the basis of morphological characters. Moreover, F. spaethiana has been recognized by some researchers as a synonym of F. platypoda, which is in accordance with our results [51]. The close relatedness of F. chiisanensis to this species pair, with moderate to high support for all datasets, suggested that these three Asian species should be grouped into the same section. Given their well-supported relationship with taxa from the section Melioides, we suggest that these three species (F. chiisanensis, F. platypoda, and F. spaethiana) and F. cuspidata should be included in an expanded section Melioides, in agreement with the historical placement of F. platypoda in this section (prior to Wallander [4]; [50]). The close relationship observed between these species and the section Melioides was not in conflict with the results of Wallander [4], where a large polytomy was obtained at this level.
The section Melioides comprises 10 recognized species, although more than 28 have been described, [4]. It is the second largest section in terms of the number of species, the largest one being Ornus with 15 species. The section Melioides was poorly resolved in the analysis based on the combined dataset, as shown by the polytomy of F. americana, F. velutina, and F. pennsylvanica. Given that this polytomy included different species depending on which dataset was considered, we suggest that these species experienced rapid radiation, or that they have exchanged genes recently. Previous molecular phylogenies had outlined a polytomy involving F. pennsylvanica, F. velutina, and F. tomentosa [8], or F. caroliniana, F. pennsylvanica, F. texensis, F. velutina, F. profunda, and F. berlanderiana [4]. The observed low differentiation at the genetic level parallels similar trends at the morphological level, which resulted in either the treatment of several species as synonyms [16], [52] or in the delineation of morphological taxa of various nature (see [53], [54], [55], [56], [57], [58], [59], [60]). In the absence of morphological or molecular evidence regarding the specific status of American taxa from the section Melioides and the status of their reproductive isolation [4], it remains difficult to estimate precisely how much genetic divergence exists among these species.
Further studies that involve artificial crosses, as well as morphological and molecular characterization of trees sampled in potential zones of contact, should help clarify whether some of these species should be treated as subspecific taxa and if they are actively hybridizing. If either of these criteria applies, the correct estimation of phylogenetic relationships among species would represent a significant challenge, even if additional sequence data were obtained.
The evolution of floral morphology
When tracing the evolution of floral characters onto the present phylogeny, we observed that floral morphology varied even within taxonomic groups. As underlined by Regal [61], and as previously suggested for Fraxinus [4], floral morphology has likely been affected by ecological factors, such as the pollination system. The loss of corolla has been associated with a transition to an anemophilous pollination system, and sometimes to dioecy [4]. However, on the basis of the present phylogeny, we did not observe any systematic relationship between floral morphology and the habitat preferences of species (desert or temperate regions with predominant wind pollination vs. subtropical areas and higher diversity of pollinators); this result suggests that the type of pollination could be an auxiliary factor in the evolution of Fraxinus. Elucidation of the evolutionary relevance of these factors will require more ecological and biological studies of diverse species within the genus.
The biogeographic history of the genus Fraxinus and implications for other tree genera
Our results agree with the hypothesis of an Eocene (between 34 to 56 mya) North American origin for the genus [8], [62], [63] (Fig. 3a), as indicated by the more basal position of the species or groups of species found in North America (sections Dipetalae, Melioides, and Pauciflorae). The view of a North American origin of the genus is reinforced by evidence from the fossil record, given that the two oldest fossils that can be reliably attributed to the genus were found in the southeast of North America [35], [36], [37].
The present estimates from molecular dating are also congruent with those based on cpDNA sequences for the diversification of the family Oleaceae [63]. Fossils for which the specific status is unclear, including a putative leaflet fossil of F. juglandina from the Eocene era in China, which would be morphologically close to F. ornus [64], and samaras of F. oishii from the Miocene era in Korea, which would be morphologically close to F. mandshurica [65], could dramatically increase the inferred age of the genus if used as landmarks, and put the estimate well at odds with the conventional history of the family Oleaceae [62], [63]. These fossils might also be representative of morphological features of extinct lineages that evolved repeatedly in the genus. Owing to these uncertainties, they were not considered as landmarks in the estimation of divergence times.
The estimated diversification rates were quite consistent over time, despite large shifts in global temperature during the past 40 mya (Fig. 2b). It is possible that the diversification rate was underestimated during past periods of warmer climate, for instance, around the Mid-Miocene Climatic Optimum, given that extinct taxa could not be accounted for and that this bias should be more severe as one moves back in time. However, the number of divergence events leading to extant taxa appeared to vary extensively in each section. These sections were well supported by the phylogeny, and with the exception of the cosmopolitan section Fraxinus (which is found in Europe, East Asia, and the east of North America), all other sections of the genus are found mostly in only one of these great areas of endemism [66]. Thus, a number of factors more specific to the geographical locations of these sections have likely affected their evolution. Palaeobiogeographical inferences at the regional level are presented below; they are likely applicable to other temperate tree genera that exhibit similar geographic distributions.
A larger number of species and higher diversification rates were observed for the sections Ornus (15 species, 0.51 sp./mya) and Melioides sensu lato (10 species, 0.32 sp./mya) than in the sister sections Fraxinus and Sciadanthus (six species, 0.23 sp./mya and two species, 0.12 sp./mya, respectively). This trend is in agreement with the view that the regions where the sections Ornus and Melioides sensu lato are found, namely, East Asia and North America, respectively, were less influenced by the drying climate and glaciations that affected Europe and the west of North America, respectively, which may have led to the survival of marginally more plant species [66], including a larger number of species surviving in East Asia (for a review, see [67]).
However, other factors that have likely affected the number of species in a particular section should be considered, including the occurrence of vicariance-inducing factors, such as topographical and climatic variation that led to the fragmentation of natural distributions. For instance, it seems reasonable to assume that major geological events such as the Tibetan Plateau uplift of the early Miocene created new ecological niches that the Asian species could have invaded, or that the Mid-Miocene Climatic Optimum represented an opportunity for Asian species to colonize the newly contiguous European landmass to the west, which resulted in divergence events and new lineages. It should also be noted that, since the Miocene, major events that one would expect to have affected species in particular sections of the genus (depending of the location of these events) had no noticeable influence on the general diversification rate (see Fig. 2b).
Given that the most ancient fossils of Fraxinus representative of the morphology of European extant taxa (including pollen) date back to the Upper Miocene (11.6–5.3 mya) [68], and considering the history depicted by the phylogeny obtained herein, it is clear that the climate and conformation of inter-continental land bridges have varied extensively since then, with variations in mean annual temperature of up to 6°C [44] and variations in sea level of up to 60 meters [69]. These changing conditions likely provided opportunities for extensive shifts in natural distributions, including more or less ancient inter-continental spread. It is generally accepted that two dispersal pathways existed between North America and Eurasia during the Tertiary: the Atlantic and Pacific tracks, respectively through a North Atlantic bridge and through Beringia [67], [70]. In agreement with previous reports [8], our results suggest that the Pacific track was involved in the early emergence of both the lineage leading to F. chiisanensis, F. platypoda, and F. spaethiana and the section Ornus from a North American ancestor (Fig. 3b), because of the absence of European species in the section Melioides sensu lato and given the geographic proximity. Regarding the inferred allopatric divergence that led to the emergence of the North American F. nigra at the end of the upper Miocene (Fig. 3d), in the section Fraxinus that is otherwise composed of species from Eurasia, it is difficult to determinate whether the migration proceeded from Asia or from Europe. Because F. nigra is currently found only in the east of North America, one might argue that its ancestor is from Europe. However, given the palaeogeographical reconstruction described herein, the allopatric speciation leading to the F. nigra lineage likely proceeded from Asia through the Pacific track. Given their high levels of morphological and ecological similarity, F. mandshurica and F. nigra are also sometimes referred to as “geographical races” [71]. According to this more credible scenario, the isolation of the lineage leading to F. nigra in North America could be related to the contraction of the Northern Hemisphere boreotropical forest at the end of the Oligocene [66], [67]. Indeed, such North American - Asian disjunct taxa (e.g. Liriodendron, Liquidambar, and Magnolia, review in [72]) are thought to be relicts of the temperate forests of the Northern Hemisphere, which reached their maximum development during the warm period of the Tertiary (Axelrod, 1975 in [72]) and would have declined during the last 30 mya [66]. Such diversification patterns that suggest inter-continental allopatric speciation appear to be common in temperate forest tree genera, given that the same genera are often found in several areas of endemism around the world [66].
Whereas F. nigra is the only North American species in the section Fraxinus, all of the other three species of the section are found in Eurasia and are arranged in a distribution pattern typical of geographic speciation induced by the effects of colonization or fragmentation inducing vicariance (Fig. 3d). As a sister taxon to this Eurasian group, F. mandshurica has a large distribution in East Asia, which covers 23 degrees of latitude and 46 degrees of longitude in China and the Russian Far East [71]. It is followed westward by F. holotricha, with an endemic distribution in the Balkan Peninsula. The species F. excelsior is found further west throughout Europe, except in the Mediterranean region, to which F. angustifolia is mostly restricted. These putative speciations associated with specific geographic areas are consistent with previous reports [8] and would have been driven by climatic and geological changes [67], [70], such as the contraction of the boreotropical forest found in the Northern Hemisphere during the Tertiary [73]. For example, the Mediterranean species Quercus suber would have been affected by Tertiary geological and climatic events in Eurasia that would have lead to the early divergence of intraspecific lineages [74].
These relatively recent speciation events are likely to have been accompanied by reticulate evolution during the early divergence of these ash taxa. Such trend is still observable today where natural hybridization is occurring in zones of contact between taxa (e.g. [16]), underlining the slow evolution of reproductive isolation in ashes. Such a phenomenon has been demonstrated both in conifers and angiosperm trees in Eurasia. For instance, in the conifer genus Tsuga, T. dumosa (found in the Himalayas) has been shown to be a hybrid taxon [75], while in angiosperm trees, the central Asian species Populus tristis is thought to be a hybrid between P. nigra and another unknown Asian species [13]. In the perennial shrubs Paeonia, it has been shown that the European populations of the Asian species have been replaced by their European×Asian hybrids, probably due to differential selection promoted by Pleistocene climatic changes [76].
In addition, some species such as F. excelsior (common ash) could have expanded their southern range during periods with cooler climates, leading to the present relic populations in locations where the climate is now temperate, like in the Elburz Mountains or the adjacent mountain ranges in Turkey. These trends further suggest the occurrence of generally slow speciation process, in temperate tree genera with similar life history, reproductive and demographic attributes as well as large geographical ranges (e.g. [13], [17]). Moreover, perennial plants such as ashes have long and overlapping generation times with often large effective population sizes, which would retard reproductive isolation and speciation [77]. Some Fraxinus species (e.g., F. excelsior) have a particularly complicated mating system (trioecy: male, female, and hermaphrodite individuals), which may interfere in this respect. Despite the fact that the analysis of F. excelsior revealed almost functional dioecy [5], it is possible that the mating system plays a significant role in speciation, as shown for the transition from nondioecy to dioecy [78].
The colonization of new habitats leading to geographic speciation could further be hypothesized for several pairs of taxa in the genus, given that the species involved are phylogenetically closely related when found in parapatry or in much or less close allopatry: for instance, the pairs F. anomala and F. quadrangulata in the United States, F. papillosa and F. latifolia as well as F. xanthoxyloides and F. hupehensis in Asia, and F. angustifolia and F. excelsior in Europe. Such a repeated pattern of phylogenetic grouping of geographically close species has been noted in an increasing number of other tree genera [17], [79], which further suggests an important role for geographic speciation in promoting the past diversification of temperate and boreal tree taxa. Thereafter, these divergences may have led to ecological specialization, as each pair contains species that may have adapted to contrasting environments: dry versus more Mediterranean climates, or temperate versus warmer subtropical climates.
Thus, the repeated observations of geographical patterning at various scales in the phylogeny of the genus Fraxinus provide strong support for the importance of vicariance and geographic speciation as a central mechanism for diversification in this genus. Together with the noted slow evolution of reproductive isolation and reported instances of reticulate evolution in the genus, these processes could explain much of the patterns of interspecific variation noted at the genus-wide level in Fraxinus. These observations are in line with recent reports for other anemophilous and vastly distributed tree genera in the conifers [17] and angiosperms [80], which highlight the central role of vicariance and geographic speciation in driving the emergence of new lineages in temperate and boreal trees.
Supporting Information
Left: Matrix used for the Lagrange analysis. Right: Palaeogeographic maps showing the intensity of inferred migrations. Palaeomaps redrawn from Nie et al. [81].
(EPS)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the nETS dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous report [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the nITS dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous reports [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the phantastica dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous report [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from BIM analysis of the cpDNA dataset (trnH-psbA and rpl32-trnL). Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here shown to be closely related to the incertae sedis species. An asterisk indicates a species that was not clustered with other species of the same section.
(EPS)
Phylogenetic tree resulting from BIM analysis of the nETS dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from BIM analysis of the nITS dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from BIM analysis of the phantastica dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from the BEAST analysis of the four datasets (cpDNA – trnH-psbA and rpl32-trnL, nETS, nITS and phantastica). Bars at nodes represent the 95% confidence interval; posterior probabilities above the branches. Ages in million years before present.
(PDF)
Fraxinus samples used in this study, herbarium vouchers and newly published DNA sequences. ID stands for identifier; Sample type: Origin of the sample used in this study; A, Arboretum; W, Collected in the wild; H, Herbarium. Vouchers are deposited at the National Herbarium, Muséum National d'Histoire Naturelle, Paris, France (P00729547 to P00729694), or at the Mexico Herbarium (MEXU1032796 to MEXU991880); Arb.: Arboretum.
(DOC)
Acknowledgments
We are grateful to the staff of all arboreta and herbaria listed in Table S1, Kazuya Iizuka (Utsunomiya Univ.) and Naoko Miyamoto (Forestry and Forest Products Research Institute) who provided samples from wild Japanese species, staff at the herbarium of the Universidad Nacional Autónoma de México, who kindly provided samples of Mexican ashes, Jérôme Laroche (Bioinformatics Platform of the Institute for Systems and Integrative Biology, Univ. Laval) for access to computer facilities and assistance with phylogenetic analyses, and Mariangela Arca for generating some of the cpDNA sequences. We also thank two anonymous reviewers for their constructive comments.
Funding Statement
D.D. Hinsinger is the recipient of a fellowship from the Ministère des Affaires Etrangères (Bourse Lavoisier) and received financial support from the French Ministry of Education and the Université Paris-Sud 11 (Orsay). This work was supported by the “Consortium National de Recherche en Génomique”, and the “Service de Systématique Moléculaire” of the Muséum National d'Histoire Naturelle (IFR 101). It is part of agreement no. 2005/67 between Genoscope and the Muséum National d'Histoire Naturelle on the project “Macrophylogeny of Life”, directed by Guillaume Lecointre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bousquet J (2009) Inferring the past from the present phylogeographic structure of North American forest trees: seeing the forest for the genes. Canadian Journal of Forest Research-Revue Canadienne De Recherche Forestiere 39: 286–307. [Google Scholar]
- 2. Culley TM, Weller SG, Sakai AK (2002) The evolution of wind pollination in angiosperms. Trends in Ecology and Evolution 17: 361–369. [Google Scholar]
- 3. Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests 6: 95–124. [Google Scholar]
- 4. Wallander E (2008) Systematics of Fraxinus (Oleaceae) and evolution of dioecy. Plant Systematics and Evolution 273: 25–49. [Google Scholar]
- 5. Morand-Prieur M-E, Raquin C, Shykoff JA, Frascaria-Lacoste N (2003) Males outcompete hermaphrodites for seed siring success in controlled crosses in the polygamous Fraxinus excelsior (Oleaceae). American Journal of Botany 90: 949–953. [DOI] [PubMed] [Google Scholar]
- 6. Via S (2002) The ecological genetics of speciation. American Naturalist 159: S1–S7. [DOI] [PubMed] [Google Scholar]
- 7. Schluter D (2001) Ecology and the origin of species. Trends in Ecology and Evolution 16: 372–380. [DOI] [PubMed] [Google Scholar]
- 8. Jeandroz S, Roy A, Bousquet J (1997) Phylogeny and phylogeography of the circumpolar genus Fraxinus (Oleaceae) based on internal transcribed spacer sequences of nuclear ribosomal DNA. Molecular Phylogenetics and Evolution 7: 241–251. [DOI] [PubMed] [Google Scholar]
- 9. Gielly L, Taberlet P (1994) Chloroplast DNA polymorphism at the intrageneric level and plant phylogenie. Comptes-Rendus de l'Académies des Sciences de Paris, Sciences de la Vie/Life sciences 317: 685–692. [Google Scholar]
- 10. Wallander E, Albert VA (2000) Phylogeny and classification of Oleaceae based on rps16 and trnL-F sequence data. American Journal of Botany 87: 1827–1841. [PubMed] [Google Scholar]
- 11. Bouillé M, Bousquet J (2005) Trans-species shared polymorphisms at orthologous nuclear gene loci among distant species in the conifer Picea (Pinaceae): implications for the long-term maintenance of genetic diversity in trees. American Journal of Botany 92: 63–73. [DOI] [PubMed] [Google Scholar]
- 12. Willyard A, Cronn R, Liston A (2009) Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Molecular Phylogenetics and Evolution 52: 498–511. [DOI] [PubMed] [Google Scholar]
- 13. Hamzeh M, Dayanandan S (2004) Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. American Journal of Botany 91: 1398–1408. [DOI] [PubMed] [Google Scholar]
- 14. Rieseberg LH, Brouillet L (1994) Are many plant species paraphyletic ? Taxon 43: 21–32. [Google Scholar]
- 15. Fernandez-Manjarres JF, Gerard PR, Dufour J, Raquin C, Frascaria-Lacoste N (2006) Differential patterns of morphological and molecular hybridization between Fraxinus excelsior L. and Fraxinus angustifolia Vahl (Oleaceae) in eastern and western France. Molecular Ecology 15: 3245–3257. [DOI] [PubMed] [Google Scholar]
- 16. Santamour FSJ (1962) The relation between polyploidy and morphology in white and biltmore ashes. Bulletin of the Torrey Botanical Club 89: 228–232. [Google Scholar]
- 17. Huelsenbeck JP, Rannala B (1997) Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science 276: 227–232. [DOI] [PubMed] [Google Scholar]
- 18. Arca M, Hinsinger DD, Cruaud C, Tillier A, Bousquet J, et al. (2012) Deciduous trees and the application of universal DNA barcodes: a case study on the circumpolar Fraxinus . Plos ONE 7 3: doi:10.1371/journal.pone.0034089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Alexander JH, Zhang D (2002) Paraphyletic Syringa (Oleaceae): evidence from sequences of nuclear ribosomal DNA ITS and ETS regions. Systematic Botany 27: 592–597. [Google Scholar]
- 20. Nickrent DL, Schuette KP, Starr EM (1994) A molecular phylogeny of Arceuthobium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany 81: 1149–1160. [Google Scholar]
- 21. Wojciechowski MF, Sanderson MJ, Baldwin BG, Donoghue MJ (1993) Monophyly of aneuploid Astragalus (Fabaceae): Evidence from nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany 80: 711–722. [Google Scholar]
- 22. Waites R, Selvadurai HR, Oliver IR, Hudson A (1998) The phantastica gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell 93: 779–789. [DOI] [PubMed] [Google Scholar]
- 23. Kim M, McCormick S, Timmermans M, Sinha N (2003) The expression domain of phantastica determines leaflet placement in compound leaves. Nature 424: 438–443. [DOI] [PubMed] [Google Scholar]
- 24. Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 18: 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7: 544–548. [Google Scholar]
- 26. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 113 doi:10.1186/1471-2105-5-1131471-2105-5-113 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A (1996) Sequence Alignment Editor v2.0a11. Available from http://tree.bio.ed.ac.uk/software/seal/. Oxford University, Oxford, England
- 28. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 29. Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 30. Sullivan J, Joyce P (2005) Model selection in phylogenetics. Annual Review of Ecology, Evolution and Systematics 36: 445–466. [Google Scholar]
- 31. Huelsenbeck JP, Rannala B (2004) Frequentist properties of bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology 53: 904–913. [DOI] [PubMed] [Google Scholar]
- 32. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 33.Rambaut A, Drummond AJ (2005) Tracer v1.5. Available from http://tree.bio.ed.ac.uk/software/tracer/. Oxford University, Oxford, England
- 34. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Call VB, Dilcher DL (1992) Investigations of angiosperms from the Eocene of southeastern North America: samaras of Fraxinus wilcoxiana Berry. Review of Palaeobotany and Palynology 74: 249–266. [Google Scholar]
- 36. Magallon-Puebla S, Crane PR, Herendeen PS (1999) Phylogenetic pattern, diversity, and diversification of eudicots. Annals of the Missouri Botanical Garden 86: 297–372. [Google Scholar]
- 37. Manchester SR (1999) Biogeographical relationships of North American Tertiary floras. Annals of the Missouri Botanical Garden 86: 472–522. [Google Scholar]
- 38. Palamarev E (1989) Paleobotanical evidences of the Tertiary history and origin of the Mediterranean sclerophyll dendroflora. Plant Systematics and Evolution 162: 93–107. [Google Scholar]
- 39. Kay KM, Whittall JB, Hodges SA (2006) A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evolutionary Biology 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambaut A (2009) FigTree v1.3.1. Available from http://tree.bio.ed.ac.uk/software/figtree/. Oxford University, Oxford, England
- 41.R Development Core Team (2010) R: A language and environment for statistical computing v2.12.1. Available from http://www.R-project.org. Vienna, Austria
- 42. Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- 43. Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57: 4–14. [DOI] [PubMed] [Google Scholar]
- 44. Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- 45.Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis v2.75. Available from http://mesquiteproject.org.
- 46. Schluter D, Price T, Mooers AO, Ludwig D (1997) Likelihood of ancestor states in adaptive radiation. Evolution 51: 1699–1711. [DOI] [PubMed] [Google Scholar]
- 47. Pybus OG, Harvey PH (2000) Testing macro-evolutionary models using incomplete molecular phylogenies. Proceedings of the Royal Society B-Biological Sciences 267: 2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rabosky DL (2006) Likelihood methods for inferring temporal shifts in diversification rates. Evolution 60: 1152–1164. [PubMed] [Google Scholar]
- 59. Nakaike T (1972) A synoptical study on the genus Fraxinus from Japan, Korea and Formosa. Bulletin of the National Science Museum (Tokyo) 15: 475–512. [Google Scholar]
- 50.Wei Z, Green PS (1996) Fraxinus. In: Wu Z, Raven PH, editors. Flora of China. Saint-Louis, Missouri: Science Press, Missouri Botanical Garden. pp. 273–279. [Google Scholar]
- 51. Yonekura K (2005) Taxonomic notes on vascular plants in Japan and its adjacent regions (I). - New combinations and new names of japanese plants-. Journal of japanese Botany 80: 323–333. [Google Scholar]
- 52.Miller GN (1955) The genus Fraxinus, the ashes, in North America, North of Mexico.: Cornell University. 64 p. [Google Scholar]
- 53. Nesom GL (2010) Geography of unifoliolate taxa of Fraxinus (Oleaceae). Phytoneuron 17: 1–6. [Google Scholar]
- 54. Nesom GL (2010) Notes on Fraxinus profunda (Oleaceae). Phytoneuron 32: 1–6. [Google Scholar]
- 55. Nesom GL (2010) Observations on Fraxinus albicans Buckley (Oleaceae), the correct botanical name for Texas ash. Phytoneuron 33: 1–12. [Google Scholar]
- 56. Nesom GL (2010) Taxonomic notes on Fraxinus berlandieriana and F. velutina (Oleaceae). Phytoneuron 34: 1–8. [Google Scholar]
- 57. Nesom GL (2010) Taxonomic status of Fraxinus coriacea (Oleaceae). Phytoneuron 37: 1–9. [Google Scholar]
- 58. Nesom GL (2010) Notes on Fraxinus cuspidata and F. gooddingii (Oleaceae). Phytoneuron 38: 1–14. [Google Scholar]
- 59. Nesom GL (2010) Taxonomy of the water ashes: Fraxinus caroliniana, F. cubensis, and F. pauciflora (Oleaceae). Phytoneuron 39: 1–13. [Google Scholar]
- 60. Nesom GL (2010) Fraxinus biltmoreana and Fraxinus smallii (Oleaceae), forest trees of the eastern United States. Phytoneuron 51: 1–30. [Google Scholar]
- 61. Regal PJ (1982) Pollination by wind and animals: Ecology of geographic patterns. Annual Review of Ecology and Systematics 13: 497–524. [Google Scholar]
- 62. Besnard G, Rubio de Casas R, Christin P-A, Vargas P (2009) Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: Tertiary climatic shifts and lineage differentiation times. Annals of Botany 104: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee HL, Jansen RK, Chumley TW, Kim KJ (2007) Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Molecular Biology and Evolution 24: 1161–1180. [DOI] [PubMed] [Google Scholar]
- 64. Wang Q, Ferguson DK, Feng GP, Ablaev AG, Wang YF, et al. (2010) Climatic change during the Palaeocene to Eocene based on fossil plants from Fushun, China. Palaeogeography, Palaeoclimatology, Palaeoecology 295: 323–331. [Google Scholar]
- 65. Jung SH, Lee SJ (2009) Fossil-winged fruits of Fraxinus (Oleaceae) and Liriodendron (Magnoliaceae) from the Duho formation, Pohang basin, Korea. Acta Geologica Sinica-English Edition 83: 845–852. [Google Scholar]
- 66. Donoghue MJ, Smith SA (2004) Patterns in the assembly of temperate forests around the Northern Hemisphere. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences 359: 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wen J (1999) Evolution of Eastern Asian and Eastern North American disjunct distributions in flowering plants. Annual Review of Ecology and Systematics 30: 421–455. [Google Scholar]
- 68. Muller J (1981) Fossil pollen records of extant angiosperms. The Botanical Review 47: 1–142. [Google Scholar]
- 69. Miller KG, Kominz MA, Browning JV, Wright JD, Mountain GS, et al. (2005) The Phanerozoic record of global sea-level change. Science 310: 1293–1298. [DOI] [PubMed] [Google Scholar]
- 70. Donoghue MJ, Bell CD, Li J (2001) Phylogenetic patterns in Northern Hemisphere plant geography. International Journal of Plant Sciences 162: S41–S52. [Google Scholar]
- 71. Yihong W (1995) Study on ecology of Fraxinus mandshurica . Journal of Forestry Research 6: 61–64. [Google Scholar]
- 72. Nie Z-L, Sun H, Beardsley PM, Olmstead RG, Wen J (2006) Evolution of biogeographic disjunction between eastern Asia and eastern North America in Phryma (Phrymaceae). American Journal of Botany 93: 1343–1356. [DOI] [PubMed] [Google Scholar]
- 73. Wolfe JA (1975) Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Annals of the Missouri Botanical Garden 62: 264–279. [Google Scholar]
- 74. Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, et al. (2007) The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Molecular Ecology 16: 5259–5266. [DOI] [PubMed] [Google Scholar]
- 75. Havill NP, Campbell CS, Vining TF, LePage B, Bayer RJ, et al. (2009) Phylogeny and biogeography of Tsuga (Pinaceae) inferred from nuclear ribosomal ITS and chloroplast DNA sequence data. Systematic Botany 33: 478–789. [Google Scholar]
- 76. Sang T, Crawford D, Stuessy T (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120–1136. [PubMed] [Google Scholar]
- 77. Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annual Review of Ecology, Evolution and Systematics 37: 187–214. [Google Scholar]
- 78. Vamosi JC, Vamosi SM (2004) The role of diversification in causing the correlates of dioecy. Evolution 58: 723–731. [DOI] [PubMed] [Google Scholar]
- 79. Chen C-H, Huang J-P, Tsai C-C, Chaw S-M (2009) Phylogeny of Calocedrus (Cupressaceae), an eastern Asian and western North American disjunct gymnosperm genus, inferred from nuclear ribosomal nrITS sequences. Botanical Studies 50: 425–433. [Google Scholar]
- 80. Lopez de Heredia U, Jiménez P, Collada C, Simeone MC, Bellarosa R, et al. (2007) Multi-marker phylogeny of three evergreen oaks reveals vicariant patterns in the Western Mediterranean. Taxon 56: 1199–1209. [Google Scholar]
- 81. Nie Z-L, Sun H, Manchester SR, Meng Y, Luke Q, et al. (2012) Evolution of the intercontinental disjunctions in six continents in the Ampelopsis clade of the grape family (Vitaceae). BMC Evolutionary Biology 12: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left: Matrix used for the Lagrange analysis. Right: Palaeogeographic maps showing the intensity of inferred migrations. Palaeomaps redrawn from Nie et al. [81].
(EPS)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the nETS dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous report [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the nITS dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous reports [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from the maximum likelihood (ML) analysis of the phantastica dataset, including all sequences. ML bootstrap values are indicated above the branches. Sections according to a previous report [4] are indicated by vertical bars.
(PDF)
Phylogenetic tree resulting from BIM analysis of the cpDNA dataset (trnH-psbA and rpl32-trnL). Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here shown to be closely related to the incertae sedis species. An asterisk indicates a species that was not clustered with other species of the same section.
(EPS)
Phylogenetic tree resulting from BIM analysis of the nETS dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from BIM analysis of the nITS dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from BIM analysis of the phantastica dataset. Posterior probabilities ≥0.50 are indicated above the branches, ML bootstrap values are indicated below the branches. Sections according to a previous report [4] are indicated by vertical bars, and the dotted line indicates the occurrence of F. platypoda, previously placed in the section Fraxinus [4], but here found to be closely related to the incertae sedis species.
(PDF)
Phylogenetic tree resulting from the BEAST analysis of the four datasets (cpDNA – trnH-psbA and rpl32-trnL, nETS, nITS and phantastica). Bars at nodes represent the 95% confidence interval; posterior probabilities above the branches. Ages in million years before present.
(PDF)
Fraxinus samples used in this study, herbarium vouchers and newly published DNA sequences. ID stands for identifier; Sample type: Origin of the sample used in this study; A, Arboretum; W, Collected in the wild; H, Herbarium. Vouchers are deposited at the National Herbarium, Muséum National d'Histoire Naturelle, Paris, France (P00729547 to P00729694), or at the Mexico Herbarium (MEXU1032796 to MEXU991880); Arb.: Arboretum.
(DOC)